Abstract
Eukaryotic mRNAs are modified at the 5′ end with a cap structure. In fungal cells, the formation of the mRNA cap structure is catalyzed by three enzymes: triphosphatase, guanylyltransferase, and methyltransferase. Fungal capping enzymes have been proposed to be good antifungal targets because they differ significantly from their human counterparts and the genes encoding these enzymes are essential in Saccharomyces cerevisiae. In the present study, Candida albicans null mutants were constructed for both the mRNA triphosphatase-encoding gene (CET1) and the mRNA methyltransferase encoding gene (CCM1), proving that these genes are not essential in C. albicans. Heterozygous deletions were generated, but no null mutants were isolated for the guanylyltransferase-encoding gene (CGT1), indicating that this gene probably is essential in C. albicans. Whereas these results indicate that Cet1p and Ccm1p are not ideal molecular targets for development of anticandidal drugs, they do raise questions about the capping of mRNA and translation initiation in this fungus. Southern blot analysis of genomic DNA indicates that there are not redundant genes for CET1 and CCM1 and analysis of mRNA cap structures indicate there are not alternative pathways compensating for the function of CET1 or CCM1 in the null mutants. Instead, it appears that C. albicans can survive with modified mRNA cap structures.
Candida albicans infections are a serious health risk for immunocompromised patients, such as transplant recipients, cancer patients, neonates, human immunodeficiency virus-positive patients, and hospital patients with intravenous tubes or catheters. Few drugs are available for treating disseminated candidiasis, and there is a need for new antifungal compounds to treat these infections (13). There is a common presumption that antifungal agents can be developed from inhibitors of enzymes that catalyze essential functions. Although this presumption has not yet been proven, it has generated interest in the identification of essential genes in C. albicans. Enzymes involved in mRNA capping, especially the mRNA triphosphatase, have been proposed as potentially good antifungal targets because they are significantly different from the human enzymes and are reportedly essential for cell growth (32). In all eukaryotes, mRNA cap structures are purported to play critical roles in stability, processing, and nuclear export of mRNA, as well as in translation initiation (45). While cap-independent translation does occur, cap-dependent translation is believed to be the major pathway for translation initiation in eukaryotes (20).
The formation of the 7-methylguanylated mRNA cap (m7GpppN) in eukaryotes occurs in three steps (10). The first step is catalyzed by the mRNA triphosphatase which dephosphorylates the terminal nucleotide (pppN → ppN). In the second step, guanylyltransferase “caps” the diphosphate with guanosine 5′-monophosphate (GMP): ppN + GMP → GpppN. The third step is the transfer of a methyl group to the capped nucleotide by the methyltransferase (GpppN + S-adenosylmethionine → m7GpppN + S-adenosylhomocysteine). In mammals, two genes encode the three enzymatic steps of capping, whereas in fungi each of the three capping reactions is catalyzed by a distinct gene product (32). Fungal mRNA 5′-triphosphatase has a completely different structure and catalytic mechanism than the mammalian counterparts (14). The genes encoding mRNA 5′-triphosphatase (CET1), guanylyltransferase (CGT1), and methyltransferase (CCM1) have all been shown to be essential in the model fungal organism, Saccharomyces cerevisiae (references 34, 19, and 39, respectively). In a recent publication, Pei et al. (27) concluded the CET1 gene is essential in C. albicans because they were unable to construct a null deletion mutant. However, we show here that both alleles of CET1 can be deleted in C. albicans and the null mutants are viable. We were also able to delete both alleles of C. albicans CCM1.
Construction of null mutants in C. albicans is complicated because this diploid organism lacks a sexual cycle. Techniques to evaluate gene essentiality by using sequential disruption of both alleles of a target gene in C. albicans were introduced by Fonzi and Irwin (9), and more recently the Mitchell laboratory has developed additional methods for generating C. albicans null mutants (7, 42, 43). In the two-step transformation method, alleles are knocked out in sequential transformations selecting for homologous recombinations (42). An alternative deletion strategy, the single-transformation method, first selects for transformants that are Arg+ due to the insertion of the ura3Δ3′-ARG4-ura3Δ5′ (UAU1) cassette at the targeted locus (7). Transformants with the desired UAU1 cassette integration are then grown on synthetic complete (SC) medium lacking arginine and uridine to select Arg+ Ura+ cells. These cells have undergone a mitotic recombination event in which one allele of the nonessential gene is replaced with the UAU1 cassette and the other allele of the nonessential gene is replaced with a functional URA3 gene. In the case of an essential gene, insertional deletion of two alleles of the target gene can only be generated if an allelic duplication occurs such that the strain becomes triploid for the target gene locus. This maintenance of a wild-type allele under the selective pressure for homozygous deletions is strong evidence that a gene is essential.
We initially applied the single transformation method (7) to determine whether CET1, CGT1, and CCM1 are essential genes in C. albicans. After generating cet1-null mutants by this technique, we were also able to generate cet1-null mutants by using the two-step transformation knockout strategy (42), confirming that the C. albicans CET1 gene is nonessential. C. albicans ccm1-null mutants were generated by using the two-step transformation method. CGT1 heterozygous deletions were generated by both techniques, but we were unable to recover any isolates completely lacking a wild-type CGT1, indicating that this is probably an essential gene in C. albicans.
The viability of C. albicans cet1- and ccm1-null mutants raises questions about the mechanics and role of cap-dependent translation initiation in this fungal pathogen. There are several possible explanations why CET1 and CCM1 are not essential in C. albicans, e.g., redundant genes encoding mRNA capping enzymes, alternative mRNA capping pathways, or the possibility that C. albicans can survive with modified mRNA cap structures. Our analysis and characterization of mRNA cap structures indicates that the C. albicans cet1- and ccm1-null mutants do make modified mRNA-cap structures, and these modified mRNA-caps are sufficient for C. albicans to effectively translate mRNA and survive.
MATERIALS AND METHODS
Reagents.
Taq polymerase was purchased from Applied Biosystems (Foster City, Calif.). Restriction endonucleases and deoxynucleotidyl transferase were purchased from Invitrogen (Rockville, Md.). All enzymes were used according to manufacturers' protocols. PCR primers used in the present study were purchased from Sigma-Genosys and are described in Table 1. Plasmids pGEM-URA3, pGEM-HIS1, and pRS-ARG4−ΔSpeI (42), as well as the UAU1 cassette-containing plasmid pBME101 (7), were generous gifts from A. P. Mitchell. Synthetic complete (SC) media were made by using dropout base mixes available from Bio 101 (Vista, Calif.) with glucose added to a final concentration of 2%. Southern blot analyses were performed using probes labeled with the DIG High Prime kit and the DIG chemiluminescent detection kit (Roche, Indianapolis, Ind.). Amphotericin B, cycloheximide, hygromycin B, and actinomycin D were purchased from Sigma (St. Louis, Mo.), and blasticidin S was purchased from Calbiochem (Darmstadt, Germany). Tobacco acid pyrophosphatase (TAP) was obtained from Epicentre Technologies (Madison, Wis.). Nuclease P1, NaIO4, and polyethyleneimine-coated thin-layer chromatography (TLC) plates were purchased from VWR (Batavia, Ill.). The capped nucleotide analog standards, m7GpppA, m7GpppG, GpppA, and GpppG, were purchased from New England Biolabs, Inc. (Beverly, Mass.). Nucleotide standards, adenosine 5′-monophosphate (AMP) and GMP, were bought from Sigma. NaB[3H]4 (15 Ci/mmol) was purchased from American Radiolabeled Chemicals, Inc. (St. Louis, Mo.).
TABLE 1.
Oligonucleotide sequences
| Primer | Sequence (5′ - 3′)a |
|---|---|
| CaCET5′-DR | ctactgcataccataaaccttctgttcatgaacgtcattcaataacgagcatgttgaatgacacGTTTTCCCAGTCACGACGTT |
| CaCET3′-DR | cttcaattaattcctccaatctaagattatctacccctttctggattttatcaatagcagcTGAATTGTGAGCGGATA |
| CaCET1F | catcaagtgggaatgcgaatg |
| CaCET1R | cttgttcaatctattattgagag |
| CET1 probe F | ccgtcagattcaactccaac |
| CET1 probe R | cataacacccgcctccaactc |
| CET1-restore 5′ | acgcatgtcgactttaaactgtatatatttgtagc |
| CET1-restore 3′ | acgcatgtcgacgaaagtgggcgcggttc |
| CaCGT5′-DR | ctatacactttatgattcaattagaagaaagagaaattcctgtaataccaggtaataaacGTTTTCCCAGTCACGACGTT |
| CaCGT3′-DR | caatttattcgtcgtcactatcttcataagtgggtatgtcattaagtatttgttgcagtgTGAATTGTGAGCGGATA |
| CaCGT1F | caaggctagtctttagaaattc |
| CaCGT1R | ccagaaaccgagaaagaatc |
| CaCCM5′-DR | ctacaccactagcatgtctaccgattcgtacactccctcacaagagcctggctcaaagcGTTTTCCCAGTCACGACGTT |
| CaCCM3′-DR | cctataccttctcaaatacaaatcctatgtaaaatgccactgcttccttttcgtcgcctgTGAATTGTGAGCGGATA |
| CaCCM1F | gagttgaaacatgaatcattag |
| CaCCM1R | gctacacactacccagacata |
| CCM1 probe F | gacaggcgaatctgtatttgc |
| CCM1 probe R | cagcaccgtacttgccatcac |
| CCM1-restore 5′ | acgcatgtcgacgaaaagccaatgttatagatg |
| CCM1-restore 3′ | acgcatgtcgacacgatgcgtccggcgtagag |
| ARG4 primer | gaactatctattcgatgatg |
Lowercase roman letters indicate nucleotides that provide homology to C. albicans genomic DNA. Nucleotides in uppercase indicate the 5′ DR and 3′ DR sequences that anneal to flanking sequence of marker cassettes from the plasmids (42). Lowercase italic nucleotides indicate sequences added for cloning convenience.
Strains and growth conditions.
Strains used in the present study are described in Table 2. For transformation, C. albicans BWP17 was grown overnight in YEPD (1% yeast extract, 2% Bacto Peptone [Difco], 2% glucose) supplemented with uridine at 80 μg/ml, subcultured 1:100 into 50 ml of the same medium, and grown for two doublings at 30°C. Where indicated, uridine was added to a final concentration of 80 μg/ml, and arginine was added to a final concentration of 50 μg/ml. All transformants were plated onto SC lacking arginine (SC−arginine), SC plus arginine but lacking uridine (SC+arginine−uridine), or SC lacking both arginine and uridine (SC−arginine−uridine) as appropriate for the selectable marker(s) and incubated at 30°C for 2 to 3 days. For the selection of Arg+ Ura+ segregrants of CET1 heterozygotes disrupted with the UAU1 cassette, isolated colonies were grown overnight in YEPD lacking uridine (YEPD−uridine). A total of 1 ml of each culture was washed with sterile water and then resuspended in 1 ml of H2O, and a 100-μl portion of the cells was plated onto SC−arginine−uridine, followed by growth for 2 to 3 days at 30°C. Growth studies were performed in round-bottom 96-well plates with cells inoculated in 200 μl of YEPD+uridine to a final concentration of 2,000 to 3,000 cells/ml, incubated at 30 or 37°C for 60 h. Growth was monitored in a SpectraMax 250 (Molecular Dynamics) at 595 nm, with readings taken every 30 min after shaking for 5 s.
TABLE 2.
C. albicans strains used in this study
| Strain | Genotype | Description | Source |
|---|---|---|---|
| BWP17 | Δura3::λimm434/Δura3::λimm434 his1::hisG/his1::hisG arg4::hisG arg4::hisG | Parent strain for deletions, allows selection of three different markers (URA3, ARG4, HIS1) | A. P. Mitchell (42) |
| CQF125 | BWP17 Δcet1::UAU1/CET1 | Heterozygote of CET1 from single transformation method | This study |
| CQF136 | BWP17 Δcet1::UAU1/Δcet1::URA3 | Temperature-sensitive null mutant of cet1 from single transformation method | This study |
| CQF160 | BWP17 Δcet1::UAU1/Δcet1::URA3 | Null mutant of cet1 from single transformation method | This study |
| CQF142 | BWP17 Δcet1::ARG4/CET1 | Heterozygote of CET1 from two-step transformation method | This study |
| CQF141 | BWP17 Δcet1::UAU1/Δcet1::URA3/CET1 | Trisomic isolate for CET1 locus generated from single transformation method | This study |
| CQF152 | BWP17 Δcet1::ARG4/Δcet1::URA3 | Null mutant of cet1 from two-step transformation method | This study |
| CQF248 | BWP17 Δcet1::ARG4/Δcet1::URA3 his1::hisG::CET1 | CET1 restored to null in trans by recombination at his1::hisG locus | This study |
| CQF153 | BWP17 Δcgt1::ARG4/CGT1 | Heterozygote of CGT1 from two-step transformation method | This study |
| CQF170 | BWP17 Δcgt1::ARG4/Δcgt1::URA3/CGT1 | Trisomic for CGT1 locus from two-step transformation method | This study |
| CQF154 | BWP17 Δccm1::ARG4/CCM1 | Heterozygote of CCM1 from two-step transformation method | This study |
| CQF171 | BWP17 Δccm1::ARG4/Δccm1::URA3 | Null mutant of ccm1 from two-step transformation method | This study |
| CQF250 | BWP17 Δcet1::ARG4/Δcet1::URA3 his1::hisG::CET1 | CCM1 restored to null in trans by recombination at his1::hisG locus | This study |
DNA methods.
Genomic DNA was prepared from overnight cultures of C. albicans grown at 30°C in 5 ml of YEPD by using the glass bead lysis method (3). PCR amplifications of deletion cassettes and analytical PCRs with genomic DNA were carried out as described in Wilson et al. (42). Before transformation of C. albicans with insertional deletion cassettes, the PCR fragments were blocked with ddATP by using terminal deoxynucleotidyl transferase according to the protocol described by Shah-Mahoney et al. (33). PCR-amplified DNA was purified by using phenol-CHCl3-isoamyl alcohol (25:24:1), followed by ethanol precipitation before and after the blocking reaction. High-stringency Southern blot analysis (36) was performed on 1 to 3 μg of genomic DNA that was digested with BglII and separated on 0.8% agarose gels for capillary transfer to membrane (Hybond N; Amersham). Probes were PCR amplified from genomic DNA of C. albicans BWP17 with primers CET1 probe F and CET1 probe R for the CET1 probe and CCM1 probe F and CCM1 probe R for the CCM1 probe. Membranes were hybridized with 1 μg of DIG-labeled probe under high-stringency conditions (42°C), followed by stringent wash conditions (two 5-min room temperature washes in 2× SSC [0.15 M NaCl, 10 mM sodium citrate, pH 7] plus 0.1% sodium dodecyl sulfate [SDS], followed by two 20-min washes at 68°C in 0.5× SSC plus 0.1% SDS). Southern blots were developed by using anti-DIG-Fab fragments, which are conjugated to alkaline phosphatase, and CSPD chemiluminescent detection. The anti-DIG was diluted 1:10,000 in blocking buffer and the CSPD was diluted 1:100 in detection buffer as recommended by the supplier.
Genetic manipulations of C. albicans.
C. albicans competent cells were prepared as described by the method of Braun and Johnson (2). C. albicans BWP17 and derivatives were transformed with 80 μl of the PCR-amplified disruption cassettes as described previously (42).
In vitro susceptibility tests.
MICs were determined by using NCCLS standard protocols for broth microdilution assays with the colorimetric modification (28).
RNA methods.
Total RNA was isolated from C. albicans cells by hot phenol-chloroform extraction and ethanol precipitation as described by Lin et al. (18). Poly(A) mRNA was enriched from 1 mg of total RNA by using the Qiagen (Valencia, Calif.) RNeasy kit, as per the manufacturer's specification. To label the mRNA, 35 μg of precipitated mRNA was dissolved in 11 μl of 2.5 mM EDTA (pH 7.0), heated to 65°C for 1 min, and placed on ice for 5 min. Then, 5 μl of 8 mM NaIO4 was added, and the oxidation reaction was allowed to proceed at 4°C in the dark for 90 min. The samples were mixed with 4 μl of 1 M sodium acetate (pH 5.5), and the mRNA was precipitated with ethanol. The mRNA pellets were dissolved in 12 μl of 0.1 M potassium phosphate buffer (pH 6.8). A total of 2 μl of NaB[3H]4 (15 Ci/mmol; 55 mM in 0.1 M potassium hydroxide) was added to the oxidized RNA, and the reaction was allowed to proceed at room temperature in the dark for 60 min. Next, 75 μl of 0.2 M sodium acetate was added, and the samples were incubated for an additional 15 min. The RNA was ethanol precipitated, dissolved in 100 μl of water, and purified by using the RNeasy kit (Qiagen). The volume of purified radiolabeled mRNA solutions was adjusted to 50 μl with water, and the samples were split in half for nuclease P1 or TAP digestions. For P1 digestion, 25-μl samples were adjusted to a final concentration of 50 mM ammonium acetate (pH 5.3) and treated with nuclease P1 (100 μg/ml) at 37°C for 18 h. The remaining 25-μl fractions were digested with TAP by adding 2.9 μl of 10× TAP reaction buffer (500 mM sodium acetate [pH 6.0], 1% β-mercaptoethanol, and 0.1% Triton X-100) and 1.1 μl of TAP (10 U) and incubated at 37°C for 4 h.
Cap structure resolution.
Next, 20 μl of the RNA samples digested with nuclease P1 or TAP were spotted onto polyethyleneimine-cellulose, along with unlabeled nucleotides, AMP and GMP, and cap standards, m7GpppG, m7GpppA, GpppG, and GpppA. After they dried, the TLC plates were developed in 0.1 M sodium phosphate buffer (pH 6.8)-ammonium sulfate-n-propanol (100:60:2 [vol/wt/vol]) at room temperature. Markers were detected under UV light, and the positions of tritiated cap structures were identified by phosphorimaging.
RESULTS
C. albicans CET1 deletions.
Targeted deletions of the triphosphatase encoding gene, CET1, were constructed by using two different methods: the single-step transformation procedure of Enloe et al. (7), and the sequential two-step transformation method (42). In the single-step transformation method, the first allele was deleted by transforming C. albicans with a UAU1 PCR cassette-amplified from pBME101 by using primers CaCET15′-DR and CaCET13′-DR. Transformants were selected on SC−arginine. As detected by analytical PCR, three of four Arg+ transformants had the UAU1 disruption cassette integrated in the CET1 genomic locus. These three Δcet1::UAU1/CET1 heterozygotes were streaked out on SC−arginine plates to generate isolated colonies, and 15 isolated colonies from each were grown overnight in YEPD−uridine at 30°C. Overnight cultures were harvested and washed in water and plated on SC−arginine−uridine to select for mitotic recombinants. PCR analysis of genomic DNA for the 78 Arg+ Ura+ segregants from this selection showed that 12 were null mutants (Δcet1::UAU1/Δcet1::URA3) and 44 were trisomic for the CET1 locus (Δcet1::UAU1/Δcet1::URA3/CET1). The remaining 12 segregants probably had rearrangements or ectopic recombinations that we could not identify.
The two-step transformation method was also used to confirm that CET1 is not essential in C. albicans. Heterozygous CET1 deletion mutants were identified after selection on SD−arginine after transformation of C. albicans BWP17 with a PCR cassette amplified from pRS-ARG4−ΔSpeI with the primers CaCET15′-DR and CaCET13′-DR. Two of the nine Arg+ transformants had targeted insertions in one CET1 allele. One of these heterozygotes (Δcet1::ARG4/CET1) was then transformed with a URA3 marker cassette PCR amplified from pGEM-URA3 with the primers CaCET5′-DR and CaCET3′-DR. Eight Arg+ Ura+ transformants were screened, and six were null mutants (Δcet1::ARG4/Δcet1::URA3).
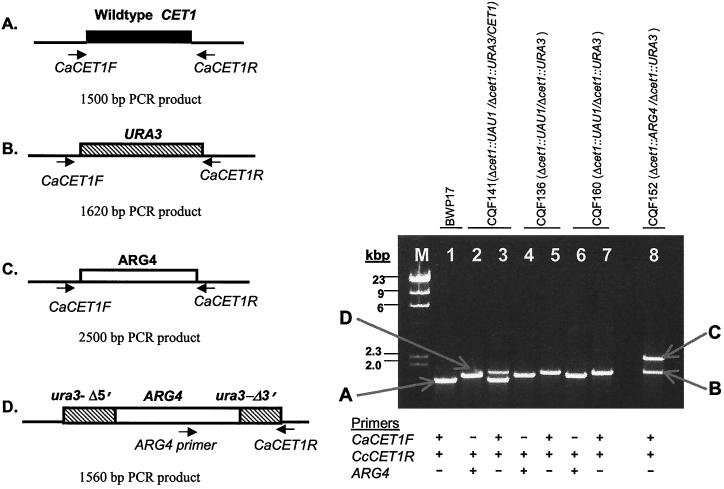
Analytical PCR of CET1 deletion strains are shown on the gel in Fig. 1. The PCR amplifications were designed to detect the four possible chromosomal constructs at this locus (depicted in Fig. 1A to D). PCR amplification of the wild-type CET1 allele (Fig. 1A) with primers CaCET1F and CaCET1R should produce a PCR product of 1,500 bp. Using the same primers, PCR amplification of DNA from isolates with the Δcet1::URA3 allele (Fig. 1B), the Δcet1::ARG4 allele (Fig. 1C), or the Δcet1::UAU1 allele produce larger products (1,620 bp for URA3, 2,600 bp for ARG4, and 4,000 bp for Δcet1::UAU1). Because the PCR product for the Δcet1::UAU1 allele obtained with the flanking primers is so large, amplification of it is not favored when the smaller alleles are present. Therefore, to determine whether this allele is definitively present, we also analyzed genomic DNA of UAU1 transformants by using the ARG4 primer paired with the CaCET1R primer. This primer pair should produce a product of 1,560 bp from the Δcet1::UAU1 allele (D). As expected, one PCR product was amplified from C. albicans BWP17 genomic DNA with the CaCET1R and CaCET1F primers (Fig. 1, lane 1), and this band corresponds to the wild-type allele (CET1; Fig. 1A). PCR analysis of genomic DNA from isolate CQF141 indicates it is trisomic, with primers ARG4 and CaCET1R producing a band corresponding to the Δcet1::UAU1 allele (lane 2) and the CaCET1R and CaCET1F primers producing two bands corresponding to the wild-type CET1 allele and the Δcet1::URA3 allele (lane 3). The PCR products for isolates CQF136 and CQF160 are the same and indicate that both are cet1-null mutants. With primers ARG4 and CaCET1R, both isolates yield the band corresponding to the Δcet1::UAU1 allele (lanes 4 and 6), whereas only the band corresponding to the Δcet1::URA3 allele is seen with CaCET1F and CaCET1R primer amplifications (lanes 5 and 7). Isolate CQF152 was generated from the two-step transformation method so only the CaCET1F and CaCET1R primers were needed for analysis. Lane 8 shows that this isolate has the two deletion alleles (Δcet1::URA3 and Δcet1::ARG4) and no wild-type CET1 allele.
FIG. 1.
Analytical PCR of genomic DNA from C. albicans BWP17 parent strain and CET1 deletion strains. (Left panel) Schematic diagrams of wild-type or insertional deletion mutants for C. albicans chromosomal DNA at the CET1 locus with oligonucleotide binding sites and predicted PCR fragments. (Right panel) PCR fragments separated on 1% agarose gel. Lane M, DNA size standards; lane 1, C. albicans BWP17 genomic DNA PCR amplified with CaCET1F and CaCET1R primers; lane 2, C. albicans CQF141 genomic DNA PCR amplified with ARG4 and CaCET1R primers; lane 3, C. albicans CQF141 genomic DNA PCR amplified with CaCET1F and CaCET1R primers; lane 4, C. albicans CQF136 genomic DNA PCR amplified with ARG4 and CaCET1R primers; lane 5, C. albicans CQF136 genomic DNA PCR amplified with CaCET1F and CaCET1R primers; lane 6, C. albicans CQF160 genomic DNA PCR amplified with ARG4 and CaCET1R primers; lane 7, C. albicans CQF160 genomic DNA PCR amplified with CaCET1F and CaCET1R primers; lane 8, C. albicans CQF152 genomic DNA genomic DNA PCR amplified with CaCET1F and CaCET1R primers.
C. albicans CGT1 and CCM1 deletions.
The single transformation method of Enloe et al. (7) was initially applied to deletion analysis of the CGT1 and CCM1 genes in C. albicans. The marker deletion cassettes were PCR amplified from pBME101 by using primers CaCGT5′-DR and CaCGT3′-DR or CaCCM15′ DR and CaCCM13′-DR. C. albicans BWP17 was transformed with the UAU1 PCR cassettes, and Arg+ transformants were selected. DNA isolated from these transformants was screened by analytical PCR to identify isolates with the correct heterozygous deletions. Heterozygous deletions were constructed for both genes. In this gene deletion procedure, the second step requires selection for a mitotic recombination event where the UAU1 cassette recombines into the second allele and the cells become Arg+ Ura+. We were unable to generate null mutants or even trisomic isolates for either of these two genes by this method.
A second method for generating C. albicans gene knockouts was also tested for CGT1 and CCM1 (42). C. albicans BWP17 was first transformed with ARG4 cassettes PCR amplified from pRS-ARG4-ΔSpeI with primers CaCGT5′-DR and CaCGT3′-DR or primers CaCCM5′-DR and CaCCM3′-DR. A heterozygous strain for each of these genes was then transformed with a URA3 marker cassette PCR amplified from pGEM-URA3 with primers CaCGT5′-DR and CaCGT3′-DR or primers CaCCM5′-DR and CaCCM3′-DR. Twelve Arg+ Ura+ transformants were screened for CCM1 deletions, and two were ccm1-null mutants (Δccm1::ARG4/Δccm1::URA3). All 24 of the Arg+ Ura+ transformants screened for CGT1 deletions retained a wild-type CGT1 allele, with 22 isolates being heterozygous (Δcgt1::ARG4/CGT1) and 2 isolates being trisomic (Δcgt1::ARG4/Δcgt1::URA3/CGT1).
The generation of ccm1-null mutants demonstrates that this is not an essential gene in C. albicans, but the CGT1 gene remains an enigma. We did not generate deletion mutants at this locus in multiple attempts by both deletion approaches. However, heterozygous deletion mutants (Δcgt1::ARG4/CGT1) can be constructed (Fig. 2), indicating that the deletion cassette can integrate into at least one allele at the desired locus. The failure to generate the double deletion could be due to preferential insertion of the deletion cassette at one allelic variant. Yesland and Fonzi (46) have reported that sequence heterology biases insertion to the allele with the best sequence homology with the transforming DNA. However, our sequence analysis of the C. albicans BWP17 CGT1 locus shows that none of the allelic variations in this gene are found in the sequences that would hybridize to the flanking homology provided by the CaCGT5′-DR and CaCGT3′-DR oligonucleotides in the PCR deletion cassette (data not shown). Therefore, we believe that we are not seeing the allelic bias and that CGT1 is likely an essential gene in C. albicans. Another group has reported failure to generate a cgt1-null mutant in C. albicans (5), supporting an essential nature for this gene.
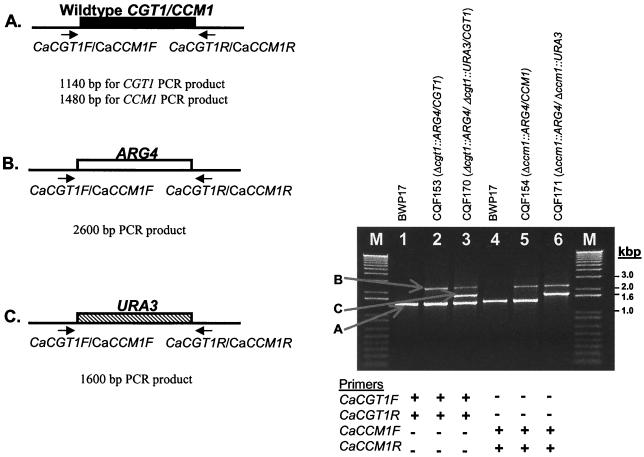
FIG. 2.
Analytical PCR of genomic DNA from C. albicans BWP17 parent strain and CGT1 or CCM1 deletion strains. (Left panel) schematic diagrams of wild-type or insertional deletion mutants for C. albicans chromosomal DNA at the CGT1 and CCM1 loci with oligonucleotide binding sites. The expected sizes for the PCR products indicated for the genomic structures are as follows: 1,140 bp for wild-type CGT1 and 1,480 bp for wild-type CCM1 (A); 2,600 bp for both Δcgt1::ARG4 and Δccm1::ARG4 (B); and 1,600 bp for both Δcgt1::URA3 and Δccm1::URA3 (C). (Right panel) PCR fragments separated on 0.8% agarose gel. Lane M, DNA size standards; lane 1, C. albicans BWP17 genomic DNA PCR amplified with CGT1F and CGT1R primers; lane 2, C. albicans CQF153 genomic DNA PCR amplified with CGT1F and CGT1R primers; lane 3, C. albicans CQF170 genomic DNA PCR amplified with CGT1F and CGT1R primers; lane 4, C. albicans BWP17 genomic DNA PCR amplified with CCM1F and CCM1R primers; lane 5, C. albicans CQF154 genomic DNA PCR amplified with CCM1F and CCM1R primers; lane 6, CQF171 genomic DNA PCR amplified with CCM1F and CCM1R primers.
Figure 2 shows the PCR analysis for the CGT1 and CCM1 deletions. PCR amplification with CaCGT1F and CaCGT1R results in PCR product sizes of 1,140 bp for the wild-type CGT1 allele (Fig. 2A), 2,600 bp for the Δcgt1::ARG4 allele (Fig. 2B), and 1,600 bp for the Δcgt1::URA3 allele (Fig. 2C). Amplification of genomic DNA with primers CaCGT1F and CaCGT1R produces a PCR product the size of the expected wild-type CGT1 allele for C. albicans BWP17 (lane 1). Two bands are seen for isolate CQF153 (lane 2), with sizes of the wild-type CGT1 allele and the size of the Δcgt1::ARG4 allele, and all three bands (A, B, and C) are seen for isolate CQF170 (lane 3). PCR amplification with CaCCM1F and CaCCM1R results in PCR product sizes of 1480 for the wild-type CCM1 allele (A), 2,600 bp for the Δccm1::ARG4 allele (B), and 1,600 bp for the Δccm1::URA3 allele (C). Amplification of genomic DNA with primers CaCCM1F and CaCCM1R produces a PCR product the size of the expected wild-type CCM1 allele for C. albicans BWP17 (lane 4), whereas two bands are seen for isolate CQF154 (lane 5) with sizes of the wild-type CCM1 allele and the size of the Δccm1::ARG4 allele. With these primers, both the deleted alleles, Δccm1::URA3 and Δccm1::ARG4, are seen for isolate CQF171 (lane 6) but no wild-type CCM1 allele.
Southern blot analysis.
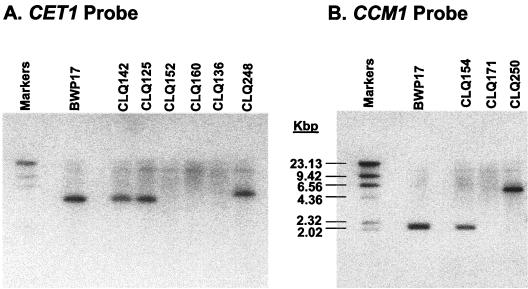
In order to eliminate the formal possibility that cet1- and ccm1-null mutants survive because a transposed copy of the wild-type CET1 allele or CCM1 allele exists in another chromosomal location, C. albicans chromosomal DNA was analyzed by Southern blot (Fig. 3). Genomic DNA from C. albicans BWP17; the CET1 heterozygotes CQF142 (Δcet1::ARG4/CET1) and CQF125 (Δcet1::UAU1/CET1); the cet1-null mutants CQF152 (Δcet1::ARG4/Δcet1::URA3), CQF160 (Δcet1::UAU1/Δcet1::URA3), and CQF136 (Δcet1::UAU1/Δcet1::URA3); the CCM1 heterozygote CQF154 (Δccm1::ARG4/CCM1); and the C. albicans ccm1-null mutant CQF171 (Δccm1::ARG4/Δccm1::URA3) was analyzed by Southern blot (Fig. 3). Genomic DNA was digested with BglII restriction endonuclease. After gel electrophoresis, the DNA was transferred to a nylon membrane for probing. For CET1 (Fig. 3A), the DNA probe encompassed codons 51 to 480 of the 520 codon CaCET1 open reading frame. For CCM1 (Fig. 3B), the DNA probe encompassed codons 18 to 455 of the 474 codon CaCCM1 open reading frame. Southern hybridization showed that, as expected, only one DNA band hybridized to the CET1 probe and one DNA band hybridized to the CCM1 probe in C. albicans BWP17 (Fig. 3). The CET1 or the CCM1 band is seen in the heterozygote isolates but not in the cet1- or ccm1-null mutants. There was no evidence that the wild-type CET1 or CCM1 was transposed in the null mutants.
FIG. 3.
Southern blot analysis of BglII-digested genomic DNA isolated from C. albicans. (A) Probe was CET1 DNA (covering codons 51 to 479). Sources of genomic DNA: BWP17 (CET1/CET1), CQF125 (Δcet1::UAU1/CET1), CQF141 (Δcet1::UAU1/Δcet1::URA3/CET1), CQF136 (Δcet1::UAU1/Δcet1::URA3), CQF142 (Δcet1::ARG4/CET1), CQF152 (Δcet1::ARG4/Δcet1:: URA3), CQF248 (Δcet1::ARG4/Δcet1::URA3 with CET1 restored). (B) Probe was CCM1 DNA (covering codons 18 to 455). Sources of genomic DNA: BWP17 (CCM1/CCM1), CQF154 (Δccm1::ARG4/CCM1), CQF171 (Δccm1::ARG4/Δccm1::URA3), CQF250 (Δccm1::ARG4/Δccm1::URA3 with CCM1 restored).
Growth of capping gene deletion mutants.
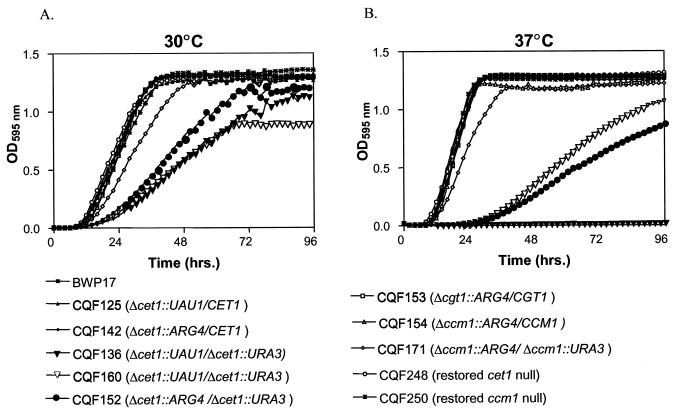
Growth in YEPD at both 30 and 37°C was measured for C. albicans BWP17 parent strain; the CET1, CGT1, and CCM1 heterozygous deletion mutants and the cet1- and ccm1-null mutants (Fig. 4). The wild-type parent and the heterozygous strains all had very similar growth curves at both temperatures. The null mutants grew slower at both temperatures, but the difference is more pronounced at 37 than at 30°C. The lag in growth was much longer for the null mutants at 37°C than for the heterozygotes or the parent. One of the null mutants constructed from the one-step transformation method was temperature sensitive, with almost no growth over 96 h at 37°C.
FIG. 4.
Growth curves for C. albicans parent strain and capping gene deletion mutants at 30 and 37°C. Cultures were grown in 200 μl of YEPD+uridine in 96-well plates with starting inoculum sizes of ca. 2,000 to 3,000 cells/ml. Growth was monitored in a Spectramax plate reader every 30 min after the plates were shaken for 5 s.
Sensitivity to antifungal compounds.
Prompted by a publication describing a C. albicans Δcgt1/CGT1 heterozygous deletion mutant that is resistant to the protein synthesis inhibitor hygromycin B (5), we decided to test the sensitivity for each of our capping gene deletion mutants to antifungal compounds (Table 3). The compounds tested included the following: amphotericin B, a polyene that interacts with ergosterol in the fungal membrane (6); cycloheximide, which inhibits peptide synthesis (24); blasticidin S, an aminohexosyl-cytosine nucleoside that inhibits ribosomal peptidyltransferase (15, 16); hygromycin B, which inhibits peptide chain elongation by disruption of EF-2-dependent translation (8); and actinomycin D, a polypeptide that inhibits DNA-dependent RNA synthesis (1). For standard yeast broth microdilution assays, drug concentrations were tested at twofold dilutions, and MIC endpoints were determined at 48 h; however, the MICs for the cet1-null strain (CQF152) were determined at 72 h because no growth was observed in the microtiter plate for this strain even in the no-drug controls at 48 h. MICs that are within two dilutions are not considered significantly different (23). All strains showed very similar sensitivities to amphotericin B and actinomycin D. The heterozygote deletion mutants for all three genes had similar sensitivities as the BWP17 parent to all drugs tested. Our C. albicans Δcgt1/CGT1 heterozygous deletion mutant was not more resistant to hygromycin B than the parent strain. However, our Δcet1/Δcet1 mutant and our Δccm1/Δccm1 mutants were slightly more sensitive to hygromycin B than the other strains tested. The Δcet1/Δcet1 mutant was more sensitive to cycloheximide than any of the other strains.
TABLE 3.
In vitro susceptibility of C. albicans parent strain and capping gene deletion mutants to antifungal compounds
| C. albicans strain (relevant phenotype) | MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|
| Amphotericin B | Cycloheximide | Blasticidin S | Hygromycin B | Actinomycin D | |
| BWP17 (parent) | 0.5 | >128 | >32 | 64 | 128 |
| CQF142 (Δcet1/CET1) | 0.5 | >128 | >32 | >128 | 128 |
| CQF152 (Δcet1/Δcet1) | 0.5 | 32 | 32 | 8 | 32 |
| CQF248 (CET1 restored null) | 0.5 | >128 | >32 | >128 | 128 |
| CQF153 (Δcgt1/CGT1) | 0.5 | >128 | >32 | >128 | 128 |
| CQF154 (ΔccmI/CCM1) | 0.5 | >128 | >32 | >128 | 128 |
| CQF171 (Δccm1/ΔccmI) | 0.5 | >128 | >32 | 32 | 64 |
| CQF250 (CCM1 restored null) | 0.5 | >128 | >32 | >128 | 128 |
All MIC values were determined after 48 h of incubation, except for strain CQF152, where they were determined after 72 h of incubation due to slow growth even in the no-drug controls.
Characterization of mRNA cap structures.
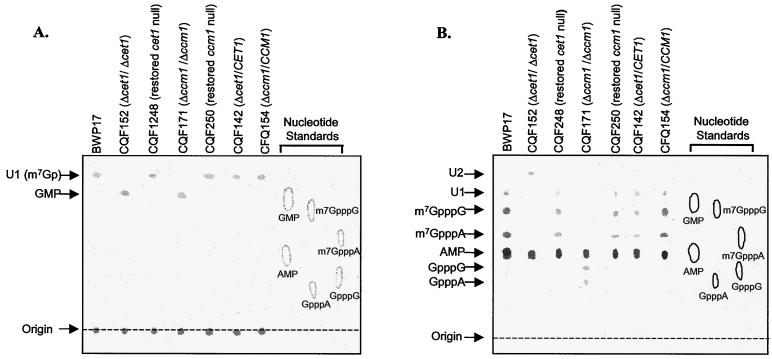
mRNA was purified from C. albicans BWP17, CQF152 (cet1-null mutant), CQF248 (the CET1 restored null), CQF171 (ccm1-null mutant), and CQF250 (the CCM1 restored null) by using hot phenol extraction (18). Treatment of mRNA with tritiated sodium borohydride labels the 2′,3′-cis-diols on the ribose rings of both mRNA cap structures and the terminal nucleotides at the 3′ end of mRNA (12). In order to separate the cap structures for TLC analysis, the radiolabeled mRNA was treated with TAP or P1 nucleases. TAP hydrolyzes a variety of nucleoside-linked pyrophosphate bonds, including those found in cap structures; however, phosphodiester bonds, which link nucleosides downstream of the 5′ terminal cap structure in mRNA, are not cleaved by TAP (35). Hence, TAP digestion of mRNA removes the 5′-terminal guanylate in capped transcripts, leaving the rest of the polynucleotide intact. In contrast, P1 nuclease possesses no pyrophosphatase activity, thus rendering cap structures impervious to its hydrolytic activity. Instead, the enzyme is a phosphodiesterase that cleaves the phosphate bond between nucleosides in mRNA, generating 5′-nucleotide monophosphates, and releasing the unhydrolyzed 5′-terminal cap structures (11, 12). TAP and P1 nuclease digests of the radiolabeled mRNA were separated by TLC and analyzed by phosphorimaging (Fig. 5). Unlabeled standards were also separated on TLC and visualized with UV light to aid in identification of the radiolabeled spots in the samples. For the TAP digestions of all samples (Fig. 5A), there was a radiolabeled spot at the origin of the TLC, corresponding to the 3′-end labeled polynucleotide. In BWP17 the cap structure migration did not correspond to any of the standards (and is identified as U1) but is consistent with a m7Gp that would be released from TAP digestion of mRNA that has the wild-type m7GpppN structure. TAP digestion of labeled mRNA that was isolated from CQF171 (the ccm1-null mutant) and CQF152 (the cet1-null mutant) did not produce the same unidentified spot as seen in the BWP17 sample (U1); instead, they both had a spot that corresponds to GMP. Nuclease P1 digestion of the samples also produced very different results for parent and null mutants (Fig. 5B). For BWP17, there were spots corresponding to AMP, m7GpppA, and m7GpppG standards, as well as an unidentified spot indicated as U2. As expected, the spot corresponding to AMP and representing the labeled AMP residue from the 3′ end of the nuclease P1-digested mRNA was seen for both null mutants. Also, as expected, there were no methylated cap structures seen for CQF171 (the ccm1-null mutant). Instead, the cap structures corresponded to the GpppG and GpppA standards. Nuclease P1 digestion of mRNA from CQF152 (the cet1-null mutant) released a product that migrated much farther than any of the standards or other digestion products (U3). We speculate that this spot also represents a cap structure since mRNA digested with TAP generates a product that migrates with GMP. This finding is consistent with TAP-mediated hydrolysis of cap structures lacking a methyl moiety at the N7 position of the 5′-terminal guanylate. Under the TLC conditions used here, U3 must represent a product with a greater polarity than any of the standards or other products. This could be the result of an extra phosphoryl group on the cap structure and would be consistent with the mutant being unable to catalyze the mRNA triphosphatase reaction.
FIG. 5.
TLC analysis shows altered mRNA cap structures for cet1- and ccm1-null mutants. TAP (A)- and P1 (B)-digested mRNA was purified from C. albicans BWP17, CQF152 (Δcet1::ARG4/Δcet1::URA), CQF248 (Δcet1::ARG4/Δcet1::URA3 with CET1 restored), CQF171 (Δccm1::ARG4/Δccm1::URA3) CQF250 (Δccm1::ARG4/Δccm1::URA3 with CCM1 restored), CQF142 (Δcet1::ARG4/CET1), andCQF154 (Δccm1::ARG4/CCM1), all labeled with NaB[3H]4. The location of the standards separated on the TLC plate were visualized by UV light and are indicated on the right side of the image.
Restoration of null mutants.
As a formal proof that the phenotypes associated with loss of cet1 and ccm1 in the null mutants are due to the loss of the respective gene functions rather than disruption of the genetic loci, we inserted wild-type copies of the genes back into the genome at the Δhis1::hisG locus (42). The wild-type genes and upstream sequence were PCR amplified from C. albicans BWP17 genomic DNA with primers for CET1 (CET1-restore 5′ and CET1-restore 3′) and primers for CCM1 (CCM1-restore 5′ and CCM1-restore 3′). After cloning the PCR fragments into the SalI site of pGEM-HIS (42), the resulting plasmids were linearized with NruI and used to transform the respective null mutants (CQF152 and CQF171). The sequence homology between the genomic HIS1 flanking sequence and pGEM-HIS1 vector provides a target sequence for recombination and insertion of the genes into the genome. Colonies with CET1 or CCM1 insertions were identified by analytical PCR of Ura+ Arg+ His+ transformants, and the genomic insertions were confirmed by Southern blot analysis (Fig. 3). These strains with CET1 or CCM1 restored in trans in the genome were phenotypically similar to the wild-type BWP17 strain in growth studies (Fig. 4), susceptibilities to antifungal compounds (Table 3), and cap structures (Fig. 5).
DISCUSSION
Two features of fungal mRNA capping enzymes indicate they might be good targets for antifungal drug development. First, mRNA capping enzymes have distinct catalytic mechanisms and gene structures from their functional human counterparts (14, 26). Second, mRNA capping is essential in S. cerevisiae (32) and is widely reported to be critical for efficient translation in all eukaryotes (for reviews, see references 17, 20, and 30). All three genes encoding the enzymes involved in mRNA capping have been shown to be essential in S. cerevisiae (19, 34, 39). Although gene essentiality in S. cerevisiae is often consistent with essentiality for the homologous genes in Candida, there are many notable exceptions. Therefore, we wanted to validate mRNA capping as a good antifungal drug target by determining whether the genes encoding the capping enzymes are essential in C. albicans. The C. albicans mRNA capping enzymes have been characterized and the three genes encoding the mRNA capping enzymes in C. albicans (CET1, CGT1, and CCM1) have been identified (31, 44, 45), but null mutants have previously not been constructed for these genes. We were able to construct null deletion mutants of both CET1 and CCM1 in C. albicans by the methods of Enloe et al. (7) and Wilson et al. (42). This contrasts with a recent report by Pei et al. (26), who were unable to delete both alleles of CET1 in C. albicans by using the UAU1 one-step transformation method (7).
We initially used the one-step transformation method of Enloe et al. (7) and were able to generate null mutants of CET1 in C. albicans. However, the recovery of null mutants was originally very low (1 of 44 initial Arg+ Ura+ segregants), and this one cet1-null mutant isolated by the UAU1 mitotic recombination procedure was temperature sensitive (Fig. 4). These two observations raised the possibility that we might have generated a compensatory mutation during the mitotic recombination allowing us to isolate a null mutant for a gene that really is essential. To test this hypothesis, 16 additional Arg+ Ura+ segregants generated from the other two CET1 heterozygotes with UAU1 targeted insertions were screened. Four of these isolates were cet1-null mutants, and none of these four mutants showed the temperature-sensitive growth phenotype seen with the first cet1-null mutant (Fig. 4). These results suggest that the temperature-sensitive phenotype was not related to the loss of CET1 function but may be due to, for example, duplication of a recessive mutation adjacent to the insertionally deleted CET1 allele when this locus was converted to homozygosity in the mitotic recombination step.
As an additional verification that the CET1 gene is not essential, we were also able to generate cet1-null mutants by a two-step transformation method for disrupting both alleles (42). This method relies on sequential transformation of ARG4 and URA3 markers into the two CET1 alleles and led to the identification of 7 null mutants in the 9 Arg+ Ura+ transformants that were screened.
Null deletion mutants of CCM1, the methyltransferase-encoding gene, were constructed in C. albicans by using the two-step transformation method (Fig. 2). However, we were unable to generate null mutants at this locus by using the single-step transformation method. The high rate of success and relative ease in generating cet1- and ccm1-null mutants with the two-step transformation method versus the one-step transformation method highlights the importance of testing multiple approaches in determining whether genes are essential in C. albicans. The reason for the differences in the two methods remains unexplained, but it could be due to a physical constraint of the DNA at specific loci that inhibits the mitotic recombination event required to select homozygous deletions in the single transformation method.
De Backer et al. (5) showed, with their C. albicans Δcgt1/CGT1 heterozygous mutant, that the mutant was temperature sensitive and resistant to the translational inhibitor hygromycin B. We did not observe temperature sensitivity for our Δcgt1::ARG4/CGT1 heterozygous strain (Fig. 4). The MICs for hygromycin B were similar for this strain and the CGT1/CGT1 parent strain (Table 3). The C. albicans cet1- and ccm1-null mutants are more sensitive to hygromycin B than the heterozygotes and the strains with capping genes added back in trans (MICs = 16 μg/ml versus >128 μg/ml). In addition, the cet1-null mutant is more sensitive to the protein translation inhibitor cycloheximide. Hypersensitivity to a compound that inhibits protein translation would be an expected phenotype for a mutant strain that has altered levels of expression for a component of protein translation. The drug sensitivity data are not conclusive but are consistent with the theory that C. albicans CCM1 and CGT1 deletion mutants may have less-efficient protein translation.
The fact that cet1- and ccm1-null mutants can be generated in C. albicans raises multiple questions about the processing of mRNA and translation in this organism. Possible explanations for why CET1 and CCM1 are not essential in C. albicans include (i) redundant genes encoding the capping functions, (ii) an alternative capping pathway, (iii) the ability to survive with cap-independent translation, or (iv) the ability to effectively translate mRNA with a modified cap. Based on the results we show here, we favor the latter hypothesis.
There are many examples of important enzymes encoded by redundant genes in C. albicans, including genes encoding chitin synthase (21); the GSC1, GSL1, and GSL2 genes involved in β-1,3-glucan synthase (22), and protein mannosyltransferases (38). However, by using sequence analysis and Southern blots we have found no evidence of redundant genes for CET1 or CCM1 in the C. albicans genome. A search of the publicly available C. albicans genomic sequence (assembly 6, Stanford Genome Technology Center, http://www-sequence.stanford.edu/group/candida/index.html) did not identify any other locus encoding an open reading frame with significant amino acid sequence homology to CET1p or CCM1p. The Southern blot analysis showed only one band hybridized to CET1 or CCM1 fragments in the parent strain and this was lost in the cet1- or ccm1-null mutants (Fig. 3). If mRNA triphosphatase or methyltransferase capping activities are essential in C. albicans, there are more likely to be nonhomologous genes encoding the functions or an alternative capping pathway. However, our data indicate that modified guanylated caps are formed in the null mutants and are sufficient for protein translation in C. albicans.
We were unable to delete, under the growth conditions used to select for deletion mutants, both alleles of CGT1 in C. albicans. This suggests that the mRNA-guanylyltransferase modification is essential even if the triphosphatase and methyltransferase activities are not. However, it is possible that the essential nature of CGT1 is due to an unknown function of this gene.
Analysis of the mRNA cap structure in the C. albicans cet1- and ccm1-null mutants indicates guanylated cap structures that are distinct from the mRNA cap structure in the wild-type cells (Fig. 5). The mRNA cap structure in the BWP17 cells migrates as expected with the m7GpppN standard. In the ccm1-null mutant, it is quite clear that the mRNA cap structures are not methylated (GpppN). Based on the pattern of labeled nucleotides seen in the TLC analyses, our hypothesis is that, in the cet1-null mutants, mRNA is guanylated without the third phosphate being removed and that it is not methylated (GppppN). Tetraphosphate linkages have been previously identified in cap structures formed by virus particles lacking RNA triphosphatase activity (4), indicating that viral guanylyltransferase activity is independent of RNA triphosphatase. If the cap structure in the cet1-null mutant contains a tetraphosphate linkage, the C. albicans guanylyltransferase activity must be independent of RNA triphosphatase. This conclusion is supported by a recent publication from Takagi et al. (37), who cite a fundamental divergence in mRNA capping machinery between S. cerevisiae and C. albicans. They report that activity of S. cerevisiae mRNA guanylyltransferase (CGT1p) depends on interaction with the RNA triphosphatase subunit (CET1p). However, their results showed that C. albicans CGT1p does not require interaction with CET1p for guanylyltransferase activity, a conclusion reinforced by our demonstration of guanylated mRNA caps in the cet1-null mutant. Further studies will be needed to determine the exact structures of the cap nucleotides in the null mutants, but they are definitely different than the cap structures in the parent strain.
The question remains as to whether any mRNA cap is essential for translation in C. albicans. The 5′-cap on mRNA binds the eukaryotic initiation factor eIF4e and is involved in the recruitment of the ribosome to form an initiation complex involving both the 5′-cap and the 3′-polyadenylated tail (41). Whereas most eukaryotic translation is thought to involve mRNA with 5′ caps (29), there are many examples of cap-independent translation in eukaryotes. Viral mRNAs have been reported to have 3′-end sequences that mimic the cap function (41), and cap-independent translation initiation that is facilitated by internal ribosome entry sites has been widely studied (47). Although eIF4e has been reported to be essential in S. cerevisiae (40), a recent report clarifies that eIF4e is not essential in starved cells (25). The proposal that mRNA capping is only essential in rapidly growing cells is supported by the growth studies with our C. albicans capping gene deletion mutants. Growth defects for the cet1- and ccm1-null mutants compared to parent strain are much more pronounced at 37°C than at 30°C (Fig. 4). The slower-growth phenotype for the cet1- and ccm1-null mutants could be due to a variety of reasons but, again, it is a phenotype expected for mutants with alterations in protein translation efficiency. The survival of the cet1 and ccm1-null mutants demonstrates that the complete m7GpppN mRNA cap structure is not absolutely required in C. albicans and the mRNA cap probably contributes to efficiency but is not essential.
Acknowledgments
We thank Aaron P. Mitchell for generous gifts of the plasmids and strains used in the C. albicans knockouts, as well as technical advice with these procedures. We also thank Ann Berger, Marek Nagiec, and Che-Shen Tomich for critical reading of the manuscript.
REFERENCES
- 1.Aivasashvilli, V. A., and R. S. Beabealashvilli. 1983. Sequence-specific inhibition of RNA elongation by actinomycin D. FEBS Lett. 160:124-128. [DOI] [PubMed] [Google Scholar]
- 2.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 3.Burke, D., D. Dawson, and T. Stearns. 2000. Yeast DNA isolations, p. 109-114. In Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Chen, D., C. L. Luongo, M. L. Nibert, and J. T. Patton. 1999. Rotavirus open cores catalyze 5′ capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology 265:120-130. [DOI] [PubMed] [Google Scholar]
- 5.De Backer, M. D., R. A. de Hoog, G. Froyen, F. C. Odds, F. Simons, R. Contreras, and W. H. M. Luyten. 2000. Single allele knockout of Candida albicans CGT1 leads to unexpected resistance to hygromycin B and elevated temperature. Microbiology 146:353-365. [DOI] [PubMed] [Google Scholar]
- 6.Elewski, B. E. 1993. Mechanisms of action of systemic antifungal agents. J. Am. Acad. Dermatol. 28:S28-S34. [DOI] [PubMed] [Google Scholar]
- 7.Enloe, B. M., A. Diamond, and A. P. Mitchell. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eustice, D. C., and J. M. Wilhelm. 1984. Mechanisms of action of aminoglycoside antibiotics in eucaryotic protein synthesis. Antimicrob. Agents Chemother. 26:53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fresco, L., and S. Buratowski. 1996. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances but is not required for splicing. RNA 2:584-596. [PMC free article] [PubMed] [Google Scholar]
- 11.Fujimoto, M., A. Kuninaka, and H. Yoshino. 1974. Substrate specificity of nuclease P1. Agric. Biol. Chem. 38:1555-1561. [Google Scholar]
- 12.Furuichi, Y., and A. J. Shatkin. 1989. Characterization of cap structures. Methods Enzymol. 180:164-176. [DOI] [PubMed] [Google Scholar]
- 13.Georgopapadakou, N. H., and T. J. Walsh. 1994. Human mycoses: drugs and targets for emerging pathogens. Science 264:371-373. [DOI] [PubMed] [Google Scholar]
- 14.Ho, C. K., Y. Pei, and S. Shuman. 1998. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J. Biol. Chem. 273:34151-34156. [DOI] [PubMed] [Google Scholar]
- 15.Kalpaxis, D. L., D. A. Theocharis, and C. Coutsogeorgopoulos. 1986. Kinetic studies on ribosomal peptidyltransferase: the behavior of the inhibitor blasticidin S. Eur. J. Biochem. 154:267-271. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita, T., N. Tanaka, and H. Umezawa. 1970. Binding of blasticidin S to ribosomes. J. Antibiot. 6:288-290. [DOI] [PubMed] [Google Scholar]
- 17.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 18.Lin, R.-J., D.-H. Kim, D. Castanotto, S. Westaway, and J. J. Rossi. 1996. RNA preparations from yeast cells, p. 43-50. In P. A. Krieg (ed.), A laboratory guide to RNA: isolation, analysis, and synthesis. Wiley-Liss, Inc., New York, N.Y.
- 19.Mao, X., B. Schwer, and S. Shuman. 1995. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 15:4167-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy, J. E. G. 1998. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 62:1492-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mio, T., T. Yabe, M. Sudoh, Y. Satoh, T. Nakajima, and M. Arisawa. 1996. Role of three chitin synthase genes in the growth of Candida albicans. J. Bacteriol. 178:2416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mio, T., M. Adachi-Shimizu, Y. Tachibana, H. Tabuchi, S. Inoue, T. Yabe, T. Yamada-Okabe, M. Arisawa, T. Watanabe, and H. Yamada-Okabe. 1997. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in β-1,3-glucan synthase. J. Bacteriol. 179:4096-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2000. Reference method for broth dilution antifungal susceptibility testing for yeasts. Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Obrig, T. G., W. J. Culp, W. L. McKeehan, and B. Hardesty. 1971. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 246:174-181. [PubMed] [Google Scholar]
- 25.Paz, I., and M. Choder. 2001. Eukaryotic translation initiation factor 4E-dependent translation is not essential for survival of starved yeast cells. J. Bacteriol. 183:4477-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei, Y., K. Lehman, L. Tian, and S. Shuman. 2000. Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 28:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pei, Y., B. Schwer, J. Saiz, J., R. P. Fisher, and S. Shuman. 2001. RNA triphosphatase is essential in Schizosaccharomyces pombe and Candida albicans. BioMed Central 1:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaller, M. A., and A. L. Barry. 1994. Evaluation of a novel colorimetric broth microdilution method for antifungal susceptibility testing of yeast isolates. J. Clin. Microbiol. 32:1992-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinkstaff, J. K., S. A. Chappell, V. P. Mauro, G. M. Edelman, and L. A. Krushel. 2001. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl. Acad. Sci. USA 98:2770-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachs, A. B., P. Sarnow, M. W. Heintze. 1997. Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell 89:831-838. [DOI] [PubMed] [Google Scholar]
- 31.Saha, N., B. Schwer, and S. Shuman. 1999. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyl transferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274:16553-16562. [DOI] [PubMed] [Google Scholar]
- 32.Schwer, B., K. Lehman, N. Saha, and S. Shuman. 2001. Characterization of the mRNA capping apparatus of Candida albicans. J. Biol. Chem. 276:1857-1864. [DOI] [PubMed] [Google Scholar]
- 33.Shah-Mahoney, N., T. Hampton, R. Vidaver, and D. Ratner. 1997. Blocking the ends of transforming DNA enhances gene targeting in Dictyostelium. Gene 203:33-41. [DOI] [PubMed] [Google Scholar]
- 34.Shibagaki, Y., N. Itoh, H. Yamada, S. Nagata, and K. Mizumoto. 1992. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J. Biol. Chem. 267:9521-9528. [PubMed] [Google Scholar]
- 35.Shinshi, H., M. Miwa, T. Sugimara, K. Shimotohno, and K. Miura. 1976. A novel phosphodiesterase from cultured tobacco cells. Biochemistry 15:2185-2190. [DOI] [PubMed] [Google Scholar]
- 36.Southern, E. M. 1975. Detection of specific sequences among DNA fragments by gel electrophoresis. J. Mol. Biol. 113:237-251. [DOI] [PubMed] [Google Scholar]
- 37.Takagi, T., E.-J. Cho, R. T. K. Janoo, V. Polodny, Y. Takase, M.-C. Keogh, S.-A. Woo, L. D. Fresco-Cohen, C. S. Hoffman, and S. Buratowski. 2002. Divergent subunit interactions among fungal mRNA 5′-capping machineries. Eukaryot. Cell 1:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timpel, C., S. Zink, S. Strahl-Bolsinger, K. Schröppel, and J. Ernst. 2000. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 182:3063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukamoto, T., Y. Shibagaki, S. Imajohmi, T. Murakoshi, M. Suzuki, A. Nakamura, H. Gotoh, and K. Mizumoto. 1997. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem. Biophys. Res. Commun. 239:116-122. [DOI] [PubMed] [Google Scholar]
- 40.Vasilescu, S., M. Ptushkina, B. Linz, P. P. Müller, and J. E. G. McCarthy. 1996. Mutants of eukaryotic initiation factor eIF-4E with altered mRNA cap-binding specificity reprogram mRNA selection by ribosomes in Saccharomyces cerevisiae. J. Biol. Chem. 271:7030-7037. [DOI] [PubMed] [Google Scholar]
- 41.Wang, S., K. S. Browning, and W. A. Miller. 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 16:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 44.Yamada-Okabe, T., O. Shimmi, R. Doi, K. Mizumoto, M. Arisawa, and H. Yamada-Okabe. 1996. Isolation of the mRNA-capping enzyme and ferric-reductase-related genes from Candida albicans. Microbiology 142:2515-2523. [DOI] [PubMed] [Google Scholar]
- 45.Yamada-Okabe, T., T. Mio, M. Matsui, Y. Kashima, M. Arisawa, and H. Yamada-Okabe. 1998. Isolation and characterization of the Candida albicans gene for mRNA 5′-triphosphatase: association of mRNA 5′-triphosphatase and mRNA 5′-guanylyl transferase activities is essential for the function of mRNA 5′-capping enzyme in vivo. FEBS Lett. 435:49-54. [DOI] [PubMed] [Google Scholar]
- 46.Yesland, K., and W. A. Fonzi. 2000. Allele-specific gene targeting in Candida albicans results from heterology between alleles. Microbiology 146:2097-2104. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, W., G. M. Edelman, and V. P. Mauro. 2001. Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl. Acad. Sci. USA 98:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]