Abstract
Fungal sphingolipids contain ceramide with a very-long-chain fatty acid (C26). To investigate the physiological significance of the C26-substitution on this lipid, we performed a screen for mutants that are synthetically lethal with ELO3. Elo3p is a component of the ER-associated fatty acid elongase and is required for the final elongation cycle to produce C26 from C22/C24 fatty acids. elo3Δ mutant cells thus contain C22/C24- instead of the natural C26-substituted ceramide. We now report that under these conditions, an otherwise nonessential, but also fungal-specific, structural modification of the major sterol of yeast, ergosterol, becomes essential, because mutations in ELO3 are synthetically lethal with mutations in ERG6. Erg6p catalyzes the methylation of carbon atom 24 in the aliphatic side chain of sterol. The lethality of an elo3Δ erg6Δ double mutant is rescued by supplementation with ergosterol but not with cholesterol, indicating a vital structural requirement for the ergosterol-specific methyl group. To characterize this structural requirement in more detail, we generated a strain that is temperature sensitive for the function of Erg6p in an elo3Δ mutant background. Examination of raft association of the GPI-anchored Gas1p and plasma membrane ATPase, Pma1p, in the conditional elo3Δ erg6ts double mutant, revealed a specific defect of the mutant to maintain raft association of preexisting Pma1p. Interestingly, in an elo3Δ mutant at 37°C, newly synthesized Pma1p failed to enter raft domains early in the biosynthetic pathway, and upon arrival at the plasma membrane was rerouted to the vacuole for degradation. These observations indicate that the C26 fatty acid substitution on lipids is important for establishing raft association of Pma1p and stabilizing the protein at the cell surface. Analysis of raft lipids in the conditional mutant strain revealed a selective enrichment of ergosterol in detergent-resistant membrane domains, indicating that specific structural determinants on both sterols and sphingolipids are required for their association into raft domains.
INTRODUCTION
Sphingolipids are ceramide-containing lipids that are highly enriched in the outer leaflet of the plasma membrane, where they exert both structural and signaling functions (for review see Dickson, 1998; Schneiter, 1999). In fungi, these lipids are composed of an inositolphosphate-containing hydrophilic head group bound to phytoceramide. The mammalian ceramide typically contains saturated acyl chains ranging from 16 to 24 carbon atoms in length (Gu et al., 1997). The fungal ceramide, on the other hand, contains a saturated C26 very-long-chain fatty acid. Synthesis of this C26 acyl chain occurs by chain elongation of saturated long-chain fatty acids (C16/C18), a reaction that requires an ER-associated acyl chain elongation complex. Elo2p and Elo3p are two components of this complex (Oh et al., 1997). They are presumed to catalyze the condensing reaction of the elongation cycle and exhibit slightly different substrate specificities. Although Elo2p elongates C16/C18 fatty acids to C22/C24, Elo3p is specifically required for the final elongation step from C22/C24 to C26. Cells with mutations in ELO3 are viable but synthesize ceramide/sphingolipids with C22/C24 instead of the natural C26 fatty acid. Synthesis of the C24/C26 fatty acid is essential as an elo2Δ elo3Δ double mutant is not viable (Oh et al., 1997). The physiological significance of this particular fatty acid requirement of the fungal ceramide is unknown, but it is interesting to note that maturation of glycerophosphatidylinositol (GPI)-anchored proteins in yeast involves acyl chain remodeling reactions that serve to introduce a C26 fatty acid into glycolipid protein anchors, suggesting that the presence of C26 is of functional importance (Sipos et al., 1997).
Synthesis of ceramide in mammals and yeast is confined to the ER membrane; subsequent maturation of ceramide to the more complex sphingolipids occurs upon transport to the Golgi apparatus (Levine et al., 2000; Funato and Riezman, 2001). The mature sphingolipids are then transported to the plasma membrane, where they are highly enriched, and constitute ∼30% of all the phosphate-containing lipids (Patton and Lester, 1991; Hechtberger et al., 1994). In addition to sphingolipids, the plasma membrane contains high levels of sterols (Lange et al., 1989; Schneiter et al., 1999). These high levels of sterols and sphingolipids are generally believed to be important for reducing the permeability of the outer most membrane, thus protecting the cell against hostile environmental conditions.
Sterols have a high affinity for lipids containing saturated acyl chains, particularly ceramide (Sankaram and Thompson, 1990; Schroeder et al., 1994). By restricting acyl chain mobility on ceramide, sterols induce the formation of dispersed liquid ordered phase domains, which are resistant to solubilization by detergents (for review see Brown and London, 2000; Rietveld and Simons, 1998). These sphingolipid/cholesterol-rich membrane domains have been proposed to serve as platforms/rafts for the lateral sorting of certain proteins, particularly for those containing a GPI anchor (Brown and Rose, 1992; Simons and Ikonen, 1997; Muniz and Riezman, 2000). Sterols and ceramides are both synthesized in the ER membrane, allowing for a possible early lipid-dependent segregation of proteins along the secretory pathway to the plasma membrane (Brown and Rose, 1992; Bagnat et al., 2000).
Sterols are the product of a complex biosynthetic pathway with more than 20 distinct reactions. The sterol-specific part of this pathway starts by the condensation of farnesyl pyrophosphate to form squalene and, in fungi, ends with the synthesis of ergosterol. The early reactions of this 11-step conversion are essential. They involve cyclization of squalene to lanosterol and subsequent removal of three methyl groups. The enzymes that catalyze the last five steps of the pathway are nonessential. They serve to modify the basic sterol backbone by introduction of a methyl group at position 24 of the side chain (ERG6), isomerization of the double bond from position 8 to position 7 (ERG2), introduction of a second double bond in the B ring (ERG3), introduction of a double bond at position 22 of the side chain (ERG5), and reduction of the methylene bond originally present at position 24 (ERG4; see Figure 1A; for review see Lees et al., 1999; Parks et al., 1999).
Figure 1.
Outline of the sterol biosynthetic pathway in yeast and comparison of sterol structure from yeast, plants, and mammals. (A) Overview of the sterol-specific part of the ergosterol biosynthetic pathway. Essential enzymes are underlined. (B) Comparison of the structure of the major sterols from fungi, mammals, and plants. The yeast sterol, ergosterol, differs from the mammalian cholesterol by i) the presence of a methyl group at position 24, ii) an additional double bond at position 22, and iii) an additional double bond at position 7 of the B ring. The plant sterols stigmasterol and sitosterol have an ethyl group at position 24, a B-ring configuration as cholesterol and the presence (stigmasterol) or absence (sitosterol) of a double bond at position 22.
Just as sphingolipids differ in structure between fungi and mammals, the sterols differ too. Ergosterol is distinct from the mammalian cholesterol by three structural modifications, two in the side chain and one in the B ring. The structure of the plant sterols is intermediate between that of the fungal ergosterol and the mammalian cholesterol. The functional significance of these structural differences, however, is not yet understood (see Figure 1B).
We were interested in investigating the physiological role of the C26 substitution on the fungal ceramide. Using a screen for mutants that are synthetically lethal with elo3Δ, genetic conditions under which the final elongation step from C22/C24 to C26 becomes essential were generated. One of the mutants isolated in this screen is defective in the sterol C-24 methyltransferase, ERG6 (Gaber et al., 1989). We show here that the interaction between ELO3 and ERG6 is highly specific and affects raft structures at the plasma membrane, indicating that fungal-specific lipid modifications in sterols and sphingolipids are codependent and of functional significance.
MATERIALS AND METHODS
Strains and Growth Conditions
Yeast strains used in this study are listed in Table 1. Strains bearing single deletions of nonessential ERG genes were obtained from EUROSCARF (see www.rz.uni-frankfurt.de/FB/fb16/mikro/euroscarf/index.html; Winzeler et al., 1999). Strains were cultivated at 24, 30, or 37°C in YPD rich media (1% Bacto yeast extract, 2% Bacto peptone [Difco Laboratories Inc., Detroit, MI], 2% glucose) or minimal media. To counterselect for the presence of URA3 containing plasmids, 5-fluoroorotic acid (5-FOA; bts BioTech Trade and Service Gmbh, St. Leon-Rot, Germany) was added to solid media at 1 g/l. Media supplemented with sterols contained 5 mg/ml Tween 80 and either 20 μg/ml or 10 ng/ml sterols (Sigma Chemical Co., St. Louis, MO). Sterol rescue experiments were performed under anaerobic conditions by placing the plates into an anaerobic jar containing an AnaeroGen sachet (Oxoid Limited, Hampshire, UK). Plates supplemented with brefeldin A contained 50 μg/ml of the drug. For DNA cloning and propagation of plasmids, E. coli strain XL1-blue (Stratagene, La Jolla, CA) was used throughout this study.
Table 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| FY1679 | MATa/MATα TRP1/trp1Δ63 HIS3/his3Δ200 LEU2/leu2Δ1 ura3-52/ura3-52 | Winston et al. (1995) |
| FY1679a | MATa his3Δ200 leu2Δ1 ura3-52 | Winston et al. (1995) |
| CH1462 | MATα ade2 ade3 his3 leu2 ura3 | Kranz and Holm (1990) |
| RH2607 | MATa his4 leu2 ura3 bar1 lcb1-100, | Sütterlin et al. (1997) |
| YRS1411 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 erg2∷kanMX | EUROSCARF; Winzeler et al. (1999) |
| YRS1256 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 erg3∷kanMX | EUROSCARF; Winzeler et al. (1999) |
| YRS1254 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 erg4∷kanMX | EUROSCARF; Winzeler et al. (1999) |
| YRS1253 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 erg5∷kanMX | EUROSCARF; Winzeler et al. (1999) |
| YRS1266 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 erg6∷kanMX | EUROSCARF; Winzeler et al. (1999) |
| YRS1410 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 erg24∷kanMX4 | EUROSCARF; Winzeler et al. (1999) |
| YRS1265 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 elo2∷kanMX4 | EUROSCARF; Winzeler et al. (1999) |
| YRS1116 | MATα ade2 ade3 his3 leu2 ura3 elo3∷his5 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1118 | MATα his3Δ200 leu2Δ1 ura3-52 elo3∷his5 | This study |
| YRS1119 | MATa his3Δ200 leu2Δ1 ura3-52 elo3∷his5 | This study |
| YRS1247 | MATa his3Δ200 leu2Δ1 ura3-52 elo3∷his5 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1513 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg2∷kanMX4 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1504 | MATa his3 leu2 ura3 lys2Δ0 elo3∷his5 erg3∷kanMX4 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1510 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg4∷kanMX4 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1506 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg5∷kanMX4 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1508 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg6∷kanMX4 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1514 | MATα his3 leu2 ura3 elo3∷his5 erg24∷kanMX4 pPS2[URA3 ADE3 ELO3] | This study |
| YRS1516 | MATa his3 leu2 ura3 elo2∷kanMX4 erg6∷kanMX4 pRS416-ERG6[URA3 ERG6] | This study |
| YRS1518 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg6∷kanMX4 pRS426-ERG6[URA3 ERG6] | This study |
| YRS1520 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg6∷kanMX4 pME1[LEU2 ERG6] | This study |
| YRS1519 | MATα his3 leu2 ura3 lys2Δ0 elo3∷his5 erg6∷kanMX4 pME1ts#6[LEU2 erg6ts] | This study |
| YRS1550 | MATa his3-Δ200 leu2-Δ1 ura3-52 trp1 elo3∷his5 pep4∷LEU | This study |
| YRS1554 | MAT his3 leu2 ura3 elo3∷his5 end4∷kanMX4 | This study |
| YRS1551 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6∷kanMX4 pME1[LEU2 ERG6] | This study |
Plasmid Constructions
To generate the ADE3-containing sectoring plasmid used for the synthetic lethal screen, ELO3 was amplified by PCR using the primer pair ELO3RegF2/ELO3RegR (see Table 2 for sequence of oligonucleotides) and genomic DNA from strain FY1679 as template. The ELO3 fragment was cloned blunt end into the unique SmaI site of pCH1122 to yield pPS2 (Table 3).
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| ELO3RegF2 | ATCGACGCGTCGACCGAAGTTTTTATTCCGTCCAC |
| ELO3RegR | ATCCCCCGGGGCACCAAGCCAATAATCTATCA |
| ERG6RegF | TATGCAGCTCAAAGGTACTACTGTGCTTAATC |
| ERG6RegR | GATTGAGGATCCTATGGAAACAGTTACGATGC |
| ELO3koF | ATGAACACTACCACATCTACTGTTATAGCAGCAGTTGCCGACCAGTTCCACGGATCCCCGGGTTAATTAA |
| ELO3koR | TTAAGCTTTCCTGGAAGAGACCTTGGTGTTAGAGGTCTTGACACCAGTAGGAATTCGAGCTCGTTTAAAC |
| Bubble-1 | GAAGGAGAGGACGCTGTCTGTCGAAGGTAAGGAACGGACGAGAGAAGGGAGAG |
| Bubble-2 | GACTCTCCCTTCTCGAATCGTAACCGTTCGTACGAGAATCGCTGTCCTCTCCTTC |
| Bubble 224 | CGAATCGTAACCGTTCGTACGAGAATCGCT |
| ERG6mutF | TGAAGAACTGTGGTTTCGAAGTCC |
| ERG6mutR | AAAATACTGGTCGTTTGCCACG |
Table 3.
Plasmids used in this study
| Designation | Marker(s) | Comments | Source |
|---|---|---|---|
| pRS315 | CEN LEU2 | Sikorski and Hieter (1989) | |
| pRS426 | 2μm URA3 | Christianson et al. (1992) | |
| pCH1122 | CEN URA3 ADE3 | Kranz and Holm (1990) | |
| mTn3 | LEU2 lacZ | Transposon-based knock-out library | Burns et al. (1994) |
| pFA6a | kanMX4 | kanMX4 template for gene replacement | Wach et al. (1994) |
| pFA6a-His3MX6 | his5 | his5 template for gene replacement | Longtine et al. (1998) |
| pIBI96 | CEN LEU2 ELO3 | ELO3 KpnI/SalI genomic fragment in pRS315 | García-Arranz et al. (1994) |
| pPS2 | CEN URA3 ADE3 ELO3 | ELO3 in SmaI-site of pCH1122, sectoring plasmid | This study |
| pPS5 | CEN LEU2 ELO3 kanMX4 | kanMX4 fragment of pFA6a into NotI/SacI of pIBI96 | This study |
| pRS426-ERG6 | 2μm URA3 | ERG6 in SacI/BamHI site of pRS426 | D. Zweytick and G. Daum, Graz University of Technology |
| pME1 | CEN LEU2 ERG6 | ERG6 in SacI/BamHI site of pRS315 | This study |
| pME1ts#6 | CEN LEU2 erg6ts | L341P, temperature-sensitive allele of ERG6 | This study |
The kanMX4-containing ELO3 tester plasmid, pPS5, was generated by cloning the NotI/SacI fragment harboring the kanMX4 gene from pFA6a (Wach et al., 1994) into the corresponding sites of pIBI96 (García-Arranz et al., 1994).
The ERG6 gene was cloned, after PCR amplification with the primer pair ERG6RegF/ERG6RegR from genomic DNA, as a SacI/BamHI fragment into the corresponding sites of pRS426 (Christianson et al., 1992) to generate pRS426-ERG6. The ERG6-containing SacI/BamHI fragment from pRS426-ERG6 was then cloned into the corresponding sites of pRS315 (Sikorski and Hieter, 1989) to generate pME1.
Restriction and ligation of DNA fragments, preparation of plasmid DNA, elution of DNA from low-melt agarose gels and transformation of lithium acetate competent yeast cells was performed according to standard procedures. Enzymes used for DNA manipulations were supplied by MBI Fermentas (St. Leon-Rot, Germany).
Synthetic Lethal Screen
To generate a host strain for the synthetic lethal screen, the ade2 ade3 parental strain CH1462 was first transformed with pPS2 [ADE3 URA3 ELO3] to render it HIS3-dependent auxotrophic for histidine (Kranz and Holm, 1990). The ELO3 gene of this strain was then replaced by the bacterial his5 gene, amplified with the primer pair ELO3koF/ELO3koR from pFA6a-His3MX6 (Longtine et al., 1998), to generate strain YRS1116. This strain formed red/white colony sectors on YPD and was used for the synthetic lethal screen using a transposon-based knockout library (m-Tn3; see http://ygac.med.yale.edu/; Burns et al., 1994). Approximately 40,000 leucine prototrophic transformants were screened for nonsectoring colonies. Plasmid independence of the nonsectoring phenotype was then assessed by transformation with pPS5. The site of transposon insertion was identified using a vectorette PCR protocol that used the oligonucleotides Bubble-1, -2, and -224, as described (http://genome-www.stanford.edu/group/botlab/protocols/vectorette.html). DNA sequencing was performed on an ABI 373A sequencer using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA Polymerase (PE Perkin Elmer-Cetus, Foster City, CA).
Generation of ERG6 Alleles That Are Temperature Sensitive for Function
To generate a strain in which the synthetic lethal interaction between ELO3 and ERG6 could be conditionally imposed, the 3′ end of ERG6 was amplified by PCR under error prone conditions (2.5 mM dGTP instead of the normal 250 μM) using the primer pair ERG6mutF/ERG6mutR. The 569-base pair fragment was then cotransformed with NheI/MscI gapped pME1 into strain YRS1508. One thousand leucine prototrophic colonies were replica plated on 5-FOA media to force loss of pPS2. Uracil auxotrophic colonies were screened for temperature-sensitive growth by replica plating to 37°C. Three plasmids conferring temperature-dependent growth to an elo3Δ erg6Δ double mutant were isolated and subject to sequence analysis. pME1ts#6 was used for all further studies.
Isolation of Detergent-insoluble Membrane Domains
Detergent-insoluble membrane domains were isolated after floatation on Optiprep gradients (Axis-Shield, Huntingdon, UK) exactly as described by Bagnat et al. (2000). Proteins were precipitated with TCA (10%), dissolved in sample buffer, and subjected to SDS-PAGE and Western blot analyses. Raft association of newly synthesized Pma1p was examined as described by Gong and Chang (2001). Lysates of labeled cells (3–4 OD600 nm equivalents) were extracted with 1% Triton X-100 for 30 min at 4°C. Samples were centrifuged at 100,000 × g for 1 h. Pellets were resuspended in 1% SDS. Detergent concentrations in aliquots of total, supernatant, and pellet samples were adjusted for immunoprecipitation.
For lipid analysis, cholesterol was added as internal standard, and lipids were extracted using n-heptane. Silylated sterol adducts were then quantified and analyzed by gas chromatography–mass spectroscopy (GC-MS) analysis as described below. Raft association of inositol containing lipids was examined after growing cells in inositol-free media containing 0.1 μCi [3H]myo-inositol (Perkin Elmer-Cetus Life Sciences, Cambridge, UK) for 16 h.
Western blot analyses using rabbit anti-Erg6p or anti-Pma1p (1:10,000; G. Daum, Graz University of Technology, Austria) or rabbit anti-Gas1p (1:2000; A. Conzelmann, University of Fribourg, Switzerland) was performed as described before by Schneiter et al. (1999).
Protein Secretion and Maturation
Secretion of invertase was assayed as follows. Cells were grown in minimal media containing 5% glucose to an OD600 nm of 0.2. Invertase expression was induced by resuspending the cells in low glucose media (0.05%), and the cultures were preshifted to 37°C for 15 min. Samples were then removed at the indicated time points, and cells were metabolically poisoned by the addition of 10 mM NaN3. Invertase activity of equal number of cells was determined as described by Goldstein and Lampen (1975). Carboxypeptidase Y and Gas1p maturation was analyzed as described by Munn et al. (1999) and quantified using NHI Image 1.61.
Lipid Analysis
Sterols were isolated as described by Quail and Kelly (1996), using cholesterol as internal standard. GC-MS analysis of silylated sterol adducts was performed on a Hewlett-Packard HP 5890 Series II gas chromatograph (Palo Alto, CA), equipped with a HP 5972 mass selective detector, and HP 5-MS column (cross-linked 5% phenyl methyl siloxane; dimensions: 30 m × 0.25 mm × 0.25 μm film thickness). The following temperature program was run: 1 min at 100°C, 10°C/min to 250°C, and 3°C/min to 300°C. Sterols were identified based on their mass fragmentation pattern and by comparison to commercially available standards.
Inositol-containing lipids were labeled by incubating 10 OD of cells with 30 μCi [3H]myo-inositol (Perkin Elmer-Cetus Life Sciences) at either 24 or 37°C for 2 h and analyzed as described by Reggiori et al. (1997). For analysis of sphingolipid precursors, 10 OD of cells were incubated with 200 μCi [3H]serine (Perkin Elmer-Cetus Life Sciences) at either 24 or 37°C for 4 h, lipids were extracted with chloroform/methanol (1:1; vol/vol), and mild-base–resistant lipids were analyzed by TLC. Mild-base treatment was performed by incubating lipids in 0.1 M NaOH at 30°C for 1 h.
RESULTS
A Synthetic Lethal Screen with elo3Δ Identifies Mutations in ERG6
To investigate the physiological function of very-long-chain fatty acid (C26)-substituted lipids in yeast, we created genetic conditions under which acyl chain elongation from C22/C24 to C26 becomes essential. Therefore, a red/white colony sectoring-based visual screen was performed to identify mutants that are synthetically lethal with mutations in ELO3. In an initial evaluation of the screening strategy, a transposon–based knockout library was used to mutagenize the genome (Burns et al., 1994). This approach has the advantage of allowing rapid identification of the mutant loci by a PCR-based protocol (see MATERIALS AND METHODS). Of ∼40,000 colonies screened, 19 mutations were isolated that could be grouped into 14 complementation groups.
One of the mutants isolated in this screen contained a transposon insertion in a gene of the ergosterol biosynthetic pathway, ERG6 (unpublished data). Erg6p catalyzes the methylation at position 24 of zymosterol to form fecosterol (Gaber et al., 1989). The synthetic lethality of mutations in ELO3 and ERG6 was independently confirmed by crossing of the respective single mutants followed by tetrad analysis (unpublished data).
The Interaction between ELO3 and ERG6 Is Specific
Mutations in ELO3 and ELO2/FEN1 have previously been isolated in screens for mutants that are resistant against the immunosupressor SR31747, an inhibitor of the sterol C-8 isomerase, ERG2 (Silve et al., 1996). In addition, mutations in ELO2/FEN1 result in resistance against the fungicide fenpropimorph, an inhibitor of the sterol C-14 reductase, encoded by ERG24 (Ladevèze et al., 1993). These ELO-dependent suppressions of inhibitors of the ergosterol biosynthetic pathway, however, have been reported only for the FL100 genetic background, in which both ERG2 and ERG24 are essential. In most other genetic backgrounds, ERG2 and ERG24 are nonessential (Crowley et al., 1996; Winzeler et al., 1999).
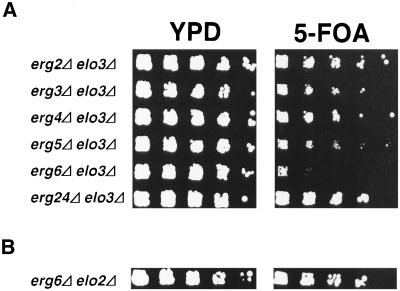
In light of this apparently complex genetic interaction between mutations in ELO2/3 and the ergosterol biosynthetic pathway, we first determined whether the synthetic lethal interaction that we uncovered between ELO3 and ERG6 is specific. Therefore, an elo3Δ mutant strain harboring a wild-type ELO3 gene on an URA3 marked plasmid (pPS2, strain YRS1247) was crossed to all viable mutants in genes of the ergosterol biosynthetic pathway, i.e., ERG2, ERG3, ERG4, ERG5, ERG6, and ERG24. Viability of the respective double mutant was then assessed by plasmid loss on media containing 5-FOA. As shown in Figure 2A, this analysis revealed that the mutation in ELO3 is viable with mutations in all the nonessential ERG genes, except for ERG6, indicating that the synthetic lethal interaction between mutations in ELO3 and ERG6 is specific.
Figure 2.
The interaction between ELO3 and ERG6 is specific. (A) Growth of the elo3Δ mutant (YRS1247) in combination with mutations in genes of the ergosterol biosynthetic pathway—ERG2 (YRS1513), ERG3 (YRS1504), ERG4 (YRS1510), ERG5 (YRS1506), ERG6 (YRS1508), and ERG24 (YRS1514)—was assessed by loss of a plasmid-borne wild-type copy of the ELO3 gene (pPS2) on media containing 5-FOA. Cells of the indicated genotype were serially diluted 10-fold and replica plated on rich media (YPD) or media containing 5-FOA. Plates were incubated for 3 d at 30°C. (B) erg6Δ is viable in combination with mutations in ELO2. Viability of an elo2Δ erg6Δ double mutant (YRS1516) was assessed by loss of a plasmid-borne copy of a wild-type ERG6 gene (pRS426-ERG6) on media containing 5-FOA.
To determine whether a mutation in ERG6 is synthetically lethal with other mutations that affect C26 metabolism, we crossed an elo2Δ mutant strain to erg6Δ, bearing a wild-type ERG6 gene on an URA3-containing plasmid. Unlike elo3Δ mutants, cells defective in ELO2 are still capable of synthesizing the C26 very-long-chain fatty acid, albeit at reduced efficiency (Oh et al., 1997). Viability of the resulting elo2Δ erg6Δ double mutant was again assessed by plasmid loss on media containing FOA. As shown in Figure 2B, this analysis revealed that the elo2Δ erg6Δ double mutant is viable, indicating that the synthetic lethality observed between ELO3 and ERG6 is specific for both partners of the respective biosynthetic pathways.
The Viability of an elo3Δ erg6Δ Double Mutant Is Rescued by Ergosterol but not by Cholesterol
Analysis of sterol auxotrophic mutants has revealed distinct cellular functions of sterols that can be fulfilled by supplementations with different concentrations of sterols. The function of sterols required for membrane integrity, termed the bulk function, requires supplementation with 5 μg/ml ergosterol and is also provided by cholesterol. A second function of ergosterol is required for cell cycle progression (Dahl et al., 1987). This sparking function of ergosterol requires 10 ng/ml ergosterol and is not provided by cholesterol but has a stringent structural requirement for ergosterol (Rodriguez and Parks, 1983).
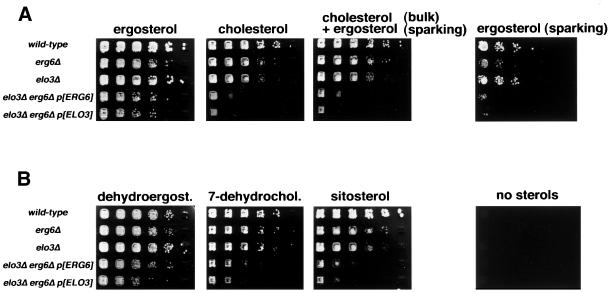
To determine which of these two essential functions of ergosterol is affected in the elo3Δ erg6Δ double mutant, cells were supplemented with different sterols and grown under anaerobic conditions to allow sterol uptake. Viability of the double mutant was again assessed by monitoring loss of a plasmid-borne copy of a wild-type ELO3 or ERG6 gene on media containing 5-FOA. As shown in Figure 3A, the elo3Δ erg6Δ double mutant was rescued on media supplemented with bulk concentrations of ergosterol. Cholesterol, a mixture containing bulk concentrations of cholesterol and sparking concentrations of ergosterol, or sparking concentrations of ergosterol alone, failed to rescue the double mutant. This observation indicates that the elo3Δ erg6Δ mutant has a stringent requirement for bulk concentrations of ergosterol that is not provided by cholesterol and that is not depending on sparking concentrations of ergosterol.
Figure 3.
Viability of an elo3Δ erg6Δ double mutant is rescued by supplementation with ergosterol but not by cholesterol. (A) Viability of the elo3Δ erg6Δ double mutant was assessed by loss of a plasmid-borne wild-type copy of either the ERG6 gene (pRS426-ERG6, p[ERG6]) or the ELO3 gene (pPS2, p[ELO3]) on media containing 5-FOA and supplemented with various sterols. Cells of indicated genotype were serially diluted 10-fold and replica plated on solid media supplemented with the indicated sterols (20 μg/ml), a combination of bulk concentrations of cholesterol (20 μg/ml) and sparking concentrations of ergosterol (10 ng/ml), or sparking concentra-tion of ergosterol only. Plates were incubated under anaerobic conditions for 5 d. (B) Structural requirements of sterols for the rescue of the elo3Δ erg6Δ double mutant was assessed by plasmid loss on media containing 5-FOA and supplemented with dehydroergosterol (dehydroergost.), 7-dehydrocholestrol (7-dehydrochol.), or sitosterol at bulk concentrations (20 μg/ml), or no sterols.
To further define the structural requirements for this bulk function-dependent rescue of the elo3Δ erg6Δ mutant, viability of this strain was assessed on media supplemented with various sterol derivatives. As shown in Figure 3B, growth of the double mutant could also be rescued by supplementation with bulk concentrations of dehydroergosterol (ergosta-5,7,9(11),22-tetraene-3-ol), a sterol with a third double bond in the ring system and a methyl group at position 24 of the aliphatic side chain, as is characteristic for ergosterol. However, dehydrocholestrol (5,7-cholestadien-3-ol), which lacks this methyl group at position 24 but has a double bond configuration of the B ring identical to that of ergosterol, failed to rescue the double mutant, indicating that the presence of the methyl group at position 24, rather than the double-bond configuration of the B ring, is essential for rescue. An ethyl group at position 24, as present in the plant sitosterol, on the other hand, was nonfunctional, because sitosterol did not rescue the double mutant. Taken together, these results indicate that the synthetic lethal interaction between mutations in ELO3 and ERG6 is likely to reflect an increased structural requirement for the methyl group at position 24 of the ergosterol molecule in an elo3Δ mutant background.
Characterization of a Strain That Is Temperature Sensitive for Erg6p Function
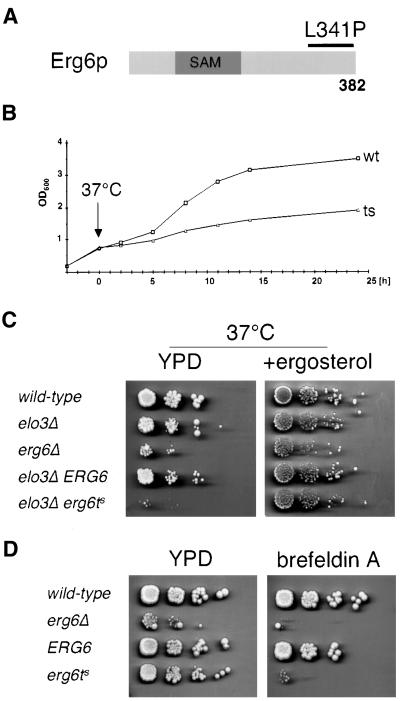
To investigate the increased structural requirement for ergosterol in the elo3Δ mutant in more detail, we generated a strain that is temperature sensitive for the function of Erg6p. Therefore, the 3′ end of the ERG6 gene, covering the carboxy-terminal 120 amino acids, was subject to mutagenesis by error prone PCR (see Figure 4A). After in vivo recombination and plasmid shuffling, three temperature-sensitive ERG6 alleles were isolated (see MATERIALS AND METHODS). Sequence analysis of one of these alleles revealed a single base substitution in codon 341 (CTA to CCA), resulting in the incorporation of a proline instead of the wild-type leucine at this position of Erg6p.
Figure 4.
Characterization of an elo3Δ erg6Δ mutant that is conditionally defective for Erg6p. (A) Structure of the Erg6p protein with the S-adenosyl-methionine (SAM)-binding domain indicated. The 3′ end of ERG6 was subject to mutagenesis using error prone PCR (black bar). The mutant used for further analysis has a leucine to proline substitution at position 341 of Erg6p. (B) Growth of the elo3Δ erg6ts strain stops at 37°C. The elo3Δ erg6Δ double mutant containing either a plasmid-borne copy of a wild-type allele of ERG6 (YRS1520) or containing the erg6ts allele (YRS1519), was assessed upon a temperature shift to 37°C by following the optical density in minimal media lacking leucine. (C) Growth of the elo3Δ erg6ts strain at 37°C is rescued by supplementation with ergosterol. Cells of indicated genotype were serially diluted 10-fold and replica-plated on solid media with ergosterol (+ergosterol) and without (YPD) and incubated at 37°C for 4 d. (D) erg6ts is sensitive to brefeldin A. Strains of the indicated genotype were serially diluted 10-fold and replica-plated on media with brefeldin A (+brefeldin A) and without (YPD) and incubated at 24°C for 4 d.
Analysis of the growth characteristics of this elo3Δ erg6ts conditional strain revealed that the strain grows like the corresponding wild-type at 24°C but that it rapidly ceases growth upon incubation at nonpermissive conditions (Figure 4B). Plating of cells that have been shifted for up to 8 h to nonpermissive conditions revealed that the capacity of the mutant to restore growth and form colonies when shifted back to permissive conditions (24°C) was not impaired, indicating that the metabolic block imposed by the temperature shift was fully reversible. In agreement with these observations, Western blot analysis with an anti-Erg6p antibody revealed that the mutant protein is stable and not subject to temperature-dependent turnover (unpublished data).
The conditional growth phenotype of the elo3Δ erg6ts mutant could be fully rescued on rich media by ergosterol supplementation, confirming the ergosterol auxotrophic phenotype of the double mutant that was so far based on plasmid loss on 5-FOA containing synthetic media (Figure 4C). Analysis of the in vivo activity of the temperature-sensitive enzyme by labeling cells with [3H]methionine revealed an ∼800-fold reduced activity of the enzyme at permissive conditions. This activity declined to undetectable levels upon incubation of cells at nonpermissive conditions (unpublished data). Consistent with this low activity of the conditional Erg6p allele, erg6Δ mutant cells bearing this allele are sensitive to brefeldin A, as is an erg6Δ null allele (Shah and Klausner, 1993; Figure 4D).
Characterization of the Secretory Pathway in the elo3Δ erg6ts Conditional Mutant
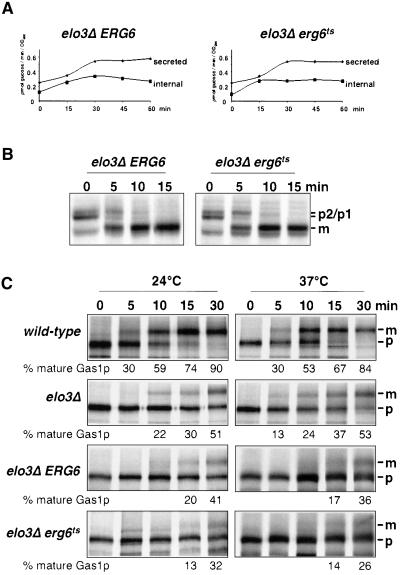
To determine whether protein maturation and secretion were conditionally affected in the elo3Δ erg6ts mutant strain, the kinetics of invertase induction and secretion was analyzed (Novick and Schekman, 1979). Induction and secretion of invertase in the elo3Δ erg6ts conditional mutant was comparable to that observed in the elo3Δ ERG6 strain, indicating that vesicular transport from the ER to the plasma membrane is not affected by the inactivation of Erg6p in the elo3Δ mutant background (Figure 5A).
Figure 5.
Characterization of the secretory pathway in the elo3Δ erg6ts mutant. (A) Secretion of invertase was analyzed in the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519). Cells were preshifted to 37°C for 15 min before invertase induction by glucose limitation. Samples were removed at the time points indicated and the activity of internal and secreted invertase was determined. (B) Maturation of CPY was analyzed in the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519). Cells were preshifted to 37°C for 15 min before labeling with [35S]methionine for 5 min and chasing for the time indicated. CPY was recovered by immunoprecipitation, subjected to SDS-PAGE, and visualized by autoradiography. The position of precursor (p1, ER; p2, Golgi) and mature CPY (m, vacuolar) is indicated. (C) Maturation of Gas1p was analyzed in wild-type (FY1679a), elo3Δ (YRS1118), and the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519). Cells were incubated at 24°C or preshifted to 37°C for 15 min before pulse-chase analysis and immunoprecipitation. The positions of ER precursor (p, 105 kDa) and mature Gas1p (m, 125 kDa) are indicated. Quantification of the degree of Gas1p maturation is shown below.
To examine whether vesicular transport between the Golgi apparatus and the vacuole was affected, transport of the soluble vacuolar hydrolase carboxypeptidase Y (CPY) was examined by pulse-chase analysis. CPY leaves the ER as a 67-kDa preprotein to mature in the Golgi apparatus to a 69-kDa form, which is cleaved to the active 61-kDa protease in the vacuole. After a preshift to 37°C for 15 min, the elo3Δ erg6ts mutant exhibited wild-type kinetics of CPY maturation, indicating that vesicular transport between ER and the vacuole was not impaired by inactivation of Erg6p in an elo3Δ mutant background (Figure 5B).
To complete the analysis of the secretory pathway, we examined whether maturation of the GPI-anchored membrane protein Gas1p was affected. Gas1p exits the ER as a 105-kDa precursor and matures in the Golgi apparatus to a 125-kDa form (Fankhauser and Conzelmann, 1991). The accumulation of the ER form of Gas1p is a sensitive assay to detect perturbations in ER-to-Golgi transport. Western blot analysis revealed wild-type maturation of Gas1p in erg6Δ but a strong accumulation of the ER precursor of Gas1p in the elo3Δ erg6ts mutant strain, irrespective of whether the mutant was incubated at permissive or nonpermissive conditions (unpublished data). Accumulation of the ER form of Gas1p was also observed in the elo3Δ mutant alone and in the elo3Δ erg6Δ double mutant bearing a wild-type copy of ERG6 on a plasmid. To determine whether there is a superimposed delay in Gas1p maturation by the inactivation of Erg6p in an ELO3 mutant background, we followed maturation of Gas1p by pulse-chase analysis and immunoprecipitation. This analysis failed to reveal a significant delay in Gas1p maturation by the reduction of Erg6p activity in the elo3Δ mutant background (Figure 5C). Taken together, these results indicate that the efficiency of the secretory pathway is not generally perturbed in the elo3Δ erg6ts conditional strain.
The elo3Δ erg6ts Mutant Has a Sphingolipid Composition Characteristic of elo3Δ
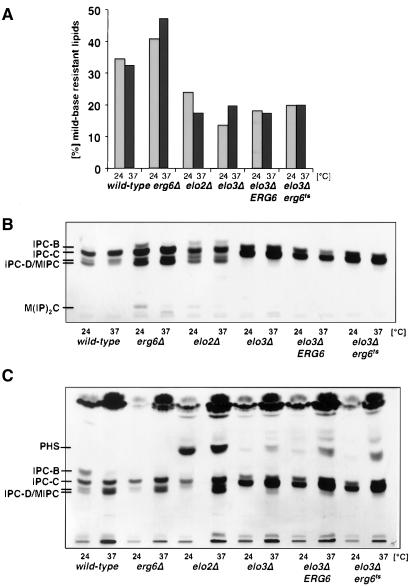
To determine whether inactivation of Erg6p affects sphingolipid synthesis in the double mutant, cells were preshifted to nonpermissive conditions and incorporation of [3H]inositol into the mild-base–resistant lipid fraction was examined. This analysis revealed an approximately twofold decrease in the incorporation of [3H]inositol into sphingolipids in cells mutant for either ELO2 or ELO3, irrespective of the activity of Erg6p (Figure 6A). Inactivation of ERG6 alone, however, resulted in a slight increased incorporation of label into sphingolipids.
Figure 6.
elo3Δ erg6ts has a sphingolipid composition characteristic for elo3Δ. (A) Sphingolipid synthesis in wild-type (FY1679a), erg6Δ (YRS1266), elo2Δ (YRS1265), elo3Δ (YRS1118), and the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519) was assessed by labeling cells at either 24 or 37°C with [3H]inositol for 2 h. Lipids were extracted, and label incorporated into the mild-base–resistant sphingolipid fraction was determined in comparison to label incorporated into the total lipid fraction. (B) Sphingolipid composition in strains of indicated genotype was analyzed by TLC and fluorography. Cells were labeled at either 24 or 37°C with [3H]inositol for 2 h, and mild-base–resistant lipids were separated by TLC. The position of different sphingolipid species is indicated to the left. MIPC, mannosyl-inositolphosphorylceramide; M(IP)2C, mannosyl-diinositolphosphorylceramide. (C) Comparison of the levels of sphingolipid precursors in strains of indicated genotype. Cells were labeled at either 24 or 37°C with [3H]serine for 4 h, and mild-base–resistant lipids were separated by TLC. The position of sphingolipid precursors and mature sphingolipid species is indicated to the left. MIPC, mannosyl- inositolphosphorylceramide.
TLC analysis of the sphingolipid classes synthesized under these conditions revealed a predominance of IPC-C (inositolphosphorylceramide-C) species at the expense of the more complex mannosylated sphingolipids in all strains with mutations in ELO3, irrespective of Erg6p activity (Figure 6B). These alterations in sphingolipid levels and composition in the elo3Δ mutant are in agreement with previous analyses (Oh et al., 1997; David et al., 1998). The IPC-C species produced in an elo3Δ mutant background has a somewhat lower mobility compared with the IPC-C species of wild-type cells, consistent with the idea that this sphingolipid is less hydrophobic because of the presence of a shorter acyl chain. Taken together, the results of the sphingolipid analysis indicate that sphingolipid synthesis/composition of the elo3Δ mutant is not affected by the activity of Erg6p.
The sphingolipid precursors, ceramide and phytosphingosine (PHS), regulate cell growth by activation of different signaling pathways (Hannun and Luberto, 2000). To determine whether accumulation of these signaling lipids results in growth arrest of the elo3Δ erg6ts conditional mutant, ceramide and PHS levels were examined by labeling cells with [3H]serine (Figure 6C). This analysis revealed a strong accumulation of PHS in the elo2Δ mutant strain and lower levels of PHS in all strains with mutations in ELO3, irrespective of Erg6p activity. These results thus indicate that the activity of Erg6p does not affect sphingolipid signaling in an elo3Δ mutant background.
The elo3Δ erg6ts Mutant Affects Raft Association of the Plasma Membrane ATPase
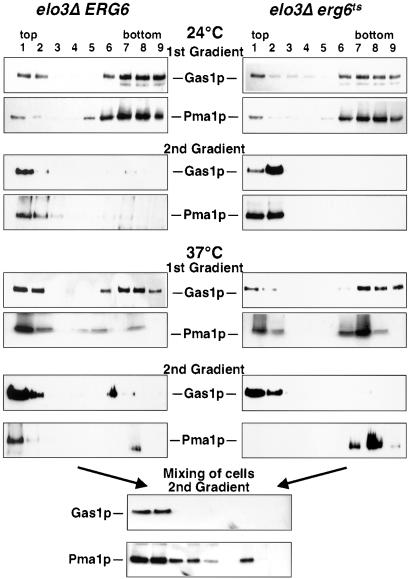
Given that sphingolipid synthesis is continuing in the conditional mutant under nonpermissive conditions, we wondered whether membrane raft formation might be impaired. Therefore, raft association of two marker proteins, the GPI-anchored Gas1p, and the plasma membrane ATPase, Pma1p, was investigated (Bagnat et al., 2000). After two successive rounds of extraction with the nonionic detergent Triton X-100 and floatation in density gradients, both marker proteins were recovered in the detergent-insoluble fraction of the gradient when membranes of cells that were cultivated at permissive conditions were analyzed. On incubation of the conditional elo3Δ erg6ts mutant at nonpermissive conditions for 2 h, however, raft association of Pma1p but not that of Gas1p was specifically impaired (Figure 7). It is important to note that unambiguous determination of raft association of Pma1p in the mutant required analysis by two successive gradients as the portion of Pma1p from the detergent-resistant fraction of the first gradient was recovered in the detergent-soluble fraction of the second gradient. Raft association of Pma1p was not impaired in the elo3Δ erg6Δ double mutant harboring a plasmid-borne wild-type copy of ERG6, indicating that steady state raft association of Pma1p is dependent on Erg6p activity in an elo3Δ mutant background.
Figure 7.
elo3Δ erg6ts affects steady state raft association of the plasma membrane ATPase. Raft association of Gas1p and the plasma membrane ATPase, Pma1p, in the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519). Cells were cultivated at 24°C or shifted to nonpermissive conditions (37°C) for 2 h, before isolation of detergent-resistant membranes by either one (1st Gradient) or two rounds (2nd Gradient) of detergent extraction and floatation on density gradients (Bagnat et al., 2000). The position of mature Gas1p and that of Pma1p is indicated. For the mixing experiment, cells of the two genotypes were mixed at a 1:1 ratio after incubation at nonpermissive conditions for 2 h, and detergent solubility of Gas1p and Pma1p was assessed after two successive rounds of detergent extraction and floatation on density gradients. All blots were processed under comparable conditions.
To examine whether raft association of Pma1p is detergent induced, an equal number of cells of the two genotypes that had been incubated at nonpermissive conditions for 2 h was mixed, and raft association of Gas1p and Pma1p was reexamined. This analysis revealed that Gas1p maintained its raft association, but Pma1p was now observed in both the detergent-resistant and the -soluble fractions of the gradient.
elo3Δ Conditionally Affects Stability of the ATPase at the Plasma Membrane
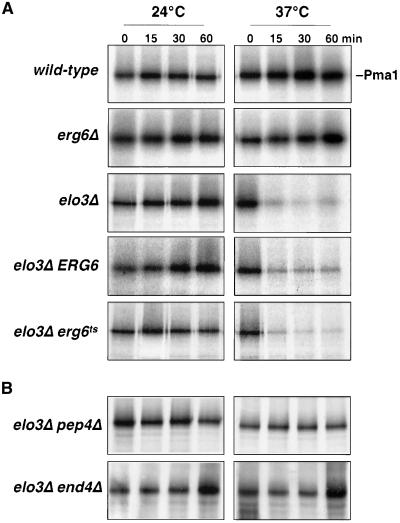
Pma1p has recently been shown to move into raft domains during biosynthetic transport to the plasma membrane (Bagnat et al., 2001; Gong and Chang, 2001; Ferreira et al., 2002; Lee et al., 2002). Raft association of Pma1p is required for proper delivery to the plasma membrane because inhibition of raft association results in rerouting of the protein to the vacuole where it becomes degraded (Bagnat et al., 2001). To examine whether raft association required for biosynthetic transport of Pma1p to the plasma membrane is affected by inactivation of Erg6p in an elo3Δ mutant background, cells were pulse-labeled and maturation of Pma1p was followed by immunoprecipitation. This analysis revealed rapid turnover of Pma1p in the elo3Δ mutant when cells were incubated at 37°C (Figure 8A). Under these conditions, 80% of newly synthesized Pma1p was rapidly degraded, irrespective of the activity of Erg6p. These results thus indicate that mutations in ELO3 affect raft association of newly synthesized Pma1p at 37°C.
Figure 8.
elo3Δ affects the stability of newly synthesized Pma1p. (A) The half-life of newly synthesized Pma1p in wild-type (FY1679a), elo3Δ (YRS1118), and erg6Δ (YRS1266), and the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519) was assessed by pulse-chase analysis and immunoprecipitation. Cells were cultivated at 24°C or preshifted to 37°C for 15 min before labeling under the conditions indicated. (B) Pma1p degradation in the elo3Δ mutant requires PEP4 and END4. Turnover of newly synthesized Pma1p in elo3Δ pep4Δ (YRS1550) and elo3Δ end4Δ (YRS1554) double mutant strains was assessed as in A.
The increased turnover of newly synthesized Pma1p in the elo3Δ mutant occurs by vacuolar degradation because Pma1p is stabilized by a mutation in the vacuolar hydrolase Pep4p (Jones, 1991). Moreover, transport of Pma1p to the vacuole appears to occur after it has reached the plasma membrane, because it is stabilized by blocking internalization from the cell surface in end4Δ cells (Raths et al., 1993; Figure 8B). Thus, Elo3p is required for stabilizing newly synthesized Pma1p at the plasma membrane at 37°C.
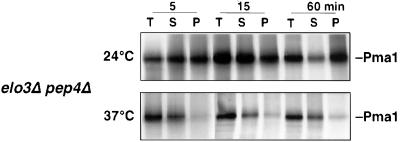
To examine at which point along the secretory pathway raft association of Pma1p is affected in elo3Δ, the detergent solubility of Pma1p was assessed by pulse labeling. To stabilize newly synthesized Pma1p, this analysis was performed in an elo3Δ pep4Δ double mutant strain. As shown in Figure 9, Pma1p becomes detergent insoluble already after 5 min in cells cultivated at 24°C. At 37°C, however, Pma1p does not acquire detergent resistance in elo3Δ, indicating that elo3Δ affects raft association of Pma1p early in its biosynthetic transport and that raft association of Pma1p is important for maintaining its stability at the plasma membrane.
Figure 9.
Newly synthesized Pma1p does not acquire detergent resistance in elo3Δ at 37°C. elo3Δ pep4Δ (YRS1550) cells were incubated at the indicated temperatures for 15 min, pulse-labeled for 5 min, and chased for 15 and 60 min. Cell lysates (T) were then extracted with Triton X-100 and separated into a soluble (S) and insoluble (P) fractions. Pma1p was immunoprecipitated and analyzed by SDS-PAGE and fluorography.
ERG6 Is Required for Maintaining Raft Domains in elo3Δ
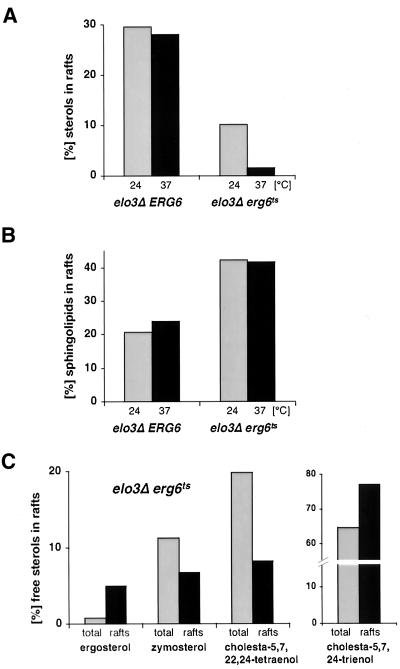
Given the observation that the conditional elo3Δ erg6ts mutant affects steady state raft association of Pma1p but not Gas1p, we examined whether lipid rafts by themselves were affected. Therefore, the sterol content of the detergent-resistant membrane fraction of mutant cells grown at permissive conditions or incubated at nonpermissive conditions for 2 h was determined by quantitative GC-MS. This analysis revealed that 30% of free sterols were raft associated in an elo3Δ ERG6 control strain at either 24 or 37°C. In the elo3Δ erg6ts conditional mutant, however, the content of raft-associated sterols was reduced threefold at permissive conditions and further declined more than sixfold upon incubation at nonpermissive conditions (Figure 10A). These results thus indicate that preexisting raft structures are affected upon inactivation of Erg6p in an elo3Δ mutant strain.
Figure 10.
The elo3Δ erg6ts conditional mutant affects membrane raft domains. (A) Comparison of free sterol levels in detergent-resistant membrane domains from the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519), cultivated at permissive conditions (24°C) or shifted to nonpermissive conditions (37°C) for 2 h. Total membranes and detergent-resistant membrane domains were isolated by two successive rounds of detergent extraction and floatation in density gradients. Sterols present in total and detergent-resistant membranes were quantified by comparison to an internal standard by GC-MS analysis, and the proportion of sterols present in the detergent-resistant membrane fraction relative to the amount of sterols in total membranes is shown. (B) Comparison of sphingolipid levels in the detergent-resistant membrane fraction from the elo3Δ erg6Δ double mutant strain bearing a plasmid-borne copy of either ERG6 (elo3Δ ERG6, YRS1520) or erg6ts (elo3Δ erg6ts, YRS1519), cultivated at permissive conditions (24°C) or shifted to nonpermissive conditions (37°C) for 2 h. Cells were labeled with [3H]myo-inositol for 16 h. Total and detergent treated membranes were isolated by floatation in density gradients. Sphingolipids were quantified after mild-base treatment of lipid extracts. The proportion of mild-base–resistant lipids present in the detergent-resistant membrane fraction relative to that of total membranes is shown. (C) Comparison of the free sterol composition of total membranes (total) and the detergent-insoluble membrane fraction (rafts) of the elo3Δ erg6ts (YRS1519) conditional mutant at permissive conditions. Total and detergent-resistant membrane domains were isolated by two successive rounds of flotation in density gradients. The sterol composition of the two membranes was analyzed by GC-MS and the proportion of different sterols present in the two membranes is shown.
To determine whether the decrease in raft lipids observed in the conditional mutant is specific for sterols, we examined the sphingolipid content of raft membranes in the two strains. Therefore, cells were labeled with [3H]myo-inositol for 16 h and raft association of mild-base–resistant lipids was determined. This analysis revealed an approximately twofold elevated level of sphingolipids in the detergent-resistant membrane fraction of the conditional mutant (Figure 10B). Unlike the sterol content, however, the sphingolipid content of raft membranes did not decline upon incubation of these cells at nonpermissive conditions. These results thus indicate that the aberrant sterols in raft membranes of the conditional mutant do not resist detergent extraction.
A quantitative analysis of total cellular sterols revealed significantly higher sterol levels in the elo3Δ and erg6Δ single mutants. Combining the two mutations results in a further increase in total sterol levels, resulting in four times elevated total saponifiable sterol levels (Table 4). A compositional analysis of the total saponifiable sterols in the conditional mutant revealed a sterol pattern characteristic of an erg6Δ single mutant with zymosterol (45.4%) and cholesta-5,7,24-trienol (33.7%) as the major sterols, which is in agreement with a previous analysis of the sterol composition of an erg6Δ mutant (Munn et al., 1999; Table 5). The low ergosterol content observed in the conditional mutant indicates that the temperature-sensitive allele of ERG6 has a greatly reduced activity already at the permissive temperature and that the plasmid-borne wild-type copy of ERG6 also does not fully complement the erg6Δ deletion.
Table 4.
Total sterol content of mutant strains at 24°C
| Wild-type | elo2Δ | elo3Δ | erg6Δ | elo3Δ ERG6 | elo3Δ erg6ts | |
|---|---|---|---|---|---|---|
| μg sterol/OD | 1.17 ± 0.13 | 1.19 ± 0.09 | 1.71 ± 0.19 | 2.46 ± 0.05 | 2.61 ± 0.24 | 3.73 ± 0.40 |
Table 5.
Total sterol composition of mutant strains at 24°C
| Wild-type | elo2Δ | elo3Δ | erg6Δ | elo3Δ ERG6 | elo3Δ erg6ts | |
|---|---|---|---|---|---|---|
| Zymosterol | 9.8 ± 1.3 | 14.6 ± 1.2 | 15.3 ± 1.8 | 35.4 ± 1.5 | 18.1 ± 2.5 | 45.4 ± 1.9 |
| Fecosterol | 9.3 ± 1.3 | 8.8 ± 0.4 | 14.1 ± 1.0 | n.d. | 14.4 ± 0.9 | n.d. |
| Cholesta-5,7,24-trienol | n.d. | n.d. | n.d. | 42.1 ± 2.5 | 10.1 ± 2.5 | 33.7 ± 2.6 |
| Cholesta-5,7,22,24-tetraenol | n.d. | n.d. | n.d. | 10.2 ± 0.1 | 2.0 ± 0.3 | 10.4 ± 0.6 |
| Ergosterol | 70.6 ± 1.4 | 63.4 ± 3.0 | 58.2 ± 2.9 | n.d. | 37.5 ± 2.3 | 1.3 ± 0.3 |
| Others | 8.7 ± 5.0 | 9.8 ± 3.4 | 7.9 ± 1.4 | 9.5 ± 1.0 | 12.2 ± 3.5 | 2.0 ± 0.6 |
n.d.; not detectable. Values are % of total. Other sterols are lanosterol, 14-desmethyl-lanosterol, 4-methyl-zymosterol, cholesta-5,8,24-trienol, episterol, cholesta-7,24-dienol, and ergosta-5,7-dienol.
Given the fact that the conditional mutant strain has a fourfold increased level of total sterols but a threefold reduced level of sterols in detergent-resistant membranes, we examined whether detergent-insoluble membrane domains exhibit preference for certain sterol species. Therefore, the composition of free (nonesterified) sterols in the detergent-insoluble membrane fraction from an elo3Δ erg6ts conditional strain cultivated at permissive conditions was examined by GC-MS. This analysis revealed a sevenfold enrichment of ergosterol in the detergent-resistant membrane fraction (Figure 10C). Zymosterol and cholesta-5,7,22,24-tetraenol, on the other hand, are deenriched in the raft fraction. Given that ergosterol accounts for only ∼1.3% of the total saponifiable sterols in an elo3Δ erg6ts conditional mutant (Table 5) in which 10% of the sterols are raft associated, a sevenfold enrichment of ergosterol in these raft structures, suggests that ergosterol synthesis and hence Erg6p activity becomes limiting for the maintenance of raft domains in this strain.
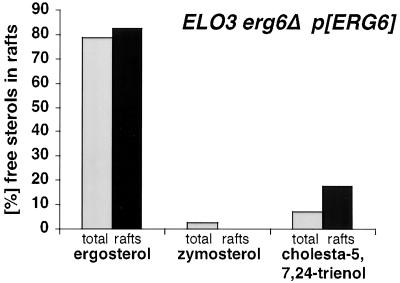
To determine whether the observed preference of membrane rafts for ergosterol is specific for elo3Δ mutant cells or a more general property of yeast membranes, the sterol composition of membrane rafts of cells making normal C26-substituted sphingolipids was analyzed. Therefore, the sterol composition of total and detergent-resistant membranes from an erg6Δ mutant that harbors a plasmid-borne wild-type allele of ERG6 was analyzed by quantitative GC-MS. This analysis revealed that both ergosterol and cholesta-5,7,24-trienol were slightly enriched in raft membranes, whereas zymosterol and other low abundant sterols such as fecosterol and 14-desmethyl-lanosterol were deenriched to undetectable levels (Figure 11). These data are consistent with the proposal that ergosterol is preferentially included in raft membranes in cells that have normal C26-substituted sphingolipids. However, given that erg6Δ mutants are viable in an ELO3 wild-type background, this preference is much less stringent.
Figure 11.
Sterol composition in raft domains of ELO3 cells. Comparison of the free sterol composition of total membranes (total) and the detergent-insoluble membrane fraction (rafts) in cells that have C26 substituted sphingolipids, but a mixture of different sterols (ELO3 erg6Δ p[ERG6]; YRS1551). Detergent-resistant membrane domains were isolated by two successive rounds of detergent extraction followed by flotation in density gradients. Sterol composition was analyzed by GC-MS.
DISCUSSION
We report a specific genetic interaction between mutations in genes required for fungal specific lipid modifications: one in the synthesis of long-chain fatty acids and the other in the synthesis of sterols. The synthetic lethality between mutations in ELO3 and ERG6 is likely due to a vital interaction of the respective lipid products, because the viability of the double mutant is rescued by supplementation with ergosterol. The fact that the double mutant is only rescued by sterols that have a methyl group at position 24 of the side chain indicates that the elo3Δ mutant has a stringent structural requirement for this otherwise nonessential sterol modification.
Cross-talk between Lipid Biosynthetic Pathways
Comparison of the lipid composition of erg6Δ, elo3Δ, and double mutant strains revealed apparently compensatory changes in sterol and sphingolipid synthesis, indicating that the two lipid biosynthetic pathways must be under coordinate control (Swain et al., 2002). First, the erg6Δ mutant displayed an increased rate of incorporation of radiolabeled inositol into mild-base–resistant lipids and a ∼2.1-fold increase in total sterol levels, suggesting that the absence of ergosterol in this strain is compensated by an increase in both sterol and sphingolipid levels (Figure 6A, Table 4). Second, mutations in ELO3 result in a reduced rate of sphingolipid synthesis and in the absence of the more complex mannosylated sphingolipid species. These changes in the sphingolipid pattern of an elo3Δ mutant appear to be compensated by a ∼1.5-fold increase in sterol levels. The sterol composition of the elo3Δ mutant, however, is comparable to that of wild-type cells, indicating that the altered sphingolipid composition affects sterol levels only but not the activity of individual sterol biosynthetic enzymes. Third, the combination of mutations in both ELO3 and ERG6 does not further affect sphingolipid synthesis, but results in an additional increase in total sterol levels, and a sterol composition characteristic of an erg6Δ mutant.
Raft Formation in the elo3Δ erg6Δ Double Mutant
A common structural function of sphingolipids and sterols is their association into membrane microdomains/rafts. Formation of these raft domains is important for lateral sorting of proteins and for signal transduction (Brown and Rose, 1992; Simons and Ikonen, 1997). Previous studies have established that depletion of either sterols or sphingolipids in yeast affects raft association of Gas1p and Pma1p (Bagnat et al., 2000, 2001; Lee et al., 2002). In our strains, sterols and sphingolipids are not depleted, but their structure and composition is altered. This affects raft association of the two marker proteins differently. Although the GPI-anchored Gas1p remains raft associated in the elo3Δ erg6ts conditional strain, Pma1p becomes detergent soluble. Analysis of the composition of the detergent-resistant lipids in this strain revealed that the sterol content is greatly reduced, whereas the sphingolipid content of raft membranes is increased, suggesting that sphingolipids compensate for the absence of appropriate sterols to maintain raft function in this strain, at least at 24°C. Under more stringent conditions (37°C), the stability of preexisting raft domains becomes impaired to a point where they can no longer resist detergent solubilization, resulting in the release of Pma1p but not Gas1p. The observation that Gas1p maintains detergent resistance may indicate that under these conditions this GPI-anchored protein partitions into a domain that has a different lipid composition than the typical raft, i.e., a sphingolipid-rich but ergosterol-poor domain.
Analysis of the sterol composition of the raft membranes in the conditional strain revealed a strong enrichment in ergosterol. This observation and the fact that the elo3Δ erg6Δ double mutant can only be rescued by sterols that have a methyl group in the aliphatic side chain suggests that C24-substituted sphingolipids have an increased structural requirement for this methyl group. This notion is supported by a recent biophysical study, which demonstrates that ergosterol is significantly more strongly domain-promoting than cholesterol (Xu et al., 2001). Moreover, in erg6Δ mutants the GPI-anchored plasma membrane protease, Yps1p, is mistargeted to the vacuole, indicating that the precise structure of the sterol is important for raft function in vivo (Sievi et al., 2001).
Raft Formation in the elo3Δ Mutant
Stable delivery of Pma1p to the plasma membrane requires functional rafts (Bagnat et al., 2001; Gong and Chang, 2001; Lee et al., 2002). The observation that ∼80% of newly synthesized Pma1p are rapidly turned over in the elo3Δ mutant at 37°C through internalization from the cell surface and delivery to the vacuole, indicates that nonraft-associated Pma1p is unstable when it reaches the plasma membrane. A similar phenotype, raft-dependent stabilization of surface delivered Pma1p has been observed with a mutant allele of Pma1p, Pma1–10 (Gong and Chang, 2001). In this case, raft association of Pma1–10 was suggested to be required but not sufficient for its subsequent stabilization at the cell surface. This is consistent with observations made with a second allele of Pma1, Pma1-G381A, which remains raft associated but nevertheless fails to become stabilized at the cell surface (Ferreira et al., 2002).
Pma1p that has entered rafts and that has been delivered to the plasma membrane in elo3Δ at 24°C, however, remains stable even if raft function is subsequently impaired by shifting cells to 37°C, as indicated by the observation that steady state raft association of Pma1p is not affected in the elo3Δ ERG6 mutant at 37°C. These observations may indicate that elo3Δ at 37°C affects raft assembly but that preexisting structures remain stable unless ergosterol is depleted.
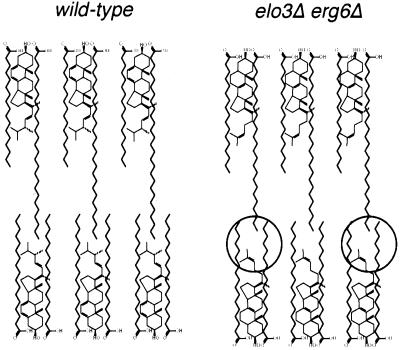
elo3Δ mutants display a number of lipid alterations that potentially could affect raft formation, particularly when combined with mutations in the sterol biosynthetic pathway. First, elo3Δ mutant cells have reduced levels of sphingolipids. A complete block in sphingolipid synthesis as achieved using a conditional allele of serine palmitoyltransferase (lcb1–100; Sütterlin et al., 1997), which catalyzes the rate-limiting step of sphingolipid synthesis, has been shown to affect raft association of both Gas1p and Pma1p (Bagnat et al., 2000; Lee et al., 2002). The observation that the elo2Δ mutant has a similar reduction in sphingolipid synthesis but is not synthetically lethal with erg6Δ indicates that reduced levels of sphingolipids cannot fully account for the defect observed in elo3Δ. Second, elo3Δ mutant cells contain IPC-C as their main sphingolipid species at the expense of the more complex mannosylated lipids. Whether a defect in head group maturation of sphingolipids could affect raft formation needs to be established. Third, the IPC-C species in the elo3Δ mutant contains a C22/C24 very-long-chain fatty acid instead of the natural C26 fatty acid. Biophysical studies indicate that the interaction of cholesterol with sphingomyelin is not affected by differences in acyl chain length (Ohvo-Rekila et al., 2002). Phase separation of lipids in model membranes, however, is favored by the mismatch of the hydrophobic interaction surface of the interacting lipids (Silvius et al., 1996). A simplified static model of the interaction of ergosterol with C26-substituted lipids, assuming interdigitation of the C26 acyl chain into the hydrophobic core of the opposing membrane leaflet (Slater and Huang, 1988; but see also McIntosh et al., 1992), is illustrated in Figure 12. According to this model, shortening of the C26 acyl chain to C22 with the simultaneous removal of the C24 methyl group on ergosterol could result in significant alterations in the hydrophobic core of the membrane that could affect the domain-forming properties of the respective membrane.
Figure 12.
Comparison of the structure of the plasma membrane of wild-type and elo3Δ erg6Δ mutant cells. Schematic model of the presumed structure of the hydrophobic core of the plasma membrane of wild-type cells compared with cells that contain C22-substituted sphingolipids and zymosterol instead of ergosterol. Alteration in the hydrophobic core of the mutant membrane is encircled. See text for details.
Taken together, the observation of a genetic interaction between mutations in genes required for fungal specific lipid modifications of sphingolipids and sterols provides a new means to study the function, biogenesis, and regulation of raft structures in vivo, using genetic approaches unique to yeast.
ACKNOWLEDGMENTS
We thank A. Conzelmann, G. Daum, J. Gerst, C. Holm, G. Loison, F. Portillo, H. Riezman, M. Rose, and M. Snyder for generously providing plasmids, yeast strains, or antibodies; P. Seiser for initial help with the synthetic lethal screen; and S. Rosenberger, D. Gaug, and S. Reiner for help in the revision phase of this manuscript. This work was supported by the Austrian Science Found (P13767 and P15210 to R.S.). R.S. acknowledges receipt of a professorial award from the Swiss National Science Foundation.
Footnotes
DOI: 10.1091/mbc.E02–02–0116.
REFERENCES
- Bagnat M, Chang A, Simons K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol Biol Cell. 2001;12:4129–4138. doi: 10.1091/mbc.12.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Nat Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Crowley JH, Smith SJ, Leak FW, Parks LW. Aerobic isolation of an ERG24 null mutant of Saccharomyces cerevisiae. J Bacteriol. 1996;178:2991–2993. doi: 10.1128/jb.178.10.2991-2993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C, Biemann HP, Dahl J. A protein kinase antigenically related to pp60v-src possibly involved in yeast cell cycle control: positive in vivo regulation by sterol. Proc Natl Acad Sci USA. 1987;84:4012–4016. doi: 10.1073/pnas.84.12.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D, Sundarababu S, Gerst JE. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Conzelmann A. Purification, biosynthesis and cellular localization of a major 125-kDa glycophosphatidylinositol-anchored membrane glycoprotein of Saccharomyces cerevisiae. Eur J Biochem. 1991;195:439–448. doi: 10.1111/j.1432-1033.1991.tb15723.x. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Mason AB, Pypaert M, Allen KE, Slayman C. Quality control in the yeast secretory pathway. J Biol Chem. 2002;277:21027–21040. doi: 10.1074/jbc.M112281200. [DOI] [PubMed] [Google Scholar]
- Funato K, Riezman H. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol. 2001;155:949–959. doi: 10.1083/jcb.200105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arranz M, Maldonado AM, Mazón MJ, Portillo F. Transcriptional control of yeast plasma membrane H+ATPase by glucose. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- Goldstein A, Lampen JO. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Gong X, Chang A. A mutant plasma membrane ATPase, Pma1–10, is defective in stability at the yeast cell surface. Proc Natl Acad Sci USA. 2001;98:9104–9109. doi: 10.1073/pnas.161282998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Kerwin JL, Watts JD, Aebersold R. Ceramide profiling of complex lipid mixtures by electrospray ionization mass spectrometry. Anal Biochem. 1997;244:347–356. doi: 10.1006/abio.1996.9915. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000;10:73–80. doi: 10.1016/s0962-8924(99)01694-3. [DOI] [PubMed] [Google Scholar]
- Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast Saccharomyces cerevisiae. Eur J Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- Jones E. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- Kranz JE, Holm C. Cloning by function: An alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladevèze V, Marcireau C, Delourme D, Karst F. General resistance to sterol biosynthesis inhibitors in Saccharomyces cerevisiae. Lipids. 1993;28:907–912. doi: 10.1007/BF02537499. [DOI] [PubMed] [Google Scholar]
- Lange Y, Swaisgood MH, Ramos BV, Steck TL. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured fibroblasts. J Biol Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- Lee MCS, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- Lees ND, Bard M, Kirsch DR. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1999;34:33–47. [PubMed] [Google Scholar]
- Levine TP, Wiggins CA, Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- McIntosh TJ, Simon SA, Needham D, Huang CH. Structure and cohesive properties of sphingomyelin/cholesterol bilayers. Biochemistry. 1992;31:2012–2020. doi: 10.1021/bi00122a017. [DOI] [PubMed] [Google Scholar]
- Muniz M, Riezman H. Intracellular transport of GPI-anchored proteins. EMBO J. 2000;19:10–15. doi: 10.1093/emboj/19.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, HeesePeck A, Stevenson BJ, Pichler H, Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol Biol Cell. 1999;10:3943–3957. doi: 10.1091/mbc.10.11.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Ohvo-Rekila H, Ramstedt B, Leppimaki P, Slotte JP. Cholesterol interactions with phospholipids in membranes. Prog Lipid Res. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- Parks LW, Crowley JH, Leak FW, Smith SJ, Tomeo ME. Use of sterol mutants as probes for sterol functions in the yeast Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1999;34:399–404. doi: 10.1080/10409239991209381. [DOI] [PubMed] [Google Scholar]
- Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J Bacteriol. 1991;173:3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Kelly SL. The extraction and analysis of sterols from yeast. Methods Mol Biol. 1996;53:123–131. doi: 10.1385/0-89603-319-8:123. [DOI] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid- phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Canivenc-Gansel E, Conzelmann A. Lipid remodeling leads to the introduction and exchange of defined ceramides on GPI proteins in the ER and Golgi of Saccharomyces cerevisiae. EMBO J. 1997;16:3506–3518. doi: 10.1093/emboj/16.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld A, Simons K. The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochem Biophys Res Commun. 1998;1376:467–479. doi: 10.1016/s0304-4157(98)00019-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, Parks LW. Structural and physiological features of sterols necessary to satisfy bulk membrane and sparking requirements in yeast sterol auxotrophs. Arch Biochem Biophys. 1983;225:861–871. doi: 10.1016/0003-9861(83)90099-1. [DOI] [PubMed] [Google Scholar]
- Sankaram MB, Thompson TE. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry. 1990;29:10670–10675. doi: 10.1021/bi00499a014. [DOI] [PubMed] [Google Scholar]
- Schneiter R. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. Bioessays. 1999;21:1004–1010. doi: 10.1002/(SICI)1521-1878(199912)22:1<1004::AID-BIES4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Schneiter R, et al. Electrospray ionization tandem mass spectrometry (E.S.I-M.S/M.S) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Klausner RD. Brefeldin A reversibly inhibits secretion in Saccharomyces cerevisiae. J Biol Chem. 1993;268:5345–5348. [PubMed] [Google Scholar]
- Sievi E, Suntio T, Makarow M. Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic. 2001;2:896–907. doi: 10.1034/j.1600-0854.2001.21205.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, et al. The immunosuppressant S. R31747 blocks cell proliferation by inhibiting a steroid isomerase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2719–2727. doi: 10.1128/mcb.16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius JR, delGiudice D, Lafleur M. Cholesterol at different bilayer concentrations can promote or antagonize lateral segregation of phospholipids of differing acyl chain length. Biochemistry. 1996;35:15198–15208. doi: 10.1021/bi9615506. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Sipos G, Reggiori F, Vionnet C, Conzelmann A. Alternative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J. 1997;16:3494–3505. doi: 10.1093/emboj/16.12.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater JL, Huang CH. Interdigitated bilayer membranes. Prog Lipid Res. 1988;27:325–359. doi: 10.1016/0163-7827(88)90010-0. [DOI] [PubMed] [Google Scholar]
- Sütterlin C, Doering TL, Schimmoller F, Schroder S, Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Swain E, Baudry K, Stukey J, McDonough V, Germann M, Nickels JT., Jr Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J Biol Chem. 2002;277:26177–26184. doi: 10.1074/jbc.M204115200. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Xu X, Bittman R, Duportail G, Heissler D, Vilcheze C, London E. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J Biol Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]