Abstract
In Saccharomyces cerevisiae, the actin cytoskeleton is depolarized by NaCl stress. In this study, the response was maximal after 30 min, and then actin patches repolarized. Rvs161p was required for actin repolarization because the rvs161Δ mutant did not repolarize actin patches after growth in a salt medium. Mutations suppressing the rvs161Δ-related salt sensitivity all occurred in genes required for sphingolipid biosynthesis: FEN1, SUR4, SUR2, SUR1, and IPT1. These suppressors also suppressed act1-1-related salt sensitivity and the defect in actin repolarization of the rvs161Δ mutant, providing a link between sphingolipids and actin polarization. Indeed, deletion of the suppressor genes suppressed the rvs161Δ defect in actin repolarization in two ways: either actin was not depolarized at the wild-type level in a set of suppressor mutants, or actin was repolarized in the absence of Rvs161p in the other suppressor mutants. Rvs161p was localized as cortical patches that concentrated at polarization sites, i.e., bud emergence and septa, and was found to be associated with lipid rafts. An important link between sphingolipids and actin polarization is that Rvs161p was required for actin repolarization and was found to be located in lipid rafts.
Membrane lipid microdomains called rafts are formed by the lateral association of sphingolipids and cholesterol (ergosterol in yeasts). In mammalian cells, rafts are involved in endocytosis and exocytosis and more generally in lipid-mediated membrane trafficking (reviewed in reference 22), in the immunological response (23), and in establishing platforms for vesicle-linked actin polymerization, visualized as comets (36).
Rafts recruit specific sets of membrane proteins and exclude others. They harbor signaling molecules (phosphatidylinositol bisphosphate), kinases (Src) involved in signal transduction (29, 40), SNAREs (10, 26), and glycophosphatidylinositol-anchored proteins (1, 22). This specific lipid-protein association forms insoluble complexes in cold, nonionic detergents such as Triton X-100 (9) that can be separated by floatation in density gradients in the form of detergent-resistant membranes (DRMs).
In the yeast Saccharomyces cerevisiae, rafts have been defined biochemically as DRMs and proven to be important for protein sorting through the endoplasmic reticulum and Golgi apparatus (1, 2). Importantly, sphingolipid biosynthetic intermediates are also signaling molecules (Fig. 1). In particular, the sphingoid base phytosphingosine is a signaling molecule required for endocytosis (21, 45) and ubiquitin-dependent proteolysis following heat stress (13).
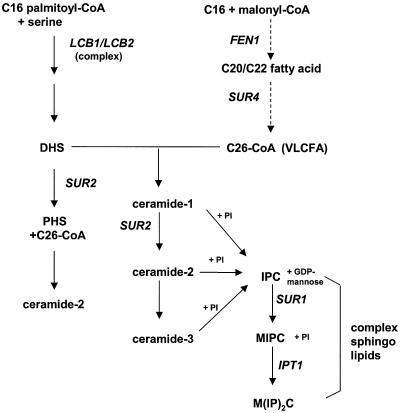
FIG. 1.
Sphingolipid biosynthetic pathway in S. cerevisiae, adapted from Dickson and Lester (19) and Schneiter (37). Genes are shown in capital italic letters. LCB1 encodes a subunit of the serine palmitoyl transferase, FEN1 and SUR4 encode the elongases and are partially redundant, SUR2 encodes a hydroxylase, SUR1 encodes a mannosyl transferase, and IPT1 encodes an inositol phosphotransferase. Abbreviations: CoA, coenzyme A; VLCFA, very long chain fatty acid; PI, phosphatidylinositol; DHS and PHS, sphingoid bases dihydrosphingosine and phytosphingosine, respectively. IPC, inositolphosphorylceramide; MIPC, mannosyl-inositolphosphorylceramide; and M(IP)2C, mannosyl-diinositolphosphorylceramide.
The yeast RVS161/END6 gene is involved in cell polarity (20), actin cytoskeleton polarization (41), endocytosis (30), and secretory vesicle trafficking (6). rvs161 mutant cells die during stationary phase and are sensitive to NaCl (3, 16). rvs161Δ, together with a specific set of actin alleles, leads to synthetic lethality (5). Furthermore, the end6-1 allele of RVS161 displays nonallelic noncomplementation with the act1-1 allele, while rvs161Δ complements act1-1 (5, 30). Therefore, the primary defect of the rvs161Δ mutant lies in actin cytoskeleton organization.
rvs161-related salt sensitivity and cell death during starvation are suppressed by mutations in the SUR1, SUR2, SUR4, and FEN1 genes (18, 34). Moreover, mutations in these suppressor genes lead to abnormal lipid composition (18), and they were recently shown to encode enzymes required for sphingolipid biosynthesis (19, 37). Therefore, we designed experiments to unravel the relationships between actin cytoskeleton organization, Rvs161p, and sphingolipids.
In this paper, we show that both Rvs161p and sphingolipids have major roles in depolarization and repolarization of actin following salt stress. We also establish that Rvs161p is associated with lipid rafts, suggesting for the first time a link between rafts and actin in S. cerevisiae. Finally, we show that Wsc1p is a sensor for actin depolarization after salt stress.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The S. cerevisiae strains used in this study are listed in Table 1. Complete medium was YPD (10 g of yeast extract, 20 g of Bactopeptone, and 20 g of glucose per liter) solidified with 20 g of agar per liter when required.
TABLE 1.
S. cevevisiae strains used in this studya
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| BY4742 | α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | EUROSCARF |
| Y13489 | α rvs161Δ leu2Δ0 lys2Δ0 ura3Δ0 his3Δ1 | EUROSCARF |
| LG973-1B | a rvs161Δ his3Δ1 leu2Δ0 ura3Δ0 | This study |
| LG742-17A | a act1-1 his4 | 5 |
| LG1041-6B | α rvs161Δ fen1Δ ura3Δ0 his3Δ1 leu2Δ0 | This study |
| LG1040-6A | a rvs161Δ sur4Δ ura3Δ0 his3Δ1 leu2Δ0 | This study |
| LG1043-6B | a rvs161Δ sur2Δ ura3Δ0 his3Δ1 leu2Δ0 | This study |
| LG1034-20C | a rvs161Δ sur1Δ ura3Δ0 his3Δ1 leu2Δ0 | This study |
| LG1035-6B | a rvs161Δ ipt1Δ ura3Δ0 his3Δ1 leu2Δ0 | This study |
| LG1039-2B | α act1-1 fen1Δ leu2Δ0 lys2Δ0 his-? | This study |
| LG1045-8C | a act1-1 sur4Δ leu2Δ0 ura3Δ0 | This study |
| LG1044-9A | a act1-1 sur2Δ his-? | This study |
| LG1036-10D | α act1-1 sur1Δ his leu2Δ0 lys2Δ0 ura3Δ0 | This study |
| LG1037-3C | α act1-1 ipt1Δ his leu2Δ0 | This study |
| RH1800 | α leu2 his4 ura3 bar1 | H. Riezman |
| RH3804 | α lcb1-100 trp1 leu2 ura3 lys2 | H. Riezman |
All strains are in the S288C background or are congenic except the lcb1-100 strain. All the EUROSCARF disruptant strains are Kanr and are derived from BY4742 and therefore bear the same auxotrophies. The disruptions, noted Δ, are kanMX4. The segregant LG973 is from Y13489 × LG811-8A (5). The fen1, sur4, sur2, sur1, and ipt1 mutants are strains from EUROSCARF. The double mutants LG1041-6B, LG1040-6A, LG1034-20C, LG1043-6B, and LG1035-6B come from a cross between LG973-1B and the EUROSCARF strains. Backcrosses were made to check for the segregation of the Kanr markers. The double mutants containing act1-1 are from a cross between LG742-17A and the EUROSCARF strains. The presence of the act1-1 allele was attested by complementation tests with plasmids containing the wild-type FEN1, SUR4, and SUR1 genes. The wsc1, wsc2, wsc3, wsc4, and mid2 disruptants are from EUROSCARF.
Minimal medium was SD (6.7 g of yeast nitrogen base [Difco] per liter) containing 20 g of glucose as the carbon source. SD-casa was SD supplemented with Casamino Acids (0.2%). To check for sodium chloride sensitivity, this medium contained 7% NaCl. To assay for the ability to grow under different conditions, droplets of cells resuspended in water containing 5 × 104 cells and serial dilutions were laid on different media, and growth was checked after 48 h.
For microscopic observations of the green fluorescent protein (GFP) fusion proteins, yeast cells were grown exponentially (<107 cells/ml) in SD-casa supplemented with tryptophan when required. For NaCl stress-induced experiments, cells were grown in liquid YPD to 3 × 106 to 5 × 106 cells/ml. Cells were collected by centrifugation and resuspended for different times in 10 ml of YPD or YPD containing 3.4% NaCl, an NaCl concentration that is sublethal for the rvs mutants.
Matings, sporulations, and dissections were carried out according to standard procedures (38). For each cross, the segregants were dissected from at least 20 asci and allowed to grow on YPD medium at 25°C. Phenotypes were assessed by replica-plating the spore colonies on different media.
DNA manipulations.
Restriction enzymes and other DNA modification enzymes were purchased from Promega and used according to the specifications of the manufacturer. Amplifications of DNA fragments were done by PCR on a DNA Thermal Cycler (Perkin Elmer Cetus). Custom oligonucleotides were provided by Genset. Transformation of Escherichia coli was carried out by the CaCl2 method (31). Yeast cells were transformed by the one-step method (11).
Plasmid construction.
The RVS161-GFP gene was obtained in two steps. First, the GFP gene was directly cloned in the pFL44 plasmid (4) after digestion of pGFP (Clontech) and pFL44 by HindIII and EcoRI. The resulting plasmid was called pFL44-GFP. A 1,281-bp PCR product containing the promoter and the open reading frame (except the stop codon) of RVS161 was obtained by using primers R161Bam (5′-GGGATCCGTAAGCAAGGTACGGTTA-3′ and R161Kpn 5′-GGGGTACCTTTATCCCGAGCGCACA-3′). R161Bam introduced a BamHI site at −470 of the RVS161 promoter, and R161Kpn introduced a KpnI site in place of the stop codon of RVS161. The PCR product was cloned in pFL44-GFP with BamHI and KnpI. The resultant RVS161-GFP fusion construct was not able to complement the rvs161Δ mutant (data not shown).
The PMA1-GFP fusion gene was obtained directly by homologous recombination in S. cerevisiae. The 400 nucleotides upstream of the ATG and the entire coding sequence without the stop codon of PMA1 were amplified by PCR with primers PMA1up (ACTCACTATAGGGCGAATTGTGACCGGTGACGAAACGTGGTCGATGGTGG) and PMA1dw (AGCCCGGGGGATCCACTAGTGGTTTCCTTTTCGTGTTGAGTAGAGACTCT). The PCR product was used to transform S. cerevisiae with a monocopy vector derived from pRS316 (39) containing the yeast GFP3 gene (15).
Visualization of polymerized actin.
Alexa-phalloidin staining of actin was carried out by a modification of the method of Kaiser et al. (25). Cells grown in liquid culture under the described conditions were fixed by addition of formaldehyde to a final concentration of 3.7%. After 30 min, cells were collected by centrifugation, washed twice in phosphate-buffered saline (PBS) (8 g of NaCl, 0.2 g of KCl, 1.14 g of Na2HPO4, and 0.2 g of KH2PO4 per liter, adjusted to pH 7.3), resuspended in 50 μl of PBS containing 0.8 U of Alexa-Fluor 594-phalloidin (Molecular Probes), and incubated at 4°C overnight. Cells were washed three times with PBS and resuspended in 20 μl of a mounting medium containing 1 mg of p-phenylenediamine (Sigma) per ml. The actin cytoskeleton organization was then observed by fluorescence microscopy.
Microscopic imaging.
Epifluorescence microscopy was carried out with a Leica DMRXA microscope fitted with a 100× immersion objective (Leica PL APO) and standard fluorescein isothiocyanate or tetramethyl rhodamine isothiocyanate filter sets. Images were captured with a cooled charge-coupled device camera (MicroMax; Princeton Instruments) controlled by Metamorph 3.5 software (Universal Imaging).
Classification of cells depending on actin polarization state.
Only cells with small buds were scored. Cells with actin patches concentrated in the small bud, with four or fewer patches in the mother cell and polarized actin cables, were classified as polarized cells. Cells with most actin patches in the bud and more than four patches in the mother cell were classified as partially depolarized cells. Cells with more actin patches in the mother cell than in the small bud were classified as totally depolarized cells. The confidence in the counts, which were done twice, was within ±2%.
Membrane and DRM association.
Membrane and DRM association of Rvs161p were done essentially as described previously (2). Briefly, cells (≈5 optical density units at 600 nm) were labeled with [35S]methionine (0.5 mCi) for 30 min at the indicated temperatures and chased for 30 min more. Then cells were lysed, treated with Triton X-100 or buffer and subjected to Optiprep density gradient centrifugation. After centrifugation, a floating fraction and a soluble fraction were obtained. The floating fraction corresponds either to membrane, when cells were treated with buffer, or to detergent-resistant membrane, when cells were treated with Triton X-100. Rvs161p was then immunoprecipitated from the membrane, detergent-resistant membrane, and soluble fractions with specific antibodies and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Phosphorimager analysis.
RESULTS
Rvs161p is required for actin repolarization following salt stress.
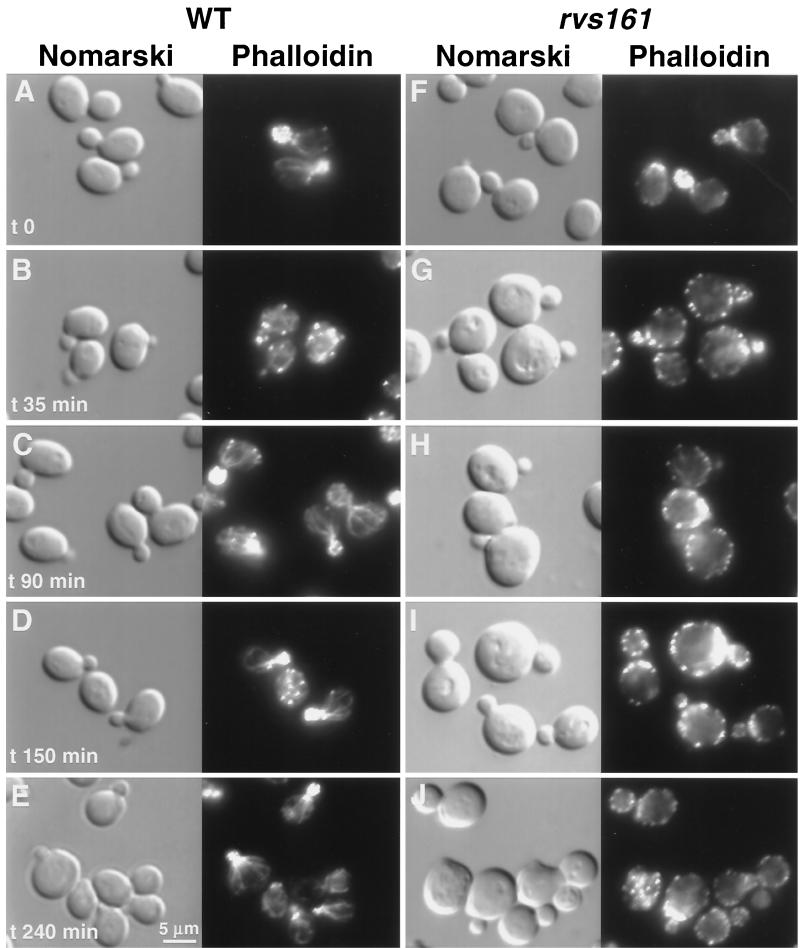
Actin patch polarization is slightly impaired in the rvs161 mutant, and this defect is dramatically enhanced by salt stress (41). Moreover, it has previously been shown that actin is transiently depolarized in wild-type cells after salt stress (12, 27). Further characterization of actin depolarization and repolarization by kinetic analysis following salt stress in the wild type revealed that after 35 min in NaCl medium, there were few cables, and when present, they appeared diffuse and the actin patches were depolarized. However, after 90 min, cables were present along the axis in the mother cell and the patches were fully repolarized (Fig. 2). In rvs161Δ mutant cells, actin patches were depolarized after 35 min in salt-containing medium, as in the wild-type strain. However, further incubation of the rvs161Δ mutant in salt-containing medium (90, 150, or 240 min) resulted in the appearance of some faint unoriented cables, while the actin patches never repolarized (Fig. 2). It should be noted that the actin patches were slightly depolarized in the rvs161 mutant cells grown in regular medium (32% of cells partially depolarized versus 0% in the wild type; see Fig. 5). This could be due to small stresses occurring during normal growth with which the rvs161Δ mutant was unable to cope.
FIG. 2.
Rvs161p is required for actin repolarization following salt stress. Wild-type (WT, A to E) and rvs161Δ mutant (F to J) strains were grown in YPD at 30°C, and at time zero, NaCl was added to a final concentration of 3.4%. Aliquots were removed at different times as indicated. Samples were fixed, stained with Alexa-phalloidin, and observed by fluorescence and Nomarski microscopy to visualize the actin cytoskeleton and whole cells, respectively.
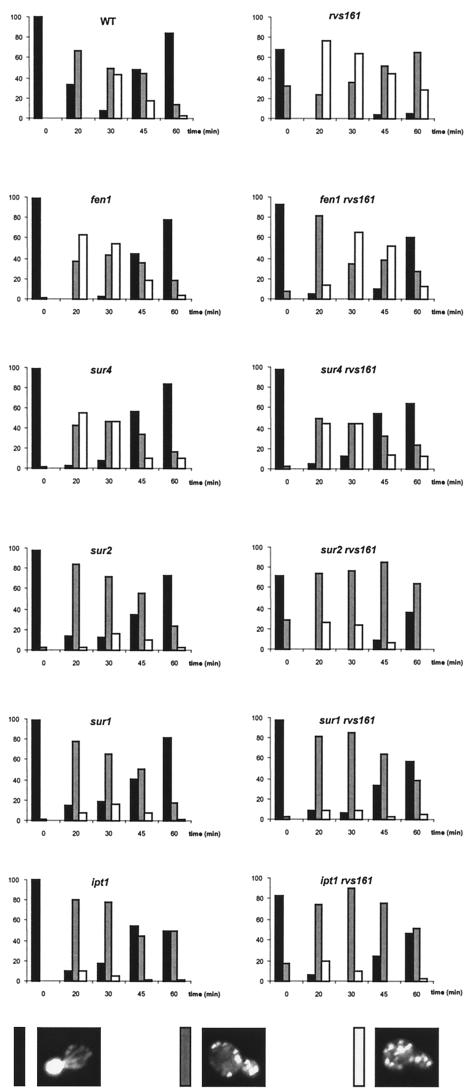
FIG. 5.
Sphingolipid mutants are impaired in actin depolarization and suppress the repolarization defect of the rvs161Δ mutant. The wild-type (WT) and the indicated mutant strains were grown as described for Fig. 2. Cells (≈100) were classified and quantified according to their polarization state as in Fig. 3.
Therefore, while the wild-type cells depolarized actin following salt stress and repolarized after a period of adaptation, the rvs161Δ mutant was able to depolarize actin in response to salt stress but was unable to repolarize afterwards. Therefore, the Rvs161p protein is required for actin repolarization following salt stress.
Search for a signal to salt stress.
Wsc1p is required for the actin patch depolarization response to heat stress (17). The timing of this heat stress response was similar to what we found above during salt stress (see also Fig. 5).
Wsc1p and Mid2p are thought to function as sensors following different stresses, such as heat and cell wall stress induced by Calcofluor or α-factor (32). Wsc1-4 proteins encompass a family of four homologous proteins with an N-terminal extracellular domain that is rich in cysteine, serine, and threonine, followed by a transmembrane domain and a C-terminal cytoplasmic tail (43). Mid2p is not homologous to Wscp but is like it structurally.
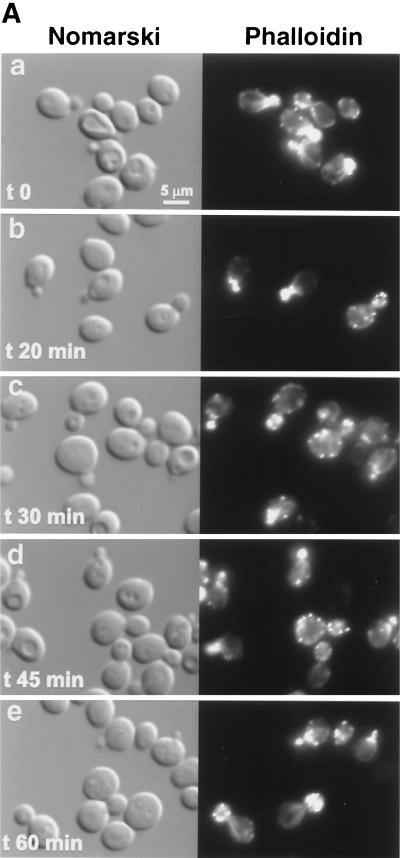
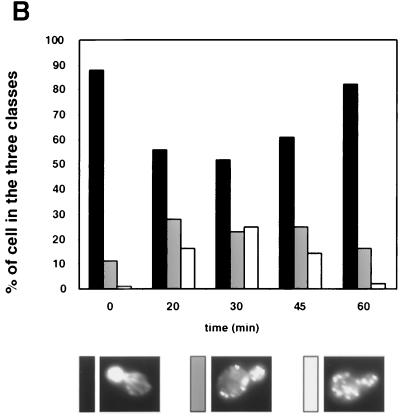
Therefore, we studied actin cytoskeleton polarization following salt stress in wsc1, wsc2, wsc3, wsc4, and mid2 mutant strains. While the wsc2, wsc3, wsc4, and mid2 mutants were able to depolarize actin patches comparably to the wild type (data not shown), the wsc1 mutant was severely impaired in depolarizing actin (Fig. 3). Indeed, 30 min after salt stress, the wsc1 mutant still presented 52% fully polarized cells (Fig. 3), compared to 8% in the wild type (Fig. 5), and only 25% fully depolarized cells, compared to 43% in the wild type. No more cells depolarized subsequently. Rather, repolarization occurred as in the wild type. Therefore, Wsc1p, a heat shock sensor for actin depolarization (17), also behaved as a sensor responding to salt stress.
FIG. 3.
wsc1 mutant impaired for actin cytoskeleton depolarization following salt stress. (A) The wsc1 strain was grown and treated as described for Fig. 2. (B) Cells (≈100) were classified and quantified according to their polarization state. Only cells with small buds were scored. Cells with actin patches concentrated in the small bud, with fewer than four patches in the mother cell, were classified as polarized cells (black bars). Cells with actin patches concentrated in the small bud but with more than four patches in the mother cell were classified as partially depolarized cells (grey bars). Cells with more actin patches in the mother cell than in the small bud were classified as totally depolarized cells (white bars).
Genetic relationships between mutations in sphingolipid metabolism genes and rvs161Δ or act1-1.
The salt sensitivity phenotype of the rvs161Δ mutant was suppressed by mutations in the FEN1, SUR4, SUR2, and SUR1 genes (Fig. 4A and 4B) (18, 34). Since all these suppressor genes encode enzymes required for sphingolipid biosynthesis (Fig. 1), we assayed whether a mutation in the IPT1 gene, also involved in this pathway (Fig. 1), behaved as a suppressor of rvs161Δ. Indeed, the ipt1 rvs161 double mutant was no longer sensitive to salt, strengthening the hypothesis of a connection between Rvs161p function and sphingolipid synthesis (Fig. 4A and B).
FIG. 4.
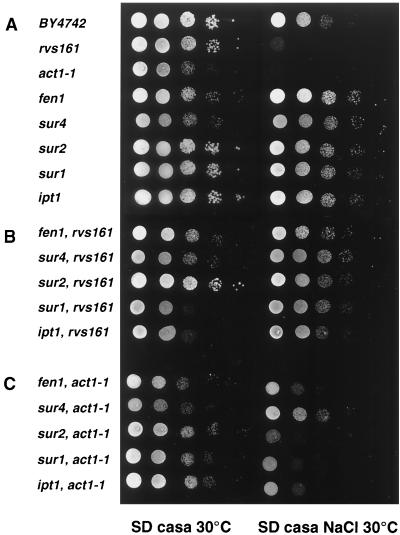
fen1, sur4, sur2, sur1, and ipt1 mutants suppress the salt sensitivity of the rvs161Δ and act1-1 mutants. The growth phenotypes were evaluated following a serial dilution drop test. The undiluted drop contained 5 × 104 cells. The complete genotype of the strains is given in Table 1.
According to growth on SD-casa at 30°C, the sphingolipid mutants fell into three classes: the fen1 rvs161 and sur4 rvs161Δ mutants grew at the same slow rate as the fen1 and sur4 mutants (class I); the sur2 rvs161Δ mutant grew at the rate of the sur2 mutant, close to the wild-type rate (class II); the sur1 and ipt1 mutants grew at the wild-type rate, but the double sur1 rvs161Δ and ipt1 rvs161Δ mutants grew much more slowly than the single mutants, showing a strong negative synergistic effect on growth between sur1 or ipt1 and the rvs161Δ mutation (class III) (Fig. 4A and B).
Since end6-1, an RVS161 allele, and act1-1 display nonallelic noncomplementation (30), Rvs161p and actin most probably belong to the same functional complex. This prompted us to test whether the fen1, sur4, sur1, sur2, and ipt1 mutations were also able to suppress the act1-1-related salt sensitivity phenotype. Indeed, all the sphingolipid biosynthesis mutations also partially suppressed the salt sensitivity of the act1-1 mutant (Fig. 4A and C). The resistance of the sur2 act1-1 mutant to salt was more obvious on YPD medium (not shown).
Therefore, these data further substantiate the involvement of Rvs161p and actin in the same function and strongly suggest that the latter involves sphingolipids.
Role of sphingolipid metabolism genes in salt stress and actin polarization.
The fact that rvs161 suppressors were also able to alleviate the act1-1-related growth defects prompted us to study actin depolarization-repolarization kinetics following salt stress in fen1, sur4, sur2, sur1, and ipt1 mutants and in the double mutants (Fig. 5). All the single fen1, sur4, sur2, sur1, and ipt1 mutants were able to lose actin cables, to depolarize actin patches, and subsequently to repolarize, although the timing and extent of depolarization varied among the different mutants. Indeed, the fen1 and sur4 mutants depolarized faster than the wild type (maximal depolarization occurred at 20 min, while it was 30 min for the wild type [Fig. 5]) and repolarized like the wild type. The sur2, sur1, and ipt1 mutants were not able to depolarize actin to the same degree as the wild type. Indeed, 30 min after salt stress, twice as many polarized cells were present in the mutants (16%, versus 8% in the wild type) and fewer fully depolarized cells were present (12%, versus 43% in the wild type), while a higher percentage of cells in an intermediate stage of depolarization (72%, versus 49% in the wild type) was observed. The sur2 and sur1 mutants repolarized to the same degree as the wild type, while ipt1 repolarization was lower and incomplete after 60 min (50%, versus 84% in the wild type).
Therefore, fen1 and sur4 mutants, carrying mutations in the genes encoding the first two enzymatic steps of the sphingolipid pathway, clearly depolarized faster than the wild-type cells, while the sur2, sur1, and ipt1 mutants, carrying mutations in the genes involved in subsequent biosynthetic steps, never fully depolarized. Roughly the same results were obtained when the double mutant strains between the rvs161Δ mutant and all these suppressor mutant genes were analyzed (Fig. 5). Thus, as expected, all the suppressive mutations were epistatic on the rvs161 disruption. The suppressors allowed the rvs161Δ mutant to grow on salt-containing medium as a consequence of suppression of the defect in actin repolarization. Therefore, sphingolipids and biosynthetic intermediates might be involved in actin depolarization and repolarization.
Rvs161p is linked to raft microdomains.
Complex sphingolipids are major components of lipid rafts. Since mutations in genes required for sphingolipid biosynthesis are suppressors of the rvs161Δ mutation, we could hypothesize that Rvs161p and other proteins required for actin depolarization and repolarization are localized in raft microdomains. In G1, Rvs161p-GFP was visualized as small cortical patches (Fig. 6) localized close to the plasma membrane, as revealed by the equatorial and top views. Later, while the patches remained cortical, Rvs161p-GFP was more concentrated at bud emergence (Fig. 6B). Finally, during cytokinesis, Rvs161p-GFP concentrated at the mother bud neck, where it could be visualized as a disk or a bar in most of the images (Fig. 6C). These results are in good agreement with previous data (7) showing that Rvs161p is found at polarization sites, in particular at the tip of the shmoos.
FIG. 6.
Rvs161p-GFP localizes as cortical patches and concentrates at polarization sites. Wild-type cells grown in SD-casa medium were observed by Nomarski and fluorescence microscopy to visualize whole cells and Rvs161p-GFP localization, respectively. The equatorial view represents the middle of the cell. For the top view, the focusing was done on the top of the cell. (A) Unbudded cell, (B) bud emergence cell, and (C) budded cell with the septum. The arrow on the Nomarski image indicates the septum between the mother cell and the bud.
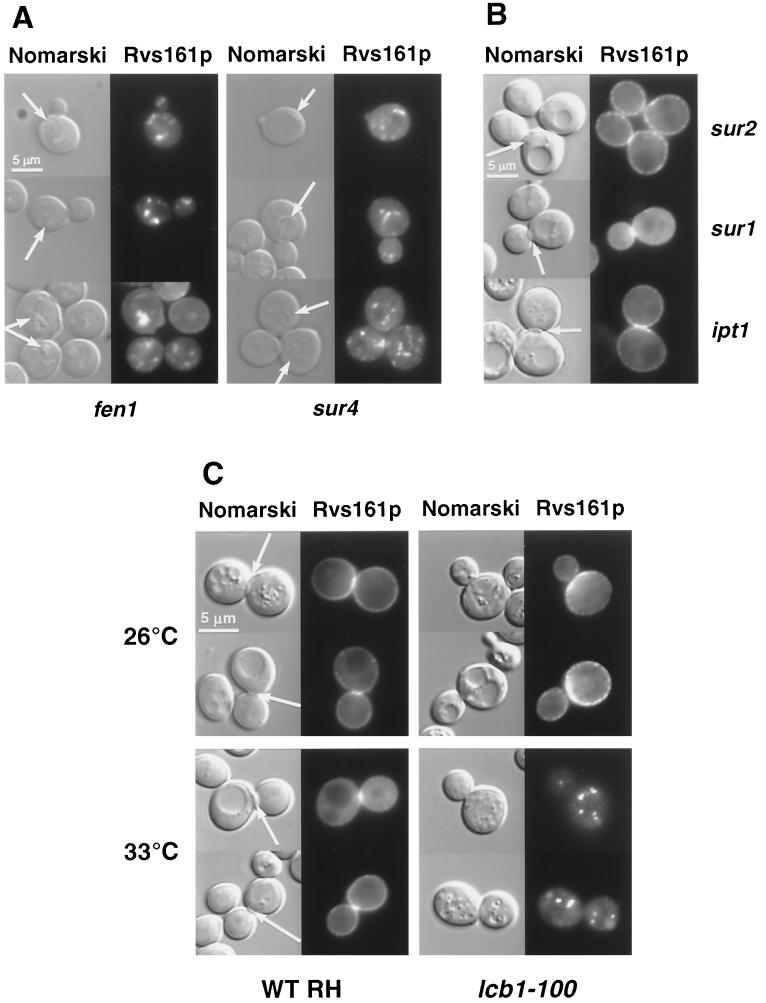
We then studied the localization of Rvs161p-GFP in the sphingolipid single fen1, sur4, sur2, sur1, ipt1, and lcb1-100 mutants; the last is a temperature-sensitive mutant impaired in the first step of sphingolipid biosynthesis (Fig. 1) that has been shown to be impaired in establishing DRMs (1). In sur2, sur1, and ipt1 deletants, the localization of Rvs161p was similar to what was seen in the wild-type strain (Fig. 7B), while Rvs161p-GFP was clearly mislocalized in the fen1 and sur4 deletants and in the lcb1-100 mutant. In the fen1 and sur4 mutants, cortical patches were still present but in a lower amount. No fluorescence could be detected in the septa, while undefined structures visible with Nomarski optics were stained (Fig. 7A). In the lcb1-100 mutant at the permissive temperature (26°C), cortical patches were present, but the septa were not stained, although the fluorescence increased in the mother bud neck area on the mother side. Importantly, when the lcb1-100 mutant was shifted to the restrictive temperature (33°C), the cortical staining disappeared, while nonmotile spots appeared in the cytoplasm (Fig. 7C).
FIG. 7.
Rvs161p-GFP localization is impaired in fen1, sur4, and lcb1-100 mutants. Different mutant cells grown in SD-casa medium were observed by Nomarski and fluorescence microscopy to visualize whole cells and Rvs161p-GFP localization, respectively. (A) In the fen1 and sur4 mutants, the arrow on the Nomarski images indicates the structures that contain Rvs161p-GFP. (B) In the sur2, sur1, and ipt1 mutants, the arrow on the Nomarski images indicates the septum between the mother cell and the bud. (C) Rvs161p-GFP localization in the lcb1-100 mutant and the corresponding wild-type (RH1800) cells grown at 26°C and shifted to 33°C for 90 min. The arrow on the Nomarski images indicates the septum between the mother cell and the bud.
Thus, Rvs161p-GFP was not properly localized in the fen1 and sur4 mutants, which are impaired in the elongation of the very long chain fatty acyl moiety of the sphingolipids, and in the lcb1-100 mutant, impaired in the synthesis of the sphingoid base. However, Rvs161p-GFP was found to be correctly localized in mutants carrying mutations affecting later steps of the sphingolipid pathway.
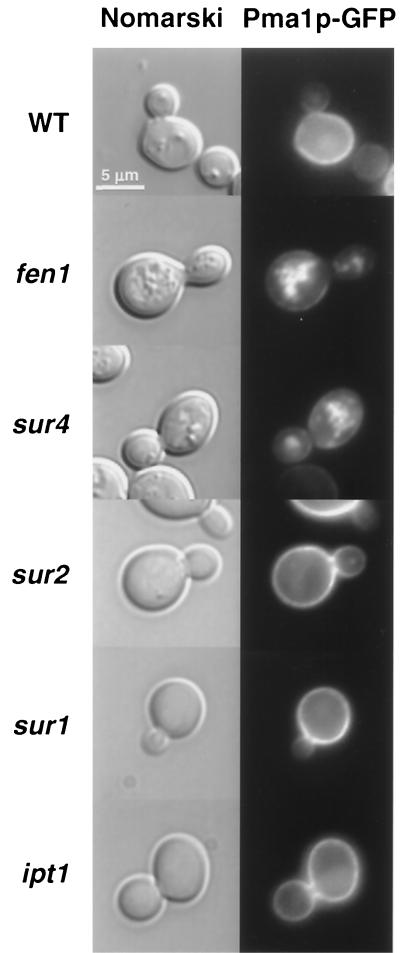
Next, we examined the localization of Pma1p, a protein that is associated with detergent-resistant membranes (DRMs) (1) and requires lipid rafts for proper sorting (2), in the same sphingolipid mutants. Like Rvs161p-GFP, Pma1p-GFP was correctly localized at the cell surface in the sur2, sur1, and ipt1 mutants, while it was delocalized in undefined structures in the fen1 and sur4 mutants (Fig. 8). Therefore, in this respect, Rvs161p behaved like Pma1p, a plasma membrane protein associated with DRMs, indicating that Rvs161p could be localized in lipid rafts.
FIG. 8.
Pma1p-GFP localization is affected in the fen1 and sur4 mutants. The indicated strains were grown in SD-casa medium and observed by Nomarski and fluorescence microscopy to visualize whole cells and Pma1p-GFP localization, respectively. WT, wild type.
This hypothesis was tested directly by searching for Rvs161p in DRMs. In wild-type cells, 63% of the protein was membrane associated (floating in the density gradient), and 40% of the protein in this membranous fraction was found in the DRM fraction (25% of total Rvs161p) (Fig. 9A). Thus, a substantial amount of Rvs161p can be recovered from membranes, further substantiating the cortical localization of Rvs161-GFP. In addition, at least some of these membranes were resistant to detergent treatment, as are the sphingolipids and ergosterol contained in raft microdomains. Consistently, in the lcb1-100 mutant, which is impaired in DRM assembly, the Rvs161p-DRM fraction dropped from 28% to 17% at the permissive temperature and from 20% to 12% at the restrictive temperature (Fig. 9B).
FIG. 9.
Rvs161p is associated with DRMs. (A) Wild-type cells were treated as described in Materials and Methods. Rvs161p was immunoprecipitated from the membrane (M), the detergent-resistant membrane (R), and the respective soluble (S) fractions with specific antibodies, followed by SDS-PAGE and Phosphorimager analysis. (B) The lcb1-100 mutant and the corresponding wild-type (WT, RH1800) strain were grown at the permissive (24°C) or the restrictive (37°C) temperature and treated as described in Materials and Methods. Rvs161p was immunoprecipitated from detergent-resistant membrane (R) and soluble (S) fractions with specific antibodies and analyzed by SDS-PAGE and Phosphorimager analysis.
In the sur4 and fen1 mutants, the Rvs161 protein was still found in DRMs at the same level as in the wild type (not shown), although Rvs161p was mislocalized in these mutants (Fig. 7A). Therefore, we hypothesize that in these mutants, Pma1p or Rvs161p accumulates in DRMs in which the sphingolipids are different from those in the wild type, such that they are improper for the protein to be routed any further. Taken together, these data show that Rvs161p is associated with lipid rafts.
DISCUSSION
We now confirm and extend previous observations on actin depolarization under salt stress (12, 27). Following the shift to NaCl-containing medium, actin patch delocalization was maximal after 30 min, and full repolarization of actin patches occurred between 60 and 90 min. These rearrangements occurred with the same time course as those following heat stress (17). Therefore, both salt and heat stress lead to the same physiological response, actin patch delocalization.
Following salt stress, the rvs161 mutant was able to depolarize actin patches even faster than the wild type, but the actin patches did not repolarize later. The rvs161 mutant responded to salt stress but was unable to recover from it. Therefore, Rvs161p is required for actin repolarization following salt stress.
From these data, we propose that rvs161Δ cells die when they are grown in NaCl-containing medium because the Rvs161 protein is required for repolarization and a polarized actin cytoskeleton is required for cell survival. Also, this could explain why the rvs161Δ mutant dies following starvation, since the actin cytoskeleton is depolarized during stationary phase (3) and repolarizes when cells enter a new cycle. Indeed, Rvs161p could be required to repolarize actin whatever the primary signal for actin depolarization. The fact that exponentially growing rvs161Δ mutant cells display a partially depolarized actin cytoskeleton further substantiates this model. Likely, actin patches are transiently depolarized during normal growth following small stresses occurring during the culture. This phenomenon is only visible in a strain that is unable to repolarize actin, such as the rvs161Δ mutant.
Also, it is interesting that the rvs161 mpk1/slt2 double mutation is lethal (6). Since Mpk1p is required for actin repolarization following heat stress (17), both Rvs161p and Mpk1p could be required for actin repolarization following a wide range of stresses.
Mutations in the suppressor genes have an effect on actin depolarization and actin repolarization: mutations in class I genes (fen1 and sur4) cause faster depolarization, while those in class II (sur2) and III genes (sur1 and ipt1) cause lesser extents of depolarization. The same suppressors of the rvs161Δ mutation suppressed the inability of the act1-1 mutant to sustain salt stress and were all able to suppress the defect of the rvs161Δ mutant in actin repolarization. This reinforces the notion that the primary function of Rvs161p lies in actin cytoskeleton repolarization. Because the Fen1p, Sur4p, Sur2p, Sur1p, and Ipt1p proteins are not likely to act by themselves in actin polarization or depolarization, sphingolipids or biosynthetic intermediates are involved in actin polarization. Indeed, sphingolipids or biosynthetic intermediates are missing or are present in greater amounts in the different suppressor mutants (14, 45) (Fig. 1). Either biosynthetic intermediates could act as signaling molecules, or complex sphingolipids could act as structural components of lipid rafts. These two mechanisms might not be exclusive.
The intermediate sphingoid base phytosphingosine has been described as a signaling molecule for resistance to heat stress (13) and protein phosphorylation required for endocytosis (21). The fen1 mutant is known to accumulate phytosphingosine (14), and phytosphingosine signals actin repolarization via the Pkh1/Pkh2-Pkc1-Mpk1 pathway (17, 21). In the class I suppressor mutants (fen1 or sur4), the excess of phytosphingosine could exacerbate the pathway in such a way that Rvs161p is no longer required for actin repolarization.
Complex sphingolipids together with cholesterol (ergosterol in yeasts) are the main components of the raft membrane microdomains. In S. cerevisiae, the three main complex sphingolipids, inositolphosphoceramide, mannosyl-inositolphosphorylceramide, and mannosyl-diinositolphosphorylceramide, together with variations in the chain lengths of the sphingoid moiety and the very long chain fatty acid and different levels in hydroxylation, make up a variety of more than 30 different molecular species (37) that potentially lead to a variety of different rafts (35). The proteins associated with specific rafts could be present in lower or higher amounts in the suppressor mutants. In the sur2, sur1, and ipt1 mutants, the proteins signaling salt stress or proteins essential for actin depolarization could be missing in their proper environment. We found that the sensor Wsc1p responds to salt stress for actin depolarization. Moreover, Wsc1p is an integral membrane protein that is likely associated with lipid rafts because it is resistant to detergent extraction (28). Wsc1p could be mislocalized in sur2, sur1, and ipt1 mutants, explaining why these mutants do not completely signal salt stress to actin patch depolarization. Here the suppression can be explained by the fact that depolarization only reaches a level at which Rvs161p is no longer required for repolarization.
Rvs161p is required for actin repolarization, and several lines of evidence show that this protein is associated with lipid rafts. Although the RVS161-GFP fusion did not complement the rvs161Δ mutant, Rvs161p-GFP was localized to very specific areas of the cell, i.e., in cortical patches that polarized to bud emergence and septum. In accordance, Rvs161p was recovered from a membranous fraction and part of it was found in DRMs. The fraction that was associated with DRMs was relatively low (40% of membrane-associated protein). Nevertheless, considering that Rvs161p has no glycophosphatidylinositol signal anchor or transmembrane domain, it cannot be directly integrated in rafts. Most probably, Rvs161p is associated with a protein(s) integrated in rafts. Moreover, Rvs161p is delocalized in the fen1 and sur4 mutants, and Fen1p and Sur4p could be involved in the establishment of rafts. Indeed, Pma1p, a raft protein, is mislocalized in the fen1 and sur4 mutants. This is consistent with previous results demonstrating a role for lipid rafts in Pma1p sorting (2). Also consistent with this is that, the rvs161Δ mutant depolarized actin patches faster than the wild type, just like the fen1 and sur4 mutants. Therefore this behavior could result from the mislocalization of Rvs161p in the fen1 and sur4 mutants.
In the raft hypothesis, Wsc1p could be required for actin depolarization and Rvs161p for actin repolarization. Therefore, in S. cerevisiae, lipid rafts could function as platforms for actin depolarization and actin repolarization.
Rafts are difficult to visualize by microscopy unless there is clustering (8, 24, 33). Therefore, the cortical patches where Rvs161p localizes could be viewed as clustered rafts. After Sur7p, this is the second report of a protein localizing as small cortical patches in S. cerevisiae (44). Interestingly, SUR7 was uncovered as a multicopy suppressor of both the rvs167 and rvs161Δ mutations. Actin patches are not delocalized in the rvs161Δ mutant overexpressing SUR7 following salt stress (42). Raft clustering could be a physical means to bring together partners for actin repolarization in S. cerevisiae.
Acknowledgments
We are greatly indebted to Bertrand Daignan-Fornier and Kai Simons for discussions and reviewing the manuscript. Special thanks to Mafalda Escobar-Henriques and Isabelle Sagot for supportive discussions. We also thank Howard Riezman for the gift of strains and Ray Cooke for editing the manuscript.
This work was supported by the Association Française pour les Myopathies (AFM grant 7314), the CNRS, and the University of Bordeaux 2.
REFERENCES
- 1.Bagnat, M., S. Keränen, A. Shevchenko, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnat, M., M. Chang, and K. Simons. 2001. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, F., M. Urdaci, M. Aigle, and M. Crouzet. 1993. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol. Cell. Biol. 13:5070-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonneaud, N., O. Ozier-Kalogeropoulos, G. Y. Li, M. Labouesse, L. Minvielle-Sebastia, and F. Lacroute. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7:609-615. [DOI] [PubMed] [Google Scholar]
- 5.Breton, A. M., and M. Aigle. 1998. Genetic and functional relationship between Rvsp, myosin and actin in Saccharomyces cerevisiae. Curr. Genet. 34:280-286. [DOI] [PubMed] [Google Scholar]
- 6.Breton, A. M., J. Schaeffer, and M. Aigle. 2001. The yeast Rvs161 and Rvs167 proteins are involved in secretory vesicles targeting the plasma membrane and in cell integrity. Yeast 18:1053-1068. [DOI] [PubMed] [Google Scholar]
- 7.Brizzio, V., A. E. Gammie, and M. D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and choloesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 9.Brown, D. A., and J. K. Rose. 1992. Sorting of glycophosphatidylinositol-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 10.Chamberlain, L. H., R. D. Burgoyne, and G. W. Gould. 2001. SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl. Acad. Sci. USA 98:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury, S., K. W. Smith, and M. C. Gustin. 1992. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol. 118:561-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung, N., G. Jenkins, Y. A., Hannun, J. Heitman, and L. M. Obeid. 2000. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 275:17229-17232. [DOI] [PubMed] [Google Scholar]
- 14.Chung, N., C. Mao, J. Heitman, Y. A. Hannun, and L. M. Obeid. 2001. Phytoshingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J. Biol. Chem. 276:35614-35621. [DOI] [PubMed] [Google Scholar]
- 15.Cormack, B. P., G. Bertram, M. Egerton, N. A. R., Gow, S. Falkow, and A. J. P. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 16.Crouzet, M., M. Urdaci, L. Dulau, and M. Aigle. 1991. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast 7:727-743. [DOI] [PubMed] [Google Scholar]
- 17.Delley, P.-A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desfarges, L., P. Durrens, H. Juguelin, C. Cassagne, M. Bonneu, and M. Aigle. 1993. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast 9:267-277. [DOI] [PubMed] [Google Scholar]
- 19.Dickson, R. C., and R. L. Lester. 1999. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1438:305-321. [DOI] [PubMed] [Google Scholar]
- 20.Durrens, P., E. Revardel, M. Bonneu, and M. Aigle. 1995. Evidence for branched pathway in the polarized cell division of Saccharomyces cerevisiae. Curr. Genet. 27:213-216. [DOI] [PubMed] [Google Scholar]
- 21.Friant, S., R. Lombardi, T. Schmelzle, M. N. Hall, and H. Riezman. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20:6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikonen, E. 2001. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13:470-477. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, K., M. Sakakibara, S. Yamasaki, A. Takeuchi, H. Arase, M. Miyasaki, N. Nakajima, M. Okada, and T. Saito. 2002. Cutting edge: negative regulation of immune synapse formation by anchoring lipid raft to cytoskeleton through Cbp-EBP50-ERM assembly. J. Immunol. 168:541-544. [DOI] [PubMed] [Google Scholar]
- 24.Janes, P. W., S. C. Ley, and A. I. Magee. 1999. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 147:447-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Lang, T., D. Bruns, D. Wenzel, D. Riedel, P. Holroyd, C. Thiele, and R. Jahn. 2001. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20:2202-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lillie, S. H., and S. Brown. 1994. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 125:825-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodder, A. L., T. K. Lee, and R. Ballester. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, T. F. J. 2001. PI(4,5)P2 regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13:493-499. [DOI] [PubMed] [Google Scholar]
- 30.Munn, A. L., B. J. Stevenson, M. I. Geli, and H. Riezman. 1995. end5, end6 and end7 mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 6:1721-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okayama, H., and P. Berg. 1982. High-efficiency cloning of full-length cDNA. Bio/Technology 24:210-219. [PubMed] [Google Scholar]
- 32.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pralle, A., P. Keller, E.-L. Florin, K. Simons, and J. K. H. Hörber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revardel, E., M. Bonneu, P. Durrens, and M. Aigle. 1995. Characterization of a new gene family developing pleiotropic phenotypes upon mutation in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1263:261-265. [DOI] [PubMed] [Google Scholar]
- 35.Röper, K., D. Corbeil, and W. B. Huttner. 2000. Retention of prominin in microvilli reveals distinct cholesterol-based lipid microdomains in the apical plasma membrane. Nat. Cell Biol. 2:582-592. [DOI] [PubMed] [Google Scholar]
- 36.Rozelle, A. L., L. M. Machesky, M. Yamamoto, M. H. E. Driessens, R. H. Insall, M. G. Roth, K. Luby-Phelps, G. Marriott, A. Hall, and H. L. Yin. 2000. Phosphatidylinositol 4,5-biphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10:311-320. [DOI] [PubMed] [Google Scholar]
- 37.Schneiter, R. 1999. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. BioEssays 21:10004-11010. [DOI] [PubMed] [Google Scholar]
- 38.Sherman, F., and J. Hicks. 1991. Micromanipulation and dissection of asci, p. 21-37. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 41.Sivadon, P., F. Bauer, M. Aigle, and M. Crouzet. 1995. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant. The Rvs161 protein shares common domains with the brain protein amphiphysin. Mol. Gen. Genet. 246:485-495. [DOI] [PubMed] [Google Scholar]
- 42.Sivadon, P., M.-F. Peypouquet, F. Doignon, M. Aigle, and M. Crouzet. 1997. Cloning of the multicopy suppressor gene SUR7: evidence for a functional relationship between the yeast actin-binding protein Rvs167 and a putative membranous protein. Yeast 13:747-761. [DOI] [PubMed] [Google Scholar]
- 43.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young, M. E., T. S. Karpova, B. Brügger, D. M. Moschenross, G. K. Wang, R. Schneiter, F. T. Wieland, and J. A. Cooper. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanolari; B., S. Friant, K. Funato, C. Sütterlin, B. J., Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19:2824-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]