Abstract
Alterations in the genetic content of a cell are the underlying cause of many human diseases, including cancers. We have developed a method, called digital karyotyping, that provides quantitative analysis of DNA copy number at high resolution. This approach involves the isolation and enumeration of short sequence tags from specific genomic loci. Analysis of human cancer cells by using this method identified gross chromosomal changes as well as amplifications and deletions, including regions not previously known to be altered. Foreign DNA sequences not present in the normal human genome could also be readily identified. Digital karyotyping provides a broadly applicable means for systematic detection of DNA copy number changes on a genomic scale.
Somatic and hereditary variations in gene copy number can lead to profound abnormalities at the cellular and organismal levels. In human cancer, chromosomal changes, including the deletion of tumor suppressor genes and the amplification of oncogenes, are hallmarks of neoplasia (1). Single copy changes in specific chromosomes or smaller regions can result in a number of developmental disorders, including Down, Prader Willi, Angelman, and cri du chat syndromes (2). Current methods for the analysis of cellular genetic content include comparative genomic hybridization (CGH) (3), representational difference analysis (4), spectral karyotyping/multiplex-fluorescence in situ hybridization (M-FISH) (5, 6), microarrays (7–10), and traditional cytogenetics. Such techniques have aided in the identification of genetic aberrations in human malignancies and other diseases (11–14). However, methods employing metaphase chromosomes have a limited mapping resolution (≈20 Mb; ref. 15), and therefore cannot be used to detect smaller alterations. Recent implementation of CGH to microarrays containing genomic or transcript DNA sequences provides improved resolution, but is currently limited by the number of sequences that can be assessed (16) or by the difficulty of detecting certain alterations such as homozygous deletions (9).
To circumvent these limitations, we have developed a method that permits the comprehensive examination of cellular DNA content based on the quantitative analysis of short fragments of genomic DNA. This method is based on two concepts. First, short sequence tags (21 bp each) can be obtained from specific locations in the genome. These tags generally contain sufficient information to uniquely identify the genomic loci from which they were derived. Such tags are in principle related to those obtained in the serial analysis of gene expression (SAGE) approach (17, 18), but are obtained from genomic DNA, rather than from mRNA, and are isolated by using different methods. Second, populations of tags can be directly matched to the assembled genomic sequence, allowing observed tags to be sequentially ordered along each chromosome. Digital enumeration of tag observations along each chromosome can then be used to quantitatively evaluate DNA content with high resolution.
Materials and Methods
Digital Karyotyping Library Construction.
Digital karyotyping was performed on DNA from colorectal cancer cell lines DiFi and Hx48, and from the lymphoblastoid cells of a normal individual (GM12911, obtained from Coriell Cell Repositories, Camden, NJ). Genomic DNA was isolated by using DNeasy or QIAamp DNA blood kits (Qiagen, Valencia, CA) and following the manufacturer's protocols. For each sample, 1 μg of genomic DNA was sequentially digested with mapping enzyme SacI, ligated to 20–40 ng of biotinylated linker (5′-biotin-TTTGCAGAGGTTCGTAATCGAGTTGGGTGAGC-3′, 5′-phosphate-CACCCAACTCGATTACGAACCTCTGC-3′; Integrated DNA Technologies, Coralville, IA) by using T4 ligase (Invitrogen), and then digested with the fragmenting enzyme NlaIII. DNA fragments containing biotinylated linkers were isolated by binding to streptavidin-coated magnetic beads (Dynal, Oslo). The remaining steps were similar to those described for LongSAGE of cDNA (18). In brief, linkers containing MmeI recognition sites were ligated to captured DNA fragments, and tags were released with MmeI (University of Gdansk Center for Technology Transfer, Gdansk, Poland, and New England Biolabs). The tags were ligated to form ditags, and the ditags were isolated, and then ligated to form concatemers, which were cloned into pZero (Invitrogen). The sequencing of concatemer clones was performed by using the Big Dye terminator v3.0 kit (Applied Biosystems) and analyzed with an SCE-9610 192-capillary electrophoresis system (SpectruMedix, State College, PA) or by contract sequencing at Agencourt (Beverly, MA). Digital karyotyping sequence files were trimmed by using phred sequence analysis software (CodonCode, Dedham, MA), and 21-bp genomic tags were extracted by using the SAGE2000 software package, which identifies the fragmenting enzyme site between ditags, extracts intervening tags, and records them in a database. Detailed protocols for performing digital karyotyping and software for the extraction and analysis of genomic tags are available at www.digitalkaryotyping.org.
Simulations.
The theoretical sensitivity and specificity of digital karyotyping for copy number alterations was evaluated by using Monte Carlo simulations. For each alteration type, 100 simulations were performed as follows: Either 100,000 or 1,000,000 experimental tags were randomly assigned to 730,862 equally spaced virtual tags in a genome containing a single randomly placed copy number alteration of a predefined size and copy number. Moving windows containing the same number of virtual tags as the simulated alteration were used to evaluate tag densities along the genome. Tag density values of >4.9, <0.1, <0.6, and >1.4 located within the area of amplifications, homozygous deletions, heterozygous losses, and subchromosomal gains, respectively, were considered true positives. Tag densities of these values in areas outside the altered region were considered false positives.
Data Analysis.
All tags adjacent to the NlaIII fragmenting enzyme (CATG) sites closest to SacI mapping enzyme sites were computationally extracted from the human genome sequence (University of California, Santa Cruz, June 28, 2002 Assembly, http://genome.ucsc.edu/). Of the 1,094,480 extracted tags, 730,862 were obtained from unique loci in the genome and were termed virtual tags. The experimentally derived genomic tags obtained from NLB, DiFi, and Hx48 cells were electronically matched to these virtual tags. The experimental tags with the same sequence as virtual tags were termed filtered tags and were used for subsequent analysis. The remaining tags corresponded to repeated regions, sequences not present in the current genome database release, polymorphisms at the tag site, or sequencing errors in the tags or in the genome sequence database. Tag densities for sliding windows containing N virtual tags were determined as the sum of experimental tags divided by the average number of experimental tags in similar sized windows throughout the genome. Tag densities were dynamically analyzed in windows ranging from 50 to 1,000 virtual tags. For windows of 1,000 virtual tags, DiFi tag densities were normalized to evaluated NLB tag densities in the same sliding windows to account for incomplete filtering of tags matching repetitive sequences, and visualized by using tag density maps. For windows <1,000 virtual tags, a bitmap viewer was developed that specifically identified tag densities above or below defined thresholds.
Quantitative PCR.
Genome content differences between DiFi and normal cells were determined by quantitative real-time PCR performed on an iCycler apparatus (Bio-Rad). DNA content was normalized to that of Line-1, a repetitive element for which copy numbers per haploid genome are similar among all human cells (normal or neoplastic). Copy number changes per haploid genome were calculated by using the formula 2(Nt−Nline)−(Dt−Dline) where Nt is the threshold cycle number observed for an experimental primer in the normal DNA sample, Nline is the threshold cycle number observed for a Line-1 primer in the normal DNA sample, Dt is the average threshold cycle number observed for the experimental primer in DiFi, and Dline is the average threshold cycle number observed for a Line-1 primer in DiFi. Conditions for amplification were as follows: one cycle of 94°C for 2 min, followed by 50 cycles of 94°C for 20 sec, 57°C for 20 sec, and 70°C for 20 sec. Threshold cycle numbers were obtained by using ICYCLER V 2.3 software. PCRs for each primer set were performed in triplicate and threshold cycle numbers were averaged. For analysis of homozygous deletions, the presence or absence of PCR products was evaluated by gel electrophoresis. PCR primers were designed by using Primer 3 (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) to span a 100- to 200-bp nonrepetitive region and were synthesized by GeneLink (Hawthorne, NY). Primer sequences for each region analyzed in this study are included in Table 4, which is published as supporting information on the PNAS web site, www.pnas.org.
Karyotyping and Comparative Genomic Hybridization.
CGH was performed as described (19), and hybridization data were analyzed with Leica Microsystems (Deerfield, IL) imaging software. Karyotyping was performed with conventional procedures.
Results
Principles of Digital Karyotyping.
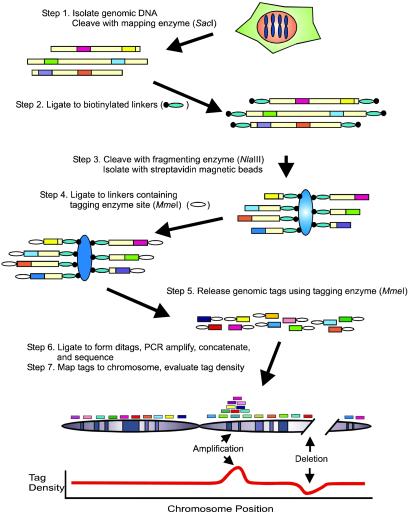
The basic concepts of digital karyotyping have been implemented as described in Fig. 1. Genomic DNA is cleaved with a restriction endonuclease (mapping enzyme) that is predicted to cleave genomic DNA into several hundred thousand pieces, each, on average, <10 kb in size (step 1). A variety of different endonucleases can be used for this purpose, depending on the resolution desired. In the current study, we used SacI, with a 6-bp recognition sequence. Biotinylated linkers are ligated to the DNA molecules (step 2) and then digested with a second endonuclease (fragmenting enzyme) that recognizes 4-bp sequences (step 3). As there are, on average, 16 fragmenting enzyme sites between every 2 mapping enzyme sites (46/44), the majority of DNA molecules in the template are expected to be cleaved by both enzymes and, thereby, be available for subsequent steps. DNA fragments containing biotinylated linkers are separated from the remaining fragments by using streptavidin-coated magnetic beads (step 3). New linkers, containing a 6-bp site recognized by MmeI, a type IIS restriction endonuclease (18), are ligated to the captured DNA (step 4). The captured fragments are cleaved by MmeI, releasing 21-bp tags (step 5). Each tag is thus derived from the sequence adjacent to the fragmenting enzyme site that is closest to the nearest mapping enzyme site. Isolated tags are self-ligated to form ditags, PCR-amplified en masse, concatenated, cloned, and sequenced (step 6). As described for SAGE (17), the formation of ditags provides a robust method to eliminate potential PCR-induced bias during the procedure. Current automated sequencing technologies identify up to 30 tags per concatemer clone, allowing for the analysis of ≈100,000 tags per day by using a single 384-capillary sequencing apparatus. Finally, tags are computationally extracted from sequence data and matched to precise chromosomal locations, and tag densities are evaluated over moving windows to detect abnormalities in DNA sequence content (step 7).
Fig 1.
Schematic of the digital karyotyping approach. Colored boxes represent genomic tags. Small ovals represent linkers. Large blue ovals represent streptavidin-coated magnetic beads. See text for details.
The sensitivity and specificity of digital karyotyping for detecting genome-wide changes were expected to depend on several factors. First, the combination of mapping and fragmenting enzymes determines the minimum size of the alterations that can be identified. For example, the use of SacI and NlaIII as mapping and fragmenting enzymes, respectively, was predicted to result in a total of 730,862 virtual tags (defined as all possible tags that could theoretically be obtained from the human genome). These virtual tags were spaced at an average of 3,864 bp, with 95% separated by 4 bp to 46 kb. Practically, this resolution is limited by the number of tags actually sampled in a given experiment and the type of alteration present (Table 1). Monte Carlo simulations confirmed the intuitive concept that fewer tags are needed to detect high-copy-number amplifications than are needed to detect homozygous deletions or low-copy-number changes in similar sized regions (Table 1). Such simulations were used to predict the size of alterations that could be reliably detected given a fixed number of experimentally sampled tags. For example, the analysis of 100,000 tags would be expected to reliably detect a 10-fold amplification ≥100 kb, homozygous deletions ≥600 kb, or a single gain or loss of regions ≥4 Mb in size in a diploid genome (Table 1).
Table 1.
Theoretical detection of copy number alterations by using digital karyotyping
| Size of alteration | Amplification, % (copy number = 10) | Homozygous deletion, % (copy number = 0) | Heterozygous loss, % (copy number = 1) | Subchromosomal gain, % (copy number = 3) | |||||
|---|---|---|---|---|---|---|---|---|---|
| No. of base pairs | No. of virtual tags | 100,000 | 1,000,000 | 100,000 | 1,000,000 | 100,000 | 1,000,000 | 100,000 | 1,000,000 |
| 100,000 | 30 | 100 | 100 | 0.06 | 100 | 0.008 | 0.02 | 0.006 | 0.08 |
| 200,000 | 50 | 100 | 100 | 1 | 100 | 0.01 | 3 | 0.01 | 0.7 |
| 600,000 | 150 | 100 | 100 | 96 | 100 | 0.07 | 100 | 0.05 | 100 |
| 2,000,000 | 500 | 100 | 100 | 100 | 100 | 11 | 100 | 3 | 100 |
| 4,000,000 | 1,000 | 100 | 100 | 100 | 100 | 99 | 100 | 97 | 100 |
Copy number alteration refers to the gain or loss of chromosomal regions in the context of the normal diploid genome, where the normal copy number is 2. The limiting feature of these analyses was not sensitivity for detecting the alteration, as this was high in every case shown (>99% for amplifications and homozygous deletions and >92% for heterozygous losses or subchromosomal gains). What was of more concern was the positive predictive value (PPV), that is, the probability that a detected mutation represents a real mutation. PPVs were calculated from 100 simulated genomes, using 100,000 or 1,000,000 filtered tags, and are shown in the table as percentages.
Size of alteration refers to the approximate size of the genomic alteration assuming an average of 3,864 bp between virtual tags.
Analysis of Whole Chromosomes.
We characterized 210,245 genomic tags from the lymphoblastoid cells of a normal individual (NLB) and 171,795 genomic tags from the colorectal cancer cell line (DiFi) by using the mapping and fragmenting enzymes described above. After filtering to remove tags that were within repeated sequences or were not present in the human genome (see Materials and Methods), we recovered a total of 111,245 and 107,515 filtered tags from the NLB and DiFi libraries, respectively. Tags were ordered along each chromosome, and average chromosomal tag densities, defined as the number of detected tags divided by the number of virtual tags present in a given chromosome, were evaluated (Table 2). Analysis of the NLB data showed that the average tag density for each autosomal chromosome was similar, ≈0.16 ± 0.04. The small variations in tag densities were likely because of the incomplete filtering of tags matching repeated sequences that were not currently represented in the genome databases. The X and Y chromosomes had average densities about half this level, 0.073 and 0.068, respectively, consistent with the normal male karyotype of these cells. Analysis of the DiFi data revealed a much wider variation in tag density, ranging from 0.089 to 0.27 for autosomal chromosomes. In agreement with the origin of these tumor cells from a female patient (20), the tag density of the Y chromosome was 0.00. Estimates of chromosome number by using observed tag densities normalized to densities from lymphoblastoid cells suggested a highly aneuploid genetic content, with ≤1.5 copies of chromosomes 1, 4, 5, 8, 17, 21, and 22, and ≥3 copies of chromosomes 7, 13, and 20 per diploid genome. These observations were consistent with CGH analyses (see below) and the previously reported karyotype of DiFi cells (20).
Table 2.
Chromosome number analysis
| Chromosome
|
No. of virtual tags
|
NLB | DiFi | |||
|---|---|---|---|---|---|---|
| No. of observed tags | Tag density | No. of observed tags | Tag density | Chromosome content | ||
| 1 | 61,694 | 10,090 | 0.16 | 6,991 | 0.11 | 1.4 |
| 2 | 61,944 | 9,422 | 0.15 | 9,545 | 0.15 | 2.0 |
| 3 | 46,337 | 6,732 | 0.15 | 7,379 | 0.16 | 2.2 |
| 4 | 41,296 | 5,581 | 0.14 | 3,666 | 0.089 | 1.3 |
| 5 | 43,186 | 6,216 | 0.14 | 4,136 | 0.10 | 1.3 |
| 6 | 41,633 | 6,120 | 0.15 | 7,291 | 0.18 | 2.4 |
| 7 | 38,928 | 5,836 | 0.15 | 9,875 | 0.25 | 3.4 |
| 8 | 35,033 | 5,009 | 0.14 | 3,260 | 0.093 | 1.3 |
| 9 | 30,357 | 4,909 | 0.16 | 4,861 | 0.16 | 2.0 |
| 10 | 37,320 | 6,045 | 0.16 | 4,865 | 0.13 | 1.6 |
| 11 | 37,868 | 6,081 | 0.16 | 5,432 | 0.14 | 1.8 |
| 12 | 30,692 | 4,631 | 0.15 | 4,056 | 0.13 | 1.8 |
| 13 | 22,313 | 3,012 | 0.13 | 5,197 | 0.23 | 3.5 |
| 14 | 23,378 | 3,658 | 0.16 | 3,171 | 0.14 | 1.7 |
| 15 | 22,409 | 3,581 | 0.16 | 4,159 | 0.19 | 2.3 |
| 16 | 23,028 | 4,119 | 0.18 | 3,201 | 0.14 | 1.6 |
| 17 | 22,978 | 4,298 | 0.19 | 3,145 | 0.14 | 1.5 |
| 18 | 18,431 | 2,712 | 0.15 | 2,389 | 0.13 | 1.8 |
| 19 | 16,544 | 3,271 | 0.20 | 3,589 | 0.22 | 2.2 |
| 20 | 20,585 | 3,573 | 0.17 | 5,460 | 0.27 | 3.1 |
| 21 | 9,245 | 1,465 | 0.16 | 1,036 | 0.11 | 1.4 |
| 22 | 12,579 | 2,476 | 0.20 | 1,655 | 0.13 | 1.3 |
| X | 30,737 | 2,249 | 0.073 | 3,147 | 0.10 | 1.4 |
| Y | 2,347 | 159 | 0.068 | 9 | 0.00 | 0.06 |
| Total | 730,862 | 111,245 | 0.15 | 107,515 | 0.15 | 2.0 |
DiFi chromosomal content is calculated for autosomal chromosomes as 2 times the ratio of DiFi tag densities to corresponding NLB tag densities, and for the X chromosome as the ratio of DIFI tag density to NLB tag density. Underlined values represent autosomal chromosome content <1.5, while boldface values represent autosomal chromosome content >3.
Analysis of Chromosomal Arms.
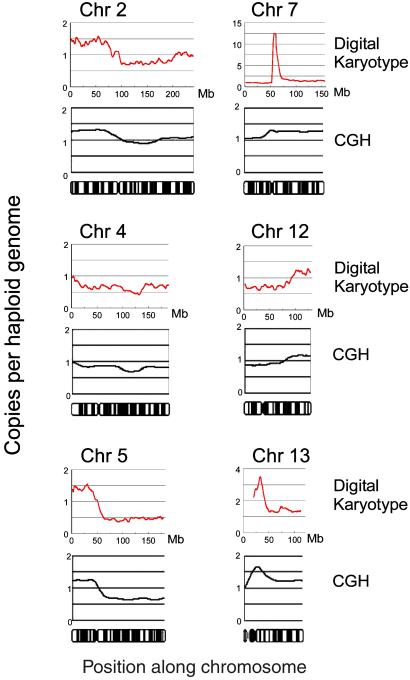
We next evaluated the ability of digital karyotyping to detect subchromosomal changes, particularly gains and losses of chromosomal arms. Tag densities were analyzed along each chromosome by using sliding windows containing 1,000 virtual tags (≈4 Mb), as windows of this size were predicted to reliably detect such alterations (Table 1). For the NLB sample, tag density maps showed uniform content along each chromosome, with small variations (<1.5-fold) present over localized regions, presumably because of the overrepresentation of tags matching repeated sequences (data not shown). In contrast, the DiFi tag density map (normalized to the NLB data) revealed widespread changes, including apparent losses in large regions of 5q, 8p, and 10q, and gains of 2p, 7q, 9p, 12q, 13q, and 19q (Fig. 2 and Fig. 5, which is published as supporting information on the PNAS web site). These changes included regions of known tumor suppressor genes (21) and other areas commonly altered in colorectal cancer (11, 12, 22). These alterations were confirmed by chromosomal CGH analyses, which revealed aberrations that were largely consistent with digital karyotype analyses in both location and amplitude (Fig. 2 and Fig. 5).
Fig 2.
Low-resolution tag density maps reveal many subchromosomal changes. The upper graph in each set corresponds to the digital karyotype, while the lower graph represents CGH analysis. An ideogram of each normal chromosome is present under each set of graphs. For all graphs, values on the y axis indicate genome copies per haploid genome, and values on the x axis represent positions along the chromosome (Mb for the digital karyotype; chromosome bands for CGH). Digital karyotype values represent exponentially smoothed ratios of DiFi tag densities, using a sliding window of 1,000 virtual tags normalized to the NLB genome. Chromosomal areas lacking digital karyotype values correspond to unsequenced portions of the genome, including heterochromatic regions. Note that using a window of 1,000 virtual tags does not permit accurate identification of alterations less than ≈4 Mb, such as amplifications and homozygous deletions, and smaller windows need to be used to accurately identify these lesions (see Fig. 3 for an example).
Analysis of Amplifications.
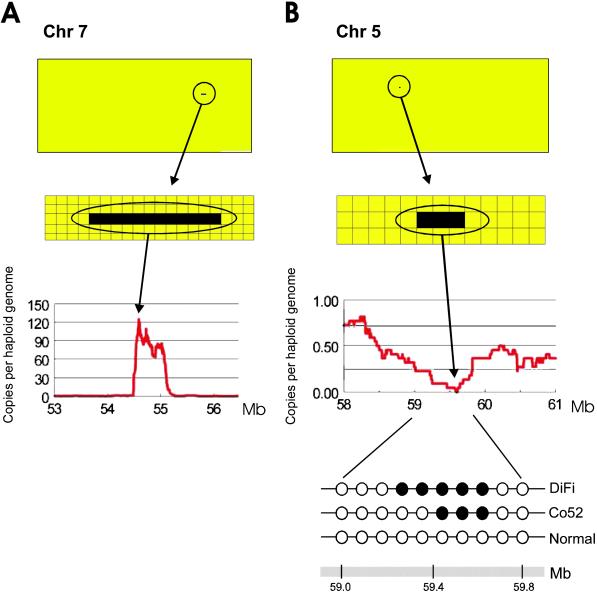
To identify amplifications, which typically involve regions much smaller than a chromosomal arm, average tag densities were dynamically calculated and visualized over sliding windows of different sizes. Although some relatively small alterations could be detected by using a 1,000 virtual tag window (Fig. 2), a window size of 50 virtual tags (≈200 kb) was used for the detailed analyses of amplifications because it would be expected to provide a relatively high resolution and sensitivity for experimental data consisting of ≈100,000 filtered tags (Table 1). To visualize small alterations, we designed a bitmap-based viewer that allowed much higher resolution views than were possible with the standard chromosome maps such as commonly used for CGH. By using this strategy, three amplification events were observed in the DiFi genome, whereas none were observed in the lymphoblastoid DNA (Table 3). The most striking was a 125-fold amplification located at position 54.54–55.09 Mb on chromosome 7p (Fig. 3A). Analysis of tags in this area resolved the boundaries of the amplified region to within 10 kb. Three genes were harbored within the amplicon, a predicted gene with no known function (DKFZP564K0822), the bacterial lantibiotic synthetase component C-like 2 (LANCL2) gene, and the epidermal growth factor receptor (EGFR) gene, an oncogenic tyrosine kinase receptor known to be amplified in DiFi cells (23). The second-highest amplification was a 6-fold change at position 30.36–32.72 Mb on chromosome 13 (Fig. 2). This area, containing eight genes, represents the apex of a broad region on 13q that is coamplified. Finally, a <300-kb region within 2 Mb of the telomere of chromosome 20q appeared to be increased >5-fold. Independent evaluation of the 7p, 13q, and 20q amplified regions by using quantitative PCR analyses of genomic DNA from DiFi cells revealed copy number gains similar to those observed by digital karyotyping (Table 3). CGH underestimated the fold amplification on 13q (Fig. 2). More importantly, CGH completely failed to identify the amplification of chromosome 7p and 20q because the <0.5-Mb amplicons were below the level of resolution achievable with this technique.
Table 3.
Quantitative analysis of amplifications and deletions
| Type of alteration
|
Location
|
Copy number | |
|---|---|---|---|
| Digital karyotyping | Quantitative PCR | ||
| Amplifications | Chromosome 7: 54.54–55.09 Mb | 125 | 139 |
| Chromosome 13: 30.36–32.72 Mb | 6.4 | 5.7 | |
| Chromosome 20: 60.54–60.83 Mb | 5.4 | 2.8 | |
| Deletions | Chromosome 18: 49.34–51.67 Mb | 0 | 0 |
| Chromosome 5: 59.18–59.92 Mb | 0 | 0 | |
| Chromosome X: 106.44–107.25 Mb | 0 | 0.4 | |
Copy number values are calculated per haploid genome as described in Materials and Methods.
Fig 3.
High-resolution tag density maps identify amplifications and deletions. (A) Amplification on chromosome 7. (Top) A bitmap viewer with the region containing the alteration encircled. The bitmap viewer is comprised of ≈39,000 pixels representing tag density values at the chromosomal position of each virtual tag on chromosome 7, determined from sliding windows of 50 virtual tags. Yellow pixels indicate tag densities corresponding to copy numbers of <110, while black pixels correspond to copy numbers ≥110. (Middle) An enlarged view of the region of alteration. (Bottom) A graphical representation of the amplified region with values on the y axis indicating genome copies per haploid genome and values on the x axis representing positions along the chromosome in Mb. (B) Homozygous deletion on chromosome 5. Top, Middle, and Bottom are similar to those for A except that the bitmap viewer for chromosome 5 contains ≈43,000 pixels, tag density values were calculated in sliding windows of 150 virtual tags, and yellow pixels indicate copy numbers >0.1 while black pixels indicate copy numbers ≤0.1. (Bottom) Below the graph is a detailed analysis of the region containing the homozygous deletion in DiFi and Co52. For each sample, white dots indicate markers that were retained, while black dots indicate markers that were homozygously deleted. PCR primers for each marker are listed in Table 4.
Analysis of Deletions.
When a homozygous deletion occurs in a cancer cell, there are zero copies of the deleted sequences, compared with two copies in normal cells. This difference is far less than that observed with amplifications, wherein 10–200 copies of the involved sequences are present in cancer cells compared with two copies in normal cells. Detection of homozygous deletions was therefore expected to be more difficult than the detection of amplifications. To assess the potential for detecting deletions, we first performed digital karyotyping on DNA from a cancer cell line (Hx48) known to have a homozygous deletion encompassing the SMAD4 and DCC genes on chromosome 18q (24). From a library of ≈116,000 filtered tags, we were able to clearly identify this deletion on chromosome 18 (Table 3). The size of this deletion was estimated to be 2.33 Mb from digital karyotyping and 2.48 Mb from PCR-based analysis of markers in the region.
We next attempted to determine whether any deletions were present in DiFi cells. Using a window size of 150 virtual tags (600 kb), we found evidence for four homozygous deletions in the DiFi genome but none in the NLB cells. These apparent deletions were on chromosomes 4p, 5q, 16q, and Xq, and were 782, 743, 487, and 814 kb in size, respectively. Assessment of the regions on 4p and 16q by quantitative PCR did not confirm the deletions, either because they were located between the markers used for PCR analyses or because there were no genuine homozygous deletions. This latter possibility was not unexpected, given the positive predicted value (PPV) estimated for a window size of 150 virtual tags (Table 1) and the expectation that the PPV would be even lower in an aneuploid genetic background. However, similar analyses did confirm the homozygous deletion at the 5q locus and showed a substantial reduction in genomic content at the chromosome X region in DiFi DNA (Fig. 3B; Table 3). Neither of these deletions was detected by conventional CGH analysis (Fig. 2). Further examination of the 5q locus by sequence-tagged site (STS) mapping demonstrated that the homozygous deletion was completely contained within the 59.18–59.92 Mb area identified by digital karyotyping and was ≈450 kb in size (Fig. 3B). Analysis of 180 additional human colorectal tumors revealed an additional cell line (Co52) with an ≈350-kb homozygous deletion of the same region, suggesting the existence of a previously unknown tumor suppressor gene that may play a role in a subset of colorectal cancers.
Detection of Foreign DNA Sequences.
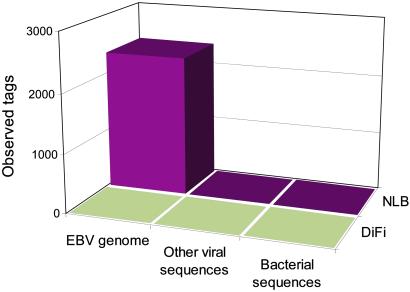
Digital karyotyping can, in principle, reveal sequences that are not normally present in human genomic DNA. The analysis of the library from NLB cells provided support for this conjecture. Like all lymphoblastoid lines, the NLB cells were generated through infection with Epstein–Barr virus (EBV) (25). EBV sequences persist in such lines in both episomal and integrated forms (26). To identify potential viral sequences in NLB cells, 210,245 unfiltered NLB tags were compared with virtual tags from the human genome, and to unfiltered DiFi tags. These comparisons yielded a subset of tags that had no apparent matches to the human genome and these were searched against virtual tags from all known viral or bacterial sequences. A total of 2,368 tags perfectly matched EBV or EBV-related primate herpes viruses, but no tags matched other viral or bacterial sequences (Fig. 4). Of the 100 virtual tags predicted to be found in the EBV genome, 94 (94%) were found among the NLB tags. A similar analysis of 171,795 unfiltered DiFi tags showed no matches to EBV or other microbial sequences (Fig. 4)
Fig 4.
Identification of EBV DNA in NLB cells. NLB, genomic tags derived from NLB cells after the removal of tags matching human genome sequences or tags matching DiFi cells. DiFi, genomic tags derived from DiFi cells after the removal of tags matching human genome sequences, or tags matching NLB cells. The number of observed tags matching EBV, other viral, or bacterial sequences is indicated on the vertical axis.
Discussion
Our data demonstrate that digital karyotyping can accurately identify regions whose copy number is abnormal, even in complex genomes such as that of humans. Whole chromosome changes, gains or losses of chromosomal arms, and interstitial amplifications or deletions were detected. All known genomic alterations in DiFi cells, including the amplification of EGFR on chromosome 7 and other gross chromosomal changes, were identified through digital karyotyping. Moreover, our analysis identified specific amplifications and deletions that had not been, to our knowledge, previously described by CGH or other methods in any human cancer. These analyses suggest that a potentially large number of undiscovered copy number alterations exist in cancer genomes and that many of these could be detected through digital karyotyping.
Like all genome-wide analyses, digital karyotyping has limitations. First, the ability to measure tag densities over entire chromosomes depends on the accuracy and completeness of the genome sequence. Fortunately, >94% of the human genome is available in draft form, and 95% of the sequence is expected to be in a finished state by the year 2003. Second, a small number of areas of the genome are expected to have a lower density of mapping enzyme restriction sites and could be incompletely evaluated by our approach. We estimate that <5% of the genome would be incompletely analyzed by using the parameters used in the current study. Moreover, this problem could be overcome through the use of different mapping and fragmenting enzymes. Finally, digital karyotyping cannot reliably detect very small regions, on the order of several thousand base pairs or less, that are amplified or deleted.
Nevertheless, it is clear from our analyses that digital karyotyping provides a heretofore unavailable picture of the DNA landscape of a cell. The approach should be immediately applicable to the analysis of human cancers, wherein the identification of homozygous deletions and amplifications has historically revealed genes important in tumor initiation and progression. In addition, one can envisage a variety of other applications for this technique. First, the approach could be used to identify previously undiscovered alterations in hereditary disorders. A potentially large number of such diseases are thought to occur because of deletions or duplications too small to be detected by conventional approaches. These diseases may be detectable with digital karyotyping, even in the absence of any linkage or other positional information. Second, mapping enzymes that are sensitive to DNA methylation (e.g., NotI) could be used to catalog genome-wide methylation changes in cancer, or diseases thought to be affected by genomic imprinting. Third, the approach could be as easily applied to the genomes of other organisms to search for genetic alterations responsible for specific phenotypes, or to identify evolutionary differences between related species. Finally, as the genome sequences of increasing numbers of microorganisms and viruses become available, the approach could be used to identify the presence of pathogenic DNA in infectious or neoplastic states. Our detection of EBV sequences through the digital karyotyping of NLB DNA provides proof of principle for this application.
Note Added in Proof.
Dunn et al. have independently reported a genomic tag-based method that can be used to analyze bacterial genomes (27).
Supplementary Material
Acknowledgments
We thank Bruce Boman for generously providing the DiFi cell line. This work was supported by the Benjamin Baker Scholarship Fund, the Clayton Fund, National Institutes of Health Grants CA 43460, CA 57345, and CA 6292, and Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie Grant NGNF KB P06T5.
Abbreviations
CGH, comparative genomic hybridization
EBV, Epstein–Barr virus
SAGE, serial analysis of gene expression
References
- 1.Vogelstein B. & Kinzler, K. W., (2002) The Genetic Basis of Human Cancer (McGraw–Hill, New York).
- 2.Scriver C. R., Beaudet, A. L., Sly, W. S. & Valle, D., (2001) The Metabolic and Molecular Bases of Inherited Disease (McGraw–Hill, New York).
- 3.Kallioniemi A., Kallioniemi, O. P., Sudar, D., Rutovitz, D., Gray, J. W., Waldman, F. & Pinkel, D. (1992) Science 258, 818-821. [DOI] [PubMed] [Google Scholar]
- 4.Lisitsyn N., Lisitsyn, N. & Wigler, M. (1993) Science 259, 946-951. [DOI] [PubMed] [Google Scholar]
- 5.Schrock E., du Manoir, S., Veldman, T., Schoell, B., Wienberg, J., Ferguson-Smith, M. A., Ning, Y., Ledbetter, D. H., Bar-Am, I., Soenksen, D., et al. (1996) Science 273, 494-497. [DOI] [PubMed] [Google Scholar]
- 6.Speicher M. R., Gwyn Ballard, S. & Ward, D. C. (1996) Nat. Genet. 12, 368-375. [DOI] [PubMed] [Google Scholar]
- 7.Solinas-Toldo S., Lampel, S., Stilgenbauer, S., Nickolenko, J., Benner, A., Dohner, H., Cremer, T. & Lichter, P. (1997) Genes Chromosomes Cancer 20, 399-407. [PubMed] [Google Scholar]
- 8.Pinkel D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., Zhai, Y., et al. (1998) Nat. Genet. 20, 207-211. [DOI] [PubMed] [Google Scholar]
- 9.Pollack J. R., Perou, C. M., Alizadeh, A. A., Eisen, M. B., Pergamenschikov, A., Williams, C. F., Jeffrey, S. S., Botstein, D. & Brown, P. O. (1999) Nat. Genet. 23, 41-46. [DOI] [PubMed] [Google Scholar]
- 10.Cai W. W., Mao, J. H., Chow, C. W., Damani, S., Balmain, A. & Bradley, A. (2002) Nat. Biotechnol. 20, 393-396. [DOI] [PubMed] [Google Scholar]
- 11.Knuutila S., Bjorkqvist, A. M., Autio, K., Tarkkanen, M., Wolf, M., Monni, O., Szymanska, J., Larramendy, M. L., Tapper, J., Pere, H., et al. (1998) Am. J. Pathol. 152, 1107-1123. [PMC free article] [PubMed] [Google Scholar]
- 12.Knuutila S., Aalto, Y., Autio, K., Bjorkqvist, A. M., El-Rifai, W., Hemmer, S., Huhta, T., Kettunen, E., Kiuru-Kuhlefelt, S., Larramendy, M. L., et al. (1999) Am. J. Pathol. 155, 683-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter N. J. (2001) Semin. Pediatr. Neurol. 8, 135-146. [DOI] [PubMed] [Google Scholar]
- 14.Hodgson G., Hager, J. H., Volik, S., Hariono, S., Wernick, M., Moore, D., Nowak, N., Albertson, D. G., Pinkel, D., Collins, C., et al. (2001) Nat. Genet. 29, 459-464. [DOI] [PubMed] [Google Scholar]
- 15.Gray J. W. & Collins, C. (2000) Carcinogenesis 21, 443-452. [DOI] [PubMed] [Google Scholar]
- 16.Snijders A. M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A. K., Huey, B., Kimura, K., et al. (2001) Nat. Genet. 29, 263-264. [DOI] [PubMed] [Google Scholar]
- 17.Velculescu V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484-487. [DOI] [PubMed] [Google Scholar]
- 18.Saha S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508-512. [DOI] [PubMed] [Google Scholar]
- 19.Speicher M. R., Prescher, G., du Manoir, S., Jauch, A., Horsthemke, B., Bornfeld, N., Becher, R. & Cremer, T. (1994) Cancer Res. 54, 3817-3823. [PubMed] [Google Scholar]
- 20.Olive M., Untawale, S., Coffey, R. J., Siciliano, M. J., Wildrick, D. M., Fritsche, H., Pathak, S., Cherry, L. M., Blick, M., Lointier, P., et al. (1993) In Vitro Cell. Dev. Biol. 29A, 239-248. [DOI] [PubMed] [Google Scholar]
- 21.Kinzler K. W., Nilbert, M. C., Vogelstein, B., Bryan, T. M., Levy, D. B., Smith, K. J., Preisinger, A. C., Hamilton, S. R., Hedge, P., Markham, A., et al. (1991) Science 251, 1366-1370. [DOI] [PubMed] [Google Scholar]
- 22.Platzer P., Upender, M. B., Wilson, K., Willis, J., Lutterbaugh, J., Nosrati, A., Willson, J. K., Mack, D., Ried, T. & Markowitz, S. (2002) Cancer Res. 62, 1134-1138. [PubMed] [Google Scholar]
- 23.Dolf G., Meyn, R. E., Curley, D., Prather, N., Story, M. D., Boman, B. M., Siciliano, M. J. & Hewitt, R. R. (1991) Genes Chromosomes Cancer 3, 48-54. [DOI] [PubMed] [Google Scholar]
- 24.Thiagalingam S., Lengauer, C., Leach, F. S., Schutte, M., Hahn, S. A., Overhauser, J., Willson, J. K., Markowitz, S., Hamilton, S. R., Kern, S. E., et al. (1996) Nat. Genet. 13, 343-346. [DOI] [PubMed] [Google Scholar]
- 25.Pelloquin F., Lamelin, J. P. & Lenoir, G. M. (1986) In Vitro Cell. Dev. Biol. 22, 689-694. [DOI] [PubMed] [Google Scholar]
- 26.Cho M. S. & Tran, V. M. (1993) Virology 194, 838-842. [DOI] [PubMed] [Google Scholar]
- 27.Dunn J. J., McCorkle, S. R., Praissman, L. A., Hind, G., van der Lelie, D., Bahou, W. F., Gnatenko, D. V. & Krause, M. K. (2002) Genome Res. 12, 1756-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.