Abstract
Spindle orientation is critical for accurate chromosomal segregation in eukaryotic cells. In the yeast Saccharomyces cerevisiae, orientation of the mitotic spindle is achieved by a program of microtubule–cortex interactions coupled to spindle morphogenesis. We previously implicated Bud6p in directing microtubule capture throughout this program. Herein, we have analyzed cells coexpressing GFP:Bud6 and GFP:Tub1 fusions, providing a kinetic view of Bud6p–microtubule interactions in live cells. Surprisingly, even during the G1 phase, microtubule capture at the recent division site and the incipient bud is dictated by Bud6p. These contacts are eliminated in bud6Δ cells but are proficient in kar9Δ cells. Thus, Bud6p cues microtubule capture, as soon as a new cell polarity axis is established independent of Kar9p. Bud6p increases the duration of interactions and promotes distinct modes of cortical association within the bud and neck regions. In particular, microtubule shrinkage and growth at the cortex rarely occur away from Bud6p sites. These are the interactions selectively impaired at the bud cortex in bud6Δ cells. Finally, interactions away from Bud6p sites within the bud differ from those occurring at the mother cell cortex, pointing to the existence of an independent factor controlling cortical contacts in mother cells after bud emergence.
INTRODUCTION
Spatial coordination between the axis of the mitotic spindle and the division plane is critical for chromosomal segregation in eukaryotic cells as well as the generation of cell diversity during metazoan development (Rhyu and Knoblich, 1995). These principles can be studied even in unicellular organisms dividing asymmetrically such as the budding yeast Saccharomyces cerevisiae. Indeed, budding yeast couples spindle orientation to the division axis, ultimately dictating the segregation of one pole of the spindle to the daughter cell while retaining the second pole within the mother cell (Segal and Bloom, 2001).
The yeast mitotic spindle pathway begins at bud emergence with the duplication of the spindle pole body (SPB), the counterpart of the centrosome of animal cells (Byers, 1981; Lew et al., 1997). SPBs organize both astral (cytoplasmic) and intranuclear microtubules (MTs) during the cell cycle. SPBs later separate to generate a short intranuclear spindle. Coupled to these events, a precise program of astral MT–cortex interactions dictates positioning of the spindle (Segal and Bloom, 2001). First, astral MTs are selectively captured at the bud cortex. Coincident with spindle assembly, new interactions occur with the bud neck region. Finally, the preanaphase spindle orients along the mother-bud axis (Carminati and Stearns, 1997; Shaw et al., 1997; Segal et al., 2000b). By virtue of this configuration, the daughter-bound pole, the SPBd, can translocate into the bud in the course of spindle elongation in anaphase, whereas the remaining pole, SPBm, is retained in the mother cell. Cyclin-dependent kinases regulate SPB function to coordinate establishment of polarity with spindle assembly (Segal et al., 1998, 2000b) by enforcing the correct response of astral MTs to temporally regulated spatial cues emanating from the cell cortex (Segal and Bloom, 2001).
We previously implicated the actin-interacting protein Bud6p in directing cortical MT capture to enforce spindle polarity. Bud6p/Aip3 (Amberg et al., 1997) follows a temporal program of cortical localization that parallels MT–cortex interactions during spindle morphogenesis (Segal et al., 2000a). Bud6p initially localizes to the prebud site and remains at the bud tip after bud emergence (Amberg et al., 1997). Concomitant with spindle assembly, Bud6p accumulates at the bud neck (Segal et al., 2000a). Finally, Bud6p mobilizes from the bud cortex to the neck and gives rise to a double ring at cytokinesis (Amberg et al., 1997; Segal et al., 2000a).
Spindle orientation is sensitive to perturbation of the actin cytoskeleton (Palmer et al. 1992; Theesfeld et al., 1999). Yet, shortly before anaphase, orientation becomes actin independent (Theesfeld et al., 1999). A candidate for a link between actin and spindle orientation is Kar9p, which is transported to the bud along actin cables (Beach et al., 2000; Yin et al., 2000). Kar9p participates in cortical capture by interacting with MTs via the EB1 homologue Bim1p (Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000; Tirnauer and Bierer, 2000). According to current models (Schuyler and Pellman, 2001), the roles of Bud6p and the formin Bni1p in MT capture may be limited to positioning Kar9p at the bud tip cortex, based on their function in actin organization (Miller et al., 1999; Evangelista et al., 2001; Sagot et al., 2001). This view, however, is inconsistent with genetic analysis of spindle orientation (Theesfeld et al., 1999; Segal et al., 2000a; Yeh et al., 2000). Indeed, spindle orientation phenotypes are very different in a bni1Δ vs. a bud6Δ mutant and unrelated to Kar9p function.
Bni1p is critical for correct retention of Bud6p at the bud tip cortex, after bud emergence. A bni1Δ mutation, which in itself is insufficient to abolish actin cables (Evangelista et al., 2001; Sagot et al., 2001), causes displacement of Bud6p from the bud cortex to the bud neck. MTs then follow mislocalized Bud6p, resulting in abnormally enhanced capture at the bud neck. In contrast, a bud6Δ mutation abrogates the majority of cortical interactions with the bud or the bud neck (Segal et al., 2000a). Neither deletion, however, precludes Kar9p-dependent MT capture (Miller et al., 1999; Segal et al., 2000a). Finally, Bud6p is critical for spindle insertion into the bud neck past the actin-sensitive step (Segal et al., 2000a). Together, these data suggest that the role of Bud6p in MT capture is not mediated via Kar9p function.
Herein, we present digital imaging microscopy analysis of cells coexpressing GFP:Bud6 and GFP:Tub1 fusion constructs to evaluate MT–Bud6p dynamic interactions in vivo. This study supports a direct participation of Bud6p in MT capture. The analysis highlights additional, unanticipated roles of Bud6p in cueing MT–cortex interactions during stages of the cell cycle beyond spindle morphogenesis (Segal et al., 2000a) in a Kar9p-independent manner. In addition, Bud6p dictates differential modes (Carminati and Stearns, 1997; Adames and Cooper, 2000) and duration of MT–cortex interactions throughout the cell cycle. Finally, disruption of BUD6 particularly eliminates the specific events observed to occur at Bud6p sites in wild-type cells.
MATERIALS AND METHODS
Yeast Strains, Genetic Procedures, Media, and Growth Conditions
All strains used in this study were isogenic to 15Dau, a derivative of BF264-15D (Segal et al., 1998). Deletion mutations were constructed by replacing the entire open reading frames by using KANR cassettes amplified by polymerase chain reaction according to Wach et al. (1994). Deletions were confirmed in all final strains by PCR analysis. Derivatives expressing a GFP:Tub1 and GFP:Bud6 fusion were obtained by transformation with pAFS72 (Straight et al., 1997) and pRB2190 (Amberg et al., 1997), respectively. Standard yeast media and genetic procedures were used (Sherman et al., 1986). Yeast cultures were grown at 23°C unless indicated.
Digital Imaging Microscopy in Live Cells Expressing GFP:TUB1 and GFP:BUD6
Cells were grown to ∼5 × 106 cells/ml in selective glucose-containing medium and then mounted in the same medium containing 25% gelatin to perform time-lapse recordings at room temperature as described previously (Shaw et al., 1997; Maddox et al., 1999; Segal et al., 2000a). Briefly, a total of five fluorescence images were acquired at a Z-distance of 0.75 μm between each plane. A single differential interference contrast (DIC) image was taken in the middle focal plane. This acquisition regime was repeated at 15-, 30-, or 60-s intervals. Although this resolution may have limited the accuracy of estimates of duration of cortical interactions lasting under 1 min, it still provided sufficient dynamic range for interactions ranging between 1 and 7 min. Moreover, data derived for MT dynamic measures (our unpublished data) and scoring of bulk cortical interactions were in good agreement with values obtained previously based on dynamic studies undertaken with similar or higher resolution (Carminati and Stearns, 1997; Shaw et al., 1997; Maddox et al., 1999; Adames and Cooper, 2000). Images were processed as described previously (Shaw et al., 1997; Maddox et al., 1999) by using MetaMorph (Universal Imaging, Downington, PA) software. Quantitation of cytoplasmic MT interactions was carried out by scoring all possible contacts observed by following the history of individual MTs. Interactions were categorized as described previously (Carminati and Stearns, 1997), except that results were expressed as percentage of a particular type of interaction over the total cortical interactions scored at each cell cycle stage, rather than as percentage of cells in which a MT showed a particular behavior (Carminati and Stearns, 1997). Briefly, interaction categories were as follows: 1) MTs “hitting” the cortex, i.e., transient contacts with the cortex during MT cycles of growth and shrinkage; 2) MTs growing at the cell cortex; 3) MTs shortening at the cell cortex (2 and 3 occurred while the MT plus end remained in contact with the cell cortex and were accompanied by corresponding movements of the spindle pole and nucleus toward or away from the cortex, respectively); and 4) MTs displaying “sweeping” movements on the cortex. This category included rare MT sliding movements as defined by Adames and Cooper (2000). In contrast to the previous study by Carminati and Stearns (1997), data corresponding to small-budded cells was not pooled with that corresponding to unbudded cells during our analysis. Duration of cortical interactions was also determined by following the history of individual MTs. Mean values correspond to the total time for each contact event (in minutes) divided by the number of contacts scored, n is number of MTs. Results were expressed as mean ± SD.
The operational definition of neck region, for the purpose of microscopy, was the cell cortex area within a 0.5-μm distance from the point of constriction between the mother and the bud. Cortical association was defined according to Carminati and Stearns (1997).
Single still cell images were captured using 100% incident light intensity and 500-ms exposures (Segal et al., 1998). Quantitation of orientation of MT attachments relative to the division site was based on scoring at least 500 cells at each cell cycle stage from an asynchronous population in two independent counts. Spindle measurements and SPB distance in digital images were carried out as described previously (Segal et al., 2000b). The kinetics of spindle elongation was determined for each time-lapse series analyzed to separately score MT–cortex interactions during the “fast” and “slow” phases of anaphase B.
RESULTS
Microtubule–Bud6p Dynamic Interactions during Cell Cycle
We have previously correlated Bud6p cortical distribution with the program of MT–cortex interactions during the cell cycle (Segal et al., 2000a). Herein, we undertook the characterization of yeast cells coexpressing GFP:Bud6p and GFP:α tubulin (Tub1p) fusions to establish whether cortical interactions indeed coincided with sites of Bud6p localization in live cells. The use of time-lapse digital imaging microscopy further provided information on the dynamic properties of such interactions. A summary of the cell cycle program of MT–Bud6p interactions emerging from this study is presented in Figure 1. Interactions were studied along the complete cell cycle (Figure 1A) and categorized according to defined cortical areas in the bud or mother cell (Figure 1B).
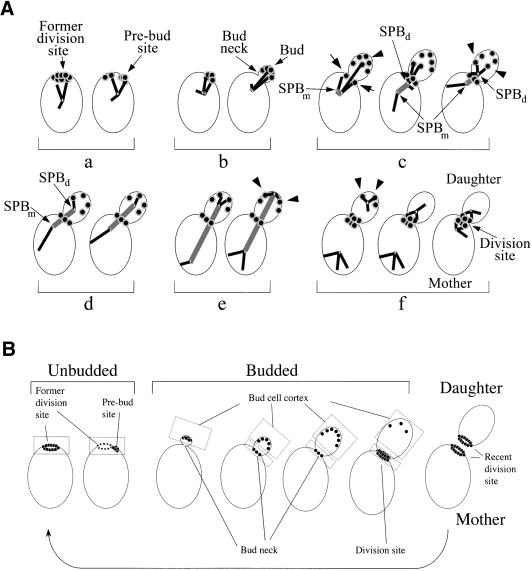
Figure 1.
Cell cycle program of Bud6p-driven microtubule–cortex interactions revealed by real-time imaging microscopy of live cells. (A) Cartoon depicting a summary of the cortical interactions documented in the present study. (a) MTs emanating from the SPB primarily interacted with Bud6p decorating the cytokinesis site (former division site in a G1 cell) after spindle disassembly. These contacts tethered the SPB near the recent division site. (b) Accumulation of Bud6p at the prebud site (the future budding site) was immediately followed by new MT contacts at this new area of capture. Interactions continued with the emerging bud directing the duplicated SPBs to face the bud neck as MTs underwent capture at the bud tip cortex. Bud6p initially decorates the distal portion of the bud cortex. The constriction between the mother and the bud, the bud neck, was free of label at this stage. (c) Bud6p began to partition at the neck shortly before spindle assembly. This was followed by interactions occurring within the bud (arrowhead) or neck (arrows) as the SPBs separated. The SPBd maintained a tight dynamic association with the bud neck and the proximal bud cortex by a combination of pulling and pushing contacts until the SPBd was inserted at the neck. Contacts with the bud tip were rare and it was the interactions with a broad bud neck region that seem to maintain the spindle in place at this stage. The SPBm was essentially prevented from interacting with the bud cell cortex. (d) Once the SPBd translocated into the bud it continued to interact with the daughter face of the neck until the fast phase of spindle elongation began. At this point MTs reached further into the bud and interacted precisely at Bud6p sites, often close to the bud tip in a series of short-lived contacts. (e) During the slow phase of spindle elongation, cortical contacts lengthened and the SPBd often interacted with several cortical Bud6p sites. (f) These contacts continued after spindle disassembly. Once Bud6p abandoned the distal daughter cell cortex, MT interactions concentrated at the Bud6p ring. As a result, the SPBd became tethered to the division site. This generally preceded a similar MT-mediated movement of the SPBm. (B) Spatial definitions for the scoring of cortical interactions in this study. Interactions were classified by compartment (mother vs. bud) in budded cells. Bud6p localization is confined to the regions highlighted by boxes. Data in Tables 1 and 2 were collected for cortical interactions within these regions exclusively. Table 3 summarizes all interactions taking place within the mother cell cortex of budded cells (excluding the neck region).
After cytokinesis, both mother and daughter cells inherited a Bud6p ring marking the recent division site. At this point, each SPB established interactions with this Bud6p ring, thus causing the SPBs to approach and be retained at the recent division site. Once selection of a new polarity axis and Bud6p accumulation at the prebud site began, MTs became tethered to this site (Figure 1A, a and b). MT interactions continued with the bud tip after bud emergence. Slightly preceding spindle assembly, Bud6p began to redistribute to the bud neck region, directing a subset of the MT interactions to this new area of capture (Figure 1A, c). As the bud continued to grow, the GFP:Bud6p label further resolved into discrete dots over the bud surface. As a result, it was easier to visualize MTs interacting with defined dots of cortical Bud6p in the later portion of the cell cycle. During anaphase, interactions occurred with remarkable precision at Bud6p sites (Figure 1A, d and e). Finally, spindle disassembly coincided with the formation of a double Bud6p ring at the bud neck (Figure 1A, f).
Time-lapse analysis revealed a correlation between the interaction of MTs at Bud6p sites and the relative duration of this cortical contact (Table 1). Astral MT–cortex interactions lasted two- to fourfold longer at Bud6p sites relative to contacts in the same areas at sites devoid of Bud6p. Yet, duration was also dependent on the cell cycle position, indicating that astral MT dynamic behavior is not solely determined by interaction with Bud6p. On average, interactions at Bud6p sites lasted 2 min in G1, 2.6 min during bud emergence, 1.5 min throughout spindle assembly, <1 min in early spindle elongation, and 2 min toward late mitosis. In contrast, interactions away from Bud6p sites lasted 0.5–0.7 min (Table 1).
Table 1.
Average duration of MT–cortex interactions during the cell cycle
| At Bud6p cortical sites | Away from Bud6p sites | |
|---|---|---|
| Spindle disassembly | ||
| Daughter cell cortex | 2.25 ± 1.45 min (n = 22) | |
| Daughter cell, cytokinesis site | 1.91 ± 1.08 min (n = 60) | 0.55 ± 0.30 min (n = 35)a |
| Mother cell, cytokinesis site | 2.10 ± 1.10 min (n = 51) | |
| Bud emergence | ||
| Bud cell cortex | 2.65 ± 1.05 min (n = 13) | 0.68 ± 0.20 min (n = 15)b |
| Spindle assembly | ||
| Bud neck | 1.14 ± 1.03 min (n = 25) | |
| 0.65 ± 0.50 min (n = 12)c | ||
| Bud cell cortex | 1.55 ± 1.08 min (n = 55) | |
| Preanaphase orientation | ||
| Bud neck | 0.96 ± 0.74 min (n = 47) | |
| 0.62 ± 0.30 min (n = 14)d | ||
| Bud cell cortex | 1.43 ± 0.68 min (n = 7) | |
| Spindle elongation | ||
| Fast phase, bud cortex | 0.79 ± 0.51 min (n = 81) | |
| 0.58 ± 0.20 min (n = 20)e | ||
| Slow phase, bud cortex | 2.01 ± 1.17 min (n = 39) |
For all spatial definitions, see Figure 1B.
Events in unbudded cells within 1 μm of the division site.
Events in the proximal portion of the bud and bud neck region.
Events within the bud cell cortex only.
Distinct modes of MT–cortex interactions contribute to MT-mediated positioning of the spindle poles throughout the cell cycle (Carminati and Stearns, 1997; Adames and Cooper, 2000). Quantitation of these modes of interaction at or away from Bud6p cortical sites was carried out relative to cell cycle stage (Table 2). Throughout the cell cycle, MT “growth” or “shrinkage” at the cortex, which is coupled to SPB movements, was typically absent at sites devoid of Bud6p at the bud or bud neck (Table 2, boxes). In contrast, hitting or sweeping events occurred both at or away from Bud6p sites (Table 2). The following sections describe in detail these MT–cortex interactions as a function of cell cycle stage.
Table 2.
Differential microtubule interactions with cortical Bud6p during the cell cycle
| Unbudded cellsa | Bud emergenceb | Spindle assembly | Spindle elongationc
|
Spindle disassemblyd | ||
|---|---|---|---|---|---|---|
| Fast | Slow | |||||
| Interactions at Bud6p sites (%) | 67.2mc 78.3dc | 69.7 | 83.0 | 76.7 | 82.5 | 56.8 |
| Number of events | 138 | 256 | 216 | 60 | 177 | 49 |
| Hit cortex (%) | ||||||
| Coincident with Bud6p | 33.0 | 24.3 | 26.9bn 15.0b | 3.3bn 43b | 30.0 | 17.6 |
| Away from Bud6p | 16.7 | 20.8 | 6.9bn 5.6b | 13.3b | 7.9 | 25.5 |
| Grow at cortex (%)e | ||||||
| Coincident with Bud6p | 17.2 | 20.8 | 6.3bn 2.5b | 5.0b | 21.4 | 23.5 |
| Away from Bud6p | 3.6 | 3.8 | 1.1 | 13.8 | ||
| Sweep at cortex (%) | ||||||
| Coincident with Bud6pf | 1.5 | 3.8 | 3.7b | 3.3b | 9.6 | 5.9 |
| Away from Bud6p | 1.8 | 5.7 | 4.5b | 10.0b | 6.8 | 3.9 |
| Shrink at cortex (%)e | ||||||
| Coincident with Bud6p | 26.2 | 20.8 | 13.1bn 15.5b | 5.4bn 16.7b | 21.5 | 9.8 |
| Away from Bud6p | 1.7 | |||||
| Time at cortex (%)g | 53.6 | 51.6 | 42.6 | 50.2 | 58.5 | 41.0 |
Combines data from mother cells (mc) from the time of cytokinesis and daughter cells (dc) from the time when Bud6p is completely relocalized to the division site. Only interactions within 1 μm of the recent division site were considered (72% of all interactions). Cells were followed for 40 min or until Bud6p started labeling the prebud site.
Data in budded cells corresponds to events at the bud neck (bn) or the bud cell cortex (b). For interactions within the mother cell see Table 3.
Events within 20 min after bud emergence.
The kinetics of pole-pole separation was determined for each time lapse series to distinguish between the fast and slow phases of spindle elongation.
Interactions in the daughter cell cortex before complete recruitment of Bud6p to the recent division site.
Interaction modes highly correlated with cortical Bud6p sites.
Sweeping interactions ending at a Bud6p site.
At least 30 MTs were followed at each stage to determine the total cortical association expressed as percentage of total time elapsed.
Orientation of MT Interactions and Bud6p-mediated Capture during G1 through S Phases
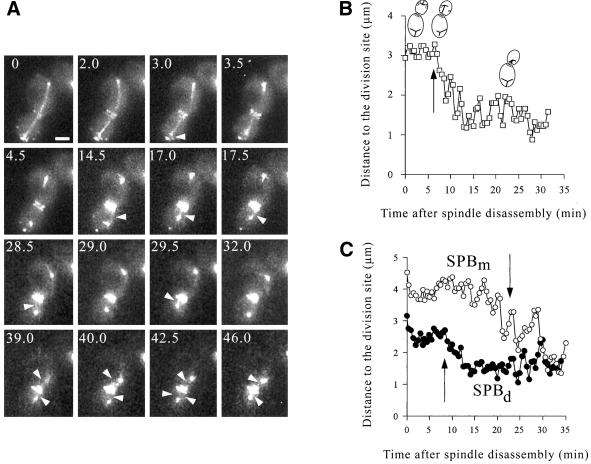
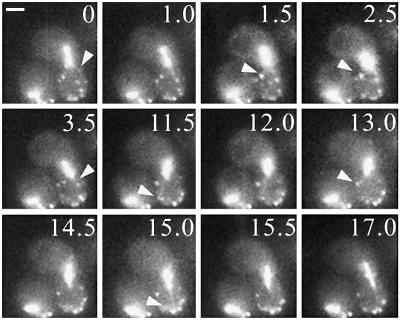
Formation of a double ring containing Bud6p at the site of division was coincident with spindle disassembly (100%, n = 17 time-lapse series). As cells entered G1, the former SPBm and SPBd were typically positioned in proximity to the distal cell cortex of the mother or daughter cell, respectively (Figure 2A, 0–3.5 min). Astral MTs emerging from the SPBd still interacted with the cell cortex at remaining Bud6p sites (Figure 2A arrowhead, 3.0 min). As Bud6p became fully displaced from the distal daughter cell cortex, labeling of the recent division site increased and the distal cortex apparently failed to attract additional MT contacts. This caused a shift of cortical interactions toward the recent division site. Approximately 15 min after spindle disassembly (Figure 2A, arrowhead 14.5–17 min), the SPBd established contacts with the ring, bringing about the movement of the SPBd toward the recent division site (Figure 2A, 17 min). Once in contact with the ring, the SPBd continued to interact preferentially with this region (∼72% of all possible contacts in the cell; see legend to Figure 2).
Figure 2.
Effect of Bud6p-mediated MT capture on SPB position during early G1. (A) Time-lapse series illustrating events after mitotic exit. Coincident with spindle disassembly Bud6p forms a double ring at the cytokinesis site (2–4.5 min). The SPBd interacts with the distal portion of the cell cortex at Bud6p sites (3 min, arrowhead). As Bud6p concentrates at the ring, MTs encounter this Bud6p area and initiate persistent dynamic interactions with the recent division site (14.5–28.5 min, arrowheads). Finally the SPBm also approaches the division site after MT contacts to the Bud6p ring (40–46 min, arrowheads). Scale bar, 2 μm. (B) SPBd movement relative to the former division site after spindle disassembly. Plot showing the shortest SPBd distance to the division site as a function of time elapsed after spindle disassembly. Data were collected for the complete time-lapse series shown in Figure 2A, as described in MATERIALS AND METHODS. The arrow indicates the first time-lapse frame in which MT contacts to the ring at the division site were first observed. Before this point, MTs established 70% of total interactions with the distal daughter surface (arbitrarily defined as the surface beyond a perpendicular line intersecting the center of the axis of polarity in the daughter cell). Of those, 56.8% occurred coincident with Bud6p. Once contacts occurred with the ring, MTs oriented 72% of all attachments toward the division site. Contacts away from this area became short-lived and only 5% of total contacts occurred with the distal portion of the bud. As a result, the SPBd positioned near the division site. (C) SPBd vs. SPBm movement after spindle disassembly. The plot shows the behavior of SPBs after spindle disassembly. Data were collected as in B for a time-lapse series in which both SPBs remained in focus throughout the series. Arrows indicate the first time frame in which contacts with the division site occurred. SPBd displayed 42% of MT contacts to the distal cell cortex at Bud6p sites until MTs redirected toward the ring (∼7 min). From that moment 69% of total contacts occurred with the ring. The SPBm displayed 62% of total cortical contacts with the distal cell surface which translated in 42% of cortical retention per total time elapsed. Once contacts were established with the division site (∼24 min), MTs spent 62% of the time associated with the ring. By 32 min, the SPBm already contacted the prebud site.
Throughout this phase, particular modes of interaction contributed toward SPBd positioning. After spindle disassembly, MTs preferentially grew or hit the distal cell cortex, at or away from Bud6p (Table 2, spindle disassembly). Once Bud6p relocalized to the division site, prevalent interactions were MTs hitting the division site or MTs growing and shrinking while interacting with the Bud6p ring (Table 2, unbudded cells). The interaction at Bud6p sites (67.2–78.3% of contacts; Table 2, unbudded cells) and the effect of Bud6p on duration of cortex retention (Table 1) can explain the apparently nonrandom distribution of MT contacts. Thus, once the distal cortex was devoid of Bud6p, net retention shifted toward the recent division site (Figure 2, A and B). The SPBm, however, continued to contact the distal cell cortex for an additional 10–25 min but finally reached the division site by interacting with the Bud6p ring (Figure 2, 28.5–39 min). This trend turned out to be the rule (90%, n = 20 time-lapse series). Thus, daughter cell cortical behavior mirrored the redistribution of Bud6p. In contrast, MT-mediated retention of the SPB by the mother cell cortex seemed to involve alternative factors in early G1. Data from a representative time-lapse series illustrates this differential behavior (Figure 2C). In this case, the SPBm repositioned ∼18 min after the SPBd became tethered to the division site (Figure 1C).
A role of Bud6p in driving MT-mediated movement of the SPBs toward the recent division site was further supported by the fact that contacts with the ring preceded SPB movement toward the division site both in the mother and daughter cells (Figure 2, B and C). Thus, MT shortening while in contact with Bud6p sites produced associated movement toward the contact points positioning the SPBs near the recent division site.
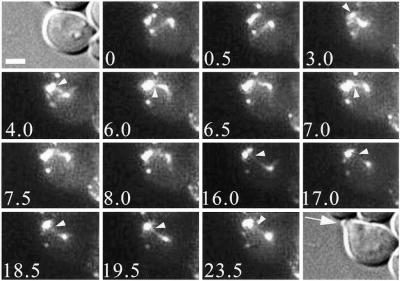
As soon as Bud6p began to accumulate at the prebud site (cortical region where a bud will emerge), the proximity of the SPB tethered to the nearby former division site allowed the immediate establishment of new interactions before bud emergence without further probing of the mother cell cortex. MTs spent the majority of time associated with Bud6p sites, suggesting that MT-based search and capture are actually assisted by the cell cycle-regulated presence of Bud6p at the recent and future division sites (Figures 2A and 3). As shown in Figure 3, dynamic contacts initiated at the prebud site (an unbudded cell at time 0 min) were followed by continued interactions as the bud emerged (Figure 3, 3–17 min). MTs continued to interact with the bud tip cortex where Bud6p was highly concentrated (Figure 3, 17–23.5 min arrowheads; notice the DIC image highlighting the newly formed bud, arrowhead). Although interactions with individual dots could not be resolved during this phase, 69.7% of all contacts by MTs facing the bud occurred at the distal bud region decorated by Bud6p (Table 2, bud emergence). During bud emergence, MT growth and shrinkage at the bud tip reached a balance (24.6 vs. 20.8%; Table 2, bud emergence) to maintain the duplicated SPBs facing the bud neck within a 1.5-μm range. In rare instances, MT-based search proceeded beyond the time of Bud6p association to the prebud site (2 of 18 series analyzed). Nevertheless, MT capture at the bud cortex occurred at most within 10 min after bud emergence. This was in contrast with the lack of processivity of these events observed in bud6Δ cells (see below).
Figure 3.
Orientation of MT interactions toward the prebud site persists during bud emergence. During G1, cortical interactions at Bud6p remnants of the old division site are evident (0–3 min). Progressively, MTs undergo selective capture at the prebud site where Bud6p becomes concentrated (4 min). Notice how the GFP:Bud6p label becomes focused to the new budding site, the prebud site, which lies slightly above the recent division site. Dynamic MT interactions persist throughout bud emergence (16–23.5 min). The presence of Bud6 at high concentration within the bud encourages cortical retention within a relatively small area, resulting in the positioning of duplicated SPBs facing the bud neck as soon as a bud forms. For reference, DIC images corresponding to the first and last frames (arrowhead points at the newly formed bud) in the sequence are also shown. Numbers indicate time elapsed in minutes relative to the first frame shown. Scale bar, 2 μm.
Bud6p–MT Interactions Establish Spindle Polarity and Contribute to Preanaphase Positioning
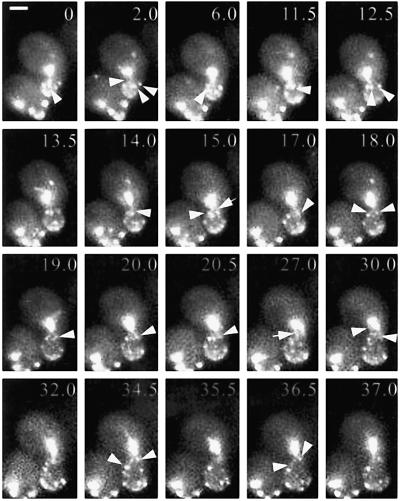
Duplicated SPBs remained facing the bud neck while the bud continued to grow and Bud6p label became more dispersed over the bud surface (Figure 4). Before initiation of SPB separation, GFP:Bud6p began to associate with the bud neck (Figure 4, 2–6 min). From that time, precise contacts occurred at Bud6p sites within the bud or neck region (Figure 4, arrowheads). These continued during spindle assembly (Figure 4, 11.5–27 min). The combined set of interactions with the bud and bud-neck drove the SPBd close to the bud neck within the mother (Figure 4, 32 min). At this stage, hitting interactions (54.4% of all interactions during spindle assembly; Table 2) seemed to antagonize SPB translocation into the bud while the spindle oriented along the mother-bud axis. These hitting events occurred preferentially at Bud6p sites (26.9% at the bud neck and 15% in the bud, or 41.9% of all interactions modes; Table 2) and prevented MTs emerging from the SPBm from reaching the cortex beyond the bud-neck (only 8 of 49 MTs). Moreover, MTs emanating from the SPBm always underwent catastrophe, if they did contact the bud (Figure 4, 27 min, arrowhead), suggesting that dynamic properties of MTs organized after accumulation of Bud6p at the neck (Segal et al., 2000a) might be inherently different than those of MTs undergoing early capture at the bud cell cortex (Segal et al., 2000b). As a result, the SPBm was always drawn away from the bud neck by additional MT dynamic contacts with the mother cell cortex, as the SPBs continued to separate, causing the spindle to align. The progressive penetration of MTs emanating from the SPBd across the neck to Bud6p sites within the bud (Figure 5) caused the SPBd to insert within the bud neck enabling an increased number of processive contacts with the daughter cell cortex (Figure 5, 0–13 min) at or slightly before initiation of spindle elongation (Figure 5, 14.5–17 min).
Figure 4.
Interactions at Bud6p sites during SPB separation enforce spindle polarity. Duplicated SPBs already face the bud neck at time 0. At 2 min, the first interactions with Bud6p at the bud neck region occur (arrowheads). As the spindle assembles, MTs from the SPBd reach into the bud at Bud6p sites near the neck (11.5–12.5 min, arrowheads). These interactions contribute to position the SPBd near the bud neck within the mother (14–20 min). At 15 min, an MT from the SPBm also encounters Bud6p near the neck (arrow). An MT from the SPBm enters the bud at 27 min (arrow). After shortening this MT hits at Bud6p at the bud neck (30 min, left arrowhead) and the SPBm moves further away from the neck. The spindle aligns and the SPBd continues to interact with Bud6p at the neck and bud (34.5–37 min, arrowheads). Numbers indicate time elapsed in minutes relative to the first frame shown. Scale bar, 2 μm.
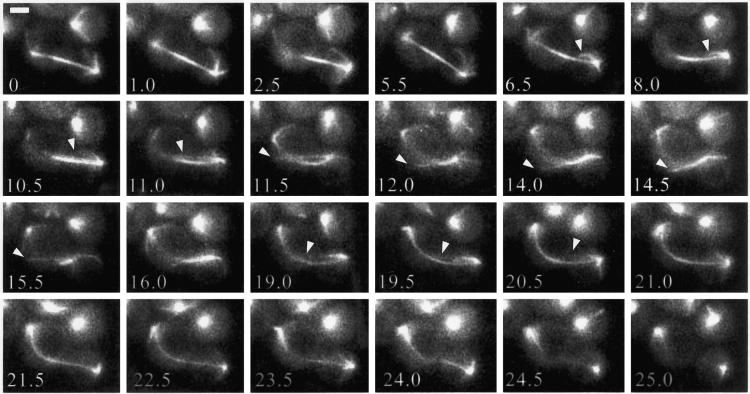
Figure 6.
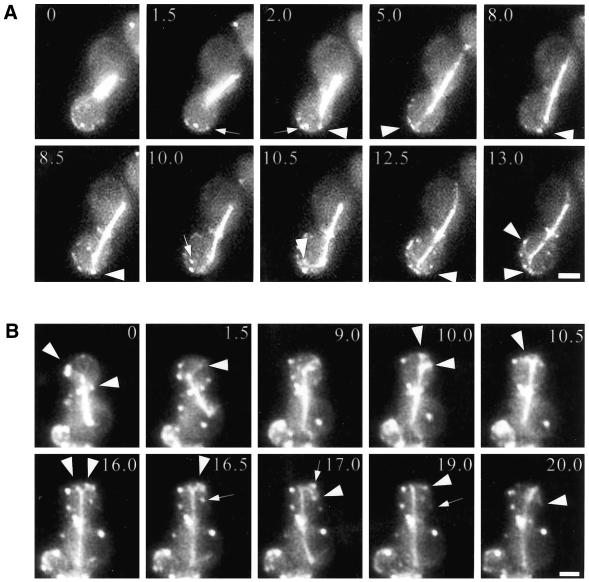
Microtubule–Bud6p interactions accompanying spindle elongation. Two representative time-lapse series are shown. Individual Bud6p–MT interactions are depicted by an arrowhead pointed at the MT contacting the Bud6p site indicated by an arrow in the preceding frame. (A) Interaction between MTs and Bud6p during the fast phase of spindle elongation. MTs precisely interacted with Bud6p dots (arrow, 1.5 min and arrowhead, 2 min; arrow, 2 min and arrowhead, 5 min). Toward the transition to the slow phase of spindle elongation, the SPBd interacts at two sites with MT curving along the cell cortex (arrow, 10 min and arrowheads, 10.5–13 min). (B) Interactions during the fast phase of spindle elongation occur with the bud neck and the bud cortex (arrowheads, 0–1.5 min). During the slow phase the SPB establishes multiple contacts (10.5–16.5, arrowheads). An MT interacts with Bud6p dots with slight curving (16.5 min, arrow and 17 min, arrowhead; 17 min arrow and 19 min, arrowhead; 19 min, arrow and 20 min, arrowhead). These interactions are accompanied by the SPB moving toward the cell cortex. Numbers indicates time elapsed in minutes relative to the first frame. Scale bar, 2 μm.
Figure 5.
Cortical interactions mediating preanaphase spindle insertion at the bud neck. The SPBd progressively gains access to the bud when MTs interact with Bud6p sites at the bud surface (arrowheads). At onset of spindle elongation (15 min), an MT reaches for a Bud6p dot ∼2 μm beyond. Numbers indicate time elapsed in minutes relative to the first frame shown. Scale bar, 2 μm.
Bud6p–MT Interactions at Distal Portion of Bud Resume during Anaphase
Spindle elongation during anaphase exhibits a two-step kinetics. During the fast phase of elongation, the spindle reaches ∼6 μm in 10 min (Yeh et al., 1995; Straight et al., 1998). This is followed by a slow phase during which the spindle continues to elongate to 10–12 μm over >25–30 min. During the fast phase, MTs emerging from the SPBd interacted with Bud6p sites with remarkable precision (76.7% of total contacts; Table 2, spindle elongation). We observed MTs establishing interactions with a Bud6p site roughly 3.5 μm distal to the SPBd (Figure 6A, 1–5 min, arrow–arrowhead pairs). These interactions were relatively short lived and accompanied the movement of the SPB into the bud as the spindle elongated (Figure 6). MTs also displayed sweeping movements while apparently interacting with the cortex devoid of Bud6p (13.3%; Table 2). These movements, however, ended when a Bud6p site was encountered, which occurred in 25% of all sweeping events (Table 2).
MT dynamic behavior at Bud6p sites distinctly changed during the slow phase. Contacts lasted on average two- to threefold longer than during the fast phase (Table 1), and MTs frequently grew or shortened at Bud6p sites (21.4 or 21.5%, respectively; Table 2). The SPBd was often engaged in several interactions combining these dynamic behaviors, apparently causing transient MT curving along the cortex (Figure 6A, 10–13 min, and 6B, 10–17 min). These interactions were coupled to spindle pole movements toward the cortex as described previously (Carminati and Stearns, 1997). Virtually no MT shortening at the cortex was apparent at sites devoid of Bud6p (Table 2). The biological importance of these interactions is demonstrated by the impact of a bud6Δ mutation on spindle dynamics in the latter portion of the cell cycle (see below).
Properties of Mother Cell Cortex of Budded Cells Are Distinct from Those of Bud Cortex Devoid of Bud6p
We observed that the relative prevalence of different modes of cortical interactions at or away from Bud6p sites changed throughout the cell cycle (Table 2). During the G1 interval, cortical retention and particular forms of interaction such as shrinkage at the cortex were dramatically reduced and constrained to sites of Bud6p accumulation. In fact, as stated above, MT shortening occurred very rarely away from Bud6p sites throughout all stages of the cell cycle.
Dynamic properties of the G1 cell cortex at sites lacking Bud6p were in general comparable with those of the bud cortex or neck regions devoid of Bud6p during the budded portion of the cell cycle (0.55 vs. 0.58–0.68 min; Table 1, lack of shrinking at the cortex; Table 2, box). This similarity extended during early bud emergence (Table 3, bud emergence). After bud emergence, however, the properties of the mother cell cortex, which does not recruit Bud6p, were clearly distinct from those of the bud cortex devoid of Bud6p (duration of interactions 1–3 min, Table 3 vs. 0.5–0.7 min, Table 1). Modes of cortical interaction associated with Bud6p such as growth and shrinkage at the bud cell cortex (Table 2, boxes) were well represented also within the mother cell (20.3–31.0% shrinking at the cortex with SPB coupled movement; Table 3, boxes). Because Bud6p does not localize at the mother cell cortex to contribute toward these interactions, other mechanisms must underlie mother cell MT behavior. Supporting this notion, a bud6Δ mutation differentially perturbed MT–cortex interactions within the bud relative to the mother cell, particularly during anaphase (see below). This differential regulation of the mother cell cortex seemed to continue until early G1 (Figure 1C and Table 3, spindle disassembly, boxes).
Table 3.
Microtubule interactions with the mother cell cortex of budded cells
| Bud emergence | Spindle assembly | Spindle elongation
|
Spindle disassembly | ||
|---|---|---|---|---|---|
| Fast | Slow | ||||
| Time at cortex (%) | 33.7 | 38.8 | 52.9 | 39.8 | 38 |
| Number of events | 22 | 19 | 26 | 50 | 15 |
| Hit cortex (%) | 68.8 | 42.5 | 62.5 | 13.5 | 60.3 |
| Grow at cortex (%)a | 12.5 | 35.7 | 12.5 | 26.0 | 20.2 |
| Sweep at cortex (%) | 16.7 | 1.5 | 29.5 | ||
| Shrink at cortex (%)a | 2.0 | 20.3 | 25.0 | 31.0 | 19.5 |
| Average duration (min/interaction) | 0.8 ± 0.2 | 1.3 ± 0.8 | 1.0 ± 0.7 | 2.7 ± 1.2 | 2.0 ± 0.8 |
Lines highlight modes of interaction specifically associated with sites of Bud6p in the bud. These modes of interaction occur with significant frequency at the mother cell cortex.
MT–Cortex Interactions Characteristic of Bud6p Sites Are Selectively Disrupted in bud6Δ Cells
The significance of MT–cortical Bud6p interactions for correct spindle positioning and dynamics was further evaluated by reexamining MT behavior in bud6Δ cells. This analysis extended our previous study (Segal et al., 2000a) by determining whether SPB orientation during early G1 or particular modes of cortical interaction occurring at Bud6p sites in wild-type cells were selectively perturbed in bud6Δ mutants.
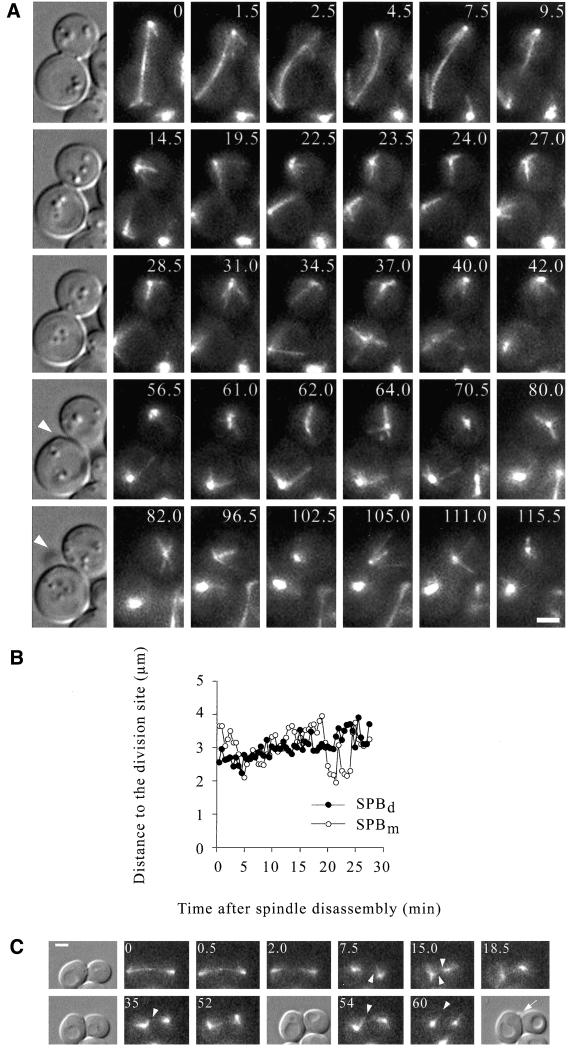
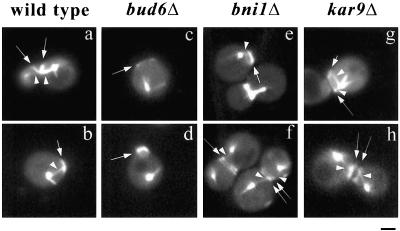
Time-lapse analysis of bud6Δ cells expressing a GFP:TUB1 fusion indicated a failure to reposition the SPBs in proximity to the recent division site (Figure 7A) after cytokinesis. MT interactions did not become restricted to the bud neck region, in contrast to wild-type cells (Figure 7A, 14.5–42 min vs. C, 7.5–35 min). Cortical interactions of MTs emerging from the SPBs were randomly distributed. In addition, contacts with the recent division site were not followed by MT-mediated SPB repositioning (Figure 7B). As cells proceeded through the cell cycle, MTs entered the bud with a significant delay in the course of spindle assembly (Figure 7A, 102.5 min), as described previously (Segal et al., 2000a). To further confirm that early SPB positioning depended on Bud6p activity at the division or prebud sites, cells coexpressing GFP:TUB1 and a septin component, GFP:CDC3 (providing an alternative landmark for the recent division site and prebud site), were used to determine the orientation of MT interactions in a population of cells (Figure 8 and Table 4). Although wild-type cells displayed a preferential orientation of contacts toward the recent division or the prebud sites (along with positioning of SPBs in proximity; Figure 8, a and b), a bud6Δ mutation decreased contacts with the cortex decorated by the septin (Figure 8, c and d). A bni1Δ mutation had no effect on the orientation of cortical contacts during the G1 interval (Figure 8, e and f) consistent with the fact that Bni1p is not critical for initial association of Bud6p with the prebud site (Segal et al., 2000a). Furthermore, loss of affinity for these structures was not observed in a kar9Δ mutant (Figure 8, g and h), indicating that MT capture at the prebud site requires Bud6p but not Kar9p.
Figure 7.
MT–cortex interactions after spindle disassembly in a bud6Δ cell. (A) Time-lapse series showing late anaphase spindle positioning followed by spindle disassembly of a bud6Δ cell expressing GFP:Tub1. Spindle poles are highly mobile (0–7.5 min). After spindle disassembly (9.5 min), MTs interact with the cell surface without preferential contacts with the division site. In the mother cell, after bud emergence (DIC image 56.5 min, arrowhead), MTs still interact randomly over the cell surface. Persistent dynamic interactions with the bud cell cortex were only established after initiation of spindle assembly. For reference, a DIC image corresponding to the first frame of each row is also shown along with fluorescence images from selected frames of the time-lapse series. Numbers indicate time elapsed in minutes. Scale bar, 2 μm. (B) SPB position relative to the division site after spindle disassembly in a bud6Δ cell. The plot represents SPB behavior corresponding to the complete time-lapse series shown in A. SPBs did not reposition near the division site even when occasional contacts with this region occurred. (C) Time-lapse series depicting late anaphase spindle behavior and MT interactions after cytokinesis in a wild-type cell expressing GFP:Tub1. The spindle was significantly less mobile than in the bud6Δ cell shown in A. After spindle disassembly, the SPBd became positioned near the recent division site at 7.5 min followed by repositioning of the SPBm at 15 min. Once cells separated, the SPBm progressively contacted the prebud site and interactions continued during bud emergence (52–60 min). For reference, the DIC images corresponding to 0, 35, 54, and 60 min (arrow points to the newly formed bud) are also provided. Numbers indicate time elapsed in minutes. Scale bar, 2 μm.
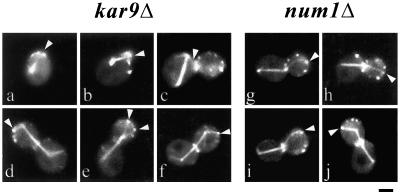
Figure 8.
Orientation of MT interactions relative to the division site in cells expressing GFP:TUB1 and GFP:CDC3 constructs. Single fluorescence images corresponding to wild-type (a and b), bud6Δ (c and d), bni1Δ (e and f), or kar9Δ (g and h) cells expressing GFP:TUB1 and GFP:CDC3 are shown. (a) Wild-type cell after cytokinesis and cell separation. MTs (arrowheads) are directed toward the septin ring (arrows). (b) Wild-type cell with MT (arrowhead) directed to the prebud site (arrow). (c) bud6Δ cell after cytokinesis, the SPB is away from the septin ring (arrow). (d) A bud6Δ cell does not direct MTs toward the prebud site (arrow). (e and f) In contrast, bni1Δ cells show MT contacts (arrowheads) to the septin ring (arrows) and position the SPBs near the division site. (g) MTs in a kar9Δ cell encounter the prebud site (short arrow) while the SPB is still in contact with remnants of the old ring (long arrow). (h) kar9Δ cells establish MT interactions (arrowheads) with septin sites (arrows). Scale bar, 2 μm.
Table 4.
Orientation of MT attachments relative to the division site
 |
Results are expressed as percentage of total attachments coincident with the septin ring in 500 cells counted for each cell cycle stage shown. In budded cells, attachments directed inside the bud (b) or coincident with the septin ring (bn) were scored separately.
A bud6Δ mutation clearly perturbed particular modes of cortical interactions throughout the cell cycle. Interestingly, this was particularly the case within the bud (Table 5). MTs continued to hit or sweep the cortex while growth and shortening at the bud cortex were dramatically impaired (Table 5, bud cell cortex, boxes). In contrast, growth and shortening were well represented in the mother cell (Table 5, mother cell cortex, boxes). Cells seemed to tolerate these defects and most carried on through the cell cycle with abnormally mobile spindles. Toward the end of anaphase, however, MTs emerging from the SPBd failed to shrink at the cell cortex and grew along the surface of the cell, suggesting a failure of (+)-end processing. Failure to productively interact with the cortex resulted in occasional MT growth beyond the bud neck (Figure 9). These MTs were still dynamic (in contrast to MTs of dhc1Δ cells; Carminati and Stearns, 1997) and could also establish interactions with the mother cell cortex (Figure 9), further pointing to differences in mother and bud cortical properties in bud6Δ cells. As shown in Figure 9, after this interaction in the mother cell, the MT shortened back into the bud. This behavior was consistently accompanied by a delay in mitotic exit until the MT was processed back past the bud neck (Figure 9). Thus, MTs emerging from the SPBd can delay mitotic exit if cortical interactions within the bud are defective or MTs grow past the bud neck irrespective of the presence of the SPBd in the bud. Indeed, bub2Δ cells proceeded to disassemble the spindle in the presence of MTs extending from the SPBd into the mother cell (our unpublished data), suggesting a role for the mitotic exit checkpoint.
Table 5.
Microtubule interactions with the cell cortex in bud6Δ cells
| Bud emergence | Spindle assembly | Spindle elongation
|
Spindle disassembly | ||
|---|---|---|---|---|---|
| Fast | Slow | ||||
| Bud cell cortex | |||||
| Time at cortex (%) | 21.3 | 42.0a | 36.9 | ||
| Number of events | 13 | 22 | 19 | 52 | 90 |
| Hit cortex (%) | 84.6 (45.1) | 91.0 (44.4) | 78.9 (61.7) | 34.6 (37.9) | 86.7 (43.1) |
| Grow at cortex (%)b | 7.7 (24.5) | 9.0 (5.0) | 5.2 (5.5) | 52.0c (22.5) | 8.9 (37.3) |
| Sweep at cortex (%) | 7.7 (9.5) | (17.5) | 15.8 (14.5) | 13.5 (16.4) | 4.4 (9.8) |
| Shrink at cortex (%)b | (20.8) | (33.1) | (18.3) | (23.2) | (9.8) |
| Duration of cortical contacts (min)d | 0.52 ± 0.1 | 0.57 ± 0.5a | 0.53 ± 0.1 | ||
| Mother cell cortex | |||||
| Time at cortex (%) | 31.5 | 38.9 | 57.0a | 41.8 | |
| Number of events | 34 | 55 | 13 | 44 | 58 |
| Hit cortex (%) | 88.0 (68.8) | 50.0 (42.5) | 53.8 (62.5) | 34.0 (13.5) | 77.6 (60.3) |
| Grow at cortex (%)b | 5.9 (12.5) | 32.0 (35.7) | 15.3 (12.5) | 25.0 (26.0) | 12.1 (20.2) |
| Sweep at cortex (%) | 5.9 (16.7) | 1.0 (1.5) | 11.3 (29.5) | 3.4 | |
| Shrink at cortex (%)b | (2.0) | 17.0 (20.3) | 30.8 (25.0) | 29.5 (31.0) | 6.9 (19.5) |
| Duration of cortical contacts (min)d | 0.54 ± 0.2 | 1.05 ± 0.46 | 1.68 ± 1.4a | 0.85 ± 0.4 | |
Numbers in small parentheses correspond to the same categories in wild-type cells provided for reference.
Data from fast and slow phase were combined.
Lines highlight the fact that modes of interaction associated with cortical Bud6p are absent in the bud of bud6Δ cells but are still present in the mother cell.
This category represents MTs growing along the cell cortex and was not accompanied by the SPB moving away from the cell cortex as in wild-type cells, in which MTs grew against the cortex.
Duration of cortical interactions represents the average of 20 independent events ± S.D.
Figure 9.
Effect of bud6Δ on cortical interactions in the bud vs. the mother cell cortex during late anaphase. Time-lapse series showing failure of MT shrinkage at the cell cortex in late anaphase in a bud6Δ cell expressing GFP:Tub1. A MT grows along the cell cortex, rather than against the cortex (arrowhead, 6.5–10.5 min). The MT continues to grow past the bud neck (11 min) until it reaches the mother cell cortex (arrowhead, 11.5–15.5 min). Notice that these interactions with the mother cell cortex, in contrast, are coupled to SPB movement toward the site of interaction, causing the spindle to curve. After these interactions, the MT shortens back, past the bud neck (19–20.5 min, arrowhead) and the spindle disassembles (23.5 min). Numbers indicate time elapsed in minutes. Scale bar, 2 μm.
In general, these phenotypes indicate that spindle orientation can be loosely achieved by MTs transiently hitting the bud cell cortex without efficient cortical interactions. However, cells may display significant checkpoint-dependent delays due to lack of these interactions.
MT Capture at Cortical Bud6p Sites in kar9Δ or num1Δ Mutants
Current models suggest that Bud6p contribution to MT capture entails positioning of Kar9p at the bud tip cortex via actin organization. The fact that MT capture during G1 and early bud emergence was not affected by a kar9Δ mutation, however, suggested that MT–cortex interactions at Bud6p sites are independent of Kar9p. Indeed, a kar9Δ mutant coexpressing GFP:Bud6 and GFP:Tub1 exhibited MTs interacting at Bud6p sites after cytokinesis and early bud emergence (Figure 10, a and b). The proportion of cells containing MTs directed into the bud decreased dramatically in midsize-budded cells. Instead, an increase in interactions with the bud neck correlated with Bud6p accumulation at the neck (Figure 10c). However, as soon as the SPBd gained access to the bud, MT contacts occurred coincident with Bud6p sites at the bud cell cortex (Figure 10, d and f). Such behavior defines a temporal window in which Kar9p is critical to maintain dynamic interactions within the bud after bud emergence through spindle assembly.
Figure 10.
MT interactions with Bud6p sites in kar9Δ or num1Δ cells. Single fluorescence images corresponding to kar9Δ (a and f) and num1Δ (g and j) cells expressing GFP:TUB1 and GFP:BUD6 are shown. Arrowheads point to MT-Bud6p contacts. Scale bar, 2 μm. MT–Bud6p interactions at the bud tip occur efficiently in kar9Δ cells until early bud emergence. After accumulation of Bud6p at the bud neck MTs also reach this region. Interactions at the bud cell cortex resume after SPBd translocation into the bud during anaphase. (a) An unbudded cell directs MTs to Bud6p decorated cortex. (b) MTs interact with Bud6p sites at bud emergence. (c) Bud neck interactions at Bud6p sites are observed in large budded cells before SPBd translocation into the bud. (d–f) Once the SPBd translocated into the bud during anaphase, MTs interacted with Bud6p cortical sites. MT–Bud6p interactions during anaphase in num1Δ cells. (g–i) MTs contact Bud6p sites at the cortex before SPB translocation (notice MT curving along the cortex, proposed to participate in dynein-dependent translocation of the SPBd; Heil-Chapdelaine et al., 2000a). (j) Once the SPBd translocated into the bud, MTs continued to interact at Bud6p sites. The behavior of interactions during G1 was comparable with that observed in wild-type cells (our unpublished data).
MT shrinkage at the cell cortex occurred characteristically at Bud6p cortical sites throughout the cell cycle (Table 2, box), a dynamic behavior known to be abolished in dynein mutants (Carminati and Stearns, 1997). Previous studies have suggested, however, that Num1p may serve as a cortical anchor for dynein, particularly, during the later portion of the cell cycle (Heil-Chapdelaine et al., 2000a). Interestingly, a num1Δ mutation did not affect MT–Bud6p contacts during anaphase before (Figure 10, g–i) or after (Figure 10j) SPBd translocation into the bud. The implications of these results are discussed below.
DISCUSSION
Bud6p Cortical Program Provides a Spatial Cue for MT Capture throughout Cell Cycle
Study of cells coexpressing GFP:Bud6 and GFP:Tub1 has enabled us to determine the dynamic properties of cortical Bud6p–MT interactions throughout the spindle pathway. This analysis offers an integrated view of the program of Bud6p cortical localization along with the orientation of dynamic astral MT interactions during the cell cycle.
Bud6p follows a characteristic cortical program during which it initially becomes concentrated at the prebud site and then the bud tip cortex. Subsequently, it accumulates at the bud neck during spindle assembly and, finally, at the site of cytokinesis (Segal et al., 2000a). The orientation of MT capture to these discrete areas kinetically follows Bud6p localization (57–83% of total cortical contacts; Table 2), which accounts for a nonrandom distribution of interactions, even during the G1 phase of the cell cycle. After cytokinesis, MT interactions shift from the cell cortex to the Bud6p ring, dictating MT-mediated positioning of the SPBs near the recent site of division. In the absence of a second landmark for the position of the division site, this MT dynamic behavior was not apparent in previous studies documenting SPB movements during early G1 (Yeh et al., 1995; Shaw et al., 1997; Lee et al., 1999). For the most part, our analysis surprisingly suggested that SPB positioning and MT-based search and capture are facilitated by the presence of Bud6p. Redistribution of Bud6p from the recent division site to the prebud site redirects MT capture as soon as a new polarity axis is established (Figures 2 and 3) independent of Kar9p function (Table 4 and Figure 10). This behavior was also apparent in diploid cells, in spite of the differences inherent to the diploid-specific bipolar budding pattern (our unpublished data). Interactions at Bud6p sites also dictate orientation of MT contacts during spindle assembly and insertion at the bud neck (Figures 4 and 5). This is a critical step in which spindle polarity becomes established and orientation along the mother-bud axis is accomplished. The behavior of MT–cortex interactions at this stage is clearly complex and paradoxically seems to be designed to offer resistance to the translocation of the SPBd into the bud. Yet, interactions at the bud cortex seem to gain momentum at the onset of anaphase. From this moment, MTs interact with Bud6p dots with remarkable precision (Figures 5 and 6). After the fast phase of spindle elongation, the SPBd maintains multiple contacts with Bud6p sites (Figure 6). These contacts continue until Bud6p becomes fully repositioned to the division site after spindle disassembly (Figure 2).
Although the spatial resolution of Bud6p dots by light microscopy may be limited, particularly during the early portion of the cell cycle, persistence of dynamic interactions with Bud6p-decorated areas occurred in preference to contacts in adjacent areas devoid of Bud6p (e.g., distal vs. proximal portion of the bud before accumulation of Bud6p at the bud neck) and was coupled to MT-mediated movements of the SPBs. Overall, however, the density of Bud6p label and its localization to discrete sites is incompatible with the frequency of MT–Bud6p interactions observed (Table 2) being solely a function of chance encounters between MTs and Bud6p dots at any site in the cortex. Indeed, we estimated that no >3.5–8.5% of the cortical surface is occupied by Bud6:GFP at anaphase onset. Moreover, the frequency of MT–Bud6p encounters was reduced to 15% for MTs contacting the bud cortex in bim1Δ cells, supporting the notion that random encounters are insufficient to explain the rate of MT–Bud6p contacts in wild-type cells. Finally, affinity for the recent division site or the prebud site and SPB-coupled movements were also abolished in bim1Δ cells (our unpublished data). Although it is difficult to determine whether these latter effects are at least partly due to the impact of the bim1Δ mutation on astral MT dynamicity during G1 (Tirnauer et al., 1999), it is important to stress that this was the only mutation inactivating a protein implicated in MT capture other than bud6Δ, that abrogated both oriented MT interactions toward the division site and SPB movements that depend on these interactions.
The functional counterpart of Bud6p in Schizosaccharomyces pombe, bud6p, participates in the spatial control of polarized cell growth characteristic of fission yeast, a process involving both MTs and actin (Glynn et al., 2001). Fission yeast bud6p is also an actin interactor and colocalizes with 62% of MTs reaching the cell's ends in the early portion of the cell cycle. It is therefore likely that Bud6p relationship to actin and MT systems is conserved but exploited to support distinct cellular processes in these two divergent yeasts.
Bud6p Promotes, in Particular, MT Shrinkage and Growth at Cortex
Detailed analysis of MT dynamic behavior throughout the yeast cell cycle demonstrated that different modes of cortical interaction prevail during the spindle pathway (Carminati and Stearns, 1997; Adames and Cooper, 2000). We further explored the possible role of Bud6p in promoting these interactions (Table 2). MTs hitting the cortex were the most prominent category at all stages, in agreement with previous studies. This type of interaction occurred equally at or away from Bud6p. In contrast, MT growth or shrinkage at the cortex of the bud and bud neck (in budded cells) or the recent division site and prebud site (in unbudded cells) was restricted to Bud6p sites. MT growth and shrinkage at the cortex was always coupled to SPB movement away or toward Bud6p sites, respectively. Finally, MTs swept the cortex in the absence of Bud6p, with these movements ending frequently at Bud6p dots, particularly during spindle elongation. In support of these conclusions, a bud6Δ mutation selectively perturbed those modes of interaction associated with Bud6p sites (Table 5 and Figure 9).
These modes of cortical interaction are well represented within the mother cell after bud emergence (Table 2 vs. 3). Based on genetic analysis (Table 5) and the fact that Bud6p is not present at the mother cell cortex (beyond the bud neck), it is likely that an alternative mechanism is responsible for promoting cortical interactions within the mother cell.
According to epistasis analysis, elements involved in nuclear migration and spindle positioning in yeast have been organized in distinct early and late pathways (Heil-Chapdelaine et al., 1999). It has been proposed, in addition, that the “early” pathway relies critically on actin organization to stage Kar9p-mediated MT capture (Beach et al., 2000; Bloom, 2000; Yin et al., 2000; Schuyler and Pellman, 2001), whereas the “late” pathway involves dynein (Adames and Cooper, 2000; Heil-Chapdelaine et al., 2000b). It is unlikely, however, that Bud6p solely participates in the early pathway as a consequence of its proposed role in organizing actin, and indirectly Kar9p, in light of the data presented herein.
Indeed, our analysis underscores the importance of Bud6p throughout the cell cycle, even beyond the actin-sensitive step. Moreover, interactions with cortical Bud6p sites occurred at the bud, bud neck cortex, or the division site rather than solely with the bud tip during early orientation. An additional level of complexity is reflected by the observation that MTs interacting with similar Bud6p sites on the bud cortex during spindle assembly led to different dynamic outcomes, depending on whether they emanated from the SPBm or SPBd (Table 2 and Figure 4).
These findings suggest that MT dynamic behavior may reflect the presence of distinct elements associating with MTs (possibly restricted by cell cycle regulation of MT organization; Segal et al., 2000) in partnership with Bud6p. For example, MT functions known to rely on dynein-driven dynamic instability during spindle insertion and anaphase (Carminati and Stearns, 1997; Heil-Chapdelaine et al., 2000b) occurred at Bud6p cortical sites in the bud (Figures 5 and 6) and were not perturbed by deleting NUM1 (Figure 10), which encodes the proposed cortical anchor for dynein (Farkasovsky and Kuntzel, 2001).
Taken together, these results indicate that Bud6p, perhaps in addition to Num1p (Heil-Chapdelaine et al., 2000; Bloom, 2001; Farkasovsky and Kuntzel, 2001) may constitute a cortical partner for part of dynein-dependent control of MT function. Further studies on MT–Bud6p dynamic interactions in various mutant contexts will likely provide greater insight into the relationship of various elements participating in MT-mediated orientation of the mitotic spindle and nuclear migration.
Implications of Bud6p-mediated Capture for Spindle Polarity and SPB Inheritance
We have previously proposed a model integrating coordinated control of SPB function with the Bud6p cortical program to enforce spindle polarity (Segal and Bloom, 2001). The key feature of this model is that an intrinsic delay in MT organization under Clb5-dependent Cdc28p kinase control (Segal et al., 2000b) prevents new MTs generated at the SPBm from establishing contacts with the bud tip once Bud6p is at the bud neck (Segal et al., 2000a). Based on our previous results, however, the model could not address whether Bud6p, in addition, may have a role in enforcing SPB inheritance. In other words, whether it could dictate not just asymmetric SPB fate, but a specific pattern of inheritance for the new vs. the old pole.
Based on the role of Bud6p during G1, it may be that contacts established by the old SPB with the division and the prebud sites underlie the mechanism that commits this SPB to a daughter-bound fate (Pereira et al., 2001). It remains to be demonstrated, however, that preexisting MTs emerging from the outer plaque of the old SPB persist during SPB duplication, a prerequisite for validation of this model. On the other hand, there are no other obvious known means for marking the old SPB with spatial information arising before SPB duplication. The involvement of Bud6p in MT capture during the G1 interval provides the most plausible basis for the selectivity of SPB inheritance. Thus, Bud6p plays a dual role in enforcing this invariant pattern of SPB segregation. First, the old SPB interacts with the prebud site, which later becomes the cortex of the growing bud. This step singles out the old SPB for daughter-bound fate before spindle assembly. As SPB separation begins, de novo MT organization in concert with accumulation of Bud6p at the bud neck forces the new SPB to a mother-bound fate. In support of this model, bud6Δ cells still exhibit intrinsic SPB asymmetry as determined by Dhc1:GFP asymmetric acquisition (Shaw et al., 1997), but delayed acquisition (at the new SPB) is no longer linked to a mother-bound fate (Yeh et al., 2000). Nevertheless, disruption of spindle polarity and Bud6p-driven capture, although delaying spindle orientation relative to anaphase, are not essential for yeast viability. This is because yeast cells are unique in that they specify the site of division at bud emergence and the orientation of the spindle can be eventually forced by the position of the bud during anaphase. Metazoan cells, in contrast, must achieve spindle orientation by anaphase to accurately specify, in turn, the cytokinesis plane. Coordination in this case is critical to produce viable cell progeny.
ACKNOWLEDGMENTS
We thank M. Longtine for generous gift of plasmids and strains and members of the Bloom laboratory for assistance with digital microscopy. This work was supported by U.S. Public Health Service grant GM-38328 (to S.I.R.).
Abbreviations used:
- DIC
differential interference contrast
- MT
microtubule
- SPB
spindle pole body
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–05–0067. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–05–0067.
REFERENCES
- Adames NR, Cooper JA. J. Cell Biol. 149, 863–874. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Bloom K. It's a kar9ochore to capture microtubules. Nat Cell Biol. 2000;2:E96–E98. doi: 10.1038/35014089. [DOI] [PubMed] [Google Scholar]
- Bloom K. Nuclear migration: cortical anchors for cytoplasmic dynein. Curr Biol. 2001;17:R326–R329. doi: 10.1016/s0960-9822(01)00176-2. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2001;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain PacI1p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Lustig RJ, Berlin A, Chang F. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr Biol. 2001;11:836–845. doi: 10.1016/s0960-9822(01)00235-4. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Adames NR, Cooper JA. Formin' the connection between microtubules and the cell cortex. J Cell Biol. 1999;144:809–811. doi: 10.1083/jcb.144.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000a;11:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Tran NK, Cooper JA. Dynein-dependent movements of the mitotic spindle in Saccharomyces cerevisiae do not require filamentous actin. Mol Biol Cell. 2000b;11:863–872. doi: 10.1091/mbc.11.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR. In: Cell cycle control in Saccharomyces cerevisiae. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Pringle JR, Broach JR, Jones EW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 607–695. [Google Scholar]
- Maddox P, Chin E, Mallavarapu A, Yeh E, Salmon ED, Bloom K. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1999;144:977–987. doi: 10.1083/jcb.144.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Cheng SC, Rose MD. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Matheos D, Rose MD. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast. Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–589. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu MS, Knoblich JA. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–526. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2001;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D. Search, capture an signal: games microtubules and centrosome play. J Cell Sci. 2001;114:247–255. doi: 10.1242/jcs.114.2.247. [DOI] [PubMed] [Google Scholar]
- Segal M, Bloom K, Reed SI. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000a;11:3689–3702. doi: 10.1091/mbc.11.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Bloom K. Control of spindle polarity and orientation in S. cerevisiae. Trends Cell Biol. 2001;11:160–166. doi: 10.1016/s0962-8924(01)01954-7. [DOI] [PubMed] [Google Scholar]
- Segal M, Clarke DJ, Reed SI. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J Cell Biol. 1998;143:135–145. doi: 10.1083/jcb.143.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Clarke DJ, Maddox P, Salmon ED, Bloom K, Reed SI. Coordinated spindle assembly and orientation requires Clb5-dependent kinase in budding yeast. J Cell Biol. 2000b;148:441–451. doi: 10.1083/jcb.148.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld CL, Irazoqui JE, Bloom K, Lew DJ. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, O”Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–766. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Brestscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]