Abstract
During cerebral cortical development, excitatory glutamatergic projection neurons are generated from neural stem cells intrinsic to the early embryonic cortical ventricular zone by a process of radial migration, whereas most inhibitory γ-aminobutyric acid (GABA)ergic interneurons and oligodendrocytes (OLs) appear to be elaborated from ventral forebrain stem cells that initially undergo tangential cortical migration before terminal lineage maturation. In contrast to the more compartmentalized developmental organization of the spinal cord, the generation of neurons and OLs from a common ventral forebrain stem cell would expose these cells to the sequential actions of ventral and dorsal gradient morphogens [sonic hedgehog (Shh) and bone morphogenetic proteins (BMPs)] that normally mediate opposing developmental programs. Here we report that Shh promotes GABAergic neuronal/OL lineage restriction of forebrain stem cells, in part, by activation of the basic helix–loop–helix transcription factors, Olig2 and Mash1. In mutant mice with a generalized defect in tangential cortical migration (Dlx1/2−/−), there is a profound and selective reduction in the elaboration of both cortical GABAergic neurons and OLs. Our studies further demonstrate that the sequential elaboration of cortical GABAergic neurons and OLs from common Shh-responsive ventral forebrain progenitors requires the spatial and temporal modulation of cortical BMP signaling by BMP ligands and the BMP antagonist, noggin, respectively. These findings suggest an integrative model for cerebral cortical GABAergic neuronal and OL lineage maturation that would incorporate the sequential contributions of the ventral and dorsal forebrain, and the potential role of regional developmental cues in modulating transcriptional codes within evolving neural lineage species.
There is an emerging consensus that two distinct developmental pathways are required for the generation of neuronal and glial lineage diversity within the evolving mammalian cerebral cortex (1–4). During early embryonic development, excitatory glutamatergic projection neurons are elaborated from neural stem cells after radial migration from the cortical ventricular zone (VZ). By contrast, more recent slice culture cell migration studies and additional analyses using mutant mice with defects in tangential cortical migration (Dlx1/2−/−) and in regional progenitor domain specification (Nkx2.1−/−) have demonstrated that most cortical inhibitory γ-aminobutyric acid (GABA)ergic interneurons are generated from stem cells located within the medial ganglionic eminence (MGE) and lateral ganglionic eminence (LGE) of the ventral forebrain after separate waves of tangential cortical migration (5). Although the majority of these migratory cells undergo rapid cortical dispersion and the elaboration of different GABAergic neuronal subtypes, a smaller proportion of these cells are retained within the late embryonic cortical subventricular zone (SVZ), where they sequentially give rise to GABAergic neurons and additional cells that become mitotically active during the phase of perinatal cortical gliogenesis (5). In fact, clonal analysis studies have suggested that the ventral forebrain may contain common GABAergic neuronal/oligodendrocyte (OL) progenitor cells (6), and in this scenario the mitotically active SVZ cells may therefore represent proliferating OL precursors. However, in these studies, the putative contribution of ventral forebrain GABAergic neuronal/OL progenitors to cerebral cortical neurogenesis and gliogenesis was inferred from the presence of Dlx-immunoreactivity in a subset of cortical OL species (6) rather than confirmed by the use of additional genetic or developmental analyses. In the current studies, we have first used comparative clonal analysis of premigratory stage ventral and dorsal forebrain and postmigratory stage cerebral cortical progenitors and complementary genetic analysis using Dlx1/2 mutant mice to demonstrate that the ventral forebrain does, in fact, contain common GABAergic neuronal/OL progenitors capable of giving rise to cerebral cortical GABAergic neurons and OLs.
The molecular mechanisms that orchestrate cerebral cortical neurogenesis and gliogenesis, particularly with regard to the distinctive contribution of the ventral forebrain tangential migratory pathway are poorly understood (2–4, 7, 8). Throughout the neuraxis, sonic hedgehog (Shh) and bone morphogenetic proteins (BMPs) are ventral and dorsal gradient morphogens, respectively (8–11). These soluble signaling molecules specify discrete regional progenitor domains by the concentration-dependent elaboration of contiguous pairs of homeodomain and basic helix–loop–helix (bHLH) transcription factors that exhibit mutually antagonistic interactions necessary to refine individual progenitor domains and to promote the timely elaboration of specific neuronal and glial subtypes (12). In the spinal cord, astrocytes are generated from dorsal domains where BMP signaling predominates, whereas OLs are generated from a circumscribed ventral progenitor domain that initially gives rise to motor neurons under the influence of specific levels of Shh signaling (10, 12). Within the motor neuron domain, Olig2, the only bHLH member of this elaborate progenitor code, is essential for linking a specific neuronal subtype to an OL specification program and for coordinating the acquisition of pan-neuronal traits and motor neuron subtype specification by promoting the expression of Neurogenin 2 (13–16). Neurogenin 2, in turn, limits the number of progenitor cell cycle divisions and initiates a downstream transcriptional cascade that reinforces the neuronal subtype specific developmental program. The later elaboration of OL species from this progenitor domain is mediated, in part, by the dorsal shift of Nkx2.2 into the Olig2+/Nkx2.2− domain through a mechanism that may involve transient Shh signaling from the floor plate (13–16). However, genetic and developmental analyses suggest that Olig1 rather than Nkx2.2 is required for OL specification, whereas Nkx2.2 is necessary for OL terminal maturation (17, 18). In addition, the neuronal/glial switch requires the extinction of Neurogenin2 before the expression of Olig1 within the Olig2+ progenitor domain (13–16). The role of soluble signaling molecules and regional progenitor transcriptional codes in orchestrating related developmental processes during forebrain lineage specification and maturation are less well defined.
The developmental organization of the forebrain differs from the spinal cord in several important respects. First, GABAergic neurons rather than motor neurons are coupled to OLs (6, 8). In addition, GABAergic neurons are known to express the bHLH transcription factor, Mash1 rather than Neurogenin2, and Mash1 persists in OL species after lineage specification (8, 19–22). Further, because Olig2 is known to couple specific neuronal subtypes to OLs (23, 24), and Olig2 and Mash1 are preferentially expressed within the ventral forebrain before tangential cortical migration (21, 22, 25), it is likely that Olig2 and Mash1 are expressed in a common GABAergic/OL progenitor before tangential cortical migration. Finally, initial ventral forebrain specification and tangential cortical migration would expose these bipotent progenitors to sequential ventral and dorsal gradient morphogens that normally mediate opposing developmental programs (8, 26). These cumulative observations suggest that Shh and BMPs may exhibit complementary and cooperative roles in cerebral cortical GABAergic neuronal and OL lineage elaboration. During nervous system development, regional neural stem cells give rise to differentiated progeny through a process of progressive lineage restriction and the formation of intermediate progenitor species that have different response profiles to environmental cues (27). Our current studies indicate that Shh is required for the generation of GABAergic neuronal/OL progenitors from ventral forebrain stem cells through the mediation of Olig2 and Mash1.
Prior studies from our laboratory and those of others have shown that BMP ligands are expressed within the early embryonic cerebral cortex at the time of GABergic neuronal lineage specification, whereas the BMP inhibitor, noggin, is selectively expressed within the late embryonic and perinatal subcortical white matter, the site of subsequent OL lineage specification and maturation (28–30). Additional studies from our laboratory have shown that BMP ligands inhibit the initial lineage specification but subsequently promote the migration and differentiation of early-born (glutamatergic) neurons from cortical VZ progenitor cells (28, 29). Further, BMP2 has been shown to promote the elaboration of striatal GABAergic neurons (31). Finally, we have demonstrated that OL lineage elaboration requires active inhibition of BMP signaling (28, 32). These cumulative observations suggest that spatiotemporal modulation of BMP signaling may be important for the sequential elaboration of cortical GABAergic neurons and OLs from lineage-restricted ventral forebrain progenitors initially specified by regional Shh signaling. Our current studies demonstrate that BMP2 and noggin potentiate the elaboration of GABAergic neurons and OLs, respectively, from ventral forebrain progenitor cells previously exposed to specific levels of endogenous Shh signaling, and also from premigratory stage dorsal forebrain cells that have undergone a dorsal-to-ventral fate switch in response to exogenous Shh signaling. Because differential modulation of BMP signaling may alter the profiles of expression of transcription factors involved in neural lineage decisions (7, 8), the precise developmental regulation of cortical BMP signaling molecules may be essential for orchestrating GABAergic neuronal and OL lineage elaboration by promoting changing profiles of transcriptional codes within sequential stage-specific progenitor species.
Materials and Methods
Neural Cell Cultures.
Dlx1/2 mutant mice were provided by J. L. R. Rubenstein (University of California, San Francisco). Timed pregnant CD1 and Dlx1/2 mutant mice were killed at embryonic days (E) 12.5 and 16.5. Tissue samples from the embryonic murine cortex, the MGE, and the LGE were carefully separated and mechanically dissociated. Primary cultures were generated by plating dissociated cells at clonal density (15–40 cells/cm2) in culture wells containing poly-d-lysine (PDL, 20 μg/ml)-coated glass coverslips and propagated in serum-free media (SFM) consisting of DMEM/F12 (Life Technologies, Grand Island, NY), l-glutamine (2 mM), penicillin (100 units/ml), N2 and B27 (Life Technologies) supplements, and basic fibroblast growth factor (bFGF, 1 ng/ml; Collaborative Biomedical Products, Bedford, MA). Primary cultures were normally propagated for 6 days in vitro (DIV), and subsequently fixed and processed for immunocytochemical analysis. For specific experimental conditions, the active N-terminal form of Shh (N-Shh, 1–250 ng/ml; Curis, Cambridge, MA), BMP2 (10 ng/ml; Genetics Institute, Cambridge, MA) or noggin (10 ng/ml; Regeneron Pharmaceuticals, Tarrytown, NY) were added at the time of initial plating. Self-renewing, multipotent stem cells were generated from E12.5 cerebral cortical cells by clonal expansion (5–15 clones per 35 cm2) in the absence of polycationic substrate in SFM containing bFGF (10 ng/ml) and heparin (2 μg/ml) and propagated for 3 DIV. For examination of the combinatorial effects of soluble factors (N-Shh, 50 ng/ml) and specific gene manipulation paradigms (Olig2 and Mash1, see below) on clonal expansion and on the profiles of lineage elaboration, Shh and/or the appropriate adenoviral vectors or antisense oligonucleotides were added concurrently at the time of initial FGF application. For examination of progenitor cell lineage potential, individual intact cellular clones were transferred to separate culture wells on PDL-coated glass coverslips and allowed to differentiate in SFM containing bFGF (1 ng/ml) for an additional 6 DIV.
Adenovirus Construction.
An adenoviral vector expressing both Olig2 and GFP under separate cytomegalovirus (CMV) promoters was constructed with the AdEasy system (33). Full-length Olig2 cDNA was generated by RT-PCR using the murine Olig2 cDNA sequence from GenBank (accession no. AB038697). All plasmids for the adenoviral construction were provided by B. Vogelstein (Johns Hopkins University, Baltimore). At the time of plating, progenitor culture were infected with control GFP-expressing and with Olig2/GFP-expressing adenoviruses. Optimal conditions were defined by a multiplicity of infection (MOI) of 50.
Antisense Oligonucleotide Inhibition of Gene Expression in Neural Progenitor Cells.
Complete inhibition of protein expression by phosphorothioate-modified antisense oligonucleotides was verified by using Western analysis. The following oligonucleotides (final concentration, 5 μM) were added at the time of plating and reintroduced when detectable levels of protein expression reemerged: Olig2 antisense: 5′-CGTCCGAGTCCATGGCCA-3′; Olig2 sense: 5′-TGGGCCATGGACTCGGACG-3′; Mash1 antisense; 5′-CCGGGCCGTACCTCTC-3′; Mash1 sense: 5′-GGCCCGGCATGGAGAG-3′.
Immunocytochemistry.
Cellular preparations were fixed and processed for immunofluorescence microscopy as described (32). Primary antibodies were diluted to optimal concentrations in PBS: nestin (Sigma), 1:1,000; β-tubulin (Sigma), 1:400; glutamate (Sigma), 1:2,000; GABA (Sigma), 1:2,500; GAD65 (Chemicon) 1:1,000; O4 supernatant from mouse hybridoma cell line (S. Pfeiffer, University of Connecticut), 1:5; Olig1 (R. Lu, D. Rowitch, and C. Stiles, Dana–Farber Cancer Institute, Boston) 1:100; Olig2 (H. Takebayashi, Okazaki National Institute, Okazaki, Japan, and M. Nakafuku, University of Tokyo, Tokyo), 1:200; glial fibrillary acidic protein (GFAP) (Sigma), 1:400; Mash1 (PharMingen), 1:250; Ngn1 (Chemicon), 1:100; Nkx2.2 (Developmental Studies Hybridoma Bank, Iowa City, IA), 1:250.
Statistical Analysis.
Clonal lineage composition was estimated by counting a minimum of 32 clones from five defined fields (cm2) from 24-well tissue culture plates and was verified a minimum of four times. Experimental values represent the proportion of total clones containing the appropriate lineage markers as a percentage with a 95% confidence interval. The absolute proportion of target clones between control and experimental conditions were compared by using χ2 analysis. Significant differences were considered at a P value <0.05.
Results and Discussion
Developmental and Genetic Evidence That Ventral Forebrain Progenitor Cells Give Rise to Cerebral Cortical GABAergic Neurons and OLs.
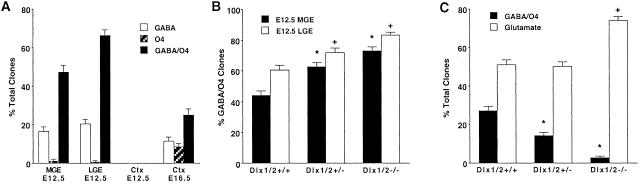
We first examined the ability of cultured progenitor cells derived from the early embryonic ventral and dorsal forebrain to give rise to GABAergic neurons and OL species. Before tangential cortical migration, individual MGE and LGE progenitors of the ventral forebrain but not age-matched cerebral cortical progenitors have the potential to generate clones containing both GABAergic neurons and OL species as well as pure GABAergic neuronal and OL clones (Fig. 1A). By contrast, after tangential migration from the MGE and the LGE, individual cerebral cortical progenitors develop the propensity to generate clones containing both GABAergic neurons and OL progenitors and also pure GABAergic neuronal and OL clones (Fig. 1A) containing cells derived from the ventral forebrain (Dlx+, not shown) (34). These observations suggest that after tangential cortical migration, individual progenitor cells derived from the ventral forebrain have the potential to give rise to both cortical GABAergic neurons as well as OLs. To further confirm the role of tangential cortical migration in the elaboration of cortical GABAergic neurons and OLs, we performed similar experimental paradigms using mutant mice that display a generalized defect in tangential cortical migration (Dlx1/2−/−). Previous studies have shown that Dlx1/2−/− mice have profound reductions in cortical GABAergic neurons, but the fate of OL species in cortical cells derived from these mutant mice have never been previously examined (5). Individual Dlx1/2−/− progenitors from the premigratory stage ventral forebrain displayed a significant increase in the elaboration of clones containing both GABAergic neurons and OL species (Fig. 1B). By contrast, there was a profound reduction in the elaboration of clones containing both GABAergic neurons and OLs and a corresponding significant increase in the generation of clones containing glutamatergic neurons from individual Dlx1/2−/− postmigratory stage cerebral cortical progenitors (Fig. 1C). Analogous postmigratory stage cortical cells from Dlx1/2+/− progenitors exhibited more limited reductions in the generation of GABAergic neurons and OLs (Fig. 1C), indicating Dlx gene dosage effects. Comparative analysis of perinatal (P0) tissue sections from Dlx1/2+/+ and Dlx1/2−/− mice confirmed the developmental stage-specific reduction in cortical GABAergic neurons and OL precursors (data not shown). These cumulative observations suggest that disruption in the process of tangential cortical migration is responsible for the profound impairment in the generation of cortical GABAergic neurons and OLs, and may result in compensatory increases in the proportion of glutamatergic neurons. These findings provide additional genetic evidence that cortical GABAergic neurons and OLs are derived from the ventral forebrain, and further suggest that a combinatorial transcriptional code is responsible for the generation of these specialized cortical neuronal and glial subtypes (8, 35).
Fig 1.
Before the stage of tangential cortical migration, individual MGE and LGE progenitors from the ventral forebrain but not the cerebral cortex give rise to both GABAergic neurons and OLs that populate the postmigratory stage cerebral cortex. (A) The proportion of total clones containing GABAergic neurons, OL species, or GABAergic neurons and OL species generated after propagation of primary E12.5 MGE and LGE and E12.5 and E16.5 cerebral cortical (Ctx) progenitors for 6 DIV. All data points represent the proportion of target clones with its 95% confidence interval. (B) The proportion of clones containing GABAergic neurons and OL species generated after propagation of primary E12.5 Dlx1/2+/+, Dlx1/2+/−, and Dlx1/2−/− MGE and LGE progenitors for 6 DIV. Statistical analysis was calculated by using χ2 analysis: *, P < 0.001 (compared with the E12.5 Dlx1/2+/+ MGE clonal condition); +, P < 0.001 (compared with the E12.5 Dlx1/2+/+ LGE clonal condition). (C) The proportion of total clones containing both GABAergic neurons and OL species or glutamatergic neurons generated after propagation of primary E16.5 Dlx1/2+/+, Dlx1/2+/−, and Dlx1/2−/− cerebral cortical progenitors for 6 DIV. Statistical analysis was calculated by using χ2 analysis: *, P < 0.001 (compared with the Dlx1/2+/+ GABA/O4 clonal condition); +, P < 0.001 (compared with the Dlx1/2+/+ glutamate clonal condition).
Shh Promotes GABAergic Neuronal/OL Lineage Restriction of Early Embryonic Forebrain Stem Cells and the Selective Induction of Transcription Factors Involved in GABAergic Neuronal and Oligodendroglial Lineage Specification and Maturation.
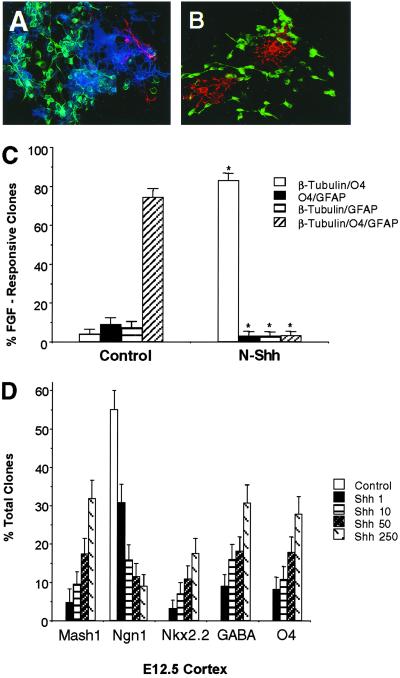
Previous studies have shown that the early embryonic cerebral cortex contains both epidermal growth factor (EGF)- and FGF-responsive stem cells (36). Because Shh is already expressed by the early gastrula stage and ventral forebrain Shh-responsive progenitors are initially derived from FGF-responsive stem cells (4, 8), we elected to examine the effects on neural lineage potential of naive dorsal forebrain FGF-responsive stem cells after exposure to Shh. When individual primary early embryonic cerebral cortical cells underwent clonal expansion in the presence of bFGF and subsequent cellular differentiation, they generated a large proportion of multipotent clones composed of neurons, OLs, and astrocytes (Fig. 2 A and C). By contrast, concurrent exposure of these cells to Shh resulted in the preferential generation of neuronal/OL clones at the expense of multipotent clones (Fig. 2 B and C). These observations suggest that the ventral gradient morphogen, Shh, promotes neuronal/OL lineage restriction of multipotent progenitors. Our studies have shown that before the stage of tangential cortical migration of ventral forebrain (MGE and LGE) progenitors, the cerebral cortex is incapable of generating OLs and GABAergic neurons (see Fig. 1A). Ventral forebrain progenitors are initially specified by the gradient morphogen, Shh, and regional dorsal-ventral patterning is mediated by the dynamic interplay between different classes of soluble signals (Shh and BMPs) and their downstream transcriptional targets (12, 37). Therefore, we next investigated whether exposure of premigratory stage cerebral cortical progenitors to Shh could induce a ventral-to-dorsal fate switch that would now allow these cells to give rise to GABAergic neurons and OL species whose sequential elaboration could be potentiated by changes in BMP signaling. Exposure of premigratory stage cortical progenitors to Shh resulted in the dose-dependent inhibition of the dorsal forebrain bHLH factor, Neurogenin1, induction of the ventral forebrain GABAergic neuronal bHLH factor, Mash1, and the OL homeodomain maturation factor, Nkx2.2 (Fig. 2D) and concomitant up-regulation of the bHLH GABAergic neuronal and OL lineage subtype factors, Olig2 and Olig1 (not shown). This ventral-to-dorsal fate switch in progenitor cell lineage effectors was also associated with the Shh-mediated dose-dependent elaboration of both GABAergic neurons and OL species but not astrocytes (Fig. 2D).
Fig 2.
Shh promotes neuronal/OL lineage restriction of early embryonic forebrain stem cells and the selective induction of transcription factors involved in GABAergic neuronal and OL lineage specification and maturation. (A and B) Immunofluorescence micrographs of individual differentiated clones derived from E12.5 cerebral cortical cells after clonal expansion under control (bFGF, 10 ng/ml, A) and N-Shh (bFGF+N-Shh, 50 ng/ml, B) conditions for 3 DIV, and subsequent plating as intact clonal populations under differentiating conditions for an additional 6 DIV. Note that FGF-responsive clones are multipotent (neurons: β-tubulin, FITC; OLs: O4, cascade blue; astrocytes: glial fibrillary acidic protein, tetramethylrhodamine B isothiocyanate), whereas FGF+Shh-responsive clones are comprised of more lineage-restricted progeny (neurons: β-tubulin, FITC; OLs:O4, tetramethylrhodamine B isothiocyanate). (C) Comparative clonal lineage profiles of bFGF- and bFGF+Shh-responsive E12.5 cortical progenitor species plated and propagated under culture conditions as noted above. All data points represent the proportion of target clones with its 95% confidence interval. Statistical analysis was calculated by using χ2 analysis: *, P < 0.001 (compared with individual clonal lineage profiles in control condition). (D) Comparative proportion of individual clones containing Mash1, Neurogenin1, Nkx2.2, GABA, and O4 derived from primary E12.5 cortical progenitors in the absence and presence of increasing concentrations of Shh after 6 DIV. Statistical analysis was calculated by using χ2 analysis: P < 0.001 [for all data points relative to their control (Ngn1), or Shh, 1 ng/ml (Mash1, Nkx2.2, GABA, O4) conditions].
Shh-Mediated Forebrain GABAergic Neuronal/OL Lineage Restriction Requires Olig2 and Mash1.
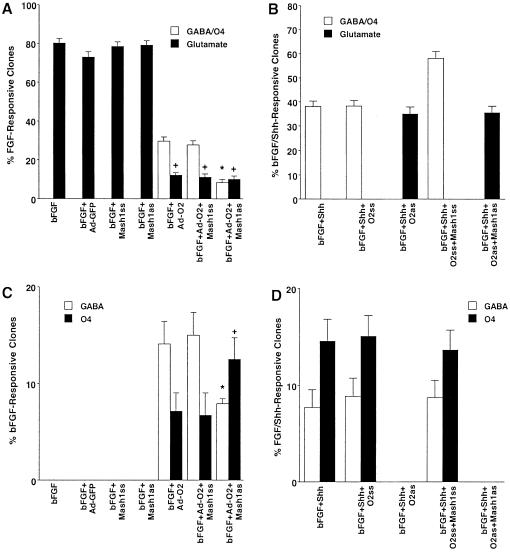
Previous studies have shown that Olig2 and Mash1 are expressed within the appropriate premigratory stage ventral forebrain progenitor domains that are exposed to endogenous levels of Shh signaling (21, 22, 25), and would therefore represent the potential sites of initial GABAergic neuronal/OL lineage restriction. In addition, these two bHLH proteins, but not Olig1 or Nkx2.2, are selectively localized to individual ventral forebrain progenitor species that give rise to cortical GABAergic neurons and OLs but not to FGF-responsive forebrain neural stem cells (S.-Y.Y., S.G., and M.F.M., unpublished observations). Therefore, to more directly examine the potential roles of Olig2 and Mash1 in Shh-mediated GABAergic neuronal/OL lineage restriction, we performed additional Olig2 and Mash1 gene manipulation paradigms. When early embryonic dorsal forebrain progenitors underwent clonal expansion in the presence of bFGF, they generated a large proportion of clones containing glutamatergic neurons but not clones containing GABAergic neurons and OL species (Fig. 3 A and C). By contrast, clonal expansion of dorsal forebrain progenitors in the presence of bFGF and Shh resulted in the generation of clones containing both GABAergic neurons and OL species as well as pure GABAergic neuronal and OL clones (Fig. 3 B and D) at the expense of clones containing glutamatergic neurons (Fig. 3B). Ectopic expression of Olig2 at the time of bFGF-mediated clonal expansion (Fig. 5A, which is published as supporting information on the PNAS web site, www.pnas.org) resulted in a conversion of the profile of differentiated clones to a pattern more similar to that seen in the bFGF and Shh clonal expansion condition (Fig. 3 A and B). Concurrent ablation of Mash1 gene expression further reduced the total complement of GABAergic neuronal/OL clones generated and altered the ratio of pure GABAergic neuronal to OL clones without additional changes in cellular viability (Fig. 3 A and C). Conversely, genetic ablation of Olig2 in the presence or absence of simultaneous inhibition of Mash1 gene expression (Fig. 5 B and C) at the time of bFGF- and Shh-mediated clonal expansion resulted in the conversion of the profile of differentiated clones to a pattern similar to that seen in the bFGF clonal expansion condition (Fig. 3 A and B). In addition, ectopic production of astrocytes was only evident in the combined Olig2 and Mash1 ablation condition (not shown), suggesting that cooperative interactions between these bHLH factors are required to actively inhibit the astroglial lineage during Shh-mediated neuronal/OL lineage restriction. These observations suggest that both Olig2 and Mash1 are required to promote Shh-mediated forebrain GABAergic neuronal/OL lineage restriction and exhibit complementary and cooperative factor interactions.
Fig 3.
Shh-mediated forebrain GABAergic neuronal/OL lineage restriction requires Olig2 and Mash1. (A) The comparative proportion of clones containing GABAergic neuronal/OL species and glutamatergic neurons generated from E12.5 FGF-responsive cortical progenitors after clonal expansion (2 DIV) in the absence or the presence of ectopic expression of Olig2 (Ad-O2) without or with concurrent ablation of Mash1 expression (Mash1as): Ad-GFP (adenoviral vector expressing GFP alone), Ad-O2 (adenoviral vector expressing Olig2 and GFP), Mash1ss (Mash1 sense oligonucleotides), and Mash1as (Mash1 antisense oligonucleotides). After clonal expansion in control and experimental conditions, intact clonal populations were plated under differentiating conditions for an additional 6 DIV. All data points represent the proportion of target clones and its 95% confidence interval. Statistical significance was calculated by using χ2 analysis: *, P < 0.001 (compared with GABA/OL clones in the bFGF+Ad-O2 clonal condition); +, P < 0.001 (compared with glutamatergic clones in the bFGF condition). (B) The comparative proportion of clones containing GABAergic neuronal/OL species and glutamatergic neurons generated from E12.5 FGF+Shh-responsive cortical progenitors in the absence or presence of ablation of Olig2 expression (Olig2as) without or with concurrent ablation of Mash1 expression (Mash1as). O2ss, Olig2 sense oligonucleotides; O2as, Olig2 antisense oligonucleotides. Control and experimental conditions were subjected to clonal expansion and differentiating conditions as described above. (C) The comparative proportion of clones containing GABAergic neurons or OL species generated under clonal culture paradigms identical to those described in A. Statistical significance was calculated by using χ2 analysis: *, P < 0.001 (compared with the bFGF+Ad-O2 GABA clonal condition); +, P < 0.001 (compared with the bFGF+Ad-O2 O4 clonal condition). (D) The comparative proportion of clones containing GABAergic neurons or OL species generated under clonal culture paradigms identical to those described in B.
Developmental Modulation of BMP Signaling Promotes the Sequential Elaboration of Cortical GABAergic Neurons and OLs from Shh-Responsive Forebrain Progenitors.
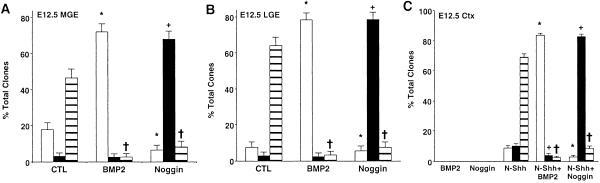
We and other groups have shown that BMP ligands inhibit the lineage specification but subsequently potentiate the migration and differentiation of early-born glutamatergic neurons from cortical VZ progenitors (28, 30, 32). By contrast, we have also demonstrated that coincident with the establishment of the cortical SVZ, BMP ligands now enhance the specification of late-born cortical (GABAergic) neurons and inhibit OL lineage elaboration (8, 32). Further, our studies have revealed that the BMP antagonist, noggin, promotes the generation of OLs and retards further neuronal lineage elaboration (8, 30, 32). These overall observations suggested that dynamic modulation of BMP signaling within the evolving cerebral cortex may regulate the elaboration of GABAergic neurons and OLs from MGE and LGE progenitor cells after tangential cortical migration. We therefore evaluated the ability of individual premigratory stage MGE and LGE progenitors to generate GABAergic neurons and OL species in response to changes in BMP signaling. Application of BMP2 to individual premigratory-stage MGE and LGE progenitors enhanced the generation of pure GABAergic neuronal clones at the expense of mixed GABAergic neuronal/OL clones, whereas exposure to noggin significantly enhanced the generation of pure OL clones also at the expense of mixed GABAergic neuronal/OL clones (Fig. 4 A and B). Our observations demonstrate that the actions of BMP2 are selective for GABAergic neurons (31). In addition, BMP ligands from the BMP2/4 factor subgroup but not from three additional BMP factor subgroups potentiated the elaboration of GABAergic neurons (S.-Y.Y., S.G., and M.F.M., unpublished observations). Different BMP ligands are expressed within the evolving cerebral cortex at the time of embryonic cortical neurogenesis, whereas noggin is selectively expressed in areas of the subcortical white matter during the late embryonic and perinatal phase of cortical OL lineage elaboration (28–30). These overall findings suggest that local modulation of BMP signaling within the evolving cerebral cortex may regulate the sequential elaboration of cortical GABAergic neurons and OLs after tangential cortical migration of ventral forebrain progenitors. Interestingly, coapplication of Shh and BMP2 or noggin to premigratory stage dorsal forebrain progenitors (Fig. 4C) now mimicked the effects of solitary application of BMP2 or noggin to ventral forebrain MGE and LGE progenitors (compare with Fig. 4 A and B) for promoting GABAergic neuronal and OL lineage elaboration, respectively. Further, the use of a neutralizing antibody to Shh confirms that endogenous Shh is not present when our clonal culture conditions are used (results not shown). Sequential exposure of these dorsal forebrain progenitors to Shh and then to BMP2 resulted in the initial induction and the subsequent inhibition of both Mash1 and Olig2 proteins (Fig. 6 A and B, which is published as supporting information on the PNAS web site). These observations suggest that exposure of early embryonic forebrain cells to Shh is required for the specification of progenitor cells capable of generating both cortical OLs and GABAergic neurons. Thereafter, tangential cortical migration is necessary to bring these lineage-restricted GABAergic neuronal/OL progenitors into contact with specific environmental signals (BMPs) whose dynamic modulation is required for the sequential developmental elaboration of cerebral cortical GABAergic neurons and OLs.
Fig 4.
Developmental modulation of BMP signaling promotes the differential elaboration of GABAergic neurons and oligodendrocytes from Shh-responsive forebrain progenitors. (A and B) The comparative proportion of clones containing GABAergic neurons (white bars), OL (O4) species (black bars), or GABAergic neurons and OL species (striped bars) generated from primary E12.5 MGE (A) or LGE (B) progenitors plated in the absence (control) or presence of BMP2 or noggin and propagated for 6 DIV. All data point represents the proportion of target clones and its 95% confidence interval. Statistical significance was calculated by using χ2 analysis: *, P < 0.001 (compared with GABA clonal control conditions); +, P < 0.05 (compared with O4 clonal control condition); †, P < 0.001 (compared with the GABA/O4 clonal control condition). (C) The comparative proportion of clones containing GABAergic neurons (white bars), OL species (black bars), or GABAergic neurons and OL species (striped bars) generated from primary E12.5 cortical (Ctx) progenitors plated in the absence or presence of N-Shh and in the absence or presence of BMP2 or noggin and propagated for 6 DIV. Statistical significance was calculated by using χ2 analysis: *, P < 0.001 (compared with GABA [N-Shh] clonal conditions); +, P < 0.001 (compared with the O4 [N-Shh] clonal condition); †, P < 0.001 (compared with the GABA/O4 [N-Shh] clonal condition).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Neurological Disorders and Stroke and the National Institute of Mental Health, National Institutes of Health (to M.F.M.).
Abbreviations
BMP, bone morphogenetic protein
OL, oligodendrocyte
Shh, sonic hedgehog
N-Shh, N-terminal Shh
VZ, ventricular zone
MGE, medial ganglionic eminence
LGE, lateral ganglionic eminence
SVZ, subventricular zone
bHLH, basic helix–loop–helix
EGF, epidermal growth factor
FGF, fibroblast growth factor
References
- 1.Tan S.-S., Kalloniatis, M., Sturm, K., Tam, P. P. L., Reese, B. E. & Faulkner-Jones, B. (2001) Neuron 21, 295-304. [DOI] [PubMed] [Google Scholar]
- 2.Parnavelas J. G. (2000) Trends Neurosci. 23, 126-131. [DOI] [PubMed] [Google Scholar]
- 3.Corbin J. G., Nery, S. & Fishell, G. (2001) Nat. Neurosci. 4, 1177-1182. [DOI] [PubMed] [Google Scholar]
- 4.Marin O. & Rubenstein, J. L. R. (2001) Nat. Neurosci. Rev. 2, 780-790. [DOI] [PubMed] [Google Scholar]
- 5.Anderson S. A., Marin, O., Horn, C., Jennings, K. & Rubenstein, J. L. R. (2001) Development (Cambridge, U.K.) 128, 358-363. [DOI] [PubMed] [Google Scholar]
- 6.He W., Ingraham, C., Rising, L., Goderie, S. & Temple, S. (2001) J. Neurosci. 21, 8854-8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehler M. F. (2002) in Cortical Development, ed. Hohmann, C. (Springer, Heidelberg), pp. 27–52.
- 8.Mehler M. F. (2002) in Cortical Development, ed. Hohmann, C. (Springer, Heidelberg), pp. 157–178.
- 9.Thomas J. L., Spassky, N., Perez Villegas, E. M., Olivier, C., Cobos, I., Goujet-Zalc, C., Martinez, S. & Zale, B. (2000) J. Neurosci. Res. 59, 471-476. [DOI] [PubMed] [Google Scholar]
- 10.Briscoe J. & Ericson, J. (1999) Cell. Dev. Biol. 10, 353-362. [DOI] [PubMed] [Google Scholar]
- 11.Miller R. H., Hayes, J. E., Dyer, K. L. & Sussman, C. R. (1999) Int. J. Dev. Neurosci. 17, 753-763. [DOI] [PubMed] [Google Scholar]
- 12.Briscoe J., Pierani, A., Jessell, T. M. & Ericson, J. (2000) Cell 101, 435-445. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Q., Choi, G. & Anderson, D. J. (2001) Neuron 31, 791-807. [DOI] [PubMed] [Google Scholar]
- 14.Mizuguchi R., Sugimori, M., Takebayashi, H., Kosako, H., Nagao, M., Yoshida, S., Nabeshima, Y., Shimamura, K. & Nakafuku, M. (2001) Neuron 31, 757-771. [DOI] [PubMed] [Google Scholar]
- 15.Novitch B. G., Chen, A. I. & Jessell, T. M. (2001) Neuron 31, 773-789. [DOI] [PubMed] [Google Scholar]
- 16.Sun T., Echelard, Y., Lu, R., Yuk, D.-I., Kaing, S., Stiles, C. D. & Rowitch, D. H. (2001) Curr. Biol. 11, 1413-1420. [DOI] [PubMed] [Google Scholar]
- 17.Qi Y., Cai, J., Wu, Y., Wu, R., Lee, J., Fu, H., Rao, M., Sussell, L., Rubenstein, J. & Qiu, M. (2001) Development (Cambridge, U.K.) 128, 2723-2733. [DOI] [PubMed] [Google Scholar]
- 18.Lu Q. R., Cai, L., Rowitch, D., Cepko, C. L. & Stiles, C. D. (2001) Nat. Neurosci. 4, 973-974. [DOI] [PubMed] [Google Scholar]
- 19.Kondo T. & Raff, M. (2000) Development (Cambridge, U.K.) 127, 2989-2998. [DOI] [PubMed] [Google Scholar]
- 20.Wang S., Sdrulla, A., Johnson, J. E., Yokota, Y. & Barres, B. A. (2001) Neuron 29, 603-614. [DOI] [PubMed] [Google Scholar]
- 21.Casarosa S., Fode, C. & Guillemot, F. (1999) Development (Cambridge, U.K.) 126, 525-534. [DOI] [PubMed] [Google Scholar]
- 22.Lo L. C., Johnson, J. E., Wuenschell, C. W., Saito, T. & Anderson, D. J. (1991) Genes Dev. 5, 1524-1537. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q. & Anderson, D. J. (2002) Cell 109, 61-73. [DOI] [PubMed] [Google Scholar]
- 24.Lu Q. R., Sun, T., Zhu, Z., Ma, N., Garcia, M., Stiles, C. D. & Rowitch, D. H. (2002) Cell 109, 75-86. [DOI] [PubMed] [Google Scholar]
- 25.Takebayashi H., Yoshida, S., Sugimori, M., Kosako, H., Kominami, R., Nakafuku, M. & Nabeshima, Y. (2000) Mech. Dev. 99, 143-148. [DOI] [PubMed] [Google Scholar]
- 26.Zhu G., Mehler, M. F., Zhao, J., Yung, S. & Kessler, J. A. (1999) Dev. Biol. 215, 118-129. [DOI] [PubMed] [Google Scholar]
- 27.McKay R. (1997) Science 276, 66-71. [DOI] [PubMed] [Google Scholar]
- 28.Mabie P. C., Mehler, M. F. & Kessler, J. A. (1999) J. Neurosci. 19, 7077-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehler M. F., Mabie, P. C., Zhang, D. & Kessler, J. A. (1997) Trends Neurosci. 20, 309-317. [DOI] [PubMed] [Google Scholar]
- 30.Li W. W., Cogswell, C. A. & LoTurco, J. J. (1998) J. Neurosci. 18, 8853-8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hattori A., Katayama, M., Iwasaki, S., Ishii, K., Tsujimoto, M. & Kohno, M. (1999) J. Neurochem. 72, 2264-2271. [DOI] [PubMed] [Google Scholar]
- 32.Mehler M. F., Mabie, P. C., Zhu, G., Gokhan, S. & Kessler, J. A. (2000) Dev. Neurosci. 22, 74-85. [DOI] [PubMed] [Google Scholar]
- 33.He T-C., Zhou, S., DaCosta, L. T., Yu, J., Kinzler, W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenstat D. D., Liu, J. K., Mione, M., Zhong, W., Yu, G., Anderson, S. A., Ghattas, I., Puelles, & Rubenstein, J. L. R. (1999) J. Comp. Neurol. 414, 217-237. [DOI] [PubMed] [Google Scholar]
- 35.Stühmer T., Anderson, S. A., Ekker, M. & Rubenstein, J. L. R. (2002) Development (Cambridge, U.K.) 129, 245-252. [DOI] [PubMed] [Google Scholar]
- 36.Martens D. J., Tropepe, V. & van der Kooy, D. (2000) J. Neurosci. 20, 1085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumsden A. & Krumlauf, R. (1996) Nat. Genet. 20, 325-326. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.