Abstract
Spectrin domains are three-helix bundles, commonly found in large tandem arrays. Equilibrium studies have shown that spectrin domains are significantly stabilized by their neighbors. In this work we show that domain:domain interactions can also have profound effects on their kinetic behavior. We have studied the folding of a tandem pair of spectrin domains (R1617) using a combination of single- and double-jump stopped flow experiments (monitoring folding by both circular dichroism and fluorescence). Mutant proteins were also used to investigate the complex folding kinetics. We find that, although the domains fold and unfold individually, there is a single rate-determining step for both folding and unfolding of the protein. This is consistent with the equilibrium observation of cooperative folding of the entire two-domain protein. The results may have important biological implications. Not only will the protein fold more efficiently during cotranslational folding, but the ability of the multidomain protein to withstand thermal unfolding in the cell will be dramatically increased. This study suggests that caution has to be exercised when extrapolating from single domains to larger proteins with a number of independently folding modules arranged in tandem. The multidomain protein spectrin is certainly more than “the sum of its parts”.
INTRODUCTION
Around 70%–80% of all proteins in eukaryotic cells are multimodular, consisting of arrays of independently folding domains (1). The general strategy employed to investigate the folding of these proteins is to study the constituent domains independently. However, the relevance of these studies to understanding the properties of the proteins as a whole will depend on whether the stability or the folding pathway change when a domain is attached to its neighbors.
One domain commonly found in tandem array is the α-helical spectrin repeat. These are 106-residue three helix coiled-coil domains (2–6). The individual domains can fold to a stable native structure (6). However, tandem repeats are more stable than the individual domains (7). Crystal structures of multiple repeats show there to be a continuous helical linker between domains (8,9). The linker extends from the last helix of the first domain to the first helix of the next domain. It has been suggested that the increase in stability in the tandem constructs is due to the presence of this linker (10).
We have studied the independent folding of two spectrin domains, domains R16 and R17 of chicken brain α-spectrin and the tandem pair R1617, which is more stable than either domain alone and folds cooperatively in an all-or-none fashion at equilibrium (11–13) (Fig. 1). Studies of a number of mutant proteins established that, in R1617, the R17 domain is stabilized by the neighboring folded R16 domain (by ∼2.8 kcal mol−1) and that the R16 domain in R1617 is stabilized both by the folded (by ∼2.3 kcal mol−1) or unfolded (by ∼0.8 kcal mol−1) R17 domain (13).
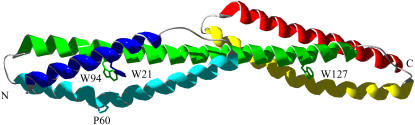
FIGURE 1.
Structure of the tandem domains R1617, showing the continuous helix formed of the helix C from R16 and helix A of R17. The A-helix of R16 is colored blue and the C-helix of R17 is colored red. This structure shows the position of the Trp residues that act as fluorescence probes. Pro60 is also shown. The N- and C-termini are labeled.
Here we investigate how inclusion in a tandem array affects the folding kinetics of the individual domains. The folding of R16 and R17 has previously been characterized (11,14). Both fold by a two-state mechanism with the population of a high-energy intermediate leading to curvature in the unfolding arms of the chevron plot for wild-type R16 (12) and some mutants of R17 (K. Scott and J. Clarke, unpublished data). Here we describe the complex kinetics of the folding of the tandem construct of spectrin domains, R1617. We have used mutant proteins to assign all folding and unfolding phases that are observable in single- and double-jump stopped-flow experiments followed by both fluorescence and circular dichroism. We show that R1617 folds via a stable intermediate that is not a species with one domain folded and the other unfolded and that, although both folding and unfolding occur in two steps, only one phase is observable (at most denaturant concentrations) due to the relative rates of the reactions. These results demonstrate that some multidomain proteins are not a simple sum of their parts. The fully folded spectrin protein is more stable both thermodynamically and kinetically than the isolated domains.
MATERIALS AND METHODS
Protein expression and purification
The phasing of the R16 and R17 domains of spectrin that we have analyzed in our laboratory have been described previously (9,12). They include the entire 106-residue region considered to define the spectrin domain, plus additional residues at either end. The R1617 construct was designed to include all residues that are in the individual domains plus extensions (13). Site directed mutagenesis was performed using a QuikChange kit from Stratagene (La Jolla, CA). Proteins were purified as has already been described (12). The purified proteins were dialyzed into water, flash frozen, and stored at −80°C.
Kinetics studies
Single- and double-jump kinetics experiments were carried out on an Applied Photophysics (Leatherhead, UK) SX.18V stopped-flow fluorimeter. An excitation wavelength of 280 nm was used for all spectrin individual and tandem constructs. The change in fluorescence above 320 nm was monitored. The final protein concentration for all fluorescence measurements was ∼1 μM. The kinetics of R1617 were also followed by the change in CD signal at 222 nm, using an Applied Photophysics π* −180 instrument, with a final protein concentration of ∼5 μM. In both cases the temperature was maintained at 25°C ± 0.1°C and experiments were carried out in 50 mM sodium phosphate buffer pH 7. Between 10 and 15 traces were averaged at every urea concentration. The data from the single-jump experiments were fitted using Kaleidagraph (Synergy Software, Reading, PA), and the data from sequential mixing experiments were fitted using global values for the rate constants with Prism (GraphPad, San Diego, CA). If individual traces were fitted, the results were the same within error as the global fits.
RESULTS
Kinetic studies of R1617
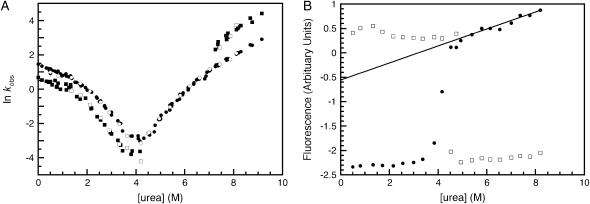
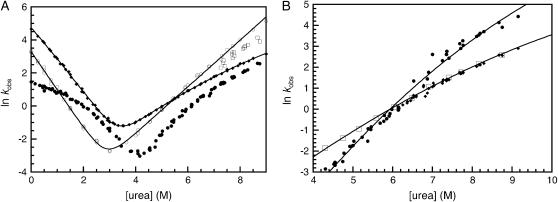
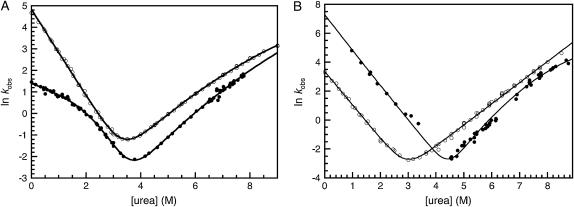
The folding and unfolding of R1617 was followed by a change in the fluorescence above 320 nm and the change in CD signal at 222 nm. The rate constants observed by both probes were the same within error (Fig. 2 A). The relative amplitudes of the observed unfolding rates differ in CD and fluorescence experiments.
FIGURE 2.
R1617 kinetics. (A) Rate constants for the folding of R1617. Data points measured by fluorescence are shown in closed symbols; data points measured using CD at 222 nm are shown in open symbols. The major amplitudes are shown as circles and the minor amplitudes as squares. (B) R1617 fluorescence amplitudes; the initial (solid circles) and final (open squares) fluorescence measurements of kinetic traces are shown. The solid line represents an extrapolation of the fluorescence of the denatured state to 0 M urea.
Refolding of R1617 was initiated by denaturant dilution and pH jump. The rates were the same within error for both methods. Refolding of R1617 was well described by a double exponential process at all concentrations of urea. The faster of the two rate constants is the major phase with 75%–90% of the amplitude by both fluorescence and CD. The observed refolding rate constants of both phases have nonlinear urea dependences and are protein concentration independent at 0 M urea (protein concentration range = 0.1–20 μM).
The unfolding of R1617 fits well to two exponential processes above 6.5 M urea and to one exponential process below 6.5 M urea. Both observed unfolding rates have a nonlinear urea dependence. The lower of the two rate constants reflects the major phase when followed by fluorescence, with ∼70% of the amplitude. However, when using CD as the probe, the two phases have approximately equal amplitudes.
Stopped-flow dead-time amplitude change in R1617
A plot of the start and end values of fluorescence and CD in stopped-flow measurements can identify dead-time loss or gain in signal, which, in turn, can indicate the presence of a burst-phase species. R1617 shows an initial gain in fluorescence at low urea concentrations (Fig. 2 B). This is accompanied by a small (∼10%) increase in CD signal at 222 nm (data not shown). This suggests that there is a collapsed intermediate forming in the dead-time of the experiments. The formation of the intermediate is associated with a small increase in helical content and a change in the fluorescence properties. A similar burst phase increase in the fluorescence was also seen in the single domain R16 and has been attributed to the formation of a collapsed denatured state involving burial of structure around Trp94 (14). In R16, however, there was no accompanying rollover in the refolding kinetics.
Interrupted refolding experiments
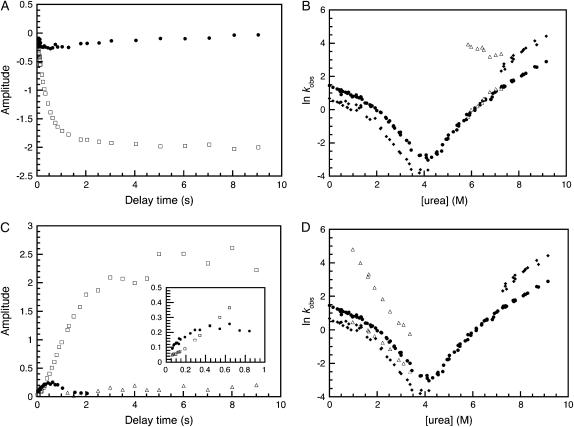
Interrupted refolding experiments indicate that both refolding phases result in the formation of the native protein. Folding was initiated at 0 M urea, by pH jump, then interrupted at various delay times by mixing to a final concentration of 6 M urea and the resulting kinetic trace recorded. At all delay times the data were well described by a double exponential process with unfolding rate constants of 41 s−1 and 0.73 s−1.
The rate constant of 0.73 s−1 is in good agreement with that of 0.8 s−1 seen in single-jump unfolding experiments under the same conditions and thus corresponds to the unfolding of native R1617. The variation of the amplitude of this rate with delay time (Fig. 3 A) also gives information about whether just one or both of the refolding phases form the native state. The rate of appearance fits well to a double exponential process with rate constants of 2.6 s−1 (with ∼90% of the total amplitude) and 0.3 s−1 (with ∼10% of the total amplitude). This is in good agreement with the refolding rates at 0 M urea from single-jump experiments of 3.5 s−1 and 1 s−1, indicating that both refolding phases form the native state.
FIGURE 3.
R1617 double-jump kinetic experiments. (A) The time course of the appearance of the two unfolding phases from interrupted refolding experiments. The 41 s−1 rate constant (solid circles) increases to a maximum amplitude at ∼500 ms and then decreases to zero. The amplitude of the 0.73 s−1 rate constant (open squares) appears with two apparent rate constants. (B) The urea dependence of the unfolding of the folding intermediate. The major (solid circles) and minor (solid diamonds) phases from the single-jump kinetic experiments are shown. The unfolding rates from the interrupted refolding experiment are shown in open triangles. (C) The time course of the appearance of the three refolding rates from interrupted unfolding experiments. The 82 s−1 phase (solid circles) builds up to a maximum amplitude at around 500 ms before decaying to an amplitude of zero. The major phase, 2.3 s−1 (open squares), has an initial lag (see inset). The minor, 0.8 s−1, phase (open triangles) accumulates with an apparent rate constant of 0.05 s−1, which is consistent with the rate of proline isomerization in the denatured state. (D) The urea dependence of the folding of the unfolding intermediate. The major (solid circles) and minor (solid diamonds) phases from the single-jump kinetic experiments are shown. The refolding rate constants from the interrupted unfolding experiment are shown in open triangles. In double-jump experiments, the amplitudes are small so it is difficult to separate the two refolding phases observed in single jump.
The amplitude associated with the 41 s−1 rate constant increases until a delay time of around 500 ms and then decreases toward zero at longer delay times. This is typical of an intermediate species. Note that there is no apparent lag in the buildup of the native species. This may suggest that the folding intermediate is not obligatory; but if the lag time is short, it may simply not be detectable.
To further explore the properties of this refolding intermediate, the denaturant dependence of its unfolding was studied. Unfolding rates were measured in the range 5.8–8.0 M urea and fit well to a double exponential process. The slower of the two observed rate constants corresponds to the major unfolding rate constant seen in single-jump experiments and represents the unfolding of the native state (Fig. 3 B). The faster of the two observed rate constants, corresponding to the unfolding of the intermediate species, is ∼40 s−1 and appears to show no urea dependence. Note that the unfolding rates of the intermediate are significantly faster than the unfolding of either R16 or R17 alone. We infer that the intermediate is a collapsed unstable species and, importantly, that it does not correspond to a species with one complete domain folded.
Interrupted unfolding experiments
Interrupted unfolding experiments allow partially unfolded states (intermediates) and cis-trans isomerization-limited species to be observed. Folded R1617 was unfolded in 6 M urea for a variety of delay times before refolding to a final urea concentration of 1 M. Single-jump refolding experiments at 1 M urea show two rate constants, 2.4 s−1 and 1 s−1.
At short delay times (<2500 ms), two exponential processes with rate constants of 82 s−1 and 2.3 s−1 were observed. For longer delay times, two exponential processes with rate constants of 2.3 s−1 and 0.8 s−1 were necessary to fit the data. A plot of the amplitudes of these phases against delay time is shown in Fig. 3 C.
The slowest of these rates is in good agreement with the rate of 1 s−1 seen for the slower of the two refolding phases. This refolding phase appears at a rate of 0.05 s−1, consistent with the rate of proline isomerization in the denatured state. The intermediate rate constant is in good agreement with that of the major folding phase seen in single-jump experiments.
The rate constant of 82 s−1 builds up to a maximum amplitude at 500 ms after which it disappears to an amplitude of zero. This is typical of an intermediate species. There is a concomitant lag in the appearance of the amplitude of the 2 s−1 phase (inset to Fig. 3 C) suggesting that the unfolding intermediate is an obligatory on-pathway intermediate (15), as the intermediate has to form before the fully unfolded state can be reached.
To further explore the properties of this unfolding intermediate, the denaturant dependence of its folding was studied in a series of interrupted unfolding experiments with a delay time of 500 ms (corresponding to maximum accumulation of the intermediate). Kinetic traces were measured in the range 1–3.5 M urea and were well described by a double exponential process. A plot of the natural logarithm of the observed folding rates is shown in Fig. 3 D. The folding of the unfolding intermediate is fast and shows a linear dependence on urea concentration. When extrapolated to 0 M urea the 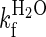 is 1000 ± 200 s−1. The second refolding rate in the double-jump experiment appears to be between the two rates observed in the single-jump experiments, showing the difficulty in separating two rates that are so similar, in particular where the amplitude of the second phase is small.
is 1000 ± 200 s−1. The second refolding rate in the double-jump experiment appears to be between the two rates observed in the single-jump experiments, showing the difficulty in separating two rates that are so similar, in particular where the amplitude of the second phase is small.
Mutant studies: slow phase in refolding
Interrupted unfolding experiments indicate that the slower of the two folding rates observed in single-jump refolding reactions has the characteristics of a proline-limited folding phase. To verify the assignment of this rate to a proline isomerization-limited process, the single proline (P60 in the R16 domain) was mutated to alanine. The folding of P60A was followed by the change in the fluorescence above 320 nm. Two refolding rates are still observed, but the major rate now has >95% of the amplitude, and the slower phase is associated with <5% of the amplitude and is hard to fit due to the very small amplitude (data not shown). The residual, low amplitude, slow phase observable by fluorescence is likely due to nonprolyl isomerization events (16). Wild-type R16 was shown to exhibit two different isomerization-limited events assigned to prolyl and nonprolyl isomerization (12), and as is the case in R1617, the P60A mutant in R16 alone shows a slow refolding phase reduced in amplitude from 15% to 4%. These data, along with the interrupted unfolding experiments, are consistent with the assignment of the slow phase in R1617 principally to a proline-limited species, plus minor nonprolyl isomerization events.
Mutant studies: deconvoluting the unfolding phases
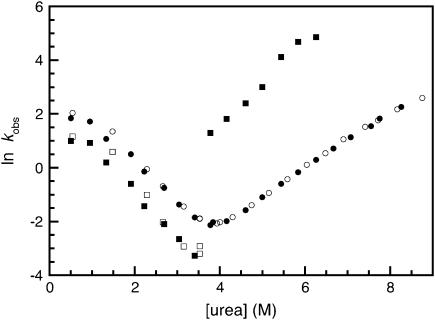
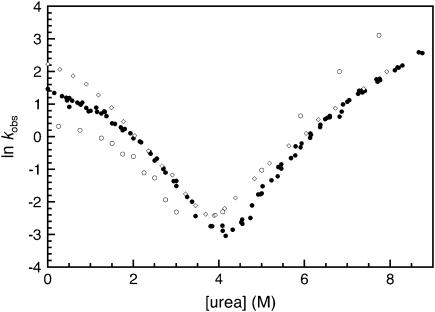
The faster of the two unfolding rates, which only appears above 6.5 M urea, accounts for ∼30% of the total fluorescence amplitude change but 50% of the total CD amplitude change. This corresponds to the relative amplitude observed for the unfolding of the R17 domain alone in the equilibrium studies of R1617 (R17 has only one Trp, whereas R16 has two). This suggests that the faster of the unfolding rates can be attributed to the unfolding of R17. To test this hypothesis the mutant W127F with the fluorescence probe in R17 removed was constructed. The R17 domain in W127F will be invisible to fluorescence measurements, but CD experiments will follow the unfolding of both domains.
The unfolding of W127F had only one unfolding phase when followed using fluorescence (Fig. 4). When followed by CD there were two observable unfolding rate constants, the slower of which was the same as the single observable phase followed by fluorescence experiments.
FIGURE 4.
The kinetics of the W127F mutant. Rate constants for the folding of W127F. Data points measured by fluorescence are shown in open symbols; data points measured using CD at 222 nm are shown in closed symbols.
The faster of the two unfolding rate constants, which is only visible by CD, is clearly the unfolding of the R17 domain within the R1617 construct. When compared with the faster unfolding phase in wild-type R1617, there is a significant increase in the unfolding rate of the R17 domain in the W127F mutant. This is consistent with the large destabilization expected upon the mutation of tryptophan to phenylalanine. (The same mutation in the R17 domain alone results in fully unfolded protein, S. Batey and J. Clarke, unpublished data.) The slow phase, unaffected by the mutation, must reflect the unfolding of the R16 domain.
The refolding followed by fluorescence and CD produced only two phases, i.e., no new “R17-only” phases were observed in CD experiments. This is consistent with the observation that both refolding phases reflect the refolding of the entire protein. The major folding rate was the faster of the two and was ∼80% of the total amplitude, as in wild type.
DISCUSSION
Comparison with the single domains
The chevrons of R1617 and R16 and R17 are shown in Fig. 5 A. (Note that the slow refolding phases of R16 and R1617, which have been assigned to cis-trans isomerization-limited folding events, have been omitted for clarity.) R1617 folds more slowly at 0 M denaturant than either R16 or R17 alone. Outside the rollover region (below ∼2.5 M urea), R1617 folds at rates similar to those of R16 alone and significantly faster than the folding rate of R17 (Fig. 5 A).
FIGURE 5.
A comparison of the observed rate constants of R1617 to the individual domains R16 and R17. Proline isomerization phases have been omitted for clarity. (A) Folding rate constants for wild-type R16 (solid diamonds), R17 (open circles), and R1617 (solid circles represent the major amplitude by fluorescence and the open squares represent the minor amplitude by fluorescence). The R16 data are fitted to a three-state (high energy intermediate) equation; the R17 data are fitted to a two-state equation. (B) Unfolding rate constants for R1617 (solid circles and solid diamonds) and W127F (open squares) (see text). The solid lines are quadratic fits to guide the eye only.
R1617 unfolds more slowly than either R16 or R17. At high denaturant concentrations (>7 M urea) the unfolding plot has a similar slope to R16, and below 6.5 M urea the R1617 unfolding kinetics have a similar denaturant dependence to R17.
Refolding kinetics
There are two refolding phases that report on the folding of R1617 as a single unit
Refolding of R1617 shows two phases by both CD and fluorescence, both of which show protein-concentration independent rollover. The obvious explanation for two refolding phases in a two-domain protein would be that each phase is reflecting the folding of an individual domain. However, this can be ruled out because of the unique spectroscopic properties of R1617. It has been shown that the fluorescence amplitude change on folding of R16 is twice that of R17, whereas the changes in CD amplitude are equal. In R1617, fluorescence and CD have the same amplitude profiles, strong evidence that both folding phases reflect the folding of the entire R1617 molecule. A second line of evidence comes from the folding of the mutant W127F, a mutant that removes the only Trp residue from the R17 domain in R1617. In the folding kinetics both phases are still apparent by both fluorescence and CD, i.e., both phases report on the folding of the fluorescent R16 domain. Further, the CD and fluorescence data coincide—there is no “extra” phase that reports the folding of the R17 domain alone.
The minor, slower phase reflects proline isomerization in the denatured state
There are two lines of evidence that suggest that this phase reflects the refolding of molecules that are limited mainly by prolyl cis-trans isomerization. Both interrupted unfolding experiments and the data for the mutant P60A are consistent with this hypothesis. Similar results have been observed for R16 alone.
The folding intermediate is not a species with one domain folded and the other unfolded
Kinetic studies of the individual domains show neither R16 nor R17 has a stable refolding intermediate, yet there is clear rollover in the refolding of R1617. Rollover in folding kinetics is evidence that a folding intermediate is formed in the dead-time of the stopped-flow experiment.
What is the nature of this intermediate? One obvious hypothesis is that this intermediate represents an R1617 molecule with one domain (either R16 or R17) folded. To check this possibility we examined the interrupted refolding of R1617. The protein was jumped from unfolding conditions into refolding conditions (at 1 M, where I is populated) for a short time and then unfolded, and the data examined to find the unfolding rate constant for I. If the intermediate were indeed a species with one domain folded, then we would expect to see it unfolding at a rate characteristic of the unfolding of R16 or R17. This is not what we see. A fast rate constant is seen at short refolding times, which represents the unfolding of I, but at rates that are significantly faster than the unfolding of either domain alone. The rate of unfolding of the intermediate at 6 M urea is 41 s−1 compared to 2.4 s−1 for R16 and 3.6 s−1 for R17. I is apparently a kinetically unstable state, which buries one or more Trp residues. Interestingly, the rollover is reduced in the W127F mutant. This implies that W127 is involved in this intermediate. Of a selection of 18 further mutants, the only proteins that show a reduction in the rollover were those from the connecting helix (L97A, R104A, F117L, A126G, W127F, and I128V found in the helix C of R16 and helix A of R17) (see, for example, Fig. 6). This indicates that the intermediate has some degree of structure/collapse in this region.
FIGURE 6.
Representative chevrons for mutants in R1617. Mutants in the connecting helix (helix C of R16 and helix A of R17) lead to a reduction in rollover, an example of such a protein is I128V, shown in open diamonds. Mutations outside of the linker region do not affect the degree of rollover, an example of such a protein is L51A shown in open circles. The wild-type data are shown in solid circles.
Unfolding kinetics
It is most straightforward to consider the unfolding kinetics in two sections: first, the unfolding at high denaturant concentrations and second, the unfolding at lower denaturant concentrations. Fig. 5 A shows the rates of the observed unfolding phases of R1617 compared to the unfolding phases of the individual domains R16 and R17.
High denaturant concentrations (≥6.5 M urea)
At high denaturant concentrations there are two unfolding phases, observable by both CD and fluorescence. In fluorescence measurements the slower phase is associated with a larger amplitude change than the fast phase, but in CD both phases have approximately the same amplitude. The comparison of the fluorescence amplitudes would infer that the faster phase represents the unfolding of the R17 domain, and the slower phase the unfolding of the R16 domain. To confirm this, the unfolding of the W127F mutant was examined. In this case only one unfolding phase was observed by fluorescence, corresponding exactly to the slower phase in the wild type at denaturant concentrations above 6.5 M. CD reveals an additional, much faster unfolding phase, which must correspond to the unfolding of the R17 domain. Thus the faster of the two unfolding phases that we observe in wild-type R1617 at high urea concentration reflects the unfolding of the R17 domain.
Low denaturant concentrations (≤5.5 M urea)
At lower denaturant concentrations, in R1617 wild type, only one unfolding phase can be observed. This unfolding rate is slower than the unfolding of R16 or R17 alone and, importantly, is slower than the unfolding of the R16 domain in the W127F mutant of R1617 (Fig. 5 B). We infer that the unfolding rate constant observed at low denaturant concentrations must reflect the unfolding of the wild-type R17 domain in R1617 and not the unfolding of the R16 domain.
Unfolding proceeds via an intermediate that is a species with the R16 domain folded and the R17 domain unfolded
The faster unfolding phase at concentrations of urea greater than 6.5 M represents the unfolding of the R17 domain. In the interrupted unfolding experiments, where the protein was unfolded at 7.5 M urea then refolded at 1.25 M, three refolding phases were observed, including a fast refolding phase that had not been previously detected in the single-jump experiments. This refolding phase represents the refolding of an intermediate with the R17 domain unfolded and the R16 domain folded. This refolding phase is significantly faster than the refolding of R17 (46 s−1 compared to the 1.6 s−1 at 1.25 M urea), reflecting the fact that R17 is stabilized by folded R16 (13). In these interrupted unfolding experiments the fast R17-only phase appeared and then disappeared, and the two slower “true” refolding phases appeared with a lag, reflecting the disappearance of the R17-unfolded intermediate. This shows that the partially folded species with an unfolded R17 domain is an obligatory on-pathway intermediate. Importantly, the unfolding intermediate is present at all concentrations of urea; we still observe this intermediate species at 6 M urea where there is only one unfolding rate observed.
Making sense of the unfolding phases
The unfolding of R16 in the presence of unfolded R17 is monitored by the fluorescence phase of the unfolding of W127F. When we plot these data along with the wild-type data, it is clear that the W127F and wild-type data are identical above 6.5 M urea (Fig. 5 B); i.e., the slower phase at high concentrations of urea represents the unfolding of R16 in R1617 (in the presence of unfolded R17). However, at concentrations below 6.5 M, the unfolding of the R16 domain (as observed in W127F) is faster than the single unfolding phase observed for R1617 wild type. We infer that the single unfolding phase observed at low urea concentrations for R1617 wild type is monitoring the unfolding of the R17 domain in the presence of folded R16. Note that this phase is continuous with the fast unfolding phase (Fig. 5 B).
At high urea concentrations the unfolding of R17 (with R16 folded) is faster than the subsequent unfolding of R16. Thus we see two kinetic phases. At lower urea concentrations, however, the unfolding of R16 (once R17 has unfolded) is faster and is thus kinetically silent in the experiments.
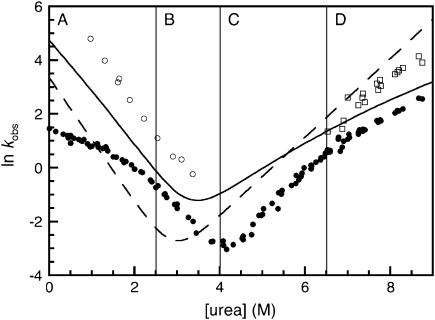
Model for the folding kinetics of R1617
In this model we consider the folding behavior of R1617 in four regions of the chevron separately. A plot of all the observed rate constants in R1617, except for the proline phases, from single- and double-jump experiments as well as the rate constants from R16 and R17 are shown in Fig. 7. The figure is divided into the four separate regions considered in this model.
FIGURE 7.
The kinetics for the folding of R16, R17, and R1617. The major folding phase of the individual domains of R16 (solid line) and R17 (dashed line) are shown. The major phase of R1617 (solid circles), the minor unfolding phase (open squares), and the folding of the unfolding intermediate (open circles) are shown. The plot is divided into four sections as discussed in the text.
Region A: folding kinetics at low urea, <2.5 M
In this region there is a rapid collapse, or preequilibrium, between the denatured state, D, and the folding intermediate, I1. The formation of I1 slows the refolding rate, as under these conditions I1 is more stable than D. The rapid preequilibrium between I1 and D may be on- or off-pathway; we cannot distinguish these alternatives.
Region B: folding kinetics at >2.5M urea
In this region, R1617 folds at a rate approaching that of the individual R16 domain. The folding rate of R16 is ∼10-fold faster than that of R17, so R16 folds first to form a second intermediate I2. This is the intermediate detected in the interrupted unfolding experiments, a species with R17 unfolded and R16 folded. Once the R16 domain is folded, the folding rate of the R17 domain is significantly increased due to the stabilizing effect of folded R16. The R17 domain now folds rapidly, at a rate much greater than the folding of R16. This second, fast step cannot be observed in the single-jump experiments. Folding is a two-step process, but we only observe the folding of the R16 domain since this is the rate-limiting step; the second step, the folding of R17, is kinetically silent.
Region C: unfolding kinetics at urea concentrations <5.5 M
In this region there is only one observable unfolding rate constant, which is slower than that of either of the individual domains. R17 is unfolding first; the interrupted unfolding experiments indicate that the intermediate with R17 unfolded forms before any fully unfolded molecules. Presumably the rate of unfolding of R16 domains with folded R17 domains is significantly slower, since we can never detect molecules with unfolded R16 and folded R17 domains. Once R17 has unfolded, the stability that folded R17 confers on R16 is lost and now R16 unfolds at a rate faster than that of R17. The fast step cannot be observed in the wild-type protein; again the unfolding is a two-step process but the second, faster phase is kinetically silent.
Region D: unfolding kinetics at high urea concentrations, >5.5 M
In this region there are two observable unfolding phases, the faster of which corresponds to the unfolding of R17. The rate constant and m-value of this phase approach that of the individual domain of R17, but is slightly lower. The presence of a neighboring folded R16 domain stabilizes the R17 domain. The slower phase corresponds to the unfolding of R16 in the presence of unfolded R17. In these conditions the faster unfolding of R17 precedes the slow unfolding of R16, so both kinetic rates are observable. Interestingly, R16, with unfolded R17 attached, also unfolds more slowly than R16 alone. This reflects the fact that R16 is stabilized by unfolded R17 (13). A schematic representation of the folding and unfolding pathways of R1617 is shown in Fig. 8.
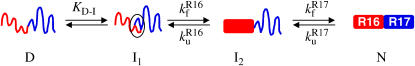
FIGURE 8.
A schematic representation of the folding pathway of R1617. The first step on the folding pathway is the rapid (dead-time) formation of an unstable intermediate (I1). The R16 domain then folds to form I2. Finally, the R17 domain folds. The first folding step (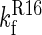 ) is slower than the second step (
) is slower than the second step (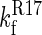 ); therefore, only the folding of R16 in the presence of unfolded R17 can be observed in the single-jump experiments. The first step on the unfolding pathway is the formation of an intermediate with R17 unfolded and R16 folded (I2). R16 then unfolds. Above 6.5 M urea the unfolding of R17 (
); therefore, only the folding of R16 in the presence of unfolded R17 can be observed in the single-jump experiments. The first step on the unfolding pathway is the formation of an intermediate with R17 unfolded and R16 folded (I2). R16 then unfolds. Above 6.5 M urea the unfolding of R17 (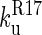 ) is faster than the unfolding of R16 (
) is faster than the unfolding of R16 (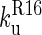 ), so both phases can be observed. Below 6.5 M urea,
), so both phases can be observed. Below 6.5 M urea, 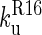 >
> 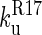 and so the only observable unfolding phase is that of R17 in the presence of folded R16.
and so the only observable unfolding phase is that of R17 in the presence of folded R16.
A chevron plot can be constructed that shows the kinetics of R16 within R1617 in the presence of unfolded R17. The folding rate constant is the major phase from regions A and B of the wild-type, and the unfolding data are the W127F unfolding rate constants combined with the wild-type data for the slower unfolding rate in region D (Fig. 9 A). Compared to R16 alone, the folding is slightly slower but the unfolding is significantly slower. We have previously demonstrated that R16 is stabilized by unfolded R17 (13), and it appears that this stabilization results largely from a decrease in the unfolding rate constant. As has been seen in wild-type R16, there is curvature in the unfolding data. We have shown that this is consistent with a folding pathway that proceeds via a high-energy kinetic intermediate (11). The R16 data from R1617 were, therefore, fitted to a four-state equation that accounts for a rapid preequilibrium between D and I1 plus a high-energy intermediate (note that this fit assumes I1 is on-pathway, which has not been confirmed). This gives folding and unfolding rate constants in water and an estimate for the stability of I1 (∼2.6 kcal mol−1) (Table 1). R16 is significantly stabilized by folded R17. Since we have not been able to observe the unfolding of R16 in the presence of folded R17 (even in mutants with a destabilized R16 domain, S. Batey and J. Clarke, unpublished data) we can assume that this stabilization comes, at least in part, from significant slowing of the unfolding rate.
FIGURE 9.
The folding of R16 and R17 within R1617. (A) Rate constants for the folding of R16 (open circles) and the rate constants for the folding of R16 within R1617 (solid circles). The data for R16 in R1617 are in the presence of unfolded R17 and are taken from the major folding rate from the single-jump experiments, the W127F unfolding rates by fluorescence and the slower of the two unfolding rates from R1617 wild-type above 6.5 M urea. (B) Rate constants for the folding of R17 (open circles) and the folding rates of R17 within R1617 (solid circles) are shown. The data for R17 in R1617 are in the presence of folded R16 and are taken from the folding of the unfolding intermediate of wild-type R1617, the unfolding rate of R1617 wild type below 6.5 M urea, and the faster unfolding rate above 6.5 M urea. See Table 1 for details of the fits.
TABLE 1.
Kinetic properties of R16, R17, and R1617
| Protein |
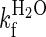 (s−1) (s−1) |
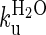 (s−1) (s−1) |
KUI |
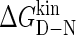 (kcal mol−1) (kcal mol−1) |
|---|---|---|---|---|
| R16* | 125 (± 3) | 2.6 × 10−3 (±3 × 10−4) | – | 6.3 (± 0.3) |
| R17 | 30 (± 2) | 4.0 × 10−4 (±3 × 10−5) | – | 6.6 (± 0.3) |
| R16 (R1617)† | 4.2 (± 0.2) | 9.2 × 10−4 (±1.8 × 10−4) | 85 (±30) | 7.5 (± 0.3) |
| R17 (R1617)‡ | 1000 (± 200) | 1.3 × 10−5 (±5.8 × 10−6) | – | 10.8 (± 0.4) |
Data for R16 and R17 are taken from Scott et al. (12). The errors are from the errors of the fits. Note that the true error in the estimate of kinetic values of ΔG are likely to be higher.
Values from a fit to a sequential transition state model. Note that the values obtained from a fit to a broad transition state are the same within error.
R16 in the presence of unfolded R17. The data are fit using a four-state model assuming an on-pathway preequilibrium between D and I and with a high energy intermediate (sequential transition state model) to account for curvature in the unfolding arm.
R17 in the presence of folded R16. The data are fit using a two-state model in the region of 0 M urea to 5 M urea where no curvature was observed. Although it would be preferable, the data could not be accurately fitted to a three-state fit assuming a high energy intermediate (sequential transition state model) to account for curvature in the unfolding arm.
A chevron can also be plotted for the kinetics of R17 within R1617. In this case the species that is folding and unfolding is I2; the R17 domain is folding and unfolding in the presence of folded R16. The folding data are those from the interrupted unfolding experiments. The unfolding data are the wild-type unfolding data from region C and the fast unfolding rate constants from region D of the wild-type chevron (Fig. 9 B). Compared to R17 domain alone, R17 folds significantly faster and unfolds more slowly; i.e., the significant stabilization of R17 by folded R16 can be attributed to both an increase in the folding rate and a decrease in the unfolding rate. These data have been fit to a system with two rate-limiting transition states separated by a high energy intermediate to account for the curvature in the unfolding arm (Fig. 9 B and Table 1). This was not necessary for R17 wild type, but a number of R17 mutants showed significant curvature in the unfolding arms of their chevrons (K. A. Scott and J. Clarke, unpublished data). Note that the stability of R17 is unaffected by the presence of unfolded R16, suggesting that its folding and unfolding rates are likely to be unaffected (13).
The effect of domain:domain interactions in R1617
The equilibrium and kinetic data both show that there are stabilizing interdomain effects in the tandem construct R1617. Both domains in the tandem construct are stabilized relative to the individual domains. The kinetics are somewhat complex at first glance. Between 0 M denaturant and ∼6.5 M denaturant (if we ignore cis-trans isomerization phases), we can observe only a single folding and unfolding rate constant for formation of R1617 although the two domains fold individually. This reflects the fact that there is a single rate-determining step for the folding and unfolding of R1617. For both folding and unfolding, the first step is a slow step and this is followed by a second rapid step. This is consistent with our equilibrium data, which suggested that folding is a cooperative all-or-none event.
Equilibrium data suggested that R16 was stabilized by ∼0.8 kcal mol−1 (±0.2 kcal mol−1) by unfolded R17. This is the same, within error, as our estimate from the kinetic data (1.2 ± 0.3 kcal mol−1). However, we suggested using the equilibrium data that R17 was stabilized by ∼2.8 kcal mol−1 (± 0.2 kcal mol−1) by folded R16 in R1617, but the kinetic data suggest the stabilization is larger (∼4 ± 0.4 kcal mol−1). This is likely to reflect the complexities of fitting the chevron (with uncertainties from the fit, and the long extrapolation to 0 M for the unfolding rate constant). Thus, although we cannot be absolutely certain of the magnitude of the stabilization, the kinetic and equilibrium data are entirely consistent.
Forced unfolding
There are two folding/unfolding regimes that all proteins in the cell will experience. The first is cotranslational folding as the protein is synthesized (17). The second is the thermal unfolding and refolding events that occur naturally without external denaturants during the protein's lifetime. In the case of proteins that experience force in the cell, there is a third regime: unfolding induced by the addition of a denaturant in the form of an externally applied force. Spectrin, as a constituent of the cytoskeleton, is likely to experience force, and atomic force microscopy experiments have shown that spectrin domains unfold at relatively low forces (18,19). If a spectrin domain unfolds due to mechanical stress, our data suggest it will recover (refold) more rapidly in the presence of folded neighboring domains once the force is released.
Our data cannot, however, be used to infer that the forced unfolding of spectrin domains will be slowed by folded neighbors, even though there is experimental evidence for cooperative unfolding in some spectrin domains (18,20), since we have no information as to whether the forced unfolding pathway is the same as that observed in the absence of force. Indeed, there is evidence for other proteins to suggest that the mechanical unfolding pathway (the pathway sampled in the presence of an external force) may not be the same as that sampled in chemical denaturation studies (21,22), and kinetic unfolding rate constants are a poor predictor of mechanical stability (23,24). Moreover, simulations of mechanical and thermal unfolding of spectrin domains have suggested that the unfolding pathways are “very different” (25). We are currently undertaking some experiments to specifically address this question.
CONCLUSION
There have been few studies of the kinetics of folding of multidomain proteins in which each domain can fold independently (26–33). In the muscle protein titin, the immunoglobulin domains are essentially independent of each other; neighboring domains have no effect on either the stability or the kinetics (27). In contrast, domain:domain interactions in the α-helical spectrin repeats not only stabilize each other but have profound effects on their kinetic behavior. This may have important biological implications.
In R1617, the N-terminal R16 domain, once folded, significantly speeds the folding of the subsequent domain. If this is a common effect, then this may be of significance during folding of spectrin as it is synthesized in the cell. Cotranslational folding of multiple domain proteins is thought to provide a first line of defense against misfolding in the cell (17). The catalyzed folding of downstream domains may also be important. Sanchez et al. (28) have recently suggested that “Rapid structure formation in the N-terminal domain (of the capsid protein of the Semliki Forest virus) might provide a template for efficient and rapid formation of the complete three-dimensional structure”. It is not clear from our results whether there is a specific structural “template” effect in spectrin domains. Further investigation will be needed to establish whether and how the pathway of folding is affected by domain:domain interactions.
Kinetic stability (maintaining the folded structure) is important for a protein to function in vivo. Once folded, the effect of domain:domain interactions is to slow the unfolding of the protein significantly. The R17 domain unfolds ∼2 orders of magnitude more slowly when in tandem with R16. The effect of this on the half-life is dramatic. The half-life of R17 alone is ∼30 min. In R1617 the half-life of the R17 domain is extended to ∼15 h. (The lifetime of a human red blood cell is estimated to be ∼120 days, although it may be less in birds.) We have also established that the R16 domain must also unfold significantly more slowly when attached to R17. Again, if these results are typical of spectrin domains and if each domain is stabilized by both its N- and C-terminal neighbors, it is clear that the ability of the protein to withstand thermal unfolding in the cell will be dramatically increased.
This study shows that caution has to be exercised when extrapolating from single domain proteins to larger multidomain proteins with a number of independently folding modules arranged in tandem. Unlike titin, spectrin is certainly more than “the sum of its parts”.
Acknowledgments
We thank Thomas Kiefhaber and Annett Bachmann for helpful discussions.
This work was supported by the Wellcome Trust and the Medical Research Council, UK. J.C. is a Wellcome Trust Senior Research Fellow.
Kathryn A. Scott's present address is Dept. of Medicinal Chemistry, University of Washington, Seattle, WA 98195-7610.
References
- 1.Teichmann, S. A., C. Chothia, and M. Gerstein. 1999. Advances in structural genomics. Curr. Opin. Struct. Biol. 9:390–399. [DOI] [PubMed] [Google Scholar]
- 2.Winograd, E., D. Hume, and D. Branton. 1991. Phasing the conformational unit of spectrin. Proc. Natl. Acad. Sci. USA. 88:10788–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speicher, D. W., and V. T. Marchesi. 1984. Erythrocyte spectrin is comprised of many homologous triple helical segments. Nature. 311:177–180. [DOI] [PubMed] [Google Scholar]
- 4.Yan, Y., E. Winograd, A. Viel, T. Cronin, S. C. Harrison, and D. Branton. 1993. Crystal structure of the repetitive segments of spectrin. Science. 262:2027–2030. [DOI] [PubMed] [Google Scholar]
- 5.Pascual, J., M. Pfuhl, G. Rivas, A. Pastore, and M. Saraste. 1996. The spectrin repeat folds into a three-helix bundle in solution. FEBS Lett. 383:201–207. [DOI] [PubMed] [Google Scholar]
- 6.Pascual, J., M. Pfuhl, D. Walther, M. Saraste, and M. Nilges. 1997. Solution structure of the spectrin repeat: a left-handed antiparallel triple-helical coiled-coil. J. Mol. Biol. 273:740–751. [DOI] [PubMed] [Google Scholar]
- 7.Macdonald, R. I., and E. V. Pozharski. 2001. Free energies of urea and of thermal unfolding show that two tandem repeats of spectrin are thermodynamically more stable than a single repeat. Biochemistry. 40:3974–3984. [DOI] [PubMed] [Google Scholar]
- 8.Ylanne, J., K. Scheffzek, P. Young, and M. Saraste. 2001. Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure 9:597–604. [DOI] [PubMed] [Google Scholar]
- 9.Grum, V. L., D. Li, R. I. Macdonald, and A. Mondragon. 1999. Structures of two repeats of spectrin suggest models of flexibility. Cell. 98:523–535. [DOI] [PubMed] [Google Scholar]
- 10.MacDonald, R. I., and J. A. Cummings. 2004. Stabilities of folding of clustered, two-repeat fragments of spectrin reveal a potential hinge in the human erythroid spectrin tetramer. Proc. Natl. Acad. Sci. USA. 101:1502–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott, K. A., and J. Clarke. 2005. Spectrin R16: broad energy barrier or sequential transition states? Protein Sci. 14:1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott, K. A., S. Batey, K. A. Hooton, and J. Clarke. 2004. The folding of spectrin domains I: wild-type domains have the same stability but very different kinetic properties. J. Mol. Biol. 344:195–205. [DOI] [PubMed] [Google Scholar]
- 13.Batey, S., L. G. Randles, A. Steward, and J. Clarke. 2005. Cooperative folding in a multi-domain protein. J. Mol. Biol. 349:1045–1059. [DOI] [PubMed] [Google Scholar]
- 14.Scott, K. A., L. G. Randles, and J. Clarke. 2004. The folding of spectrin domains II: phi-value analysis of R16. J. Mol. Biol. 344:207–221. [DOI] [PubMed] [Google Scholar]
- 15.Bieri, O., and T. Kiefhaber. 2000. Kinetic models in protein folding. In Mechanisms of Protein Folding, 2nd ed. R. H. Pain, editor. Oxford University Press, Oxford. 34–64.
- 16.Pappenberger, G., H. Aygun, J. W. Engels, U. Reimer, G. Fischer, and T. Kiefhaber. 2001. Nonprolyl cis peptide bonds in unfolded proteins cause complex folding kinetics. Nat. Struct. Biol. 8:452–458. [DOI] [PubMed] [Google Scholar]
- 17.Netzer, W. J., and F. U. Hartl. 1997. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 388:343–349. [DOI] [PubMed] [Google Scholar]
- 18.Law, R., P. Carl, S. Harper, P. Dalhaimer, D. W. Speicher, and D. E. Discher. 2003. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys. J. 84:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. 1999. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561. [DOI] [PubMed] [Google Scholar]
- 20.Law, R., G. Liao, S. Harper, G. Yang, D. W. Speicher, and D. E. Discher. 2003. Pathway shifts and thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys. J. 85:3286–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng, S. P., R. W. Rounsevell, A. Steward, C. D. Geierhaas, P. M. Williams, E. Paci, and J. Clarke. 2005. Mechanical unfolding of TNfn3: the unfolding pathway of a fnIII domain probed by protein engineering, AFM and MD simulation. J. Mol. Biol. 350:776–789. [DOI] [PubMed] [Google Scholar]
- 22.Best, R. B., S. B. Fowler, J. L. Herrera, A. Steward, E. Paci, and J. Clarke. 2003. Mechanical unfolding of a titin Ig domain: structure of transition state revealed by combining atomic force microscopy, protein engineering and molecular dynamics simulations. J. Mol. Biol. 330:867–877. [DOI] [PubMed] [Google Scholar]
- 23.Best, R. B., B. Li, A. Steward, V. Daggett, and J. Clarke. 2001. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 81:2344–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forman, J. R., S. Qamar, E. Paci, R. N. Sandford, and J. Clarke. 2005. The remarkable mechanical strength of polycystin-1 supports a direct role in mechanotransduction. J. Mol. Biol. 349:861–871. [DOI] [PubMed] [Google Scholar]
- 25.Paci, E., and M. Karplus. 2000. Unfolding proteins by external forces and temperature: the importance of topology and energetics. Proc. Natl. Acad. Sci. USA. 97:6521–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Head, J. G., A. Houmeida, P. J. Knight, A. R. Clarke, J. Trinick, and R. L. Brady. 2001. Stability and folding rates of domains spanning the large A-band super-repeat of titin. Biophys. J. 81:1570–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott, K. A., A. Steward, S. B. Fowler, and J. Clarke. 2002. Titin; a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 315:819–829. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, I. E., M. Morillas, E. Zobeley, T. Kiefhaber, and R. Glockshuber. 2004. Fast folding of the two-domain semliki forest virus capsid protein explains co-translational proteolytic activity. J. Mol. Biol. 338:159–167. [DOI] [PubMed] [Google Scholar]
- 29.Kirkitadze, M. D., D. T. Dryden, S. M. Kelly, N. C. Price, X. Wang, M. Krych, J. P. Atkinson, and P. N. Barlow. 1999. Co-operativity between modules within a C3b-binding site of complement receptor type 1. FEBS Lett. 459:133–138. [DOI] [PubMed] [Google Scholar]
- 30.Kirkitadze, M. D., M. Krych, D. Uhrin, D. T. Dryden, B. O. Smith, A. Cooper, X. Wang, R. Hauhart, J. P. Atkinson, and P. N. Barlow. 1999. Independently melting modules and highly structured intermodular junctions within complement receptor type 1. Biochemistry. 38:7019–7031. [DOI] [PubMed] [Google Scholar]
- 31.Martin, A., and F. X. Schmid. 2003. The folding mechanism of a two-domain protein: folding kinetics and domain docking of the gene-3 protein of phage fd. J. Mol. Biol. 329:599–610. [DOI] [PubMed] [Google Scholar]
- 32.Rudolph, R., R. Siebendritt, G. Nesslauer, A. K. Sharma, and R. Jaenicke. 1990. Folding of an all-beta protein: independent domain folding in gamma II-crystallin from calf eye lens. Proc. Natl. Acad. Sci. USA. 87:4625–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker, M. J., J. Spencer, G. S. Jackson, S. G. Burston, L. L. Hosszu, C. J. Craven, J. P. Waltho, and A. R. Clarke. 1996. Domain behavior during the folding of a thermostable phosphoglycerate kinase. Biochemistry. 35:15740–15752. [DOI] [PubMed] [Google Scholar]