Abstract
HIV entry inhibitors include coreceptor antagonists and the fusion inhibitor T-20. T-20 binds the first helical region (HR1) in the gp41 subunit of the viral envelope (Env) protein and prevents conformational changes required for membrane fusion. HR1 appears to become accessible to T-20 after Env binds CD4, whereas coreceptor binding is thought to induce the final conformational changes that lead to membrane fusion. Thus, T-20 binds to a structural intermediate of the fusion process. Primary viruses exhibit considerable variability in T-20 sensitivity, and determinants outside of HR1 can affect sensitivity by unknown mechanisms. We studied chimeric Env proteins containing different V3 loop sequences and found that gp120/coreceptor affinity correlated with T-20 and coreceptor antagonist sensitivity, with greater affinity resulting in increased resistance to both classes of entry inhibitors. Enhanced affinity resulted in more rapid fusion kinetics, reducing the time during which Env is sensitive to T-20. Reduced coreceptor expression levels also delayed fusion kinetics and enhanced virus sensitivity to T-20, whereas increased coreceptor levels had the opposite effect. A single amino acid change (K421D) in the bridging sheet region of the primary virus strain YU2 reduced affinity for CCR5 and increased T-20 sensitivity by about 30-fold. Thus, mutations in Env that affect receptor engagement and membrane fusion rates can alter entry inhibitor sensitivity. Because coreceptor expression levels are typically limiting in vivo, individuals who express lower coreceptor levels may respond more favorably to entry inhibitors such as T-20, whose effectiveness we show depends in part on fusion kinetics.
HIV enters cells by membrane fusion mediated by the envelope (Env) glycoprotein (reviewed in ref. 1). Env is a homotrimer in which each subunit contains surface gp120 and transmembrane gp41 glycoproteins (2). CD4 binding induces conformational changes in gp120 that allow binding to a cellular coreceptor, generally either the CCR5 or CXCR4 chemokine receptors (3, 4). Coreceptor binding is thought to trigger exposure of the hydrophobic fusion peptide at the amino terminus of gp41, which inserts into the membrane of the host cell (1). The fusion peptide and transmembrane domain of each gp41 subunit are then brought into close proximity by interactions between two helical regions (HR1 and HR2) in the ectodomain of gp41. The HR1 domain forms a triple-stranded coiled-coil with the adjoining HR1 domains in each gp41 subunit, whereas the HR2 domains bind with high affinity to grooves on the outside of the triple-stranded coiled-coil, resulting in a six-helix bundle in which the fusion peptides and transmembrane domains are at the same end of the molecule (5, 6). The change in free energy associated with formation of the six-helix bundle is thought to provide the force needed to elicit a fusion pore (7).
Inhibitors that block HIV entry are under clinical development, including small molecule antagonists that target CCR5 or CXCR4 (reviewed in ref. 8) and the fusion inhibitor T-20 that can reduce viral load by 1–2 logs in vivo (9, 10). T-20 is a peptide based on the sequence of the HR2 domain in gp41 and inhibits fusion by binding to the HR1 domain of gp41, preventing six-helix bundle formation. CD4 binding appears to make Env sensitive to T-20, whereas coreceptor binding triggers formation of the six-helix bundle, at which point T-20 can no longer bind (7, 11). Thus, T-20 targets a structural intermediate of the fusion process and factors that impact the kinetics of membrane fusion might affect T-20 sensitivity.
Mutations in the HR1 region of gp41 can affect viral sensitivity to T-20, presumably by altering the affinity of T-20 for HR1 (12, 13). Changing the V3 loop in otherwise isogenic viruses can also modulate T-20 sensitivity (14, 15). Furthermore, primary virus strains exhibit considerable variability in their sensitivity to T-20, with determinants outside of the HR1 domain being responsible for these differences in some cases.¶,∥ How changes in gp120 impact T-20 sensitivity is not obvious, nor is it known whether viral resistance to T-20 in vivo will involve mutations outside of HR1. To investigate the mechanism by which alterations in gp120 sequence impact T-20 sensitivity, we studied Env chimeras bearing different V3-loop sequences as well as the impact of a mutation in the bridging sheet region of a primary R5 virus Env that reduces gp120 affinity for CCR5 (14–16). We found that Envs that bound to coreceptor with high affinity were more resistant to T-20 than those that bound to coreceptor with reduced affinities. Coreceptor affinity also correlated with sensitivity of these viruses to the coreceptor antagonist TAK-779. Mechanistically, we found that increased coreceptor affinity resulted in faster fusion kinetics. Because fusion is a cooperative process requiring multiple Env trimers and coreceptor binding events, we propose that enhanced coreceptor affinity accelerates formation of the six-helix bundles, reducing the kinetic window during which Env is sensitive to T-20. Our finding that coreceptor expression levels also influenced sensitivity to fusion inhibitors and fusion kinetics is consistent with this hypothesis. Thus, receptor expression levels and Env/receptor affinity are cellular and viral determinants, respectively, that impact viral sensitivity to different classes of entry inhibitors. Therefore, mutations that result in drug resistance may do so directly by altering inhibitor binding sites or indirectly by affecting the rate of membrane fusion. Individuals who express lower levels of CCR5, such as Δ32-CCR5 heterozygotes, may consequently respond more favorably to T-20, and viruses that exhibit enhanced affinity for coreceptor may respond less well.
Materials and Methods
Cells.
QT6, 293T, U87/CD4, U87/CD4/CXCR4, U87/CD4/CCR5, NP2/CD4, 3T3/CD4/CCR5, and HeLa cell lines were cultured in DMEM supplemented with 10% FCS, 60 μg/ml penicillin, 100 μg/ml streptomycin (DMEM/10/PS) and G418 or puromycin where appropriate. T-REx/CCR5 cells, which allow tetracycline-regulated expression of CCR5, were generated by transfecting the T-REx cell line (Invitrogen) with the pcDNA4/TO mammalian expression vector (Invitrogen) encoding CCR5. Cells were maintained in DMEM/10/PS supplemented with 200 μg/ml zeocin and 5 μg/ml blasticidin to maintain ccr5 and Tet-repressor genes, respectively. Variable levels of CCR5 expression were induced by addition of different concentrations (0.1–100 ng/ml) of doxycycline (Sigma) to the culture medium. CCR5 expression levels were determined by flow cytometric analysis of cells immunostained with a phycoerythrin-conjugated CCR5-specific antibody (PharMingen).
Plasmids.
Env genes from NLHX, NLHXSF162-V3, and NLHXADA-V3B proviral clones (16) (provided by L. Ratner, Washington University School of Medicine, St. Louis) were excised by SalI/XhoI restriction enzyme digestion and cloned into the XhoI site of the pSI mammalian expression vector (Promega) to generate gp160 expression constructs. YU2 Env was cloned into the pCI expression construct (Promega). Stop codons were introduced at the gp120/gp41 cleavage junctions of NLHX, NLHXSF162-V3, and NLHXADA-V3B gp160 expression constructs by using the Quikchange site-directed mutagenesis kit to generate gp120 expression constructs. The YU2 K421D mutant was also generated by using the Quikchange site-directed mutagenesis kit.
Cell/Cell Fusion and Virus Infection Assays.
QT6 “effector” cells, transfected with Env expression plasmids and infected with a vaccinia virus encoding T7 polymerase (vTF1.1) (17), were added to QT6 “target” cells cotransfected with a luciferase reporter construct under the control of a T7 promotor (T7-luc; Promega), CD4 and CCR5, CXCR4 or pcDNA3.1 (control) expression plasmids. Cell/cell fusion, resulting from a functional interaction between Env-expressing effector cells and receptor-expressing target cells, was detected by assaying for T7 polymerase driven luciferase expression. This assay has been described in detail (18). Luciferase reporter pseudotype viruses bearing NLHX, NLHXSF162-V3, NLHXADA-V3B, and BaL Envs were generated by cotransfection of 293T cells with gp160 and pNL-luc expression constructs as described (19, 20). Viruses were titrated on U87/CD4, U87/CD4/CCR5, and U87/CD4/CXCR4 cells, which were assayed for luciferase expression 2–3 days postinfection, to determine coreceptor use and virus titer.
Env/Receptor Binding Assays.
gp120s were produced from 293T cells transfected with gp120 expression constructs and infected with vTF1.1 vaccinia virus encoding T7 polymerase to drive expression from the T7 promotor. Cell culture supernatants were harvested 24 h posttransfection/infection, and gp120 concentrations were determined by ELISA as described (21) with the exception that gp120 was detected with an HIV-1 Env-specific rabbit sera and an horseradish peroxidase-conjugated anti-rabbit antibody (Amersham Pharmacia Life Science) followed by 3,3′,5,5′-tetramethyl-benzidine substrate (Kirkegaard & Perry Laboratories). Receptor binding efficiencies of gp120s were determined by using a cell surface binding assay in which bound protein is detected by Western blot analysis (22). We also measured gp120/cell surface receptor binding by immunostaining and flow cytometry analysis as described (21) by using CD4-negative T-REx/CCR5 cells induced to express high levels of CCR5, or NP2/CD4 cells, with and without 5 μg/ml sCD4 to induce coreceptor binding site exposure or as a specificity control to compete for cell surface CD4 binding. TAK-779 was included as a further specificity control for CCR5 binding. Bound gp120 was detected with an HIV Env-specific rabbit serum and a phycoerythrin-conjugated anti-rabbit antibody (PharMingen).
Env Fusion Kinetics.
The fusion kinetics of NLHXSF162-V3 and NLHXADA-V3B Envs were determined in a dye transfer content mixing assay (11). QT6 effector cells were transfected with Env expression plasmids and infected with vTF1.1 to drive expression. Twenty four hours posttransfection, effector cells were labeled with 10 μM calcein acetoxymethyl ester (AM) (Molecular Probes) and 3T3/CD4/CCR5 target cells were labeled with 20 μM 5- and 6-([(4-chloromethyl)benzoyl]-amino) tetramethylrhodamine (CMTMR; Molecular Probes) for 1 h at 37°C. Effector cells were detached with cell dissociation buffer (GIBCO), washed, and cocultured for different periods of time with CMTMR-labeled target cells at 37°C. Dye redistribution was monitored microscopically (11) and the extent of fusion calculated as follows: percent fusion = 100 × number of fused cells positive for both dyes/number of bound or fused cells positive for CMTMR. The effect of CCR5 concentration on fusion kinetics was determined by using CD4-transfected T-REx/CCR5 target cells induced to express low, medium, or high levels of CCR5 and labeled with calcein AM. HeLa effector cells were infected with a recombinant vaccinia virus construct expressing BaL Env (VCB43; gift from Christopher Broder, Uniformed Services University of the Health Sciences, Bethesda) and labeled with CMTMR.
Results
Modifications in the V3 Loop Affect Coreceptor Use and T-20 Sensitivity.
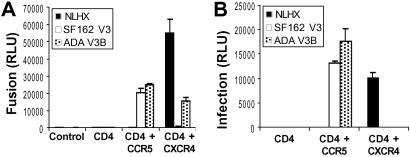
Replacement of the V3 loop of the X4 HIV-1 strain NLHX with the V3 loop of the R5 virus strain SF162 (NLHXSF162) results in a chimeric virus that uses CCR5 and is ≈6-fold more resistant to the fusion inhibitor T-20 (14, 15). When part of the NLHX V3 loop is replaced with the corresponding region of the R5 virus strain ADA (NLHXADA-V3B), CCR5 use as well as a modest 2- to 4-fold decrease in T-20 sensitivity was observed (14, 15). To confirm that these V3-loop modifications were solely responsible for altered T-20 sensitivity and to study these mutations in a context that enables measurement of fusion kinetics and receptor interactions, Env genes were excised from molecular clones of these viruses, inserted into the pSI expression plasmid, and assayed for functionality and coreceptor use in fusion and pseudotype virus infection assays. All Envs were expressed at equivalent levels and processed into gp120 and gp41 subunits with equal efficiency (data not shown). NLHX Env used CXCR4, but not CCR5, for fusion and infection, whereas NLHXSF162-V3 and NLHXADA-V3B used CCR5 (Fig. 1). NLHXADA-V3B retained partial X4 tropism for fusion but not for pseudotyped Env-mediated infection (Fig. 1).
Fig 1.
Coreceptor use of NLHX and NLHX V3 chimeric Envs. Coreceptor use of Envs determined in a cell/cell fusion luciferase reporter assay, using QT6 effector cells expressing Env and QT6 target cells expressing indicated receptors (control; pcDNA3.1-transfected cells) (A), and by infection using luciferase reporter pseudotype viruses to infect U87 cells expressing indicated receptors (B). Results, presented from representative experiments, are expressed in relative light units (RLU) ± SD, determined on replicate wells.
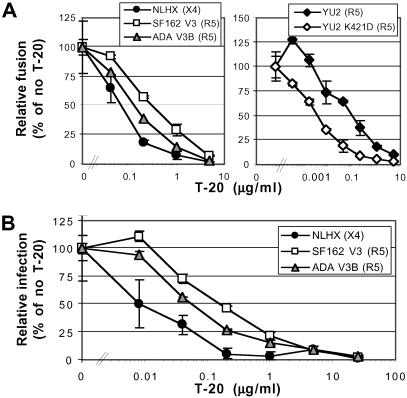
The T-20 sensitivity of this panel of Envs was measured by using cell/cell fusion and pseudotype virus infection assays, with results comparable to those obtained previously with replication-competent virus particles (14, 15). NLHX exhibited the greatest T-20 sensitivity, and NLHXADA-V3B had an intermediate T-20-sensitive phenotype, whereas NLHXSF162-V3 was most resistant to T-20 inhibition (Fig. 2). T-20 IC50 values for NLHX, NLHXADA-V3B, and NLHXSF162-V3 were 0.07, 0.13, and 0.31 μg/ml, respectively (equivalent to 16, 29, and 70 nM) for Env-mediated fusion and 0.009, 0.06, and 0.18 μg/ml (2, 13, and 40 nM), respectively, for pseudotype virus infection (Fig. 2). In humans, plasma concentrations of T-20 typically exceed 1 μg/ml (9, 10). Differences in IC50 values obtained from fusion and infection assays may be caused by differences in Env and/or receptor expression levels. Thus, changes in the V3 loop can alter sensitivity to T-20, even though this fusion inhibitor binds to gp41.
Fig 2.
T-20 sensitivity of NLHX, NLHXSF162-V3, NLHXADA-V3B, YU2, and YU2 K421D Envs. Relative T-20 sensitivities of Envs determined in a cell/cell fusion luciferase reporter assay, using QT6 effector cells expressing Env and QT6 target cells expressing CD4 and indicated coreceptors (A), and by infection using luciferase reporter pseudotype viruses to infect U87/CD4 cells expressing indicated coreceptors (B). Results are from a representative experiment ± SD determined on replicate wells.
V3-Loop Modifications Influence Virus Sensitivity to Coreceptor Antagonists.
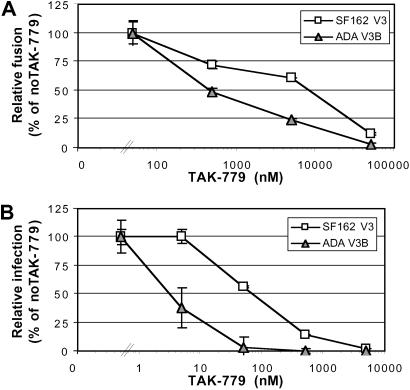
To determine whether variations in V3-loop sequence impact sensitivity of virus to coreceptor antagonists as well, we examined the sensitivity of NLHX, NLHXSF162-V3, and NLHXADA-V3B to inhibition by the CXCR4 and CCR5 antagonists AMD3100 and TAK-779. When cells expressing CD4 and CXCR4 were used, NLHXADA-V3B was 9-fold more sensitive to AMD3100 than the parental NLHX Env in a fusion inhibition assay (data not shown). This finding may reflect a weak interaction of ADAV3B with CXCR4 given that this receptor is unable to mediate infection of this virus (Fig. 1). When cells expressing CD4 and CCR5 were used, NLHXADA-V3B was ≈20-fold more sensitive to TAK-779 than NLHXSF162-V3 Env protein in fusion and infection assays (Fig. 3). An ≈2-log higher concentration of TAK-779 was required to block fusion compared with infection. Thus, modifications to the V3 loop can impact sensitivity to T-20 as well as to CCR5 and CXCR4 antagonists.
Fig 3.
TAK-779 sensitivity of NLHXSF162-V3 and NLHXADA-V3B Envs determined in a cell/cell fusion luciferase reporter assay, using QT6 effector cells expressing Env and QT6 target cells expressing CD4 and CCR5 (A), and by infection using luciferase reporter pseudotype viruses to infect U87/CD4/CCR5 cells (B). Results are representative of at least three experiments ± SD determined on replicate wells.
Coreceptor Binding Efficiency Correlates with Entry Inhibitor Sensitivity.
Differential sensitivity of NLHX and V3 chimeras to CCR5 or CXCR4 antagonists could result from differences in how each Env interacts with coreceptor. Alternatively, these varied sensitivities may result from altered coreceptor-binding affinities of these otherwise isogenic Envs. Relatively few Env proteins have been carefully tested for coreceptor affinity, with values ranging from 4 nM for an R5 Env (4, 23) to ≈200–500 nM in the case of lab-adapted X4 gp120 proteins (24, 25). This variability in Env/coreceptor binding constants, coupled with the fact that coreceptor expression levels can be limiting for virus infection both in vitro and in vivo (26, 27) and the requirement of multiple coreceptor binding events to support membrane fusion (28), prompted us to determine whether the V3-loop alterations studied here affected coreceptor affinity in a manner that would correlate with entry inhibitor sensitivity.
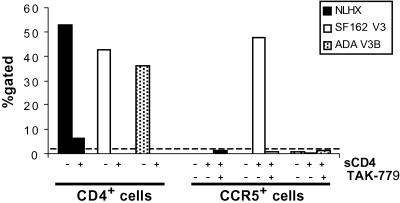
Stop codons were introduced into the NLHX, NLHXSF162-V3, and NLHXADA-V3B env genes at the gp120-gp41 cleavage junction to generate gp120 expression constructs. Equivalent amounts of gp120 proteins were examined for the ability to bind to cells expressing CD4, CCR5, or CXCR4. sCD4 was used to trigger the necessary conformational changes in gp120 to allow coreceptor binding. Bound gp120 was detected by SDS/PAGE and Western blot and also by immunostaining and flow cytometry analysis (data not shown and Fig. 4). Comparable results were obtained with both techniques. We used subsaturating amounts of gp120 so that differences in binding efficiencies could be differentiated. As a result, not all cells were in the positive gate. NLHX, NLHXSF162-V3, and NLHXADA-V3B gp120s all bound to cell surface expressed CD4 with similar efficiencies (Fig. 4). In addition, the NLHXSF162-V3 gp120 bound to cell surface expressed CCR5 after CD4 triggering and this binding could be blocked with the CCR5 antagonist TAK-779 (Fig. 4). In contrast, binding of NLHXADA-V3B to CCR5 (Fig. 4) or CXCR4 (data not shown) and NLHX to CXCR4 (data not shown) was undetectable in these equilibrium binding assays, indicating that the relative affinities of these proteins to their specific coreceptors is substantially reduced compared with NLHXSF162-V3 for CCR5.
Fig 4.
Receptor binding efficiencies of NLHX, NLHXSF162-V3, and NLHXADA-V3B Envs determined by immunostaining and flow cytometry analysis of gp120 binding to the surface of NP2/CD4 cells and T-REx/CCR5 cells. % gated, percentage of cells immunostained for bound Env. Dotted line, assay background. Results are representative of at least four experiments.
To further investigate the relationship between coreceptor affinity and T-20 sensitivity, we introduced a single amino acid change in the bridging sheet region (K421D) that reduces the affinity of the primary virus strain YU2 for CCR5 (29), a finding we confirmed by using our gp120 binding assays (data not shown). We found that the T-20 IC50 for YU2 was 0.1 μg/ml whereas the IC50 for YU2 K421D was 0.0033 μg/ml, a 30-fold difference (Fig. 2A). Thus, a single amino acid change in this primary virus strain reduces coreceptor affinity and enhances sensitivity to T-20.
Coreceptor Affinity Affects Membrane Fusion Kinetics.
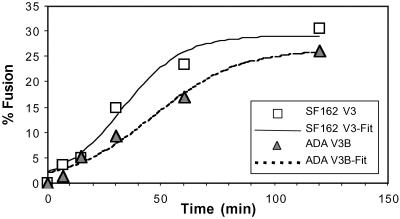
Because multiple coreceptor binding events are needed to support fusion (28), we hypothesized that enhanced coreceptor affinity would result in more rapid coreceptor binding and trigger formation of the six-helix bundle in gp41 more quickly, resulting in membrane fusion and also rendering Env resistant to the inhibitory effects of T-20. In contrast, reduced coreceptor affinity would delay fusion kinetics and maintain Env bound to CD4 in a T-20-sensitive state for a longer period. To monitor fusion kinetics, cells expressing CD4 and a coreceptor were labeled with CMTMR, and cells expressing Env were labeled with calcein AM. Dye redistribution was monitored at different times after cell mixing to measure the rate and extent of membrane fusion. We found that 2 h after mixing cells expressing NLHXSF162-V3 Env fused with CD4/CCR5-positive cells with approximately equivalent efficiency as cells expressing NLHXADA-V3B Env (Fig. 5; 31% and 27% of cell pairs fused, respectively). However, fusion mediated by NLHXSF162-V3 Env occurred more rapidly with half-maximal fusion observed at 33 min compared with 46 min for NLHXADA-V3B Env (Fig. 5). Thus, enhanced affinity for CCR5 correlated with an increased rate of membrane fusion and resistance to T-20. This finding is further supported by the observation that when T-20 was added to pseudotype infection cultures at various times postinfection then NLHXSF162-V3 infection was sensitive to inhibition by T-20 for a shorter period compared with NLHXADA-V3B infection (data not shown).
Fig 5.
Fusion kinetics of NLHXSF162-V3 and NLHXADA-V3B Envs determined in a dye transfer content mixing assay using QT6 effector cells expressing Env and labeled with calcein AM and 3T3/CD4/CCR5 target cells labeled with CMTMR. Results represent the average fusion observed in five fields photographed at ×10 magnification. T-distribution tests showed the rate differences to be statistically significant (P < 0.05).
Coreceptor Density Can Impact T-20 Sensitivity, TAK-779 Sensitivity, and Fusion Kinetics.
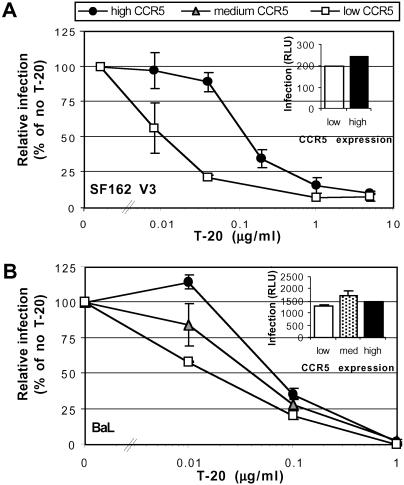
If coreceptor affinity impacts fusion kinetics and T-20 sensitivity, we reasoned that coreceptor expression levels might do so as well. If true, and because coreceptor expression levels are often limiting for virus infection of primary cell types (27), differences in coreceptor expression levels between individuals could help predict treatment success and in selecting optimal doses of entry inhibitors. To study the relationship between coreceptor expression levels, TAK-779 sensitivity, T-20 sensitivity and fusion kinetics, we used CD4+ T-REx/CCR5 cells that express CCR5 under control of a tetracycline-regulated promoter, making it possible to vary CCR5 expression levels by incubating cells with different concentrations of doxycycline. Cells expressing low, medium, or high levels of CCR5 as judged by flow cytometry analysis (mean fluorescent intensity of 14, 110, and 195, respectively) were infected with NLHXSF162-V3 or BaL Env virus pseudotypes in the presence of different concentrations of TAK-779 or T-20. In the absence of inhibitors, infection efficiencies were approximately the same regardless of coreceptor expression levels (Fig. 6 Insets). However, low CCR5 expression resulted in increased sensitivity of NLHXSF162-V3 and BaL pseudotypes to TAK-779 and T-20 inhibition (Fig. 6 and data not shown). Therefore, coreceptor density can impact both TAK-779 and T-20 sensitivity. To determine whether differences in coreceptor expression also affected fusion kinetics, we used a dye transfer assay. Cells expressing BaL Env protein were incubated with T-REx cells expressing either low, medium, or high levels of CCR5, and the rate of fusion was determined. Half-maximal fusion occurred at 102, 86, and 46 min for cells expressing low, medium, or high levels of CCR5 respectively (Table 1), with the differences between cells expressing low/medium and high levels of CCR5 being statistically significant. The percent of cell pairs fused was approximately equivalent for cells expressing low and medium levels of CCR5 and was higher for cells expressing high levels of CCR5 (Table 1). Thus, increased coreceptor expression levels resulted in more rapid membrane fusion, reducing the period that Env is sensitive to T-20. As a result, higher concentrations of T-20 were needed to inhibit virus infection.
Fig 6.
Impact of CCR5 expression on the T-20 sensitivity of NLHXSF162-V3 (A) and BaL (B) luciferase reporter pseudotype virus infection. (Insets) Absolute infection levels. RLU, relative light units.
Table 1.
Impact of CCR5 expression on fusion kinetics
| CCR5 level (MFI) | t1/2 (min) | Ymax | b | R2 |
|---|---|---|---|---|
| Low (13) | 103.2 ± 12.9 | 20.85 ± 1.85 | 0.51 ± 0.13 | 0.96 |
| Medium (127) | 85.8 ± 16.4 | 21.05 ± 2.12 | 0.65 ± 0.19 | 0.94 |
| High (1,132) | 46.2 ± 13.3 | 31.27 ± 3.4 | 0.49 ± 0.23 | 0.86 |
CCR5 level determined by flow cytometry. MFI, mean fluorescent intensity; Ymax, maximal fusion; b, exponential rate constant; R2, correlation coefficients of fitted curves.
Time of half-maximal fusion of BaL Env on cells expressing variable levels of CCR5. Fusion was assayed from 30 min to 5 h, and data were fitted to the equation: Y = Ymax/(1 + exp(−(t−t1/2)/b)). The coefficients extracted from these curves are presented.
Discussion
Several coreceptor antagonists and fusion inhibitors are currently in clinical development, and some significantly reduce virus load in infected individuals (9, 10).** However, a major challenge to the development of potent, broadly crossreactive entry inhibitors is that these agents target directly or indirectly the highly variable HIV-1 Env protein. It is therefore not surprising that there is often considerable variability in the sensitivity of virus strains to entry inhibitors. With increasing numbers of entry inhibitors entering clinical trials, it will be important to characterize cellular and viral determinants that govern entry inhibitor efficacy as this may help predict treatment success and guide inhibitor selection and dosage.
Our study reveals that Env/coreceptor affinity and coreceptor expression levels represent viral and cellular determinants, respectively, that can significantly impact entry inhibitor efficacy. In the case of T-20, these determinants operate through a common mechanism: by modulating the kinetics of membrane fusion. T-20 is in advanced clinical trials and has potent antiviral activity both in vitro and in infected individuals (9, 10). Nonetheless, there is considerable variability in the sensitivity of primary virus isolates to T-20, and drug-resistant viruses have been selected for in vitro and in vivo (12, 13). The mechanism by which T-20 operates is unique as it targets a structural intermediate of the fusion process. Its binding site in the HR1 region of gp41 becomes available only after Env binds to CD4. Multiple coreceptor binding events then enable several Env trimers (28) to undergo the final conformational changes that lead to fusion, with six-helix bundle formation occurring rapidly after a full repertoire of coreceptors are engaged. Membrane fusion occurs coincident with six-helix bundle formation or shortly thereafter (7). Thus, T-20 can target Env only during a kinetic window that appears to be opened by CD4 binding and closed by coreceptor engagement (11), and factors that influence the kinetics of this process would logically impact T-20 sensitivity.
Given the above working model for the mode of action of T-20, we sought to identify viral and cellular determinants that could impact T-20 sensitivity. Because T-20 binds to the HR1 domain of gp41, mutations in HR1 can affect T-20 sensitivity (12, 13). However, other determinants in Env can also influence T-20 sensitivity. The concentration of T-20 needed to inhibit primary virus isolates can vary by at least two logs and this can be independent of mutations in HR1.¶∥ Additionally, chimeric viruses containing V3 loops from various R5 tropic strains were about 2- to 6-fold more resistant to T-20 inhibition compared with the parental virus encoding a TCLA X4 tropic Env (14, 15), and a single amino acid change in the bridging sheet region of HIV-1 YU2 enhanced T-20 sensitivity by 30-fold. How mutations in the V3 loop, the bridging sheet, or other regions of Env outside of the HR1 domain influence viral sensitivity to an inhibitor that binds to HR1 is not immediately apparent. By using receptor binding assays and measuring fusion kinetics, we found that for the Envs tested these differences correlated with alterations in Env/coreceptor affinity, which in turn correlated with fusion kinetics. Envs that bound to coreceptor with high affinity fused more quickly than nearly identical Envs that bound to coreceptor with reduced affinity. The time during which Env was sensitive to T-20 could be modulated not only by Env/coreceptor affinity, but also by coreceptor density. Higher levels of coreceptor resulted in more rapid membrane fusion and increased resistance to T-20, whereas lower levels of coreceptor resulted in slower membrane fusion and enhanced sensitivity to T-20. Changes in coreceptor affinity and coreceptor expression could alter T-20 sensitivity between 2- and 30-fold, depending on assay conditions. We speculate that viruses that exhibit high affinity for coreceptor, or viruses in individuals who express high levels of coreceptor either naturally or because of immune activation, would more easily acquire resistance to T-20.
Our results have several implications for antiviral therapy. HIV-infected individuals who have one copy of the Δ32-ccr5polymorphism have a significant survival advantage over individuals who are WT at the CCR5 locus most likely due to a relatively modest reduction in ccr5 levels on CD4-positive T cells and macrophages (30–33). The efficiency of virus infection in vitro is clearly related to coreceptor density (26), and CCR5 is typically present at <10,000 copies on freshly isolated peripheral blood mononuclear cells, whereas more than 50,000 copies of CD4 are usually expressed (27). Because T-20 sensitivity can be modulated to a significant degree by CCR5 expression levels, we predict that individuals who express lower levels of CCR5 will on average respond better to T-20 than individuals with higher levels, and that higher doses of T-20 will be needed to achieve an adequate antiviral response in individuals who have relatively high levels of CCR5. Under the conditions used in our experiments, differences in CCR5 expression levels can alter the T-20 sensitivity of a given virus by ≈1 log. It will be important to determine to what extent differences in CCR5 expression, and perhaps even CXCR4 expression, on primary cell types influence T-20 sensitivity. If naturally occurring variations in CCR5 expression significantly impact T-20 sensitivity, it may be useful to genotype individuals for the Δ32-ccr5 polymorphism, and perhaps even to measure CCR5 levels to help predict treatment success. In addition, Envs with high affinity might be better able to use the low levels of CCR5 present on some primary cells, effectively broadening viral tropism, fusogenicity, and perhaps pathogenicity as well. Thus, the relationship between coreceptor expression levels and the affinity with which an individual's predominant virus type binds to coreceptor are likely to influence the effectiveness of entry inhibitors in vivo. It will be important to more fully characterize the range of affinities with which primary virus strains engage CCR5 and CXCR4 and to determine to what extent differences in affinity influence sensitivity to both fusion inhibitors and coreceptor antagonists.
The impressive ability of HIV to evolve in the face of both immunologic and pharmacologic selective pressures likely means that combination therapy will be needed to suppress viral replication sufficiently so as to prevent or delay evolution of resistant virus strains. Our results provide a theoretical basis for the design of clinical trials that use specific combinations of entry inhibitors. Coreceptor antagonists reduce coreceptor levels on the cell surface, either by directly inhibiting Env/coreceptor binding or by inducing coreceptor down-regulation. Reductions in coreceptor expression levels in a setting in which these levels are already limiting for virus infection will prolong fusion kinetics, keeping Env in a T-20-sensitive state for a longer period of time. Thus, the use of a coreceptor antagonist in conjunction with T-20 may act to additively or synergistically inhibit HIV infection. Indeed, the CXCR4 ligand AMD3100 and CCR5 ligand SCH-C have been shown to act synergistically with T-20 to inhibit infection of peripheral blood mononuclear cells in vitro (34, 35).
In summary, we have identified viral and cellular determinants that can significantly affect the ability of HIV to be inhibited by T-20 and coreceptor antagonists. A given virus can exhibit differential sensitivity to T-20 depending on the availability of coreceptor on the surface of a target cell. Because coreceptor levels are a limiting factor for virus infection in vivo, this finding has important implications for the use of fusion inhibitors such as T-20. Between virus strains, differences in T-20 sensitivity can result from mutations in the T-20 binding site in the HR1 domain or by modifications in gp120 that alter the affinity with which Env binds to coreceptor. Formally, we have shown that mutations in the V3 loop and in the bridging sheet can impact coreceptor binding constants, fusion kinetics, and T-20 sensitivity, but we anticipate that changes elsewhere in the viral Env that affect coreceptor binding will have the same effect. Together, receptor affinity and coreceptor expression levels modulate the kinetic window during which Env is sensitive to T-20, raising the possibility that prolonging this state through the use of coreceptor antagonists will make the application of entry inhibitors more effective and the evolution of multidrug resistant viruses more difficult.
Acknowledgments
We thank George Lin for advice on the Western blot cell-surface receptor-binding assay, Igor Sidorov for statistical analyses, and Melissa Sanchez for help generating T-REx/CCR5 cells. We also thank Lee Ratner for NLHX and V3 chimeric molecular clones, Ted Pierson for the YU2 Env expression construct, Chris Broder for VCB43, Trimeris for T-20, Takeda for TAK-779, AnorMED for AMD3100, and Hiroo Hoshino for NP2/CD4 cells. This work is supported by National Institutes of Health Grant AI40880, a Burroughs Wellcome Fund Translational Research Award, and an Elizabeth Glaser Scientist Award (to R.W.D.). S.P. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
Abbreviations
HR, helical region
Env, envelope
AM, acetoxymethyl ester
CMTMR, 6-([(4-chloromethyl)benzoyl]-amino)-tetramethylrhodamine
This paper was submitted directly (Track II) to the PNAS office.
Reeves, J., Puffer, B., Ahmad, N., Derdeyn, C., Sharron, M., Edwards, T., Carlin, D., Harvey, P., Pierson, T., Hunter, E. & Doms, R., Ninth Conference on Retroviruses and Opportunistic Infections, Feb. 24–28, 2002, Seattle, abstr. 82.
Heil, M., Decker, J., Sfakianos, J., Shaw, G., Hunter, E. & Derdeyn, C., Ninth Conference on Retroviruses and Opportunistic Infections, Feb. 24–28, 2002, Seattle, abstr. 392.
Reynes, J., Rouzie, R. R., Kanouni, T., Baillat, V., Baroudy, B., Keung, A., Hogan, C., Markowitz, M. & Laughlin, M., Ninth Conference on Retroviruses and Opportunistic Infections, Feb. 24–28, 2002, Seattle, abstr. 1.
References
- 1.Doms R. W. & Moore, J. P. (2000) J. Cell Biol. 151, F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Center R. J., Leapman, R. D., Lebowitz, J., Arthur, L. O., Earl, P. L. & Moss, B. (2002) J. Virol. 76, 7863-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trkola A., Dragic, T., Arthos, J., Binley, J. M., Olson, W. C., Allaway, G. P., Cheng-Meyer, C., Robinson, J., Maddon, P. J. & Moore, J. P. (1996) Nature 384, 184-187. [DOI] [PubMed] [Google Scholar]
- 4.Wu L., Gerard, N. P., Wyatt, R., Choe, H., Parolin, C., Ruffing, N., Borsetti, A., Cardoso, A. A., Desjardin, E., Newman, W., et al. (1996) Nature 384, 179-183. [DOI] [PubMed] [Google Scholar]
- 5.Weissenhorn W., Wharton, S. A., Calder, L. J., Earl, P. L., Moss, B., Aliprandis, E., Skehel, J. J. & Wiley, D. C. (1996) EMBO J. 15, 1507-1514. [PMC free article] [PubMed] [Google Scholar]
- 6.Chan D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263-273. [DOI] [PubMed] [Google Scholar]
- 7.Melikyan G. B., Markosyan, R. M., Hemmati, H., Delmedico, M. K., Lambert, D. M. & Cohen, F. S. (2000) J. Cell Biol. 151, 413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pohlmann S. & Doms, R. W. (2002) Curr. Drug Targets Infect. Disorders 2, 9-16. [DOI] [PubMed] [Google Scholar]
- 9.Kilby J. M., Lalezari, J. P., Eron, J. J., Carlson, M., Cohen, C., Arduino, R. C., Goodgame, J. C., Gallant, J. E., Volberding, P., Murphy, R. L., et al. (2002) AIDS Res. Hum. Retroviruses 18, 685-693. [DOI] [PubMed] [Google Scholar]
- 10.Kilby J. M., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302-1307. [DOI] [PubMed] [Google Scholar]
- 11.Gallo S. A., Puri, A. & Blumenthal, R. (2001) Biochemistry 40, 12231-12236. [DOI] [PubMed] [Google Scholar]
- 12.Wei X., Decker, J. M., Liu, H., Zhang, Z., Arani, R. B., Kilby, J. M., Saag, M. S., Wu, X., Shaw, G. M. & Kappes, J. C. (2002) Antimicrob. Agents Chemother. 46, 1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rimsky L. T., Shugars, D. C. & Matthews, T. J. (1998) J. Virol. 72, 986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derdeyn C. A., Decker, J. M., Sfakianos, J. N., Wu, X., O'Brien, W. A., Ratner, L., Kappes, J. C., Shaw, G. M. & Hunter, E. (2000) J. Virol. 74, 8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derdeyn C. A., Decker, J. M., Sfakianos, J. N., Zhang, Z., O'Brien, W. A., Ratner, L., Shaw, G. M. & Hunter, E. (2001) J. Virol. 75, 8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung C. S., Vander Heyden, N. & Ratner, L. (1999) J. Virol. 73, 8216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander W. A., Moss, B. & Fuerst, T. R. (1992) J. Virol. 66, 2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rucker J., Doranz, B. J., Edinger, A. L., Long, D., Berson, J. F. & Doms, R. W. (1997) Methods Enzymol. 288, 118-133. [DOI] [PubMed] [Google Scholar]
- 19.Chen B. K., Saksela, K., Andino, R. & Baltimore, D. (1994) J. Virol. 68, 654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connor R. I., Chen, B. K., Choe, S. & Landau, N. R. (1995) Virology 206, 935-944. [DOI] [PubMed] [Google Scholar]
- 21.Reeves J. D. & Schulz, T. F. (1997) J. Virol. 71, 1453-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edinger A. L., Blanpain, C., Kunstman, K. J., Wolinsky, S. M., Parmentier, M. & Doms, R. W. (1999) J. Virol. 73, 4062-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doranz B. J., Baik, S. S. & Doms, R. W. (1999) J. Virol. 73, 10346-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman T. L., Canziani, G., Jia, L., Rucker, J. & Doms, R. W. (2000) Proc. Natl. Acad. Sci. USA 97, 11215-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babcock G. J., Mirzabekov, T., Wojtowicz, W. & Sodroski, J. (2001) J. Biol. Chem. 276, 38433-38440. [DOI] [PubMed] [Google Scholar]
- 26.Platt E. J., Wehrly, K., Kuhmann, S. E., Chesebro, B. & Kabat, D. (1998) J. Virol. 72, 2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee B., Sharron, M., Montaner, L. J., Weissman, D. & Doms, R. W. (1999) Proc. Natl. Acad. Sci. USA 96, 5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhmann S. E., Platt, E. J., Kozak, S. L. & Kabat, D. (2000) J. Virol. 74, 7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzuto C. D., Wyatt, R., Hernandez-Ramos, N., Sun, Y., Kwong, P. D., Hendrickson, W. A. & Sodroski, J. (1998) Science 280, 1949-1953. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y., Paxton, W. A., Wolinsky, S. M., Neumann, A. U., Zhang, L., He, T., Kang, S., Ceradini, D., Jin, Z., Yazdanbakhsh, K., et al. (1996) Nat. Med. 2, 1240-1243. [DOI] [PubMed] [Google Scholar]
- 31.Dean M., Carrington, M., Winkler, C., Huttley, G. A., Smith, M. W., Allikmets, R., Goedert, J. J., Buchbinder, S. P., Vittinghoff, E., Gomperts, E., et al. (1996) Science 273, 1856-1862. [DOI] [PubMed] [Google Scholar]
- 32.Michael N. L., Chang, G., Louie, L. G., Mascola, J. R., Dondero, D., Birx, D. L. & Sheppard, H. W. (1997) Nat. Med. 3, 338-340. [DOI] [PubMed] [Google Scholar]
- 33.Stewart G. J., Ashton, L. J., Biti, R. A., Ffrench, R. A., Bennetts, B. H., Newcombe, N. R., Benson, E. M., Carr, A., Cooper, D. A. & Kaldor, J. M. (1997) AIDS 11, 1833-1838. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay C. L., Giguel, F., Kollmann, C., Guan, Y., Chou, T. C., Baroudy, B. M. & Hirsch, M. S. (2002) Antimicrob. Agents Chemother. 46, 1336-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tremblay C. L., Kollmann, C., Giguel, F., Chou, T. C. & Hirsch, M. S. (2000) J. Acquired Immune Defic. Syndr. 25, 99-102. [DOI] [PubMed] [Google Scholar]