Abstract
Ycf1p is the prototypical member of the yeast multidrug resistance-associated protein (MRP) subfamily of ATP-binding cassette (ABC) transporters. Ycf1p resides in the vacuolar membrane and mediates glutathione-dependent transport processes that result in resistance to cadmium and other xenobiotics. A feature common to many MRP proteins that distinguishes them from other ABC transporters is the presence of a hydrophobic N-terminal extension (NTE), whose function is not clearly established. The NTE contains a membrane spanning domain (MSD0) with five transmembrane spans and a cytosolic linker region (L0). The goal of this study was to determine the functional significance of the NTE of Ycf1p by examining the localization and functional properties of Ycf1p partial molecules, expressed either singly or together. We show that MSD0 plays a critical role in the vacuolar membrane trafficking of Ycf1p, whereas L0 is dispensable for localization. On the other hand, L0 is required for transport function, as determined by monitoring cadmium resistance. We also examine an unusual aspect of Ycf1p biology, namely, the posttranslational proteolytic processing that occurs within a lumenal loop of Ycf1p. Processing is shown to be Pep4p dependent and thus serves as a convenient marker for proper vacuolar localization. The processed fragments associate with each other, suggesting that these natural cleavage products contribute together to Ycf1p function.
INTRODUCTION
ATP-binding cassette (ABC) proteins comprise a ubiquitous protein superfamily, present in microbes, plants, and animals, whose members play key roles in a variety of cellular transport processes (Higgins, 1992; Decottignies and Goffeau, 1997; Taglicht and Michaelis, 1998; Dean et al., 2001a,b; Sanchez-Fernandez et al., 2001). One group of ABC transporters, the multidrug resistance-associated protein (MRP) subfamily (also designated the ABCC subfamily) has recently been the focus of numerous studies. MRP proteins function in normal physiological processes, such as the leukotriene C4-mediated inflammatory response and bile pigment elimination, and can mediate cellular detoxification by excreting drugs or cellular metabolites in the form of glutathione-, glucuronide-, or sulfate-conjugates (Cole and Deeley, 1998; Hipfner et al., 1999; Konig et al., 1999; Borst et al., 2000; Gottesman et al., 2002). Alternatively, certain MRP substrates may be cotransported or noncovalently complexed with glutathione during their transport.
Members of the MRP subfamily share a conserved architecture with other ABC transporters (Bakos et al., 1996; Loe et al., 1996). The ABC “core” domain consists of two homologous halves, each half containing a membrane spanning domain (MSD) with six transmembrane spans, and a nucleotide binding domain (NBD) that includes three conserved motifs designated the Walker A, Walker B, and signature motifs. The halves are connected by a linker (L1). For many ABC transporters, including the well-characterized human multidrug-resistance protein P-glycoprotein (encoded by MDR1), the ABC core domain is sufficient for proper localization, ATP hydrolysis, and substrate recognition. A distinguishing feature of many members of the MRP subfamily is an additional N-terminal extension (NTE) that is very hydrophobic (Bakos et al., 1996; Hipfner et al., 1997; Borst et al., 2000). The NTE is surprisingly substantial (∼275 aa) and is comprised of a membrane spanning domain (MSD0) with five predicted transmembrane helices, plus a cytoplasmic linker region (L0). The functional significance of the N-terminal extension is not clearly understood and remains a major issue in the MRP field. Several recent studies have focused on understanding the roles of the MSD0 and L0 subdomains of the NTE in human MRP1 (Bakos et al., 1998; Gao et al., 1998; Bakos et al., 2000; Qian et al., 2001). These studies showed that the linker region (L0) is necessary for the plasma membrane localization and functional activity of MRP1. Surprisingly, however, no role for the membrane spanning domain (MSD0) of MRP1 has yet been determined, because constructs missing solely MSD0 show a pattern that is indistinguishable from wild-type in terms of basolateral membrane localization and transport activity (Bakos et al., 1998). In contrast, it was recently reported for human MRP2 that MSD0 plays a critical role in the proper routing to or stable association of MRP2 with the apical membrane (Fernandez et al., 2002).
The product of the YCF1 gene, the yeast cadmium factor Ycf1p, is the best-studied MRP subfamily member in yeast. Ycf1p, which resides in the vacuolar membrane, detoxifies the yeast cell of heavy metals and xenobiotics by transporting glutathione-conjugates and complexes into the vacuole where they are sequestered and/or further metabolized (Szczypka et al., 1994; Li et al., 1996; Chaudhuri et al., 1997; Li et al., 1997). Human MRP1 can complement the biochemical transport and cadmium resistance defects of a ycf1 mutant, providing compelling evidence of mechanistic conservation (Tommasini et al., 1996). Ycf1p therefore serves as an excellent model protein to dissect the functional roles of distinct domains of members of the MRP subfamily. Like human MRP1 and MRP2, the domain arrangement of Ycf1p consists of a typical ABC core domain, which includes two MSDs, and two NBDs separated by a linker (L1) as well as the additional NTE (Figure 1A).
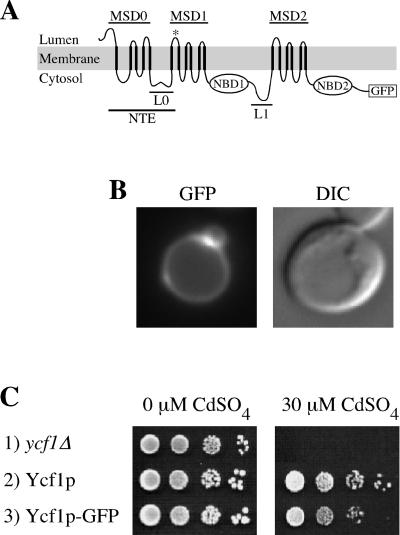
Figure 1.
GFP-tagged Ycf1p localizes to the vacuolar membrane and is functional. (A) Proposed topology of yeast Ycf1p is shown. The core region contains two membrane spanning domains (MSD1 and MSD2) and two nucleotide binding domains (NBD1 and NBD2), separated by a linker (L1) (note, for simplicity sake L1 is omitted in subsequent figures). The NTE is comprised of MSD0 and L0. The proposed topology for Ycf1p is modeled after the experimentally determined topology of the Ycf1p human homologue MRP1 (Bakos et al., 1996; Hipfner et al., 1997). Any small differences between Ycf1p and MRP1 would be unlikely to influence the conclusions of this study. The GFP coding sequence is fused immediately before the stop codon of Ycf1p. (B) GFP fluorescence pattern (left) and corresponding DIC image (right) of cells expressing Ycf1p-GFP are shown. The vacuole appears as an indentation in the DIC images. The brighter area of fluorescence appears to correspond to a region where two vacuolar lobes intersect. (C) Cadmium resistance was tested by spotting an aliquot (5 μl) of cells at 0.1 OD600 and serial 10-fold dilutions thereof onto plates containing 0 or 30 μM CdSO4 and incubating at 30°C for 3 or 6 d, respectively. The strains used are SM4516 (row 1), SM4517 (row 2), and SM4518 (B and row 3).
The goal of the present study was to assess the function of the N-terminal extension in Ycf1p. To do so, we constructed partial molecules of Ycf1p and determined their functional ability by assaying tolerance to cadmium. We show that MSD0 is required for the efficient localization of Ycf1p to the vacuolar membrane. This is the first demonstration of a role for MSD0 in the trafficking of a yeast MRP transporter and parallels the finding that MSD0 is important for the trafficking of human MRP2 (Fernandez et al., 2002). On the other hand, our results indicate that L0 is not required for localization of Ycf1p but is instead essential for Ycf1p-mediated cadmium resistance (a similar functional role has been previously demonstrated for L0 of MRP1). In this study, we also reexamined an intriguing aspect of Ycf1p biology, namely, its posttranslational proteolytic processing that was first reported by Wemmie and Moye-Rowley (1997). Our studies show that cleavage of Ycf1p occurs within a lumenal loop of MSD1 and is dependent on the master vacuolar protease Pep4p, thus providing a convenient marker for successful trafficking of Ycf1p to the vacuolar membrane. Furthermore, our analysis indicates that once processing occurs, the N- and C-terminal cleavage products remain associated with one another, suggesting that both cleavage products contribute to activity of Ycf1p.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions
Yeast strains used in this study are listed in Table 1. Plate and liquid drop-out media were prepared as described previously (Michaelis and Herskowitz, 1988). The plates used for the cadmium spot tests were prepared by adding the indicated concentration of CdSO4 to the minimal medium immediately before pouring the plates. Cultures were grown at 30°C except where indicated. All yeast transformations were performed as described previously (Elble, 1992).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Reference/source |
|---|---|---|
| BY4741 | MATa leu2Δ0 met15Δ0 ura3Δ0 his3Δ1 | (Brachmann et al., 1998) |
| MS10 | MATa ura3-52 leu2-3112 ade2-101 | M. Rose |
| MS1907 | MATa ura3-52 leu2-3112 ade2-101 pep4∷LEU2 | M. Rose |
| RH144-3D | MATa ura3 his4 leu2 bar1-1 | (Raths et al., 1993) |
| RH266-1D | MATa end3ts ura3 his4 leu2 bar1-1 | (Raths et al., 1993) |
| SM3851 | MATa trp1 leu2 ura3 his4 can1 ycf1Δ | (Sharma et al., 2002) |
| SM4516 | SM3851 [2μ URA3] | Transformant of SM3851 with pSM217 |
| SM4517 | SM3851 [2μ URA3 YCF1] | Transformant of SM3851 with pSM1752 |
| SM4518 | SM3851 [2μ URA3 YCF1-GFP] | Transformant of SM3851 with pSM1753 |
| SM4523 | MS10 [2μ URA3 YCF1-GFP] | Transformant of MS10 with pSM1753 |
| SM4524 | MS1907 [2μ URA3 YCF1-GFP] | Transformant of MS1907 with pSM1753 |
| SM4529 | RH144-3D [2μ URA3 YCF1-GFP] | Transformant of RH144-3D with pSM1753 |
| SM4531 | RH266-1D [2μ URA3 YCF1-GFP] | Transformant of RH266-1D with pSM1753 |
| SM4542 | SM3851 [2μ URA3 YCF1-HA-GFP] | Transformant of SM3851 with pSM1774 |
| SM4575a | MATa leu2Δ0 met15Δ0 ura3Δ0 his3Δ1 pep4∷KanMX4 | Research Genetics (Huntsville, AL) |
| SM4576a | MATa leu2Δ0 met15Δ0 ura3Δ0 his3Δ1 prb1∷KanMX4 | Research Genetics (Huntsville, AL) |
| SM4605 | BY4741 [2μ URA3 YCF1-GFP] | Transformant of BY4741 with pSM1753 |
| SM4606 | SM4575 [2μ URA3 YCF1-GFP] | Transformant of SM4575 with pSM1753 |
| SM4607 | SM4576 [2μ URA3 YCF1-GFP] | Transformant of SM4576 with pSM1753 |
| SM4735 | SM3851 [2μ URA3 ycf1Δ209-1515-HA] | Transformant of SM3851 with pSM1869 |
| SM4736 | SM3851 [2μ URA3 ycf1Δ272-1515-HA] | Transformant of SM3851 with pSM1870 |
| SM4741 | SM3851 [2μ URA3 ycf1Δ209-1515-HA] [2μ LEU2 ycf1Δ2-208-GFP] | Transformant of SM3851 with pSM1869, pSM1873 |
| SM4742 | SM3851 [2μ URA3 ycf1Δ272-1515-HA] [2μ LEU2 ycf1Δ2-271-GFP] | Transformant of SM3851 with pSM1870, pSM1874 |
| SM4758 | SM3851 [2μ URA3] [2μ LEU2] | Transformant of SM3851 with pSM217, pSM218 |
| SM4760 | SM3851 [2μ URA3 ycf1Δ209-1515-HA] [2μ LEU2] | Transformant of SM3851 with pSM1869, pSM218 |
| SM4761 | SM3851 [2μ URA3 ycf1Δ272-1515-HA] [2μ LEU2] | Transformant of SM3851 with pSM1870, pSM218 |
| SM4763 | SM3851 [2μ URA3] [2μ LEU2 ycf1Δ2-208-GFP] | Transformant of SM3851 with pSM217, pSM1873 |
| SM4764 | SM3851 [2μ URA3] [2μ LEU2 ycf1Δ2-271-GFP] | Transformant of SM3851 with pSM217, pSM1874 |
| SM4766 | SM3851 [2μ LEU2 ycf1Δ2-208-GFP] | Transformant of SM3851 with pSM1873 |
| SM4767 | SM3851 [2μ LEU2 ycf1Δ2-271-GFP] | Transformant of SM3851 with pSM1874 |
| SM4777 | SM3851 [2μ URA3] [2μ LEU2 YCF1-HA-GFP] | Transformant of SM3851 with pSM217, pSM1881 |
| SM4778 | SM3851 [2μ URA3 ycf1Δ209-1515-HA] [2μ LEU2 ycf1Δ2-271-GFP] | Transformant of SM3851 with pSM1869, pSM1874 |
| SM4876 | SM3851 [2μ URA3] [2μ LEU2 ycf1Δ223-235-HA-GFP] | Transformant of SM3851 with pSM217, pSM1890 |
This strain is a derivative of the strain BY4741 and was provided by J. Boeke (Johns Hopkins University School of Medicine, Baltimore, MD).
Plasmid Constructions
Plasmids used in this study are listed in Table 2. To epitope tag the N-terminal region of Ycf1p, we used a triply iterated epitope from influenza hemagglutinin (HA) as a NotI fragment in the vector GTEP1 (Tyers et al., 1992). First, a NotI restriction site was introduced after amino acid 63 of Ycf1p in pSM1753 (2μ URA3 YCF1-GFP) (Sharma et al., 2002) to yield pSM1772 (2μ URA3 YCF1-GFP), which is identical to pSM1753 except for the addition of the NotI site. The triple HA epitope tag was subcloned into NotI-digested pSM1772. The resulting plasmid, pSM1774 (2μ URA3 YCF1-HA-GFP), contains a triple HA epitope tag in the first cytosolic loop of Ycf1p and GFP fused to the C terminus of Ycf1p (Figure 4A). The addition of the triple HA epitope tag was confirmed by DNA sequence analysis.
Table 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Reference/source |
|---|---|---|
| pJAW50 | 2μ TRP1 YCF1 | Wemmie et al., 1994 |
| pRS425 | 2μ LEU2 | Sikorski and Hieter, 1989 |
| pSM217 | 2μ URA3 | Chen et al., 1997 |
| pSM218 | 2μ LEU2 | Berkower and Michaelis, 1991 |
| pSM1752 | 2μ URA3 YCF1 | Sharma et al., 2002 |
| pSM1753 | 2μ URA3 YCF1-GFP | Sharma et al., 2002 |
| pSM1772 | 2μ URA3 YCF1-GFP | This study |
| pSM1774 | 2μ URA3 YCF1-HA-GFP | This study |
| pSM1869 | 2μ URA3 ycf1Δ209-1515-HA (MSD0) | This study |
| pSM1870 | 2μ URA3 ycf1Δ272-1515-HA (MSD0-L0) | This study |
| pSM1871 | 2μ URA3 ycf1Δ2-208-GFP (L0-ΔYcf1p) | This study |
| pSM1872 | 2μ URA3 ycf1Δ2-271-GFP (ΔYcf1p) | This study |
| pSM1873 | 2μ LEU2 ycf1Δ2-208-GFP (L0-ΔYcf1p) | This study |
| pSM1874 | 2μ LEU2 ycf1Δ2-271-GFP (ΔYcf1p) | This study |
| pSM1881 | 2μ LEU2 YCF1-HA-GFP | This study |
| pSM1889 | 2μ URA3 ycf1Δ223-235-HA-GFP | This study |
| pSM1890 | 2μ LEU2 ycf1Δ223-235-HA-GFP | This study |
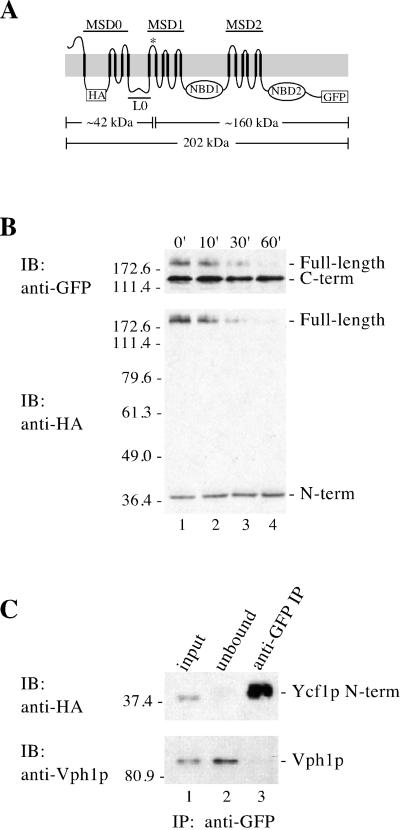
Figure 4.
Proteolytic processing of Ycf1p yields two stable products that associate with each other. (A) Schematic of Ycf1p shows the location of the potential cleavage site (*), the triple HA epitope, the GFP tag, and the approximate molecular weight of the N- and C-terminal epitope-tagged cleavage products based on SDS-PAGE migration. (B) Stability of the cleavage fragments was examined by cycloheximide chase analysis. After addition of 10 μg of cycloheximide to 10 OD600 units of mid-log cells to stop protein synthesis, aliquots were removed at the indicated times. Crude yeast cell extracts (0.5 OD600 cell equivalents per lane) were resolved by 10% SDS-PAGE and transferred to nitrocellulose. The same immunoblot was probed sequentially with two different antibodies to detect the N- and C-termini of Ycf1p-HA-GFP by using rat anti-HA monoclonal antibodies (bottom) and rabbit anti-GFP polyclonal antibodies (top), respectively. (C) Association between the N- and C-terminal cleavage products of Ycf1p was examined by coimmunoprecipitation. Crude membranes were prepared after a 60-min cycloheximide chase as described in MATERIALS AND METHODS and subjected to IP with rabbit anti-GFP polyclonal antibodies under nondenaturing conditions to pull down the C-terminal cleavage product and any associated proteins. Immunoprecipitated proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose. The presence of the N-terminal Ycf1p cleavage product in the anti-GFP IP was determined by probing the immunoblot with rat anti-HA monoclonal antibodies (top, lane 3). The N-terminal Ycf1p band detected by immunoblotting is specific, because it was not detected in a control which contains Ycf1p-GFP, but lacks the HA tag (our unpublished data). The presence or absence of an unrelated vacuolar protein, Vph1p, was assessed using mouse anti-Vph1p monoclonal antibodies (bottom, lane 3). Lanes 1, 2, and 3 contain ∼1, ∼1, and 50% of the total, respectively. The strain used for B and C is SM4542.
The Ycf1p partial molecules were constructed by recombinational cloning (Oldenburg et al., 1997). Each N-terminal partial molecule has a triple HA epitope tag at residue 63 in the first cytosolic loop. The C-terminal partial molecules have green fluorescent protein (GFP) fused immediately before the stop codon. To construct the plasmids pSM1869 (2μ URA3 ycf1Δ209-1515-HA; MSD0) and pSM1870 (2μ URA3 ycf1Δ272-1515-HA; MSD0-L0), fragments of YCF1 were amplified by polymerase chain reaction (PCR) from pSM1753 and then recombined into AgeI-PmlI-digested pSM1774. The corresponding C-terminal partial molecules of Ycf1p containing amino acids 209-1515 or 272-1515 were constructed by PCR amplification of a fragment of YCF1 from pJAW50 (2μ TRP1 YCF1) (Wemmie et al., 1994), followed by recombination of each product into AatII-digested pSM1774. The resulting plasmids were pSM1871 (2μ URA3 ycf1Δ2-208-GFP; L0-ΔYcf1p) and pSM1872 (2μ URA3 ycf1Δ2-271-GFP; ΔYcf1p). To construct LEU2 versions of these C-terminal partial molecules, PvuI fragments from pSM1871 and pSM1872 were subcloned into PvuI-digested pRS425 (2μ LEU2) (Sikorski and Hieter, 1989) generating pSM1873 (2μ LEU2 ycf1Δ2-208-GFP; L0-ΔYcf1p) and pSM1874 (2μ LEU2 ycf1Δ2-271-GFP; ΔYcf1p). A LEU2 version of full-length YCF1-HA-GFP was constructed by subcloning a PvuI fragment from pSM1774 into PvuI-digested pRS425 to yield pSM1881 (2μ LEU2 YCF1-HA-GFP).
A deletion of the 13-amino acid region predicted to form a helical wheel within L0 was created by recombinational cloning of a YCF1 PCR product containing an engineered deletion of amino acids 223–235 into AgeI-linearized pSM1774, thereby yielding pSM1889 (2μ URA3 ycf1Δ223-235-HA-GFP). To construct a LEU2 version of ycf1Δ223-235-HA-GFP, an AatII-StuI fragment from pSM1889 was subcloned into AatII-StuI–digested pSM1881, generating pSM1890 (2μ LEU2 ycf1Δ223-235-HA-GFP). The partial molecules and helical wheel deletion described above were confirmed by DNA sequence analysis. Note that sequence analysis revealed a conservative substitution of K270R in pSM1870 that was presumably generated during the plasmid construction.
A detailed description of plasmid constructions have been described previously (Mason, 2002) and can be furnished upon request.
Antibodies
The rabbit anti-GFP polyclonal antibody was a gift from R. Jensen (Johns Hopkins University School of Medicine, Baltimore, MD). The mouse anti-GFP monoclonal antibody (mAb) was purchased from CLONTECH (Palo Alto, CA). The 3F-10 rat anti-HA mAb was purchased from Roche Applied Sciences (Indianapolis, IN) and the 10D7-A7-B2 mouse anti-Vph1p mAb was purchased from Molecular Probes (Eugene, OR). The horseradish peroxidase-conjugated secondary antibodies (donkey anti-rabbit Ig, sheep anti-mouse Ig, and sheep anti-rat Ig) used for immunoblotting were purchased from Amersham Biosciences (Piscataway, NJ).
Fluorescence Microscopy
To examine the localization of Ycf1p-GFP, cells were grown overnight to saturation in minimal medium, and then subcultured at a 1:1000 dilution in minimal medium and grown overnight at 30°C to an OD600 of ∼0.7. Log phase cells were examined at 100× magnification on poly-lysine–coated slides by using an Axioskop microscope equipped with fluorescence and Nomarski optics (Carl Zeiss, Thornwood, NY). Images were captured with a Cooke charge-coupled device camera and IP Lab Spectrum Software (Biovision Technologies, Exton, PA).
Immunoblotting Analysis
Cell extracts and immunoblots were prepared as described previously (Fujimura-Kamada et al., 1997) except that samples were either heated at 65°C for 10 min before electrophoresis (Figure 2) or not heated to minimize aggregation (Figure 5). Crude yeast cell extracts (0.4 OD600 cell equivalents per lane) were resolved by either 8% (Figures 2 and 5B) or 10% (Figure 5C) SDS-PAGE and transferred to nitrocellulose. The primary antibodies used were rabbit anti-GFP (1:5000), mouse anti-GFP (1:670), and rat anti-HA (1:400).
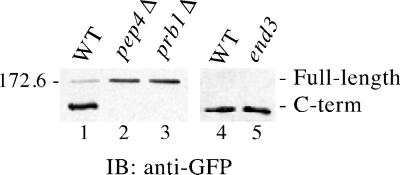
Figure 2.
Ycf1p processing is dependent on the vacuolar proteases Pep4p and Prb1p and is End3p independent. The steady-state level of Ycf1p-GFP in wild-type (WT) and mutant strain backgrounds was examined by immunoblot (IB) analysis as described in MATERIALS AND METHODS. Unprocessed Ycf1p is indicated as full-length and the C-terminal cleavage product as C-term. The end3 mutant strain (lane 5) was shifted to 37°C for 1 h to impose the endocytosis block before preparing the cell extracts, along with its isogenic WT counterpart (lane 4). Crude yeast cell extracts (0.4 OD600 cell equivalents per lane) were resolved by 8% SDS-PAGE and transferred to nitrocellulose. Ycf1p-GFP was detected using rabbit anti-GFP polyclonal antibodies. A molecular weight marker is indicated. The strains used are SM4605 (lane 1), SM4606 (lane 2), SM4607 (lane 3), SM4529 (lane 4), and SM4531 (lane 5).
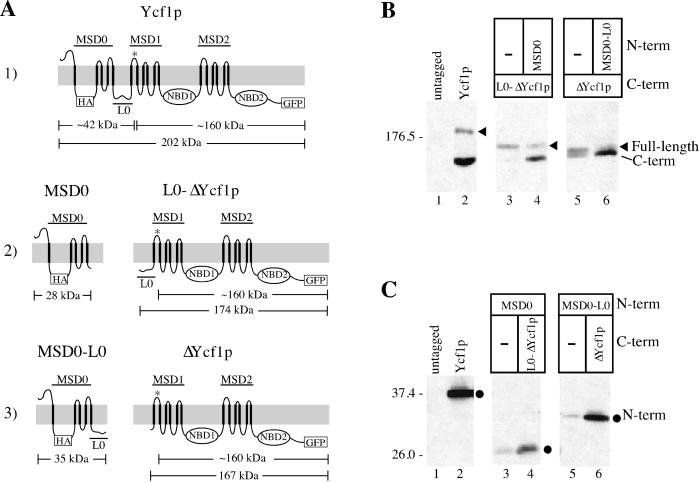
Figure 5.
Coexpression of Ycf1p partial molecules is required for the proteolytic processing and stability of Ycf1p. (A) Schematics of the Ycf1p partial molecules are shown. The N-terminal molecules contain a 3xHA epitope tag at residue 63 in the first cytosolic loop and the C-terminal molecules have GFP fused to the C terminus immediately before the stop codon of Ycf1p. The predicted cleavage site is indicated by an asterisk (*) in all of the C-terminal molecules. The approximate molecular weight of the N- and C-terminal epitope-tagged partial molecules and cleavage products based on SDS-PAGE migration are also shown. Row 1: wild-type Ycf1p-HA-GFP. Row 2: MSD0 includes the N-terminal portion of Ycf1p from amino acids 1–208; L0-ΔYcf1p contains the core region of Ycf1p plus the linker region (L0) from amino acids 209-1515. Row 3: MSD0-L0 represents the N-terminal region from amino acids 1–271; ΔYcf1p includes the core domain from amino acids 272-1515. (B and C) Immunoblot analysis of the C- and N-terminal partial molecules, respectively, expressed alone (−) or coexpressed with their corresponding partial molecules. The full-length protein encoded by each construct is indicated by an arrowhead. As indicated by SDS-PAGE mobility, the C-terminal cleavage product seems to be the same size in all cases and is indicated. The N-terminal partial molecules are indicated by circles. Crude yeast cell extracts (0.4 OD600 cell equivalents per lane) were resolved by either 8% (B) or 10% (C) SDS-PAGE and transferred to nitrocellulose. The C- and N-terminal partial molecules were detected with mouse anti-GFP monoclonal antibodies (B) and rat anti-HA monoclonal antibodies (C), respectively. Molecular weight markers are indicated. The strains used are SM4517 (lane 1), SM4542 (lane 2), SM4741 (lane 4), and SM4742 (lane 6) (B and C); SM4766 (lane 3) and SM4767 (lane 5) (B); and SM4735 (lane 3) and SM4736 (lane 5) (C).
Cycloheximide Chase Experiments
Logarithmically growing cells (10.0 OD600 units) were harvested and resuspended in 2.0 ml of synthetic complete drop-out medium, to which 10 μl of 1 mg/ml cycloheximide was added. Aliquots (2.5 OD600 units) were collected after incubation at 30°C for 0, 10, 30, and 60 min and added to an equal volume of ice-cold 2× azide stop mix (40 mM cysteine, 40 mM methionine, 20 mM sodium azide). After harvesting the cells, proteins were precipitated and analyzed by immunoblotting as described previously (Fujimura-Kamada et al., 1997) except that samples were heated at 65°C for 10 min before electrophoresis, and 0.5 OD600 cell equivalents per lane were resolved by 10% SDS-PAGE. The primary antibodies used were rabbit anti-GFP (1:5000) and rat anti-HA (1:1000).
Metabolic Labeling and Immunoprecipitation
Proteins were metabolically labeled and immunoprecipitated as described previously (Loayza et al., 1998) except that 50 μl of extract in sample buffer was brought up to 1.0 ml with dilution buffer and 2.0 μl of the rabbit anti-GFP antibody was added to each sample of 2.5 OD600 units of cells. Immunoprecipitates were dissociated from the protein A-Sepharose beads by the addition of 30 μl of 2× Laemmli sample buffer and incubated at 37°C for 10 min before electrophoresis.
The amount of the full-length and C-terminal cleavage product of Ycf1p remaining after each time point was determined using PhosphorImager analysis and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The data were graphed using the KaleidaGraph software (Synergy Software, Reading, PA).
Yeast Cell Membrane Preparations and Coimmunoprecipitation Assay
Logarithmically growing cells (100 OD600 units) were harvested, washed once with cold 10 mM sodium azide, and resuspended to 10 OD600 units per milliliter in cold 100 mM Tris and 10 mM dithiothreitol, pH 9.6. After a 10-min incubation on ice, cells were recovered by centrifugation and resuspended to 25 OD600 units per milliliter in oxalyticase buffer (50 mM KPi, pH 7.5, 1.4 M sorbitol, 10 mM sodium azide). To generate spheroplasts, oxalyticase (Enzogenetics, Corvallis, OR) was added at 1 μg/OD600 units and the cells were incubated at 30°C in a shaking water bath for 40 min. The resulting spheroplasts were chilled on ice for 5 min, harvested by centrifugation, and resuspended to 500 OD600 units per milliliter in cold lysis buffer (10 mM HEPES, pH 7.0, 0.8 M sorbitol, 1 mM EDTA, 0.02% sodium azide) containing protease inhibitors (3 μg/ml leupeptin, pepstatin, and chymostatin; 2 μg/ml aprotinin; and 1 mM phenylmethylsulfonyl fluoride). The cells were lysed by vortexing with zirconium beads in 10 1-min pulses at 4°C. The lysate was cleared twice of unbroken cells and large cellular fragments by centrifugation (500 × g for 10 min at 4°C). Membranes were recovered from the cleared lysate by centrifugation (100,000 × g for 30 min at 4°C), resuspended in lysis buffer, and protein concentrations determined using the Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA). The concentration of the proteins was adjusted to 0.8 mg/ml in lysis buffer and frozen as aliquots (500 μl) at −80°C. To carry out the coimmunoprecipitation assay, membranes (0.4 mg) were raised to 1 ml in immunoprecipitation (IP) dilution buffer (1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 50 mM Tris, pH 7.5, 3 μg/ml leupeptin, 3 μg/ml pepstatin, 3 μg/ml chymostatin, 2 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride) and the immunoprecipitation carried out as described previously (Loayza et al., 1998) except that 4 μl of rabbit anti-GFP polyclonal antibodies was added to each sample. Immunoprecipitates were dissociated from the protein A-Sepharose beads by the addition of 40 μl of 2 × Laemmli sample buffer and incubated at 65°C for 10 min before electrophoresis. Proteins were separated by 10% SDS-PAGE and immunoblots were prepared as described previously (Fujimura-Kamada et al., 1997). The N-terminal cleavage product of Ycf1p was detected with rat anti-HA monoclonal antibodies (1:250). Nonspecific interactions with Vph1p were examined with mouse anti-Vph1p monoclonal antibodies (1:5000).
Growth Inhibition by Cadmium
To examine growth on plates, cells were grown overnight to saturation in minimal medium, and then subcultured at a 1:500 dilution in minimal medium and grown overnight at 30°C to an OD600 of ∼1.0. This overnight culture was diluted to an OD600 of 0.1, which in turn was diluted in 10-fold increments. Aliquots (5 μl) of each 10-fold dilution were spotted onto synthetic complete minimal medium containing 0 and 40 μM CdSO4 and incubated at 30°C for 3 or 6 d, respectively.
Results
Posttranslational Proteolytic Processing of Ycf1p Is PEP4 Dependent
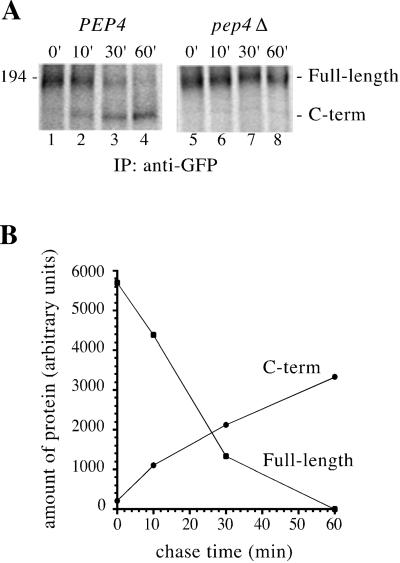
To facilitate localization and processing studies, we generated a C-terminally GFP-tagged version of Ycf1p (Figure 1A). Like untagged Ycf1p (Li et al., 1996; Wemmie and Moye-Rowley, 1997), Ycf1p-GFP localizes to the vacuolar membrane, as evidenced by the correspondence of the fluorescence pattern with the indentation of the vacuole detectable by differential interference contrast (DIC) microscopy (Figure 1B). In addition, Ycf1p-GFP is active in conferring cadmium resistance (Figure 1C). Using this construct to further examine the posttranslational processing of Ycf1p, we find that the majority of Ycf1p-GFP is in the proteolytically processed form under steady-state conditions (Figure 2, lane 1), as reported previously for untagged Ycf1p (Wemmie and Moye-Rowley, 1997).
The proteolytic processing of Ycf1p is unusual because the size of the C-terminal cleavage product (∼160 kDa) indicates that a large portion of the protein, including the hydrophobic NTE plus additional residues, is released upon cleavage. To determine whether proteolytic processing occurs once Ycf1p reaches its final destination within the cell, we examined the processing of Ycf1p in two different vacuolar protease-deficient strains, pep4Δ or prb1Δ. Pep4p, the master vacuolar protease, and the protease Prb1p play reciprocal roles in activating each other and are responsible for activating numerous other vacuolar proteases (Jones et al., 1997). In the absence of either of these proteases, Ycf1p is not cleaved (Figure 2, lanes 2 and 3). Because Ycf1p properly localizes to the vacuolar membrane in the pep4Δ and prb1Δ strains (our unpublished data), the lack of processing must not be due to mislocalization. In addition, the C-terminal proteolytic product of Ycf1p is generated in both the end3 and isogenic wild-type strains at the nonpermissive temperature (Figure 2, lanes 4 and 5). Thus, in contrast to previously published data (Wemmie and Moye-Rowley, 1997), we found that cleavage of Ycf1p is PEP4 dependent and that processing is not affected in an endocytosis-deficient end3ts mutant strain. We expect that the processing block observed in the previous study was actually due to a PEP4 deletion because the end3-1 mutant strain that was used (RH1834) was also a pep4 mutant.
We examined the kinetics of Ycf1p proteolytic processing by carrying out metabolic labeling and pulse-chase analysis. In the wild-type strain (PEP4), full-length Ycf1p chases into the C-terminal cleavage product and, as expected, no processing is observed in the pep4Δ strain at any of the time points examined (Figure 3A). As full-length Ycf1p disappears, the amount of the C-terminal proteolytic product increases, indicating a precursor–product relationship between the two molecules (Figure 3B). The apparent discrepancy between the amount of the C-terminal cleavage product accumulated by 60 min compared with the amount of full-length Ycf1p at time 0 is primarily due to the difference in the number of 35S-labeled cysteines and methionines contained in each piece, i.e., the full-length molecule has a combined total of 54 cysteines and methionines, whereas the C-terminal cleavage product has only 40. Taken together, the immunoblot and immunoprecipitation analyses indicate that Ycf1p proteolytic processing occurs at the vacuole (i.e., is dependent on vacuolar protease), and therefore cleavage can serve as a marker for vacuolar localization of Ycf1p.
Figure 3.
Metabolic pulse-chase analysis of Ycf1p-GFP processing. (A) To examine the kinetics of PEP4-dependent conversion of full-length Ycf1p-GFP into the C-terminal cleavage product, pulse-chase experiments were performed in PEP4 (wild-type) (lanes 1–4) and pep4Δ (lanes 5–8) strains. Cells were pulse labeled for 10 min with Express [35S] and the label was chased for the indicated times by addition of an excess of unlabeled cysteine and methionine. Ycf1p-GFP was immunoprecipitated with rabbit anti-GFP polyclonal antibodies and the samples were analyzed by 8% SDS-PAGE. The strains used are SM4523 (lanes 1–4) and SM4524 (lanes 5–8). (B) Bands corresponding to full-length Ycf1p and the C-terminal proteolytic product in lanes 1–4 were quantitated using PhosporImager and ImageQuant software. The data was graphed with KaleidaGraph to reveal a precursor–product relationship between full-length Ycf1p and the C-terminal cleavage product.
Proteolytic Processing of Ycf1p Yields Two Stable Cleavage Products That Associate with Each Other
To determine whether the N- and C-terminal Ycf1p cleavage products are metabolically stable and/or interact with one another, we first tagged Ycf1p-GFP with a triple HA epitope in its N-terminal cytosolic loop (Figure 4A). This construct localizes to the vacuole, retains activity (our unpublished data; Figure 8, row 2) and permits us to follow the N- and C-terminal proteolytic products with antibodies to HA and GFP, respectively. The predicted Ycf1p cleavage site, indicated by the asterisk (*) in loop 6 (Figure 4A), is based on the SDS-PAGE mobility of the N- and C-terminal cleavage products (Figure 4B) and on the requirement for a lumenal processing site that is accessible to vacuolar proteases. Thus, the N-terminal cleavage product includes MSD0, L0, and the first transmembrane span of MSD1, whereas the C-terminal cleavage product includes the majority of the core region of Ycf1p, but lacks the first transmembrane span of MSD1.
Figure 8.
Functional analysis of Ycf1p partial molecules indicates that L0 is important for activity. The partial molecules were tested for their ability to confer resistance to CdSO4 when expressed alone and in combination with their corresponding partial molecules. An aliquot of cells at 0.1 OD600, and serial 10-fold dilutions thereof, were spotted onto plates containing 0 or 40 μM CdSO4 and incubated at 30°C for 3 or 6 d, respectively. The ability to confer resistance to cadmium correlates with proper vacuolar localization of Ycf1p, as indicated at the right (rows 2, 5, and 8; Figures 6 and 7). The exceptions (rows 9 and 10) are the combination MSD0 + ΔYcf1p and Ycf1pΔ223–235 (missing L0 or containing an internal deletion of L0, respectively), which localize to the vacuole (Figure 7C; our unpublished results), but do not confer resistance to cadmium. The strains used are SM4758 (row 1), SM4777 (row 2), SM4760 (row 3), SM4763 (row 4), SM4741 (row 5), SM4761 (row 6), SM4764 (row 7), SM4742 (row 8), SM4778 (row 9), and SM4876 (row 10). NA, not applicable.
We first assessed the stability of the N- and C-terminal proteolytic products by using a cycloheximide chase experiment (Figure 4B). The fate of the Ycf1p species present at steady state (Figure 4B, lane 1) could be monitored by adding cycloheximide to stop protein synthesis and then by removing aliquots at the indicated time points. Full-length Ycf1p (detected with GFP or HA antibodies) disappears by 60 min. In contrast, the N- and C-terminal proteolytic products are stable during the chase period (Figure 4B, bottom and top, respectively). This metabolic stability of the N- and C-terminal cleavage products suggests that both are likely to contribute to the biological properties of Ycf1p.
To determine whether the N- and C-terminal cleavage products interact with each other once Ycf1p is processed, we performed a coimmunoprecipitation experiment in which immunoprecipitation with anti-GFP antibodies was followed by probing the immunoblot with anti-HA antibodies (Figure 4C). To ensure that the majority of Ycf1p in the starting material was in the processed form, we performed a 60-min cycloheximide chase before the preparation of crude membranes and immunoprecipitation of the C-terminal proteolytic product of Ycf1p. As shown in the immunoblot analysis of this immunoprecipitate, the N- and C-terminal proteolytic products of Ycf1p seem to efficiently coimmunoprecipitate, suggesting that they interact with each other after cleavage occurs (Figure 4C, top, lane 3). We also checked for nonspecific interactions in the immunoprecipitated material by examining the presence of an abundant vacuolar membrane protein, Vph1p. A comparable amount of Vph1p is detected in the input and unbound lanes and is absent from the IP lane, indicating that Vph1p does not interact with Ycf1p (Figure 4C, bottom, lanes 1–3). We conclude that the interaction between the N- and C-terminal proteolytic products of Ycf1p is specific. The interaction of the cleavage products further supports the hypothesis that both contribute to Ycf1p function.
Coexpression of Ycf1p Partial Molecules Is Required for Proper Trafficking to the Vacuole, as Assessed by Proteolytic Processing and Stability of Ycf1p
The results described above showing Pep4p-dependent proteolytic processing of Ycf1p indicate that the C-terminal cleavage product can serve as a marker for the proper vacuolar localization of Ycf1p. The localization of Ycf1p can also be assessed by its fluorescence pattern. To examine whether the MSD0 and/or L0 subregions of the NTE play a role in trafficking of Ycf1p to the vacuole, we modeled our approach on that taken by others to study human MRP1 (Bakos et al., 1998; Gao et al., 1998). This approach involves the construction and characterization of deletion mutants that we designate partial molecules. The partial molecules of Ycf1p that we generated are shown in Figure 5A. The constructs in rows 2 and 3 are distinguished by the attachment of L0 to the Ycf1p core (ΔYcf1p) or to MSD0, respectively.
We first examined the SDS-PAGE pattern of the Ycf1p partial molecules expressed singly and together. Notably, the proteolytic processing of the C-terminal partial molecules, L0-ΔYcf1p and ΔYcf1p (Figure 5A, rows 2 and 3, respectively), is dramatically increased upon coexpression with their corresponding N-terminal partial molecules that contain MSD0 (Figure 5B, lanes 3–6, bottom band). In addition, coexpression notably increases the steady-state level of these C-terminal partial molecules (Figure 5B, lanes 3–6). Reciprocally, the steady-state level of the N-terminal partial molecules, MSD0 and MSD0-L0, is also greatly enhanced by the presence of their corresponding C-terminal partial molecules (Figure 5C, lanes 3–6). Together, these results indicate that coexpression is required for efficient proteolytic processing of the Ycf1p C-terminal partial molecules and to stabilize both the N- and C-terminal partial molecules. Thus, it seems that MSD0 is critical for the targeting of L0-Ycf1p or ΔYcf1p to the vacuolar membrane. MSD0 could contain specific targeting information or contribute to the proper folding of Ycf1p.
MSD0 Is Required for Vacuolar Localization of Ycf1p, but L0 Is Not
The immunoblot analyses (Figure 5B) indicate that processing of the Ycf1p C-terminal partial molecules is greatly enhanced by coexpression with their corresponding N-terminal partial molecules, suggesting that coexpression leads to vacuolar localization whereupon proteolytic processing can occur. This hypothesis implies that the N- and C-terminal partial molecules are mislocalized when expressed alone, perhaps because they are misfolded and subjected to various cellular quality control systems that lead to degradation (Hurtley and Helenius, 1989; Chang and Fink, 1995; Hammond and Helenius, 1995; Li et al., 1999). To test this hypothesis, we visually examined the GFP fluorescence pattern of the C-terminal partial molecules when expressed alone or together with their corresponding N-terminal partial molecules (Figures 6 and 7).
Figure 6.
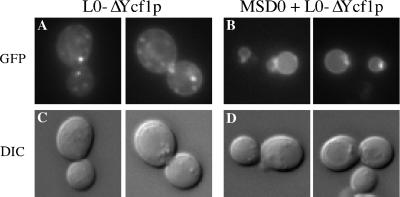
MSD0 is required for the vacuolar localization of Ycf1p. The GFP fluorescence pattern of the C-terminal partial molecule L0-ΔYcf1p is shown for cells expressing solely L0-ΔYcf1p (A) or for cells coexpressing L0-ΔYcf1p and MSD0 (B). Two examples are shown for each. In the corresponding DIC images of these cells, the vacuole appears as an indentation (C and D). The strains used are SM4763 (A) and SM4741 (B).
Figure 7.
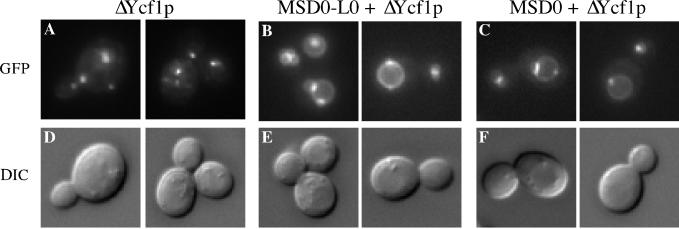
L0 is not necessary for the vacuolar localization of Ycf1p. The GFP fluorescence pattern of the C-terminal partial molecule ΔYcf1p is shown for cells expressing solely ΔYcf1p (A), for cells coexpressing ΔYcf1p and MSD0-L0 (B), or for cells coexpressing ΔYcf1p and MSD0 (C). Two examples are shown for each. The corresponding DIC images of these cells are also shown (D–F). The strains used are SM4764 (A), SM4742 (B), and SM4778 (C).
We first consider our findings with the C-terminal partial molecules expressed alone. Neither L0-ΔYcf1p nor ΔYcf1p is predominantly vacuolar (Figures 6A and 7A). Instead, both seem to mislocalize to intracellular punctate structures whose exact identity is presently unknown, but seem to be largely endosomal based on staining with the lipophilic dye FM4-64 (our unpublished data). Based on immunofluorescence with the ER marker Kar2p, we cannot exclude the possibility that some of the C-terminal partial molecules could also be endoplasmic reticulum associated. In a small percentage of cells these partial molecules showed vacuolar localization that correlates with the low level of processing observed by immunoblot analysis (Figure 5B) and could suggest that a minor population of the C-terminal partial molecules reach the vacuole on their own. By indirect immunofluorescence microscopy, we observe that the N-terminal partial molecules MSD0 and MSD0-L0 also generally mislocalize to intracellular structures when expressed alone (our unpublished data). Importantly, the mislocalization we observe for L0-ΔYcf1p shows that even though L0 is present, it is not sufficient for proper trafficking of Ycf1p in the absence of MSD0. Instead, MSD0 seems to be the key determinant for the vacuolar localization of Ycf1p.
Next, we consider the localization of the C-terminal partial molecules when coexpressed with their corresponding N-terminal partial molecules. There was a striking shift in the localization pattern of the C-terminal partial molecules from intracellular structures to the vacuolar membrane upon coexpression with their corresponding N-terminal partial molecules (Figures 6B and 7B). This shift in localization correlates precisely with the dramatic increase in proteolytic processing observed upon coexpression of the partial molecules and confirms that processing serves as a reliable marker for vacuolar localization (Figure 5B). Likewise, we observed by indirect immunofluorescence that the N-terminal partial molecules MSD0 and MSD0-L0 also shift in localization from intracellular structures to the vacuole when coexpressed with their corresponding C-terminal partial molecule (our unpublished data). Thus, the N- and C-terminal partial molecules are mutually dependent on each other for proper vacuolar localization.
To determine whether the MSD0 segment of the NTE is sufficient alone for the vacuolar localization of ΔYcf1p, we coexpressed MSD0 with ΔYcf1p, two partial molecules that both lack L0. The fluorescence pattern clearly indicates that the MSD0 portion of the NTE suffices to target the majority of ΔYcf1p to the vacuolar membrane (Figure 7C). Taken together, the results of these studies demonstrate that MSD0 is required and that L0 is dispensable for targeting Ycf1p to the vacuolar membrane.
L0 Is Required for Function of Ycf1p
Ycf1p confers cadmium resistance by transporting cadmium–glutathione complexes into the vacuole (Li et al., 1997). To test the ability of the partial molecules to mediate transport function we examined their activity by plating cells on cadmium-containing medium. Ten-fold serial dilutions of the strains were spotted onto plates lacking CdSO4 to ensure the growth properties of the strains were similar, and on plates with 40 μM CdSO4 to test their resistance to cadmium. As expected, cells lacking Ycf1p (ycf1Δ) are sensitive to cadmium, whereas cells expressing wild-type Ycf1p on a multicopy plasmid are cadmium resistant (Figure 8, rows 1 and 2, respectively). None of the partial molecules are functional when expressed alone (Figure 8, rows 3, 4, 6, and 7). However, coexpression of MSD0 with L0-ΔYcf1p or MSD0-L0 with ΔYcf1p restores cellular resistance to cadmium (Figure 8, rows 5 and 8, respectively). We could also test the cadmium phenotype of Ycf1p when L0 is missing altogether by expressing the MSD0 and ΔYcf1p combination of partial molecules. On coexpression of MSD0 with ΔYcf1p, we observe no growth in the presence of cadmium (Figure 8, row 9), even though by microscopy this combination of partial molecules localizes efficiently to the vacuole (Figure 7C). These results provide strong evidence that L0 is required for Ycf1p to confer resistance to cadmium.
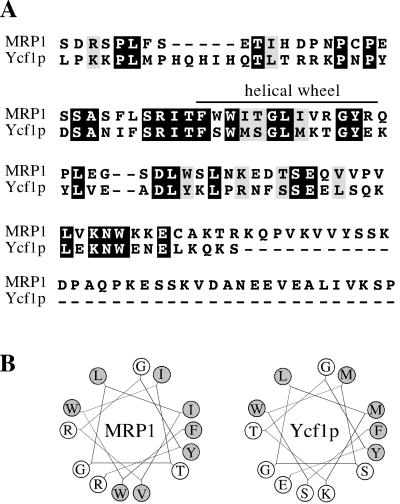
Taken together, the above-mentioned results indicate that although vacuolar localization can be achieved independently of L0, Ycf1p function is absolutely dependent on the presence of L0. This result agrees with the finding that L0 is critical for human MRP1 activity, based on studies with MRP1 partial molecules (Bakos et al., 1998). An essential role for L0 in MRP transport activity was confirmed by examining various internal deletions of L0 (Gao et al., 1998; Bakos et al., 2000; Qian et al., 2001). Bakos et al. (2000) further showed that this cytosolic linker region (L0) associates with membranes on its own, suggesting that L0 forms a distinct functional domain that interacts with the cell membrane or with hydrophobic regions of other membrane proteins. Within L0, a region predicted to form an amphipathic alpha-helix was specifically shown to be critical for the transport activity of MRP1 (Figure 9B; Bakos et al., 2000). Interestingly, based on sequence alignments we find that MRP1 and Ycf1p share a high degree of similarity in L0, particularly in the region predicted to form a helical wheel (Figure 9A). In fact, computer predictions (www.marqusee9.berkeley.edu/kael/helical.htm) indicate that this region in Ycf1p, like that of MRP1, forms an amphipathic alpha-helix as well (Figure 9B). An internal deletion of the helical wheel in Ycf1p (Δ223–235) abolishes the ability of Ycf1p to confer resistance to cadmium (Figure 8, row 10) even though it has no effect on the vacuolar localization of Ycf1p (our unpublished data). This result further demonstrates the functional significance of L0.
Figure 9.
A region within L0 of Ycf1p, like MRP1, forms a helical wheel. (A) Alignment between MRP1 and Ycf1p indicating the complete L0 region, from amino acids 192 to 305 or 189 to 271, respectively, was performed using Clustal W software (Thompson et al., 1994). Black boxes represent amino acid identity and gray boxes indicate amino acid similarity as determined with the Boxshade server. The region proposed previously (Bakos et al., 2000) to form a helical wheel in MRP1 is indicated. (B) Computer predictions indicate that residues 223–235 of Ycf1p form an amphipathic helix (right). The predicted helical wheel for MRP1 (residues 221–233) is shown for comparison and represents published data (left; Bakos et al., 2000). The hydrophobic residues are shaded in gray.
DISCUSSION
Structure–function analyses of numerous ABC proteins have revealed that the ABC “core” comprising two MSDs and two NBDs contains critical components for proper localization, ATP hydrolysis, and substrate transport. However, a universal role for the N-terminal extension (MSD0-L0) present in certain MRP subfamily members, such as Ycf1p, has not been clearly established, and indeed may differ from one MRP protein to another. In this study, we have established a role for MSD0 in targeting Ycf1p to the vacuolar membrane, and for L0 in Ycf1p cadmium resistance function. In addition, we show that Ycf1p undergoes an unusual proteolytic processing event that is mediated by vacuolar proteases and yields two stable products that associate with one another after cleavage. The functional consequences of processing, or lack thereof, are examined elsewhere (Mason and Michaelis, manuscript in preparation). Herein, we use processing as a convenient marker to signify proper trafficking to the vacuolar membrane. Below we consider our findings first in terms of Ycf1p per se and then compare our results to findings on human MRP1 and MRP2.
MSD0 Is Required for Localization of Ycf1p
The importance of MSD0 for Ycf1p localization was determined by expressing partial molecules of Ycf1p separately and together, similar to the approach used to study human MRP1. In this approach, the N-terminal partial molecules contain MSD0 and the C-terminal partial molecules contain the ABC core domain, whereas L0 is attached to either MSD0 or to the core domain. Direct and indirect fluorescence microscopy indicates that all of the Ycf1p partial molecules are mislocalized when expressed alone (Figures 6A and 7A; our unpublished data). However, we observed a striking vacuolar membrane localization when the corresponding N- and C-terminal partial molecules were coexpressed (Figures 6B and 7B). We also observed an increase in the proteolytic processing of Ycf1p by immunoblot analysis when the partial molecules were coexpressed (Figure 5B), which correlates with the increase in their trafficking to the vacuole observed by microscopy. This dramatic shift to the vacuole implies that the partial molecules can, and indeed must, interact with each other when they are coexpressed to form a properly folded molecule. Thus, our results clearly indicate a requirement for MSD0 for vacuolar localization of Ycf1p.
Importantly, the coexpression of two partial molecules that each lack L0 (MSD0 and ΔYcf1p) was sufficient for localizing Ycf1p to the vacuolar membrane (Figure 7C). These results strongly support an essential role for MSD0 in targeting Ycf1p to the vacuolar membrane and indicate that L0 is dispensable for localization of Ycf1p. Our conclusions contradict those of a previous study that suggested that the L0 region was required for vacuolar localization of Ycf1p (Wemmie and Moye-Rowley, 1997). Because the exact same amino acids and length of L0 was used in both studies, the discrepancy cannot be due to differences in the particular region chosen as L0. Several explanations could be possible, including strain differences and/or subtle physiological differences due to media or growth conditions. Whatever the explanation for the discrepancy, our data strongly supports a requirement for MSD0 for the majority of Ycf1p to reach its proper vacuolar membrane localization, under normal physiological conditions.
L0 Is Important for Activity of Ycf1p
To ask about a functional requirement for L0 we tested the cadmium resistance in a strain coexpressing partial molecules (MSD0 and ΔYcf1p) that both lack L0. We did not observe any cadmium resistance (Figure 8, row 9), even though vacuolar targeting was normal (Figure 7C). Immunoblot analysis indicated that coexpression also resulted in an increase in the steady-state protein levels of each partial molecule (our unpublished data), but not to the same dramatic extent as when either molecule contained L0, suggesting that L0 may also play a role in stabilization of Ycf1p. Although the modest destabilization of MSD0 and ΔYcf1p in the absence of L0 could make a minor contribution to reducing cadmium resistance, it is unlikely to account for the essentially complete elimination in cadmium resistance that we observed upon coexpression of MSD0 and ΔYcf1p. In contrast, coexpression of essentially the same N- and C-terminal partial molecules, but with L0 attached to either MSD0 or ΔYcf1p (Figure 8, rows 5 and 8), exhibits cadmium resistance indistinguishable from wild type. Thus, our data suggest that L0 plays a key role in modulating Ycf1p transport activity.
We further defined the region within L0 that is critical for activity by creating an internal deletion in L0. The activity of Ycf1p was abolished when we deleted a 13-amino acid region predicted to form an amphipathic alpha-helix within L0 (Figure 9 and Figure 8, row 10). Thus, similar to human MRP1 (Bakos et al., 2000; see below), L0 may form an important structural domain that could interact with other hydrophobic regions of Ycf1p. Our data suggest that this domain and/or these interactions are necessary for Ycf1p activity. Although we have shown that the L0 portion of the N-terminal extension, specifically the region predicted to form an amphipathic alpha-helix, is required for Ycf1p-mediated cadmium resistance, we were unable to determine whether L0 is sufficient for activity, because L0-ΔYcf1p, which lacks solely MSD0, is mislocalized to intracellular structures, precluding a meaningful in vivo functional test.
Comparison of Roles of MSD0 and L0 of Yeast Ycf1p and Human MRP1
Several studies have specifically examined the significance of the N-terminal extension in MRP1, the closest human homologue of Ycf1p. The most definitive of these used partial constructs, which served as the prototype for those used herein. MSD0 was not required for localization of MRP1 to the basolateral membrane, nor for its glutathione-conjugate transport activity (Bakos et al., 1998). Because only one cell type was examined, MSD0 could still be required for the localization of MRP1 to the plasma membrane in other cell types, as noted by the authors. Interestingly, a recent report examined the functional significance of the NTE in human MRP2. The results of that study strikingly parallel our findings with Ycf1p, in that MSD0 was shown to play a key role in MRP2 trafficking, in this case to the apical membrane of MDCKII cells (Fernandez et al., 2002). Taken together, these studies provide evidence that MSD0 is required for the proper trafficking of at least a subset of MRP subfamily members, both in yeast and humans.
The cytosolic linker region L0 has been directly implicated in both the localization and transport activity of human MRP1 (Bakos et al., 1998). Recently, an amphipathic helical membrane-attaching region within L0 of MRP1 was shown to be critical for the glutathione transport activity of MRP1 (Bakos et al., 2000). Likewise, we have demonstrated that the homologous amphipathic helical region within L0 of Ycf1p is critical for transport activity of Ycf1p, as determined by monitoring cadmium resistance. The striking similarity of the L0 region of yeast Ycf1p compared with that of human MRP1 further validates the utility of yeast as a model system to understand the functional domains of MRP proteins.
The N- and C-Terminal Cleavage Products of Ycf1p Associate with Each Other
Proteolytic processing of Ycf1p has been observed in a previous study (Wemmie and Moye-Rowley, 1997). An important aspect of the present study was to further examine the rather surprising posttranslational proteolytic processing event that cleaves Ycf1p within the conserved ABC core. To date, no other ABC proteins have been found to undergo an analogous processing event. Gel mobility of the cleavage product suggests that cleavage occurs within the first lumenal loop of MSD1 to liberate MSD0, L0, and the first transmembrane span of MSD1. We determined that this cleavage is PEP4 dependent in three different strain backgrounds (Figures 2 and 3; our unpublished data). It is unclear why processing was not blocked in the pep4Δ strain of Wemmie and Moye-Rowley (1997). Nevertheless, our data clearly indicate that the processing of Ycf1p is dependent on the master vacuolar proteases Pep4p and Prb1p and illustrate that cleavage can be used as a marker for vacuolar localization. Importantly, we found that this cleavage produces two stable products that directly interact (Figure 4) as assessed by coimmunoprecipitation. This result, together with our finding that L0 is required for activity and the known functional importance of the NBDs (Falcon-Perez et al., 1999, 2001), indicate that multiple regions of Ycf1p must associate with one another to form an active transporter.
ACKNOWLEDGMENTS
We thank G. Huyer for critical reading of the manuscript. S.M. was supported by a grant (DK-58029) from the National Institutes of Health.
Footnotes
DOI: 10.1091/mbc.E02–07–0405.
REFERENCES
- Bakos E, Evers R, Calenda G, Tusnady GE, Szakacs G, Varadi A, Sarkadi B. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1) J Cell Sci. 2000;113:4451–4461. doi: 10.1242/jcs.113.24.4451. [DOI] [PubMed] [Google Scholar]
- Bakos E, et al. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J Biol Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- Bakos E, Hegedus T, Hollo Z, Welker E, Tusnady GE, Zaman GJ, Flens MJ, Varadi A, Sarkadi B. Membrane topology and glycosylation of the human multidrug resistance-associated protein. J Biol Chem. 1996;271:12322–12326. doi: 10.1074/jbc.271.21.12322. [DOI] [PubMed] [Google Scholar]
- Berkower C, Michaelis S. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 1991;10:3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92:1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chang A, Fink G. Targeting of the yeast plasma membrane [H+] ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B, Ingavale S, Bachhawat AK. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics. 1997;145:75–83. doi: 10.1093/genetics/145.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Sapperstein S, Choi JD, Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SP, Deeley RG. Multidrug resistance mediated by the ATP-binding cassette transporter protein MRP. Bioessays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001a;42:1007–1017. [PubMed] [Google Scholar]
- Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001b;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Falcon-Perez JM, Martinez-Burgos M, Molano J, Mazon MJ, Eraso P. Domain interactions in the yeast ATP binding cassette transporter Ycf1p: intragenic suppressor analysis of mutations in the nucleotide binding domains. J Bacteriol. 2001;183:4761–4770. doi: 10.1128/JB.183.16.4761-4770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon-Perez JM, Mazon MJ, Molano J, Eraso P. Functional domain analysis of the yeast ABC transporter Ycf1p by site-directed mutagenesis. J Biol Chem. 1999;274:23584–23590. doi: 10.1074/jbc.274.33.23584. [DOI] [PubMed] [Google Scholar]
- Fernandez SB, Hollo Z, Kern A, Bakos E, Fischer PA, Borst P, Evers R. Role of the N-terminal transmembrane region of the multidrug resistance protein MRP2 in routing to the apical membrane in MDCKII cells. J Biol Chem. 2002;277:31048–31055. doi: 10.1074/jbc.M204267200. [DOI] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Yamazaki M, Loe DW, Westlake CJ, Grant CE, Cole SP, Deeley RG. Multidrug resistance protein. Identification of regions required for active transport of leukotriene C4. J Biol Chem. 1998;273:10733–10740. doi: 10.1074/jbc.273.17.10733. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Almquist KC, Leslie EM, Gerlach JH, Grant CE, Deeley RG, Cole SP. Membrane topology of the multidrug resistance protein (MRP). A study of glycosylation-site mutants reveals an extracytosolic NH2 terminus. J Biol Chem. 1997;272:23623–23630. doi: 10.1074/jbc.272.38.23623. [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Deeley RG, Cole SP. Structural, mechanistic and clinical aspects of MRP1. Biochim Biophys Acta. 1999;1461:359–376. doi: 10.1016/s0005-2736(99)00168-6. [DOI] [PubMed] [Google Scholar]
- Hurtley S, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- Jones EW, Webb GC, Hiller MA. Biogenesis and function of the yeast vacuole. The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 363–470. [Google Scholar]
- Konig J, Nies AT, Cui Y, Leier I, Keppler D. Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2-mediated drug resistance. Biochim Biophys Acta. 1999;1461:377–394. doi: 10.1016/s0005-2736(99)00169-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Kane T, Tipper C, Spatrick P, Jenness DD. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol Cell Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Szczypka M, Lu Y, Thiele D, Rea P. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- Loayza D, Tam A, Schmidt WK, Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe DW, Deeley RG, Cole SP. Biology of the multidrug resistance-associated protein, MRP. Eur J Cancer. 1996;32A:945–957. doi: 10.1016/0959-8049(96)00046-9. [DOI] [PubMed] [Google Scholar]
- Mason DL. Functional Analysis of MRP Transporters in Saccharomyces cerevisiae. PhD. thesis. Baltimore, MD.: The Johns Hopkins University School of Medicine; 2002. [Google Scholar]
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian YM, Qiu W, Gao M, Westlake CJ, Cole SP, Deeley RG. Characterization of binding of leukotriene C4 by human multidrug resistance protein 1: evidence of differential interactions with NH2- and COOH-proximal halves of the protein. J Biol Chem. 2001;276:38636–38644. doi: 10.1074/jbc.M107025200. [DOI] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- Sharma KG, Mason DL, Liu G, Rea PA, Bachhawat AK, Michaelis S. Localization, Regulation, and Substrate Transport Properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryotic Cell. 2002;1:391–400. doi: 10.1128/EC.1.3.391-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- Taglicht D, Michaelis S. Methods in Enzymology. ABC Transporters: Biochemical, Cellular, and Molecular Aspects. Academic Press; 1998. A complete catalogue of Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease; pp. 130–162. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R, Evers R, Vogt E, Mornet C, Zaman G, Schinkel A, Borst P, Martinoia E. The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor 1. Proc Natl Acad Sci USA. 1996;93:6743–6748. doi: 10.1073/pnas.93.13.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J, Moye-Rowley W. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol Microbiol. 1997;25:683–694. doi: 10.1046/j.1365-2958.1997.5061868.x. [DOI] [PubMed] [Google Scholar]
- Wemmie J, Szczypka M, Thiele D, Moye-Rowley W. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]