Abstract
Plasmodesmata establish a pathway for the trafficking of non-cell-autonomously acting proteins and ribonucleoprotein complexes. Plasmodesmal enriched cell fractions and the contents of enucleate sieve elements, in the form of phloem sap, were used to isolate and characterize heat shock cognate 70 (Hsc70) chaperones associated with this cell-to-cell transport pathway. Three Cucurbita maxima Hsc70 chaperones were cloned and functional and sequence analysis led to the identification of a previously uncharacterized subclass of non-cell-autonomous chaperones. The highly conserved nature of the heat shock protein 70 (Hsp70) family, in conjunction with mutant analysis, permitted the characterization of a motif that allows these Hsc70 chaperones to engage the plasmodesmal non-cell-autonomous translocation machinery. Proof of concept that this motif is necessary for Hsp70 gain-of-movement function was obtained through the engineering of a human Hsp70 that acquired the capacity to traffic through plasmodesmata. These results are discussed in terms of the roles likely played by this subclass of Hsc70 chaperones in the trafficking of non-cell-autonomous proteins.
The heat shock protein 90 (Hsp90)/Hsp70-based chaperone machinery is a dynamic, multifunctional and multicomponent system that plays a pivotal role in protecting biological organisms from environmental and genetic stresses (1–4). This chaperone system also regulates the activities and targeted distribution of proteins involved in developmental and metabolic programs (5). For example, Hsp90 controls the nuclear trafficking of steroid hormone receptors (6, 7), and Hsp70 has been implicated in the trafficking of nuclear localization signal (NLS)-containing proteins to the nuclear pore complex (8). The concept that these Hsp90/Hsp70-associated functions are highly conserved between kingdoms gained support from the finding that a plant multiprotein Hsp90/Hsp70 complex was able to correctly assemble animal steroid receptors and oncogenic protein kinases (9, 10). Further evidence consistent with this notion was afforded by the finding that an animal Hsp90-specific drug, geldanamycin, also inhibits the function of plant Hsp90s (4, 9).
The Hsp90/Hsp70 machinery also plays an important role in the life cycle of plant and animal viruses (11). During infection, the animal chaperone machinery is recruited to facilitate the formation of a complex comprised of viral protein and RNA (12, 13). The Closteroviridae family of plant viruses encodes proteins displaying homology to the Hsp superfamily (14). One such example is p65 of the Beet yellows closterovirus, which encodes a protein homologous to the plant Hsp70 chaperone family, termed Hsp70h (15). Interestingly, Hsp70h plays a direct role in the formation of movement-competent viral ribonucleoprotein (RNP) complexes (16). In plant tissues, the cell-to-cell spread of viral infection occurs via plasmodesmata (17, 18), and, consistent with the utilization of this pathway, the Hsp70h has been immunolocalized to these intercellular channels (15).
The cell-to-cell transport of plant and viral RNP complexes involves delivery to the plasmodesmata, modification of the intercellular channels, and partial unfolding of the delivered cargo (protein or RNP complex) (19–21). Given the diverse roles played by the Hsp90/Hsp70 chaperone machinery in protein folding, targeted delivery, and the formation of RNP complexes, plants may well have recruited this system to facilitate the processes involved in cell-to-cell transport of macromolecules. In the present study, we characterize a subclass of Cucurbita maxima heat shock cognate 70 (Hsc70) chaperones that have the capacity to interact with the plasmodesmal non-cell-autonomous translocation pathway. Functional analysis of mutant forms of these Hsc70s allowed us to identify a structural motif necessary and sufficient for cell-to-cell transport of plant and human Hsp70 chaperones.
Materials and Methods
Plant Materials and Protein Extraction.
Nicotiana tabacum cv. Turkish (tobacco) and C. maxima Duch. cv. Big Max (pumpkin) plants were grown in the greenhouse under natural daylight conditions with a mid-day light intensity in the range of 1,200 to 1,500 μmol m−2⋅s−1 and day/night temperatures of 26/22°C. Plasmodemal-enriched protein fractions were prepared from tobacco tissues (W2) and BY-2 suspension cells (PECP) as described (21, 22). Pumpkin proteins were extracted from stem tissues, vascular bundles, and phloem sap as described (23, 24).
Isolation of CmHsc70 and CmHsc40 Isoforms.
Degenerate primers were designed based on the highly conserved regions of the Arabidopsis and spinach Hsp70 chaperones. A primer pair designed to match the amino acid sequences, MAGKGEG and PDEAVAY, bordering the conserved ATPase domain were used to screen a pumpkin stem cDNA library (24). A resultant 1,131-bp DNA fragment was cloned, and the sequence was confirmed and then used to obtain full-length cDNAs encoding members of the CmHsp70 family present in the pumpkin stem cDNA library. Isolated cDNAs were further screened by using degenerate primers designed against the C-terminal amino acid sequence, PKIEEVD, conserved in all higher plant cytosolic Hsp70 chaperones. Three different cDNAs encoding full-length Hsp70 isoforms were identified and named kA121, kA141, and kA1251. Degenerate primers were similarly designed based on conserved regions of the Arabidopsis DnaJ-like Hsp40 proteins. A primer pair designed to match the amino acid sequences, ASQDDLKKA and PGEADEAPD, were used to screen the pumpkin stem cDNA library. When the above described procedures were used, a positive clone was identified and named kACMJ1. Based on sequence homologies, these clones were assigned the names, CmHsc70-1 (kA121), CmHsc70-2 (kA141), CmHsc70-3 (kA1251), and CmHsc40-1 (kACMJ1).
Expression and Purification of Recombinant Proteins.
Recombinant proteins were obtained by using the BAC-TO-BAC Baculovirus Expression System (Life Technologies, Grand Island, NY), the QIAexpress Protein Purification System (Qiagen, Valencia, CA), and the GST-Expression System (Amersham Pharmacia). Standard PCR cloning techniques were used to introduce the ORFs of the cDNA clones kA121, kA141, kA1251, and kACMJ1 into the pFastBac1, pQE30, and/or pGEX-KG vectors. These vectors were used to express N-terminal His-6- or GST-tagged full-length and modified forms of CmHsc70-1, CmHsc70-2, CmHsc70-3, and CmHsc40-1 in their respective systems. Plasmids expressing His-6-tagged truncated or mutant variants of CmHsc70-1 were produced by PCR-mutagenesis employing the QuikChange Site-Directed Mutagenesis kit (Stratagene) following the manufacturer's instructions. A chimeric form of CmHsc70-3–CmHsc70-1 was engineered by using the unique restriction sites, SstI and KpnI, present in CmHsc70-3, to excise the 3′ region of the ORF for replacement by the equivalent SstI–KpnI fragment excised from CmHsc70-1. Plasmids expressing His-6-tagged human Hsp70 (GenBank accession no. M11717) were obtained from Zimmer et al. (25), and the plasmid expressing His-6-tagged hHsp70–Cm Hsc70-1 tetratricopeptide-repeat protein binding (TPR–PB) fusion protein was constructed by standard PCR cloning protocols by producing a 1,851-bp NcoI–XmaI human Hsp70 cDNA fragment that was ligated with a XmaI–EcoRI digested 87-bp PCR fragment from CmHsc70-1 (bp 1873–1959, C-terminal 28 aa) into an NcoI–EcoRI digested Escherichia coli expression vector pET30a (Invitrogen). Similarily, plasmids expressing GFP–SVR (short variable region) TPR–PB or GFP–SVR TPR–PB P → T were constructed by in frame ligation of an 87-bp PCR fragment from CmHsc70-1 to a pET30a-GFP expression vector. A His-6-tagged DnaK was expressed by using a cell line provided by Suh et al. (26). Additional chromatographic protein purification of His-6- and GST-tagged recombinant proteins was carried out by using a HiTrap Q-Sepharose column as described (24). Preparation of His-6- and GST-tagged proteins and CmPP16 for use in microinjection experiments followed the protocols as described (21, 27). All microinjection experiments were performed on pumpkin cotyledons using the conditions and methods described in refs. 27 and 28.
Purification and Peptide Analysis of Phloem Sap Hsp70 Isoforms.
Phloem exudate was first dialyzed against buffer A (50 mM Tris⋅HCl, pH 7.5/50 mM β-mercaptoethanol/10% vol/vol glycerol/0.5 mM PMSF), centrifuged (27,000 × g for 10 min), and the supernatant was applied to a buffer A-equilibrated HiTrap Q-Sepharose column (Amersham Pharmacia); protein elution was achieved by using buffer A plus 1 M NaCl. The eluate was next dialyzed against buffer B (20 mM Tris⋅HCl, pH 7.5/10 mM MgCl2), centrifuged (27,000 × g for 10 min), and the supernatant was applied to a buffer B-equilibrated C8-linked ATP-agarose column (Sigma). After a wash with buffer B containing 0.5 M NaCl, bound proteins were eluted with buffer B containing 10 mM ATP. CmHsp70-containing fractions were dialyzed against buffer C (20 mM Tris⋅HCl, pH 7.0), applied to a buffer C-equilibrated Mono Q HR 5/5 column (Amersham Pharmacia) and then eluted with a 100–400 mM NaCl linear gradient. The presence of CmHsp70 homologs was confirmed by Western blot analysis using anti-wheat Hsp70 antibody.
Purified CmHsp70 was in-gel digested with trypsin, and peptide fragments were then isolated, concentrated, and subjected to electron spray/tandem mass spectrometry (ES-MS/MS) analysis with a Sciex Q STAR Hybrid Quadrupole-TOF (Applied Biosystems, Foster City, CA). In silico trypsin digestion of CmHsc70 isoforms, followed by peptide mass calculations, was carried out by using the ms-digest program from Protein Prospector (29).
ATPase Assays.
ATPase activity of recombinant His-6-tagged CmHsc70-1, CmHsc70-2, CmHsc70-3, and DnaK was assayed by using the EnzChek Phosphate Assay kit (Molecular Probes). The reaction mixture contained 20 mM Tris⋅HCl (pH 7.5), 2 mM MgCl2, 1 mM ATP, 0.2 mM 2-amino-6-mercapto-7-methyl-purine riboside, 1 unit purine nucleoside phophorylase, 4–20 μg of the recombinant protein, and an equimolar amount of GST or GST-CmHsc40-1. ATPase assays were performed in triplicate, at 25°C, as described in the manufacturer's protocol.
Results
Detection of Hsp70 Homologs Associated with Plasmodesmata.
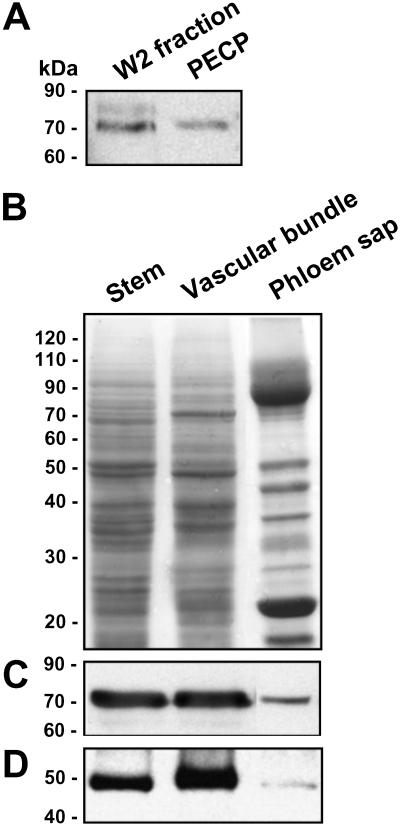
Analysis of plasmodesmal-enriched cell wall preparations [W2 (21) and PECP (22)], with a polyclonal antibody that recognizes the plant Hsp70 chaperone family, revealed the presence of at least two Hsp70 related proteins within these preparations (Fig. 1A). These results supported the concept that Hsp70-like chaperones may be involved in plasmodesmal function. Because insufficient quantities of these plasmodesmal associated Hsp70-like proteins could be obtained for a reverse genetics approach, an alternate system was tested by using the collection and fractionation protocols developed for the analysis of pumpkin phloem sap (24). The use of this system was founded on two observations; first, entry of cytosolic proteins into the phloem sap involves passage through the plasmodesmata that interconnect the companion cell–sieve tube system (30). Second, in an earlier study we established the presence of chaperones in the phloem translocation stream (31).
Fig 1.
Immunodetection of heat shock protein homologs in plasmodesmal enriched protein fractions and vascular tissues. (A) Plasmodesmal enriched protein fractions were prepared from tobacco tissue (W2) and BY-2 tobacco suspension cultured cells (PECP), and aliquots (10 μg) were separated by SDS/PAGE and subjected to Western analysis to test for the presence of Hsp70 homologs. (B–D) Immunodetection of Hsp70 and Hsp40 homologs in proteins prepared from C. maxima (pumpkin) stem tissues, vascular bundles, and phloem sap. (B) Protein samples (30 μg) were separated by SDS/PAGE and stained with GelCode Blue Reagent. (C and D) Detection of Hsp70 and Hsp40 homologs in protein fractions shown in B. Western analysis was performed with anti-wheat Hsp70 (A and C) and anti-cucumber CsDnaJ-1 polycolonal antibodies (D); control blots were performed in the absence of secondary antibodies.
Western analysis performed on protein fractions prepared from pumpkin stem tissues, vascular bundles, and the phloem sap readily detected the presence of CmHsp70-related protein(s) (Fig. 1 B and C). As expected, Western analysis performed on these same protein fractions, by using a polyclonal antibody raised against the cucumber CsDnaJ-1 (32), also identified a putative Hsp40 homolog in pumpkin tissues and phloem sap (Fig. 1D). Taken together, these results are consistent with the concept that CmHsp70 chaperones have the potential to interact with and traffic through plasmodesmata. Additionally, detection of a Hsp40-related protein in the phloem sap suggests the presence of a Hsp70-Hsp40 (DnaK–DnaJ) complex involved in the functioning of the enucleate sieve tube system. Thus, the phloem system provided us with the means to obtain analytical quantities of non-cell-autonomously acting Hsp70-related chaperones.
Identification and Characterization of CmHsp70 Isoforms.
Degenerate primers directed against concerved motifs in known cytosolic Hsp70 genes were used in the screening of a pumpkin stem cDNA library. Resultant cDNAs were sequenced and compared against the Hsp70 superfamily to select those encoding for cytosolic isoforms (33); five cDNAs encoding for different Hsp70 isoforms were identified. Based on the observed homology between the translated cDNA sequences of these five clones and the pea (34) and Arabidopsis Hsc proteins (>90% at the amino acid level), as well as using peptide analysis of Hsp70-related proteins present in the phloem sap, three cDNAs were selected for further characterization; these were assigned the names C. maxima Hsc70-1, CmHsc70-2, and CmHsc70-3 (see Fig. 5A, which is published as supporting information on the PNAS web site, www.pnas.org). The constituitive expression of these genes and their assignment as Hsc, as opposed to heat shock inducible (Hsp), chaperones was confirmed by performing semiquantitative RT-PCR (Fig. 5B) and Western analyses (data not shown) on tissue extracts collected from pumpkin plants exposed to either noninducing (25°C) or inducing (35°C) conditions.
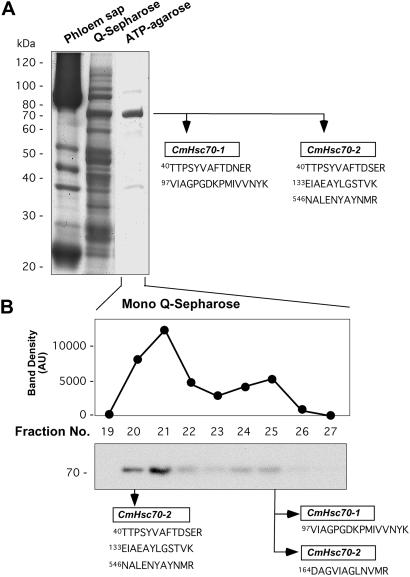
The affinity of Hsp70 for its substrate ATP was used to purify to near homogeneity the CmHsp70 related proteins present in the phloem sap (Fig. 2A). This purified CmHsp70 preparation was subjected to in-gel trypsin digestion, and the peptide fragments were isolated, concentrated, and subjected to ES-MS/MS. The duly obtained mass information (m/z values) was then compared with that of the in silico tryptic peptides predicted from the isolated CmHsc70 clones. Based on these experiments, we could identify two Hsc70-related chaperones, CmHsc70-1 and CmHsc70-2, that were present within the phloem sap (Figs. 5A and 2A).
Fig 2.
Authentication of CmHsc70-1 and CmHsc70-2 as phloem sap proteins by using peptide mass fingerprint analysis. (A) Phloem sap Hsp70 isoforms were purified to near homogeneity by chromatographic separation of phloem sap using a combination of HiTrap Q-Sepharose (Q-Sepharose) and C8-linked ATP-agarose (ATP-agarose) columns. Trypsin-digestion of the ATP-agarose purified 70-kDa protein band generated peptides that could be assigned to either CmHsc70-1 or CmHsc70-2, but not to CmHsc70-3. (B) Chromatographic separation of the ATP-agarose fractionated phloem proteins on a Mono Q-Sepharose 5/5R column. Fractions were quantified by densitometry and individual bands were excised for mass fingerprint analysis.
To further confirm the identity of these phloem proteins, the ATP-agarose purified fraction was subjected to Mono Q chromatography and western analysis of the eluted fractions identified two peaks of Hsc70 immunoreactivity (Fig. 2B). Mass spectrum analysis performed on the proteins present in these fractions indicated that the first peak contained only CmHsc70-2 peptides, whereas both CmHsc70-1 and CmHsc70-2 peptides were present in the second Mono Q peak. Western analysis performed on 2D gels of ATP-agarose purified phloem sap revealed the presence of only two immunoreactive bands (data not shown). Given that no peptides corresponding to CmHsc70-3 were identified, we concluded that, in contrast to CmHsc70-1 and CmHsc70-2, this isoform is either present at extremely low levels or more likely is unable to enter the phloem translocation stream.
Functional Assay for Recombinant Hsc70 Proteins.
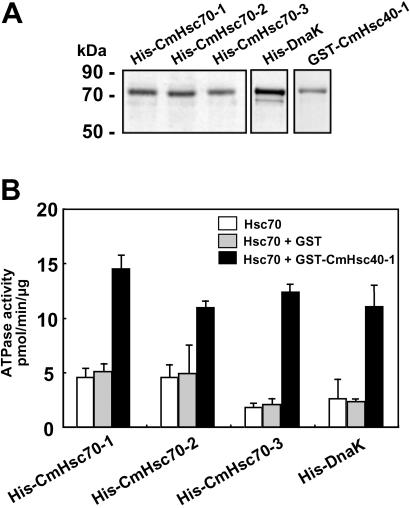
The ATPase activity of the Hsp70 chaperones was next used as a functional assay for our purified recombinant His-6-tagged proteins (Fig. 3A). Because Hsp40 (DnaJ ortholog) is a regulatory component for controlling Hsp70 ATPase activity, a member of this gene family, CmHsc40-1, was cloned from pumpkin. Recombinant GST-tagged Hsc40-1 was produced in E. coli, and purified protein was used in the Hsc70 ATPase assay (Fig. 3B). All purified recombinant CmHsc70 proteins exhibited ATPase activities that were comparable to those measured with the bacterial homolog, DnaK, and were unaffected by the addition of GST (Fig. 3B, columns 1 and 2). In the presence of GST-tagged CmHsc40-1, the CmHsc70 associated ATPase activity underwent a 2.5- to 6-fold enhancement. An equivalent stimulation was observed with DnaK when CmHsc40-1 was added to the assay medium (Fig. 3B, column 3). Similar results were obtained with the phloem-purified Hsp70 preparation (data not shown), indicating the equivalence in specific activities between the native and recombinant forms of the CmHsc70s. These experiments confirmed that our recombinant, purified, Hsc70 and DnaK proteins were in a functionally competent state.
Fig 3.
Recombinant Hsc70 proteins exhibit ATPase activity. (A) His-6-tagged CmHsc70-1, CmHsc70-2, CmHsc70-3, and DnaK purified by using Ni-based chromatography. The CmHsc40-1 cochaperone was purified as a GST fusion protein. (B) Steady-state ATP-hydrolytic activity of recombinant His-6-tagged Hsc70 isoforms (7 μg) measured alone or in the presence of GST (2.6 μg) or GST-CmHsc40-1 (7 μg). Bars represent the mean (±SD) for three independent measurements and are based on six sample points collected during the linear phase of the reaction.
Phloem CmHsc70 Chaperones Can Traffic Cell to Cell.
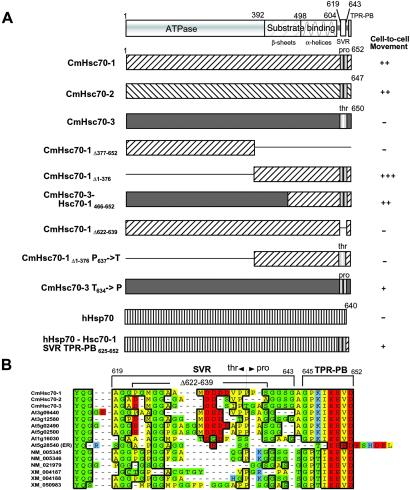
A number of characterized phloem proteins have the capacity to traffic cell to cell when introduced into pumpkin mesophyll cells by microinjection (28). This capacity of the phloem CmHsc70-1 and CmHsc70-2 chaperones was investigated to ascertain whether they have similar properties. Microinjection experiments performed with recombinant His-6-tagged proteins, earlier shown to exhibit ATPase activity, demonstrated that both CmHsc70-1 and CmHsc70-2 had the capacity to interact with plasmodesmata to increase the size exclusion limit and mediate their own cell-to-cell transport (Fig. 4 A, and Fig. 6 and Table 1, which are published as supporting information on the PNAS web site). To determine whether this property is unique to the phloem CmHsc70 isoforms, or a common activity associated with plant cytosolic Hsc70 chaperones, microinjection studies were performed by using CmHsc70-3 as a probe. The clear inability of this protein to dilate the plasmodesmal microchannels and to potentiate its own cell-to-cell movement (Fig. 4A) is consistent with its absence from the phloem sap. The ATPase activity of the CmHsc70-3 established that this recombinant protein, used in these microinjection assays, existed in the native state. Thus, the inability of CmHsc70-3 to move may reflect the absence of a motif, carried by the two phloem CmHsc70 chaperones, that imparts such a capacity.
Fig 4.
Phloem CmHsc70 isoforms contain a motif necessary and sufficient to mediate their cell-to-cell movement. (A Upper) The conserved structural organization of the Hsp70 family. The N-terminal 392 amino acid residues correspond to the ATPase domain; residues 393–604 represent the region of the protein involved in substrate/peptide binding, which is comprised of β-sandwich and α-helical subdomains. The C terminus is divided into a SVR and the TPR–PB. Movement capacity of wild-type and mutant isoforms of CmHsc70-1, CmHsc70-2, CmHsc70-3, and human Hsp70 was determined by standard microinjection methods (21, 27). The capacity of each probe to move cell to cell was evaluated against CmPP16 (27) that was injected into the same leaf tissues. Probes that moved into >20 cells were characterized as highly competent (+++); those that moved into 15–20 cells were characterized as very competent (++), those that moved into >7 cells were characterized as competent (+), and no movement was characterized as incompetent (−). (B) Comparative analysis of the C-terminal region of C. maxima, Arabidopsis thaliana, and Homo sapiens cytosolic Hsp70 chaperone families present in genomic databases (www.ncbi.nlm.nih.gov). An endoplasmic reticulum Hsp70 isoform, At5g28540, was included for comparison. Colors are as follows: green, polar; yellow, nonpolar; blue, basic; red, acidic residues.
A Motif in the C Terminus of CmHsc70-1 Facilitates Transport Through Plasmodesmata.
The highly conserved nature of the Hsp70 family afforded an opportunity to target and identify the motif(s) necessary and sufficient to potentiate cell-to-cell movement of the Hsc70 phloem isoforms. It is now well established that Hsp70 chaperones are comprised of three highly conserved functional domains (Fig. 4), the N-terminal ATPase, the C-terminal substrate/peptide binding domain, and the extreme C terminus that contains the TPR–PB (1, 35, 36). It is noteworthy that all cytosolic Hsp70 chaperones contain a SVR located between the α-helical subdomain and the extreme C-terminal TPR-PB (Fig. 4B).
To determine which of these domains is required to impart cell-to-cell movement capability, two truncated forms of the CmHsc70-1 were produced, CmHsc70-1Δ377–652 and CmHsc70-1Δ1–376, for use in microinjection assays. These studies established that the C-terminal region was necessary and sufficient to mediate cell-to-cell movement, whereas the ATPase domain appeared to be dispensable (Fig. 4A). Additional support for this finding was provided by studies carried out with a chimeric CmHsc70 chaperone, consisting of the N-terminal amino acid residues 1–465 from the cell autonomous CmHsc70-3 and the C-terminal amino acid residues 466–652 from the non-cell-autonomous CmHsc70-1. Because this chimeric Hsc70 displayed the capacity to move cell to cell, the C-terminal region of the phloem CmHsc70-1 must contain a motif that engages the non-cell-autonomous translocation machinery.
Inspection of the sequences within the C-terminal region (residues 466–652) of CmHsc70-1 identified the SVR as the most logical domain involved in protein transport through plasmodesmata. To test this hypothesis, a SVR-deletion mutant, CmHsc70-1Δ622–639, was produced and used as a probe in microinjection experiments. As was observed for CmHsc70-3, CmHsc70-1Δ622–639 lacked the capacity to mediate its own movement from cell to cell. The greatest diversity in amino acid residue composition within the CmHsc70-1 SVR was located around P637 (Fig. 4B). Consequently, site-directed mutagenesis was next used to produce a modified version of the CmHsc70-1 (P637 → T637) that reflected the structurally unique T634 residue present in CmHsc70-3. Microinjection assays performed with this P → T mutant demonstrated that this change blocked the movement capacity of the protein. Hence, the P → T substitution appears to have altered the structural integrity of a motif essential for cell-to-cell transport. A gain-of-function experiment was next performed and involved engineering a mutant form of CmHsc70-3 in which T634 was changed to P634 (Fig. 4A). Microinjection assays revealed that this mutation imparted the capacity for cell-to-cell movement, albeit of lower efficacy. Taken together, these results established that the C-terminal region of the phloem CmHsc70-1 displays a structural motif that is necessary and sufficient for its cell-to-cell transport through plasmodesmata.
Cell-to-Cell Movement of Engineered Human Hsp70.
A rigorous test for the hypothesis that the CmHsc70-1 SVR presents a motif that engages the non-cell-autonomous translocation pathway would be whether or not a heterologous protein, carrying such a motif, could move through plasmodesmata. The extreme C-terminal region, including the SVR and TPR–PB of CmHsc70-1, was used to replace the equivalent domains in the most closely related (75% identity) human Hsp70 chaperone (Fig. 4). Control experiments performed with the wild-type form of this human Hsp70 (25) established that it remained confined to the injected cells. In contrast, the engineered chaperone (hHsp701–616-Hsc70-1 SVR TPR–PB625–652) was able to move cell to cell in a manner equivalent to that displayed by the mutant CmHsc70-3 (P637 → T637).
To ascertain whether this SVR motif operates in a manner similar to that of the nuclear localization signal motif, we next fused this region to the C terminus of GFP (GFP–SVR TPR–PB625–652). In microinjection experiments, neither the wild-type nor the engineered form of GFP displayed any capacity to interact with plasmodesmata to either induce an increase in size exclusion limit or potentiate its own cell-to-cell movement (Table 1 and Fig. 6). Thus, this SVR motif does not appear to function as a simple targeting signal. These results suggest that the capacity of the SVR motif to allow protein entry into the non-cell-autonomous translocation pathway likely operates within the context of the Hsp70 chaperone machinery.
Discussion
Members of the Hsp70 chaperone superfamily play a central role in facilitating the folding, unfolding, and transport of a wide range of proteins (1, 5, 6, 37). In addition, these chaperones appear to be involved in the recognition and turnover of misfolded/destabilized proteins to thereby ensure a suitable environment for cellular function (1, 37). Recent findings have implicated members of the Hsp70 family in the targeted delivery of proteins to specific cellular domains (38–40) other than to organelles like the peroxisome, mitochondrion, chloroplast, and endoplasmic reticulum (40–46). Evidence is also accumulating that the Hsp70/Hsp90 chaperone machinery can mediate the unfolding of proteins from their native state (5). Considering the functions performed by these Hsp70 chaperones, it is easy to comprehend why in the course of plant evolution the transport machinery of the plasmodesma would have recruited cytosolic members of this family to assist in the trafficking of macromolecules.
In the present study, we demonstrated that Hsp70-related proteins could be detected in the plasmodesmal enriched cell fractions (Fig. 1A). Consistent with this finding, two pumpkin Hsp70 chaperones were identified in the phloem sap (Figs. 1C and 2), implicating their potential to enter the enucleate sieve tube system through plasmodesmata. In contrast to CmHsc70-3, a cytosolic chaperone not detected in the phloem sap, both CmHsc70-1 and CmHsc70-2 had the capacity to interact with and traffic through the companion cell-sieve element plasmodesmata (Fig. 2). Microinjection experiments conducted with enzymatically active recombinant CmHsc70 proteins (Fig. 3) provided direct evidence of their capacity for cell-to-cell movement (Figs. 4A and 6 and Table 1) and, thus, these chaperones likely represent a previously undescribed subclass of the Hsp70 family.
The proteins immunologically related to Hsp70 detected in the plasmodesmal enriched cell fractions may well be members of this new Hsc70 subclass. Alternatively, such chaperones could constitute structural components of the plasmodesmata whose function may be similar to that of organelle-associated chaperones engaged in protein unfolding/import (45, 47, 48). Although unlikely, we cannot presently discount that the detection of Hsp70-related proteins in these plasmodesmal enriched cell fractions represent the binding of cytosolic chaperones to proteins contained within this fraction.
The wide diversity of functions performed by Hsp70 chaperones, in conjunction with the high degree of structural conservation of these proteins observed across all kingdoms (33), facilitated the identification of a motif that interacts with the plant non-cell-autonomous translocation machinery. Analysis of truncated forms of CmHsc70-1 established that the highly conserved ATPase domain was dispensible in terms of protein interaction with the machinery that mediates the transport of macromolecules through plasmodesmata (Fig. 4A). The C-terminal region of CmHsc70-1, comprised of the substrate binding, SVR, and TPR–PB domains, was shown to be necessary and sufficient to mediate the cell-to-cell transport of this chaperone. Given the highly conserved structural and functional nature of both the substrate-binding and complex-forming TPR–PB domains (5, 36), the SVR was identified as the most likely site to carry a motif required for protein access to the non-cell-autonomous translocation pathway. This hypothesis gained support from the finding that deletion of the SVR, or a single amino acid substitution within the CmHsc70-1 SVR (P637 → T637), abolished the capacity for cell-to-cell movement. Consistent with this notion, an equivalent single amino acid substitution in the CmHsc70-3 SVR (T634 → P634) established the capacity for cell-to-cell movement for an otherwise cell-autonomous protein.
Analysis of this SVR, within plant and animal Hsp70 chaperones, identified the presence of an acidic region, unique to the plant Hsp70 chaperones, that is juxtaposed to a proline-rich region (Fig. 4B). The deletion engineered within the SVR (Δ622–639) of CmHsc70-1 produced a recombinant protein that closely resembled the human Hsc70 chaperone (Fig. 4). It is important to note that this region of the cytosolic Hsc70 was shown to be the only domain dispensable for chaperone activity (35). Proof of concept that this SVR region represents the site involved in the gain-of-movement functions unique to the plant kingdom was provided by our ability to impart upon a human Hsp70 the capacity to move through plasmodesmata (Fig. 4). Additional tests for this hypothesis will be performed on representative Hsc70 chaperones from a range of plant species.
Recent studies have shown that members of the Closteroviridae family of plant positive-stranded RNA viruses encode a Hsp70 homolog that, in combination with the coat proteins, functions in cell-to-cell movement of the infectious agent (15, 16). Because this viral Hsp70 was detected in association with plasmodesmata of infected tissues, a sequence analysis was conducted to ascertain whether this viral chaperone contained a similar plant-like SVR at its C terminus. No equivalent region could be detected, suggesting that this viral family may use a different pathway to interact with and traffic through plasmodesmata. Such a possibility is supported by the observation that this viral Hsp70 chaperone is more homologous to the endoplasmic reticulum-associated Bip chaperone than to the cytosolic Hsc70 subfamily (14).
The two non-cell-autonomous Hsc70 chaperones characterized in this study were shown to be present in the phloem sap of pumpkin. The observed capacity for trafficking through plasmodesmata is consistent with the notion that phloem sap proteins originate from companion cells. In the vascular system of higher plants, these companion cells are envisaged to maintain the enucleate sieve elements in a functional state. In a previous study we demonstrated that unfolding appears to be involved during protein translocation through plasmodesmata (21). Hence, CmHsc70-1 and CmHsc70-2 may play a role in the refolding of such structurally altered proteins as they enter the sieve elements. Additionally, because human Hsp70 chaperones have the capacity to bind RNA (25, 49), the phloem Hsc70 chaperones may interact in a similar manner with the RNA molecules present in the phloem sap (23, 27). Such an interaction could regulate both protein and RNA entry, long-distance movement, and exit from the phloem (50).
Supplementary Material
Acknowledgments
We thank E. Vierling for supplying anti-wheat Hsp70 antibody, H. Kindl for anti-CsDnaJ1 antibody, M. Nishimura for anti-pumpkin mitochondrial CPN60 antibody, T. Henics for providing the human Hsp70 clone, W.-C. Suh for the cell line expressing DnaK, and T. Yamada for technical assistance with the insect cell-expression system. This work was supported by National Science Foundation Grant IBN-9900539 (to W.J.L.).
Abbreviations
Hsp, heat shock protein
Hsc, heat shock cognate
RNP, ribonucleoprotein
TPR–PB, tetratricopeptide-repeat protein binding
SVR, short variable region
References
- 1.Hartl F. U. & Hayer-Hartl, M. (2002) Science 295, 1852-1858. [DOI] [PubMed] [Google Scholar]
- 2.Parsell D. A. & Lindquist, S. (1993) Annu. Rev. Genet. 27, 437-496. [DOI] [PubMed] [Google Scholar]
- 3.Rutherford S. L. & Lindquist, S. (1998) Nature 396, 336-342. [DOI] [PubMed] [Google Scholar]
- 4.Queitsch C., Sangster, T. A. & Lindquist, S. (2002) Nature 417, 618-624. [DOI] [PubMed] [Google Scholar]
- 5.Pratt W. B., Krishna, P. & Olsen, L. J. (2001) Trends Plant Sci. 6, 54-58. [DOI] [PubMed] [Google Scholar]
- 6.Pratt W. B. (1992) BioEssays 14, 841-848. [DOI] [PubMed] [Google Scholar]
- 7.Yang J. & DeFranco, D. B. (1996) Mol. Endocrinol. 10, 3-13. [DOI] [PubMed] [Google Scholar]
- 8.Shulga N., Roberts, P., Gu, Z., Spitz, L., Tabb, M. M., Nomura, M. & Goldfarb, D. S. (1996) J. Cell Biol. 135, 329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owens-Grillo J. K., Stancato, L. F., Hoffmann, K., Pratt, W. B. & Krishna, P. (1996) Biochemistry 35, 15249-15255. [DOI] [PubMed] [Google Scholar]
- 10.Harrell J. M., Kurek, I., Breiman, A., Radanyi, C., Renoir, J. M., Pratt, W. B. & Galigniana, M. D. (2002) Biochemistry 41, 5581-5587. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan C. S. & Pipas, J. M. (2001) Virology 287, 1-8. [DOI] [PubMed] [Google Scholar]
- 12.Hu J., Toft, D. O. & Seeger, C. (1997) EMBO J. 16, 59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J. & Seeger, C. (1996) Proc. Natl. Acad. Sci. USA 93, 1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karasev A. V. (2000) Annu. Rev. Phytopathol. 38, 293-324. [DOI] [PubMed] [Google Scholar]
- 15.Medina V., Peremyslov, V. V., Hagiwara, Y. & Dolja, V. V. (1999) Virology 260, 173-181. [DOI] [PubMed] [Google Scholar]
- 16.Alzhanova D. V., Napuli, A. J., Creamer, R. & Dolja, V. V. (2001) EMBO J. 20, 6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbertson R. L. & Lucas, W. J. (1996) Trends Plant Sci. 1, 260-268. [Google Scholar]
- 18.Lazarowitz S. G. & Beachy, R. N. (1999) Plant Cell 11, 535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haywood V., Kragler, F. & Lucas, W. J. (2002) Plant Cell 14, S303-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kragler F., Monzer, J., Xoconostle-Cázares, B. & Lucas, W. J. (2000) EMBO J. 19, 2856-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kragler F., Monzer, J., Shash, K., Xoconostle-Cázares, B. & Lucas, W. J. (1998) Plant J. 15, 367-381. [Google Scholar]
- 22.Lee, J.-Y., Yoo, B.-C., Rojas, M. R., Gomez-Ospina, N., Staehelin, L. A. & Lucas, W. J. (2002) Science, in press. [DOI] [PubMed]
- 23.Ruiz-Medrano R., Xoconostle-Cázares, B. & Lucas, W. J. (1999) Development (Cambridge, U.K.) 126, 4405-4419. [DOI] [PubMed] [Google Scholar]
- 24.Yoo B.-C., Aoki, K., Xiang, Y., Campbell, L. R., Hull, R. J., Xoconostle-Cázares, B., Monzer, J., Lee, J.-Y., Ullman, D. E. & Lucas, W. J. (2000) J. Biol. Chem. 275, 35122-35128. [DOI] [PubMed] [Google Scholar]
- 25.Zimmer C., von Gabain, A. & Henics, T. (2001) RNA 7, 1628-1637. [PMC free article] [PubMed] [Google Scholar]
- 26.Suh W.-C., Burkholder, W. F., Lu, C. Z., Zhao, X., Gottesman, M. E. & Gross, C. A. (1998) Proc. Natl. Acad. Sci. USA 95, 15223-15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xoconostle-Cázares B., Xiang, Y., Ruiz-Medrano, R., Wang, H.-L., Monzer, J., Yoo, B.-C., McFarland, K. C., Franceschi, V. R. & Lucas, W. J. (1999) Science 283, 94-98. [DOI] [PubMed] [Google Scholar]
- 28.Balachandran S., Xiang, Y., Schobert, C., Thompson, G. A. & Lucas, W. J. (1997) Proc. Natl. Acad. Sci. USA 94, 14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clauser K. R., Baker, P. & Burlingame, A. L. (1999) Anal. Chem. 71, 2871-2882. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Medrano R., Xoconostle-Cázares, B. & Lucas, W. J. (2001) Curr. Opin. Plant Biol. 4, 202-209. [DOI] [PubMed] [Google Scholar]
- 31.Schobert C., Baker, L., Szederkenyi, J., Grossmann, P., Komor, E., Hayashi, H., Chino, M. & Lucas, W. J. (1998) Planta 206, 245-252. [Google Scholar]
- 32.Preisig-Müller R., Muster, G. & Kindl, H. (1994) Eur. J. Biochem. 219, 57-63. [DOI] [PubMed] [Google Scholar]
- 33.Karlin S. & Brocchieri, L. (1998) J. Mol. Evol. 47, 565-577. [DOI] [PubMed] [Google Scholar]
- 34.Derocher A. & Vierling, E. (1995) Plant Mol. Biol. 27, 441-456. [DOI] [PubMed] [Google Scholar]
- 35.Wu S. J., Liu, F. H., Hu, S. M. & Wang, C. (2001) Biochem. J. 359, 419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheufler C., Brinker, A., Bourenkov, G., Pegoraro, S., Moroder, L., Bartunik, H., Hartl, F. U. & Moarefi, I. (2000) Cell 101, 199-210. [DOI] [PubMed] [Google Scholar]
- 37.Frydman J. (2001) Annu. Rev. Biochem. 70, 603-647. [DOI] [PubMed] [Google Scholar]
- 38.Pishvaee B., Costaguta, G., Yeung, B. G., Ryazantsev, S., Greener, T., Greene, L. E., Eisenberg, E., McCaffery, J. M. & Payne, G. S. (2000) Nat. Cell Biol. 2, 958-963. [DOI] [PubMed] [Google Scholar]
- 39.Tsai M. Y., Morfini, G., Szebenyi, G. & Brady, S. T. (2000) Mol. Biol. Cell 11, 2161-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pratt W. B., Silverstein, A. M. & Galigniana, M. D. (1999) Cell. Signalling 11, 839-851. [DOI] [PubMed] [Google Scholar]
- 41.Crookes W. J. & Olsen, L. J. (1998) J. Biol. Chem. 273, 17236-17242. [DOI] [PubMed] [Google Scholar]
- 42.Harano T., Nose, S., Uezu, R., Shimizu, N. & Fujiki, Y. (2001) Biochem. J. 357, 157-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishikawa S. I., Fewell, S. W., Kato, Y., Brodsky, J. L. & Endo, T. (2001) J. Cell Biol. 153, 1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang P. & MacRae, T. H. (1997) J. Cell Sci. 110, 1431-1440. [DOI] [PubMed] [Google Scholar]
- 45.Jackson-Constan D., Akita, M. & Keegstra, K. (2001) Biochim. Biophys. Acta 1541, 102-113. [DOI] [PubMed] [Google Scholar]
- 46.Hendershot L. M. (2000) Nat. Cell Biol. 2, E105-E106. [DOI] [PubMed] [Google Scholar]
- 47.Gaume B., Klaus, C., Ungermann, C., Guiard, B., Neupert, W. & Brunner, M. (1998) EMBO J. 17, 6497-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim J. H., Martin, F., Guiard, B., Pfanner, N. & Voos, W. (2001) EMBO J. 20, 941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson G. M., Sutphen, K., Bolikal, S., Chuang, K. Y. & Brewer, G. (2001) J. Biol. Chem. 276, 44450-44456. [DOI] [PubMed] [Google Scholar]
- 50.Lucas W. J., Yoo, B.-C. & Kragler, F. (2001) Nat. Rev. Mol. Cell. Biol. 2, 849-857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.