Abstract
Ustilago maydis, a pathogen of maize, is a useful model for the analysis of mating, pathogenicity, and the morphological transition between budding and filamentous growth in fungi. As in other fungi, these processes are regulated by conserved signaling mechanisms, including the cyclic AMP (cAMP)/protein kinase A (PKA) pathway and at least one mitogen-activated protein kinase (MAP kinase) pathway. A current challenge is to identify additional factors that lie downstream of the cAMP pathway and that influence morphogenesis in U. maydis. In this study, we identified suppressor mutations that restored budding growth to a constitutively filamentous mutant with a defect in the gene encoding a catalytic subunit of PKA. Complementation of one suppressor mutation unexpectedly identified the ras2 gene, which is predicted to encode a member of the well-conserved ras family of small GTP-binding proteins. Deletion of the ras2 gene in haploid cells altered cell morphology, eliminated pathogenicity on maize seedlings, and revealed a role in the production of aerial hyphae during mating. We also used an activated ras2 allele to demonstrate that Ras2 promotes pseudohyphal growth via a MAP kinase cascade involving the MAP kinase kinase Fuz7 and the MAP kinase Ubc3. Overall, our results reveal an additional level of crosstalk between the cAMP signaling pathway and a MAP kinase pathway influenced by Ras2.
Many fungi are capable of alternating between budding and filamentous growth (dimorphism), and competence for this morphological switch can be an important factor in the virulence of several fungal pathogens of plants and animals (19, 33, 34). Morphological transitions are involved in the formation of three cell types in the maize pathogen Ustilago maydis, including budding haploid cells, filamentous dikaryons, and diploid teliospores. Haploid cells grow saprophytically and can participate in mating interactions when compatible partners exchange pheromone signals and form the conjugation tubes required for cell fusion. The product of fusion is an infectious dikaryotic cell type that proliferates with a filamentous morphology in host tissue and eventually forms melanized teliospores within tumors of the maize plant. Haploid cells are then produced meiotically following teliospore germination. Pigmented, asexual spores (chlamydospores) that form in response to nutritional deprivation have been reported for U. maydis but are less well characterized (30).
Two unlinked mating type loci designated a and b play a major role in regulating dimorphism in U. maydis. A successful mating interaction is observed only when two cells have different specificities at both the a and b loci. The a locus encodes a pheromone (mfa1 or mfa2) and pheromone receptor (pra1 or pra2) and is responsible for cell recognition and cell fusion (6). The b locus encodes two homeodomain proteins, bE and bW, which are responsible for maintenance of the infectious dikaryon and completion of the life cycle (24, 28, 48). Pheromone response is regulated through a mitogen-activated protein (MAP) kinase cascade that leads to activation of the pheromone response factor Prf1 (21, 36, 40). Prf1 regulates the transcription of genes located at the a and b loci. Several components of MAP kinase cascades have been identified in U. maydis, and these include the MAP kinase kinase kinase Ubc4, the MAP kinase kinase Fuz7, the MAP kinase Ubc3, and the putative adaptor protein Ubc2 (2, 4, 36, 37). Mutation in any of these MAP kinase components results in faulty pheromone signaling.
In addition to the mating type loci, other factors such as the cyclic AMP (cAMP)/protein kinase A (PKA) pathway, nutrient availability, exposure to air, and acidic pH influence the switch from budding to filamentous growth in U. maydis (19, 27, 45). In general, high PKA activity leads to a budding phenotype in U. maydis, while low PKA activity results in filamentous growth. This conclusion is based on observations that mutants deficient in the regulatory subunit of PKA (encoded by the ubc1 gene) display a multiple budding phenotype, while those lacking the enzyme required to produce cAMP (adenylyl cyclase, encoded by uac1) or the catalytic subunit of PKA (encoded by adr1) are constitutively filamentous (5, 14, 19). In addition to their defects in morphogenesis, mutants deficient in the components of the cAMP/PKA pathway are unable to induce tumor formation in planta, demonstrating that the cAMP pathway also plays an important role in virulence.
Interestingly, the ubc2, ubc3, ubc4, and fuz7 genes were identified by their ability to complement mutations that suppressed the filamentous phenotype of adenylyl cyclase mutants (2, 36, 37). This result suggests that the MAP kinase and cAMP pathways are linked. Further evidence for a connection between the pathways has been provided by Kruger et al. (29). These authors showed that the mfa1 pheromone gene transcript was more abundant in ubc1 mutants and in wild-type cells grown in the presence of 6 M cAMP than in wild-type cells grown without exogenous cAMP (29).
In an attempt to identify additional downstream components of the cAMP pathway, suppressor mutations that restored budding growth to the otherwise filamentous adr1 mutant were identified. Complementation of one of these mutations led to identification of the hgl1 gene (13). Hgl1 is thought to act as a repressor of budding growth, and in vitro experiments indicate that Hgl1 serves as a target for phosphorylation by PKA. In addition, hgl1 mutants are severely compromised in their ability to form melanized teliospores.
In this report, we continued the genetic suppression approach that identified the hgl1 gene and found that an ortholog of the ras family of small GTP-binding proteins (designated ras2) is required for budding growth, pathogenicity, and mating in U. maydis. Our analysis also revealed that Ras2 promotes filamentous growth through a MAP kinase cascade and regulates mfa1 pheromone gene transcription.
MATERIALS AND METHODS
Strains.
All strains are listed in Table 1. U. maydis strains were grown at 30°C in potato dextrose broth (Difco), potato dextrose agar (Difco), or complete medium (CM) (23). For selection on CM plates, 250 μg of hygromycin B or 50 μg of nourseothricin was added per ml. Transformants requiring selection in liquid CM broth were grown with 150 μg of hygromycin B or 100 μg of nourseothricin per ml. Escherichia coli strain DH5α (Bethesda Research Laboratories) was used for plasmid construction, and E. coli strain DH10B (Bethesda Research Laboratories) was used for transformation by electroporation.
TABLE 1.
U. maydis strains
| Strain (alternative designation) | Genotype | Source or reference |
|---|---|---|
| 521 (002) | a1b1 | R. Holliday |
| 518 (001) | a2b2 | R. Holliday |
| 001/prV16Hyg | a2b2(prV16Hyg) | This work |
| 002/prV16Hyg | a1b1(prV16Hyg) | This work |
| 001/pHyg101 | a2b2(pHyg101) | This work |
| 002/pHyg101 | a1b1(pHyg101) | This work |
| FB1 | a1b1 | F. Banuett |
| FB2 | a2b2 | F. Banuett |
| FB1/prV16Hyg | a1b1(prV16Hyg) | This work |
| FB2/prV16Hyg | a2b2(prV16Hyg) | This work |
| FB1/pHyg101 | a1b1(pHyg101) | This work |
| FB2/pHyg101 | a2b2(pHyg101) | This work |
| FB1/prV16Sat | a1b1(prV16Sat) | This work |
| FB2/prV16Sat | a2b2(prV16Sat) | This work |
| FB1/pSat112 | a1b1(pSat112) | This work |
| FB2/pSat112 | a2b2(pSat112) | This work |
| d132 | a1/a2 b1/b2 | 28 |
| d132/prV16Hyg | a1/a2 b1/b2(prV16Hyg) | This work |
| d132/pHyg101 | a1/a2 b1/b2(pHyg101) | This work |
| 001-12 | a2b2 Δadr1::phl | 14 |
| 001 Δras2 | a2b2 Δras2-2 hygr | This work |
| 002 Δras2 | a1b1 Δras2-2 hygr | This work |
| 001 Δras2(pSat112) | a2b2 Δras2-2 hygr(pSat112) | This work |
| 001 Δras2(prV16Sat) | a2b2 Δras2-2 hygr(prV16Sat) | This work |
| 002 Δras2(pSat112) | a2b2 Δras2-2 hygr(pSat112) | This work |
| 002 Δras2(prV16Sat) | a2b2 Δras2-2 hygr(prV16Sat) | This work |
| 001 Δras2(pX696S) | a2b2 Δras2-2 hygr(pX696S) | This work |
| FB1-26 | a1b1 Δfuz7 hygr | 3 |
| FB1 Δubc3 | a1b1 Δubc3 natr | 36 |
| FB2 Δubc3 | a1b1 Δubc3 natr | 36 |
| 001 Δprf1 | a2b2 Δprf1 hygr | Y. Kohno, A. De Maria, and N. Lee, unpublished data |
| 002 Δprf1 | a1b1 Δprf1 hygr | Y. Kohno, A. De Maria, and N. Lee, unpublished data |
| 001 Δprf1(pSat112) | a2b2 Δprf1 hygr(pSat112) | This work |
| 001 Δprf1(prV16Sat) | a2b2 Δprf1 hygr(prV16Sat) | This work |
| 002 Δprf1(prV16Sat) | a1b1 Δprf1 hygr(prV16Sat) | This work |
| 002 Δprf1(pSat112) | a1b1 Δprf1, hygr(pSat112) | This work |
| 0606 | a2b2 Δubc1 phleor | 13 |
| 0606(prV16Hyg) | a2b2 Δubc1 phleor(prV16Hyg) | This work |
| 0606(pHyg101) | a2b2 Δubc1 phleor(pHyg101) | This work |
| 3020 | Δhgl1 natr | 13 |
| 3020(prV16Hyg) | Δhgl1 natr(prV16Hyg) | This work |
| 3020(pHyg101) | Δhgl1 natr(pHyg101) | This work |
| 33-1(pHyg101) | a2b2 Δadr1 phleor(pHyg101) | This work |
| 33-1(prV16Hyg) | a2b2 Δadr1 phleor(prV16Hyg) | This work |
| 33-1(pX6-9) | a2b2 Δadr1 phleor(pX6-9) | This work |
| 33-1(pX696rh) | a2b2 Δadr1 phleor(pX696rh) | This work |
| P6D | a2b2 (a1b1 loci ectopically integrated) | 17 |
| P6D Δras2 | a2b2 Δras2-2 (a1b1 loci ectopically integrated) | This work |
| P6D ras2Val16 | a2b2 ras2Val16 (a1b1 loci ectopically integrated) | This work |
DNA manipulations.
Standard procedures were followed for molecular cloning as well as Southern and Northern analysis (46). For the identification of ras family homologs, total genomic DNA was isolated from wild-type strain 518 (22), digested with several enzymes, transferred to a membrane, and analyzed by hybridization at 37°C. The blot was washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate twice for 15 min at 25°C for low-stringency conditions. For more stringent conditions, the blot was further washed with 0.1× SSC-0.1% sodium dodecyl sulfate for 15 min at 48°C.
Plasmids pX696 and pX6-9 carry a 6.2-kb genomic XbaI fragment containing the ras2 gene in pBluescript KS and pHyg101, respectively. Primers prras7 and prras10 (AAGCTTGTGGTGCTGGGAGATGTAGGTGTAGGAAAGACG and GGGCTCGAGGAGCCAGAGCG, respectively) were used to amplify the 3′ portion of ras2 and to introduce a mutation at codon 16 (glycine to valine). This product was digested with HindIII and XhoI and ligated to the 2.9-kb XbaI-HindIII fragment of pX696 containing the 5′ region of ras2 to make prV16. The 4.4-kb insert of prV16 containing the activated ras2Val16 allele was ligated into pHyg101 and pSat112 for transformation into U. maydis.
prVIH10 is a derivative of prV16 containing the hygromycin resistance cassette and a 0.3-kb genomic fragment downstream of the activated ras2Val16 allele for integration of the activated allele into the genome. prVKOH and prVKOP are derivatives of prV16 in which codons 9 to 55 or 9 to 75 have been replaced by the hygromycin resistance or phleomycin resistance cassettes, respectively. In plasmid pX696rh, the ras2 gene was disrupted by insertion of the hygromycin resistance cassette at codon 5. To replace the ras2 gene with the ras2-2 allele, plasmid prVKOH was digested with KpnI and NotI and transformed into U. maydis protoplasts by established methods (53). Gene replacements were confirmed by DNA hybridization with genomic DNA (data not shown).
Primers prras4 (CGAGAGAATGCAAGAGCC) and prras5 (GCACACACACAGCGCGG) were used to isolate the ras2 allele from strain 33-1. A Perkin-Elmer 480 thermal cycler was used to amplify the ras2 locus with the high-fidelity Vent polymerase (New England Biolabs) and the following program: 5-min time delay at 94°C; 30 cycles of 1 min at 94°C, 1 min at 65°C, and 1 min at 72°C; and 10 min at 72°C. The same primers were used to sequence three independent PCR products. A subclone of a cosmid containing the ras2 gene from a U. maydis genomic cosmid library (5) was used to sequence the wild-type ras2 allele. Primers pradr1 (5′-CCGCTTCTACGCGATCAAGG-3′), pradr2 (5′-GGTCGAACACACGAATTCGG-3′), pradr3 (5′-GGGAAGCGTTGTGATTTGCG-3′), and pradr4 (5′-GGTGGAGGTAGTCGATCGC-3′) were used to screen for adr1 disruption mutants.
RNA procedures.
Fungal cells were grown on charcoal-containing double complete medium agar for 48 h, and RNA was isolated essentially as described before (47). Standard molecular techniques were followed for gel electrophoresis, RNA blotting, and hybridization (46). A 680-bp EcoRV fragment was used to probe for mfa1 (6). The ras2 transcript was identified with a 0.9-bp HindIII-AvaI fragment.
Mating and pathogenicity assays.
Strains were tested for the production of aerial hyphae during mating reactions on charcoal-containing double complete medium plates (23). To investigate pheromone production and pheromone response, confrontation assays were performed essentially as described by Mayorga and Gold (28) with the additional step of concentrating the 5-ml overnight cultures by centrifugation and resuspension in 1 ml of potato dextrose broth. Infection of maize seedlings was performed as described before (28).
Microscopy.
Cells were grown in CM broth with the appropriate antibiotics to mid-logarithmic phase and photographed with a Zeiss Axiophot microscope with differential interference contrast optics.
Nucleotide sequence accession number.
The nucleotide sequence of the ras2 gene has been submitted to the GenBank database under accession number AF545586.
RESULTS
Genetic screen for suppressors of filamentous growth of a PKA mutant.
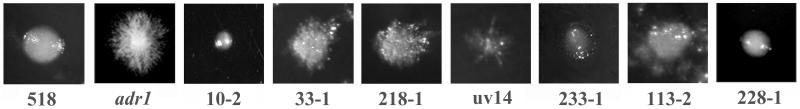
Suppressors of a constitutively filamentous PKA mutant defective in the catalytic subunit encoded by the adr1 gene were isolated to identify targets of the cAMP pathway involved in morphogenesis (14). Eighty-five yeastlike suppressor mutants were isolated, with colony morphologies ranging from reduced filamentous growth (e.g., short filaments) to a completely yeastlike morphology. The colony morphologies of six suppressor mutants chosen for further analysis are shown in Fig. 1. These mutants were screened by transformation with known genes, encoding cAMP and MAP kinase pathway components involved in morphogenesis (hgl1, ubc2, ubc3, and ubc4; Table 2). Of the six mutants, the phenotypes of three (113-2, 218-1, and 233-1) were influenced by transformation with the hgl1 gene. For example, transformation of strain 113-2 with a plasmid carrying the hgl1 gene resulted in filamentous transformants (indicative of complementation or copy number suppression). Introduction of the ubc3 gene into this strain caused filamentous growth in 20 to 30% of the transformants, suggesting that strain 113-2 may be mutated at another locus that is important for both the cAMP and MAP kinase pathways. To explore this possibility, strain 113-2 was transformed with a cosmid genomic library, and two different cosmids were isolated because of their ability to restore the filamentous phenotype to 113-2. However, DNA hybridization analysis revealed that both cosmids contained the hgl1 gene (data not shown). Two other suppressor mutants (218-1 and 233-1, Table 2) were partially complemented by the addition of a plasmid carrying the hgl1 gene, but we were unable to isolate clones from a cosmid library that completely restored filamentous growth to these strains. Complementation was attempted for the three mutants (33-1, 228-1, and uv14) that were unaffected morphologically after transformation with the plasmids containing the hgl1, ubc2, ubc3, or ubc4 gene. Successful complementation was obtained only for strain 33-1, with the identification of a cosmid, 33-4, that restored filamentous growth upon transformation (Fig. 2.) Subsequent subcloning of cosmid 33-4, retransformation of subclones into strain 33-1, and sequence analysis of the complementing region identified ras2 as the complementing gene.
FIG. 1.
Colony morphologies of U. maydis adr1 suppressor mutants. The wild-type 518 strain has a yeastlike colony morphology, and the adr1 mutant (deficient in the PKA catalytic subunit) is constitutively filamentous. The colony morphology of the selected mutants 10-2, 33-1, 218-1, uv14, 233-1, 113-2, and 228-1 range from completely yeastlike to slightly filamentous.
TABLE 2.
Complementation of suppressor mutations with known genes
| Strain | Phenotypea after transformation with:
|
||||
|---|---|---|---|---|---|
| Vector | hgl1 | ubc2 | ubc3 | ubc4 | |
| 001/002 | Yeast | Yeast | Yeast | Yeast | Yeast |
| 10-2 (hgl1) | Yeast | Filamentous | Yeast | Yeast | Yeast |
| 33-1 | Yeast | Yeast | Yeast | Yeast | Yeast |
| 113-2 | Yeast | Filamentous (100%) | Yeast | Filamentous (20-30%) | Yeast |
| 228-1 | Yeast | Yeast | Yeast | Yeast | Yeast |
| 218-1 | Yeast | Filamentous | Yeast | Yeast | Yeast |
| 233-1 | Yeast | Filamentous | Yeast | Yeast | Yeast |
| uv14 | Yeast | Yeast | Yeast | Yeast | Yeast |
Colonies displayed either a yeastlike (yeast) or filamentous phenotype.
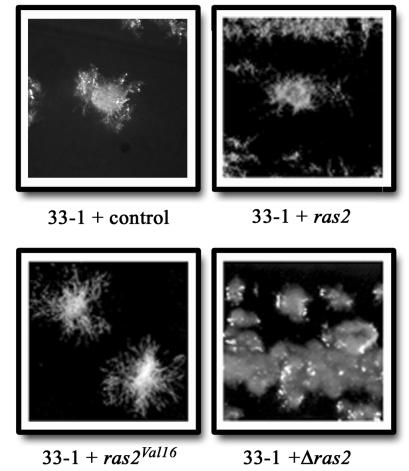
FIG. 2.
Complementation of adr1 suppressor mutant 33-1. Shown are colony morphologies of 33-1 transformed with a vector control (pHyg101, top left), a plasmid carrying the wild-type ras2 allele (pX6-9, top right), a plasmid carrying the activated ras2Val16 allele (prV16Hyg, bottom left), and a plasmid carrying a disrupted ras2 allele (pX696rh, bottom right).
Characterization of ras2 of U. maydis.
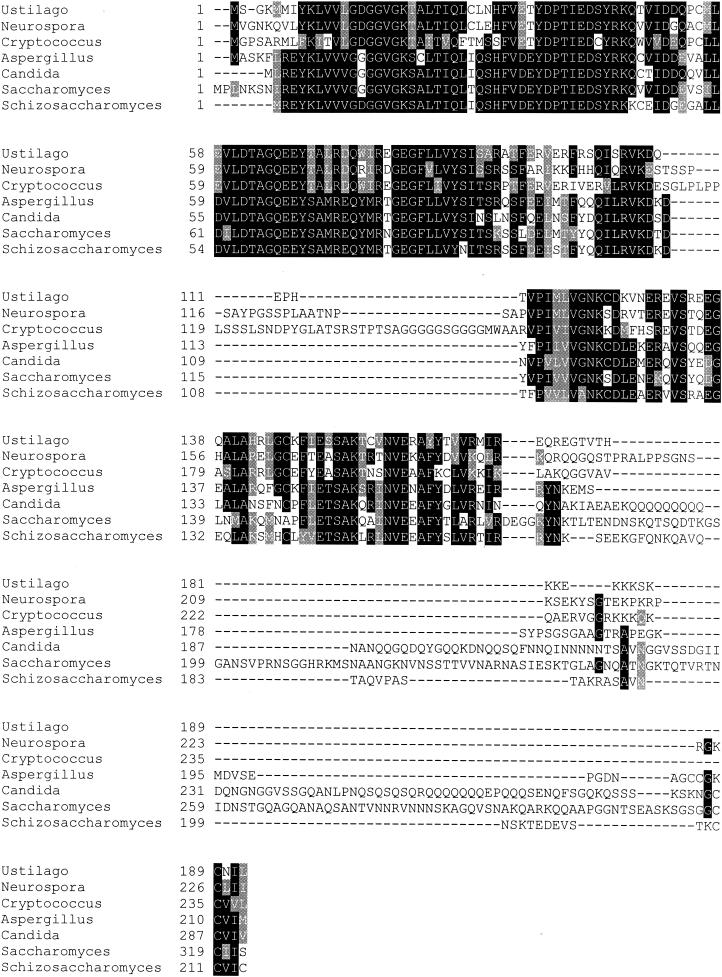
The ras2 gene of U. maydis contained an open reading frame of 576 nucleotides, encoding a predicted polypeptide of 192 amino acids. The gene had high amino acid sequence identity to other fungal Ras genes, including those of Neurospora crassa NC-ras2 (65%), Cryptococcus neoformans RAS2 (67%), Aspergillus fumigatus RAS1 (55%), Candida albicans RAS1 (55%), Schizosaccharomyces pombe ras1 (53%), Saccharomyces cerevisiae RAS2 (51%), and RAS1 from Saccharomyces cerevisiae (51%) (Fig. 3). The predicted polypeptide from the U. maydis gene did not contain the long carboxy-terminal extension found in the Ras1 and Ras2 proteins of S. cerevisiae (Fig. 3). The designation ras2 was chosen based on the higher sequence similarity to RAS2 than to RAS1 in S. cerevisiae.
FIG. 3.
ras2 encodes a member of the Ras family of GTP-binding proteins. (A) Alignment showing homology of Ras2 to other fungal Ras proteins. Identical residues are indicated by white letters on black, and similar amino acids are highlighted with a grey background. Sequence alignment was performed with ClustalW (50) and presented with Boxshade 3.21. The proteins used for comparison are Neurospora crassa NC-Ras2, Cryptococcus neoformans Ras2, Aspergillus fumigatus Ras, Candida albicans Ras1, Saccharomyces cerevisiae Ras2, and Schizosaccharomyces pombe Ras1.
To examine whether ras2 is part of a Ras gene family in U. maydis, a fragment containing the ras2 gene was used as a hybridization probe for DNA blots of genomic DNA under low-stringency conditions. Although there was a high background of weakly hybridizing bands, several distinct bands were detected, including the major band for ras2, suggesting the presence of other sequences with significant homology to ras2 in the U. maydis genome (data not shown). More stringent conditions identified a single band in each lane that represented the ras2 gene. These results suggest that more than one Ras homolog may exist in U. maydis. In this context, a second Ras gene (designated ras1) has been isolated and found to influence pheromone gene expression in U. maydis (P. Muller and R. Kahmann, personal communication).
Identification of ras2 as a multicopy suppressor.
The introduction of cosmid 33-4 and cosmid subclones containing the ras2 gene into mutant 33-1 gave rise to transformants with various phenotypes. Although we initially identified cosmid 33-4 by its ability to restore the filamentous phenotype to strain 33-1, we consistently found that some transformants remained yeastlike (despite resistance to hygromycin B, indicating the presence of the cosmid or subclones). These results prompted an examination of the mutation in the ras2 allele in strain 33-1 because the phenotypic diversity exhibited by the transformants suggested the possibility of copy number suppression rather than true complementation. The ras2 allele from 33-1 was cloned by PCR, and three independent products were sequenced; surprisingly, no mutations were found in the open reading frame of this gene (data not shown).
It was also unlikely that the ras2 gene carried a mutation in the promoter region because RNA blot analysis of 33-1 and wild-type cells revealed similar levels of the ras2 transcript (data not shown). These results suggest that the ras2 gene found on cosmid 33-4 enables filamentous growth in the yeastlike mutant 33-1 through copy number suppression. Although the nature of the mutated gene in 33-1 remains unknown, the ability of a cosmid carrying ras2 to suppress the 33-1 mutant phenotype demonstrates that Ras2 is an important factor in morphogenesis. Thus, ras2 is sufficient to promote filamentous growth upon transformation into the yeastlike suppressor mutant 33-1.
Disruption of ras2 alters cell morphology.
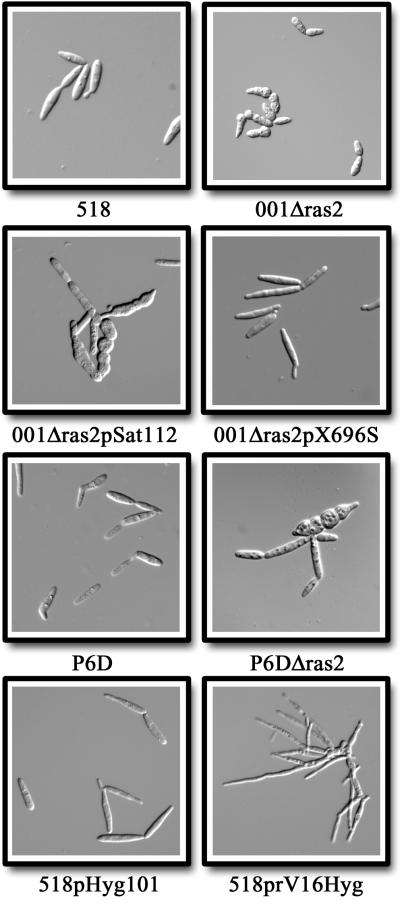
The ras2 gene was deleted from each of two mating-compatible haploid strains to further examine its role in morphogenesis. Mutants lacking ras2 were shorter and rounder than wild-type cells (Fig. 4) and exhibited a morphology reminiscent of both ukc1 mutants (12) and the chlamydospores described by Kusch and Schauz (30). The ukc1 gene encodes a protein kinase with similarity to the cot-1 product of N. crassa (58) Transformation of the wild-type ras2 allele but not the empty vector control into ras2 deletion mutants restored normal cell morphology, demonstrating that the phenotype observed was indeed due to deletion of the ras2 gene (Fig. 4). The ras2 gene was also deleted from cells of the P6D strain. This strain carries the a1 and b1 mating type sequences randomly integrated into the genome of an a2b2 haploid to construct a pathogenic haploid strain due to activated mating functions (17). The P6D Δras2 mutant also displayed a rounded cell morphology similar to that of wild-type cells deficient in ras2 (Fig. 4).
FIG. 4.
Cellular morphology of U. maydis strains carrying mutations at the ras2 locus. Wild-type strain 518 (top left), 001 Δras2 (top right), 001 Δras2 transformed with a vector control (pSat112, second from top left), 001 Δras2 transformed with a plasmid carrying the wild-type ras2 allele (pX696S, second from top right), P6D (third from top left), P6D Δras2 (third from top right), wild-type 518 transformed with a vector control (pHyg101, bottom left), and wild-type 518 transformed with a plasmid carrying the activated ras2Val16 allele (prV16Hyg, bottom right) are shown.
We were also interested in determining whether loss of Ras2 by deletion restored budding growth to an adr1 mutant, as expected from our original suppression screens. Repeated attempts to disrupt ras2 in an adr1 mutant background or adr1 in a ras2 mutant background were unsuccessful, suggesting that this combination is lethal. To explore this possibility in more detail, we exploited the fact that transformation of wild-type cells with an adr1 disruption construct results in a high frequency of filamentous transformants. For example, in a screen of 200 such transformants, 43% were filamentous, and hybridization confirmed adr1 disruption in 10 of these strains. By contrast, a screen of 200 transformants of a ras2 deletion strain with the adr1 disruption construct did not identify any filamentous strains. PCR analysis with two different primer sets confirmed that disruption of adr1 had not occurred in these strains. Overall, these results suggest that disruption of both genes results in lethality.
Ras2 promotes filamentous growth.
We constructed an activated ras2 allele (ras2Val16) by replacing the codon for glycine with that for valine at the 16th amino acid position to further investigate the role of Ras2 in morphogenesis. This dominant activating mutation is analogous to that of the S. cerevisiae ras2Val19 allele, which results in defective GTPase activity (51). We cloned the ras2Val16 allele into autonomously replicating transformation vectors containing the marker for resistance to hygromycin B or nourseothricin. Wild-type strains carrying these plasmids appeared yeastlike on solid medium, but these strains were clearly pseudohyphal when grown in liquid broth (Fig. 4). As expected, wild-type strains carrying the vector grew by budding. Interestingly, transformants of strain 33-1 with the activated ras2Val16 allele were more filamentous than those carrying the wild-type allele, while those carrying a disrupted allele (pX696rh) or the control plasmid (pHyg101) (20) remained yeastlike (Fig. 2). These results demonstrate that Ras2 acts to promote filamentous growth.
Ras2 is required for pheromone production and perception.
To determine the effect of the ras2 deletion on mating, ras2 mutants were cocultured either with compatible wild-type strains or as compatible mutant pairs on mating medium and assayed for the production of dikaryotic hyphae. Vigorous aerial hyphae were produced when ras2 mutants were cospotted with wild-type cells, indicating a positive mating reaction (Fig. 5). These mating reactions were comparable to those seen when compatible wild-type cells were mated. Interestingly, ras2 mutants were unable to induce aerial hyphae formation when cospotted with compatible ras2 strains, indicating that these mutants were defective in cell fusion and/or filamentous growth after fusion (Fig. 5).
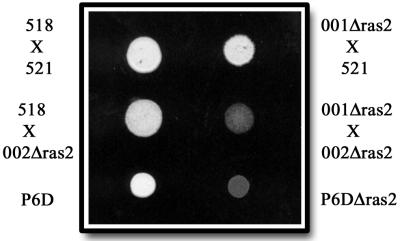
FIG. 5.
Mutants deficient in ras2 are unable to form aerial hyphae. A strong mating reaction was seen when compatible wild-type strains were cospotted on charcoal-containing medium (top left). A strong mating reaction was also observed when wild-type cells were coinoculated with ras2 mutants (top right and middle left). Coinoculation of compatible ras2 mutants resulted in a yeastlike colony (middle right). P6D cells are capable of producing aerial hyphae when inoculated without a mating partner (bottom left), but P6D cells defective in ras2 are not able to produce these hyphae (bottom right).
We also plated ras2 mutants next to compatible wild-type or ras2 mutant cells in a confrontation assay for the ability of ras2 mutants to produce and respond to pheromone. Closer inspection of the mating interaction showed that ras2 mutants were able to respond to pheromone from wild-type cells by producing conjugation tubes (Fig. 6). However, the response to pheromone exhibited by ras2 mutants was severely reduced in comparison to that of compatible wild-type cells plated next to each other. Furthermore, wild-type cells produced fewer conjugation tubes and responded less vigorously to ras2 mutants, presumably because of reduced or delayed pheromone secretion. Even when compatible ras2 mutants were spotted in very close proximity to each other, there was a complete lack of conjugation tube formation (Fig. 6). These results indicate that ras2 mutants are attenuated for pheromone response and suggest that they produce less pheromone than wild-type cells.
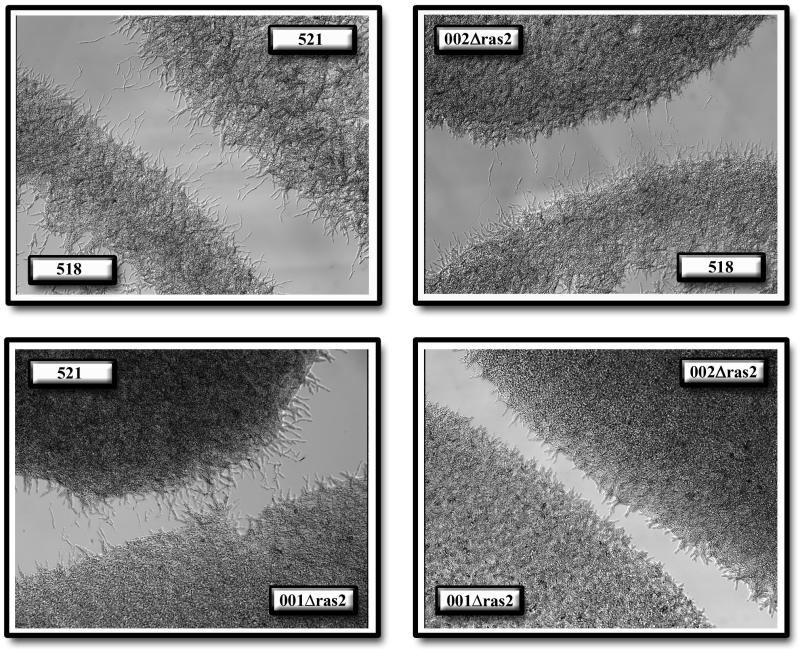
FIG. 6.
ras2 mutants produce less pheromone and are attenuated for pheromone signaling. Wild-type cells respond to pheromone from compatible cells by producing conjugation tubes that are oriented towards their mating partner (top left). Mutants deficient in ras2 produce very few conjugation tubes when spotted next to wild-type cells. Conversely, fewer conjugation tubes are formed from wild-type cells in response to pheromone produced from ras2 mutants (top right and bottom left). ras2 mutants fail to produce conjugation tubes when spotted beside compatible ras2 partners (bottom right).
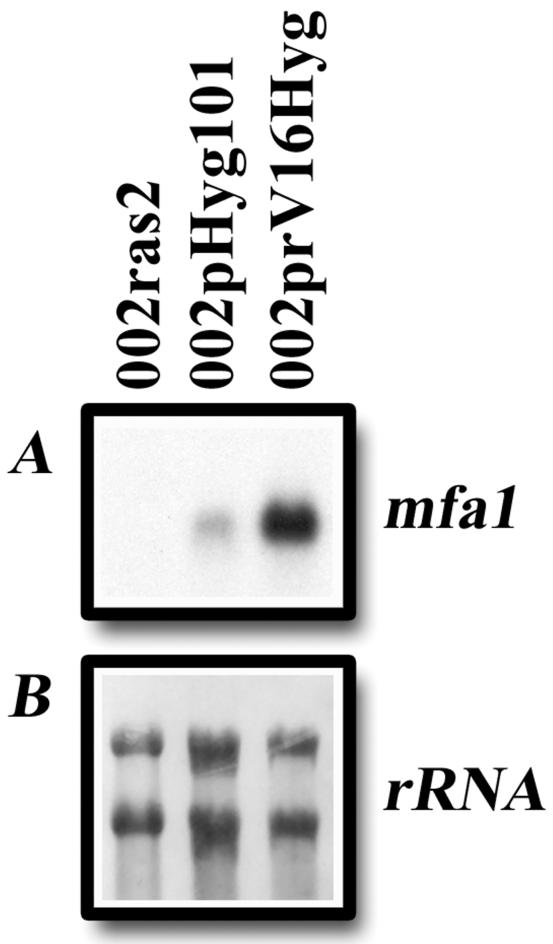
To further investigate pheromone signaling, total RNA was isolated from wild-type and ras2 mutant cells and examined for mfa1 pheromone gene transcription. Previous experiments have shown that a basal level of mating pheromone is expressed in wild-type cells (52). Similarly, hybridization with the mfa1 gene demonstrated that the mfa1 transcript was produced in wild-type cells carrying the control vector pHyg101 (Fig. 7). Interestingly, expression of the mfa1 gene was dramatically increased in wild-type cells carrying a plasmid containing the ras2Val16 allele, while mfa1 expression was completely abolished in ras2 mutants. These results show that Ras2 is necessary for signaling events leading to the production of pheromone in U. maydis.
FIG. 7.
RNA blot analysis of mfa1 transcript levels in ras2 mutants. Total RNA was isolated from 002 Δras2 cells, wild-type 002 cells carrying the control vector pHyg101, and wild-type 002 cells carrying the activated ras2Val16 allele in prV16Hyg. The RNA blot was hybridized with a probe for the mfa1 gene and exposed for 3.75 h (A) or stained with 0.04% methylene blue (B).
Ras2 is essential for postfusion filament formation and pathogenicity.
The ras2 mutant was coinoculated with wild-type cells or with another compatible ras2 mutant into maize seedlings to ascertain whether the ras2 gene plays a role in pathogenicity. Similar to the results obtained from the mating assays, ras2 mutants were pathogenic on maize when paired with wild-type cells, as expected from the positive mating reaction between those strains (Table 3). However, compatible ras2 mutants were unable to induce disease symptoms, even 4 weeks after inoculation, indicating that ras2 is required for the induction of disease symptoms on maize.
TABLE 3.
Pathogenicity of ras2 mutantsa
| Cross or strain | Total no. of plants inoculated | No. of plants producing anthocyanin | No. of plants with tumors | % of plants with tumors |
|---|---|---|---|---|
| 001 × 002 | 26 | 23 | 23 | 96 |
| 001 Δras2 × 002 | 51 | 51 | 51 | 100 |
| 001 × 002 Δras2 | 48 | 47 | 47 | 98 |
| 001 Δras2 × 002 Δras2 | 81 | 0 | 0 | 0 |
| P6D | 47 | 39 | 8 | 17 |
| P6D Δras2 | 133 | 0 | 0 | 0 |
| P6D ras2Vall16 | 53 | 45 | 40 | 76 |
These results are representative of four independent experiments.
The P6D Δras2 deletion mutant was used to determine whether the defects in mating and pathogenicity of haploid ras2 mutants were due to a defect in cell fusion. The P6D strain is solopathogenic because it can form aerial hyphae on charcoal plates and induce disease symptoms in maize seedlings in the absence of a mating partner (17). Deletion of the ras2 gene in the P6D background resulted in cells that were unable to form aerial filaments on mating medium (Fig. 5). Even though P6D is weakly pathogenic, deletion of the ras2 gene in this background further attenuated symptom formation and resulted in the complete loss of anthocyanin production and tumor formation upon injection into maize seedlings (Table 3). Interestingly, P6D cells carrying the activated ras2Val16 allele appeared to be more virulent in maize seedlings than cells carrying the vector control (Table 3). Multiple tumors were observed around the site of infection in seedlings infected with the P6D ras2Val16 mutant, whereas only a rare single small tumor could be found when the untransformed P6D strain was used as the inoculum (Fig. 8). These results indicate that Ras2 plays an essential role in postfusion events involved in filament formation and pathogenicity. Given the influence of the ras2 gene on pheromone gene transcription, it is likely that ras2 is also required for fusion during mating.
FIG. 8.
Ras2 promotes tumor formation. Anthocyanin production and the formation of very small tumors are the major symptoms of disease in maize seedlings infected with the P6D strain (left), while multiple tumors of various sizes are induced upon infection with the P6D strain carrying the activated ras2Val16 allele in prV16Hyg (right).
Ras2 and PKA regulate morphogenesis in distinct pathways.
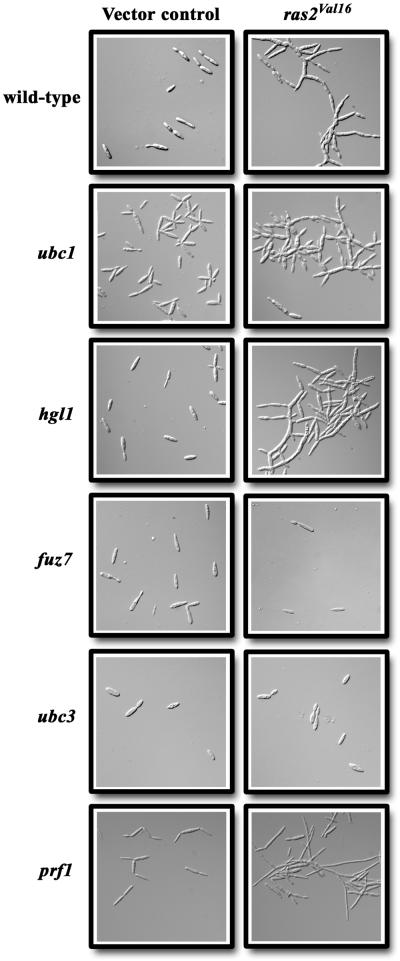
The ability of an activated ras2 allele to promote filamentation prompted an investigation into the relationship between Ras2 and pathways known to regulate filamentous growth in U. maydis. One of the factors regulating the switch between budding and filamentous growth is the activity level of PKA; mutants with low PKA activity grow filamentously, while mutants deficient in the regulatory subunit of PKA (encoded by ubc1) have a multiple budding phenotype. To examine interactions between Ras2 and cAMP signaling, we introduced perturbations in ras2 signaling into strains deficient in components of the cAMP pathway. To this end, the activated ras2Val16 allele was transformed into the constitutively budding ubc1 mutant. Interestingly, ubc1 ras2Val16 double mutants displayed a combination of the ubc1 and ras2Val16 phenotypes; multiple buds were formed at the tips of elongated cells (Fig. 9). The appearance of this novel phenotype suggests that Ras2 and Ubc1 act in different pathways to regulate morphogenesis.
FIG. 9.
Cellular phenotype of mutants with defects in Ras2 and cAMP or MAP kinase signaling. Wild-type 518 and ubc1, hgl1, and ubc3 mutant cells were transformed with the vector control pHyg101 (left column) or a plasmid containing the activated ras2Val16 allele, prV16Hyg (right column). fuz7 and prf1 mutant cells were transformed with a vector control, pSat112 (left column), or a plasmid containing the activated ras2Val16 allele, prV16Sat (right column), conferring resistance to the antibiotic nourseothricin.
Furthermore, the hgl1 gene was recently identified as an additional component of the cAMP pathway (13). The product of this gene may serve as a target for PKA and function to suppress budding growth because hgl1 mutants have a constitutive budding phenotype (13). We found that an hgl1 mutant carrying the activated ras2Val16 allele was filamentous, in marked contrast to the budding phenotype of hgl1 mutants transformed with the vector control (Fig. 9). These results illustrate that budding growth resulting from a defect in hgl1 can be bypassed by the activation of filamentous growth as a result of Ras2 activity. Overall, these results suggest that the Ras2 and cAMP pathways act antagonistically to control morphogenesis in U. maydis. That is, Ras2 appears to promote filamentous growth, and the activated cAMP pathway promotes budding growth.
Ras2 regulates morphogenesis via a MAP kinase signaling cascade.
To determine the role that Ras2 plays in filamentous growth in relation to the MAP kinase/pheromone response cascade, strains deficient in components of the pheromone signaling pathway were transformed with the ras2Val16 activated allele. The fuz7 and ubc3 genes encode a MAP kinase kinase and a MAP kinase, respectively, and mutations in these genes suppress the constitutively filamentous phenotype of a mutant lacking adenylyl cyclase (36). Strains deficient for fuz7 or ubc3, however, maintain a wild-type cellular morphology (4, 36, 40). Thus, we were interested in determining the phenotype of fuz7 and ubc3 mutants expressing the activated ras2Val16 allele.
Considering the involvement of Ras2 in pheromone signaling, it was not surprising that introduction of the ras2Val16 allele into fuz7 and ubc3 mutants resulted in strains that were no different from those transformed with the vector control (Fig. 9) (4, 21). The prf1 gene encodes a pheromone response transcription factor HMG protein that is required for transcription of genes involved in mating. However, the introduction of the activated ras2Val16 allele into the prf1 mutant resulted in cells with a filamentous cell morphology. As expected, transformation of the empty vector control did not influence the yeastlike cell morphology of the prf1 strain (Fig. 9) (21). These results indicate that Ras2 may regulate morphogenesis by signaling via a MAP kinase cascade that includes components encoded by the fuz7 and ubc3 genes but not the transcription factor encoded by prf1. We speculate that a different transcription factor may influence filamentous growth in response to signaling from Fuz7 and Ubc3.
DISCUSSION
Ras proteins are important components of signaling cascades in many organisms and act as molecular switches by alternating between GDP- and GTP-bound forms in response to environmental stimuli. The involvement of Ras proteins in fungal cell growth and differentiation has been well documented. For example, in Saccharomyces cerevisiae, an increase in Ras2 activity is correlated with sensitivity to environmental stress, growth defects on carbon sources other than glucose, the loss of carbohydrate reserves, a transient arrest in G1, a block in sporulation, and enhanced pseudohyphal growth (7-10, 35). Candida albicans mutants deficient in both copies of the RAS1 gene exhibit defects in filament formation and virulence (15, 31). The NC-ras2 gene of Neurospora crassa regulates hyphal growth, cell wall synthesis, and the formation of conidia (25). In Cryptococcus neoformans, RAS1 is required for growth at elevated temperatures, mating, filamentation, agar invasion, and sporulation (1, 49). The Schizosaccharomyces pombe ras1 gene is involved in pheromone response, morphogenesis, and sporulation (16, 57). Given these observations, it is not surprising that the Ras ortholog encoded by ras2 in U. maydis is also necessary for several processes, including morphogenesis, mating, and virulence.
Ras2 and PKA pathways have opposing effects on morphogenesis.
In S. cerevisiae, the intrinsic GTPase activity of Ras2p was reduced by specifically altering glycine 19 to eliminate GTPase activity (26, 51). We constructed a similar dominant U. maydis ras2 allele by substituting the glycine codon at the equivalent position (Gly16) with valine. Introduction of this ras2Val16 activated allele into wild-type cells resulted in transformants with a filamentous cell morphology. These cells differed from the normal unipolar, budding wild-type cells in that they were elongated and defective in cytokinesis and had multiple daughter cells growing from both ends of the mother cell.
The multiple budding phenotype was first observed in mutants with constitutively active PKA due to a defect in the ubc1 gene (19). The phenotype of ubc1 mutants resembles that of activated ras2Val16 mutants at first glance, but several lines of evidence indicate that the PKA and Ras2 pathways mediate distinct processes. While ubc1 mutant cells are most often observed in small clusters with the cells joined at a single tip, ras2Val16 mutants can be isolated as large clumps. In addition to the elongated cell size of ras2Val16 mutants, their bipolar growth pattern may account for the distinction between the ubc1 and ras2Val16 phenotypes. ubc1 mutants carrying the ras2Val16 allele display a unique phenotype: bipolar multiple budding cells that are somewhat swollen but still elongated. Thus, it appears that activation of PKA may serve to promote budding growth or repress filamentous growth by the initiation of bud sites, while the Ras2 pathway may act to promote filamentous growth through cell elongation and the inhibition of cell separation. A similar separation of morphological control by different pathways has been described for S. cerevisiae. Pseudohyphal growth in S. cerevisiae involves cell elongation, unipolar budding, mother-daughter cell adhesion, and invasive growth. The PKA pathway is thought to regulate unipolar budding and agar invasion, whereas the MAP kinase cascade regulates cell elongation and invasion (32, 39, 41, 43, 44).
In U. maydis, although the pathway regulated by PKA may appear to counter the Ras2 pathway, the processes that they regulate may not be completely disparate, because a defect in cytokinesis is associated with the activation of both pathways. Interestingly, the phenotype of wild-type cells carrying the activated ras2Val16 allele is very similar to that of uac1 ubc1, uac1 ubc2, uac1 ubc3, uac1 ubc4, and uac1 fuz7 double mutants (2, 19, 36, 37). For example, the uac1 ubc1 double mutant appears to be slightly filamentous due to an elongated cell morphology. This indicates that adenylyl cyclase may not only produce cAMP to activate PKA but may also play additional PKA-independent roles in morphogenesis. Whether these supplementary roles are associated with Ras2 activity remains to be determined.
The Ras2 pathway regulates filamentation through a MAP kinase pathway.
The activation of Ras2 failed to induce filamentous growth in mutants deficient in the MAP kinase Ubc3 or the MAP kinase kinase Fuz7, indicating that Ubc3 and Fuz7 constitute part of a MAP kinase cascade that relays signals from Ras2 to influence cell elongation and cytokinesis. The genetic interaction between ras2, ubc3, and fuz7 is consistent with the fact that both ubc3 and fuz7 were identified based on their ability to complement secondary mutations that suppressed the constitutively filamentous phenotype of uac1 mutants (2, 36).
Filamentous growth resulting from the activation of Ras proteins has been observed in a number of fungi. In response to nitrogen starvation, diploid S. cerevisiae cells undergo pseudohyphal growth, which is enhanced by the expression of the dominant active allele of RAS2 (18). Further investigation revealed that pseudohyphal growth is caused by the activation of a MAP kinase pathway by RAS2 (39, 44). Similarly, Candida albicans strains carrying the activated RAS1V13 allele formed more abundant hyphae in a shorter time period than wild-type strains (15). Under conditions of nitrogen starvation and in response to mating pheromone, certain strains of Cryptococcus neoformans are capable of forming filaments and sporulating in the absence of a mating partner (54, 56). This process, known as haploid fruiting, does not normally occur in the serotype A strain H99, but vigorous haploid fruiting was observed in cells expressing the activated RAS1Q67L allele, whose function depends on the MAP kinase cascade (1, 54).
ras2 regulates pheromone expression.
Mating and dimorphism are intricately connected in U. maydis because haploid cells must first mate before undergoing the morphological switch to filamentous growth. Therefore, it would seem appropriate that the factors controlling these processes are coordinately regulated. In fact, many of the factors mediating pheromone response are also responsible for filamentous growth. In this report, we demonstrate that Ras2 plays a central role in both mating and dimorphism.
In confrontation assays between wild-type cells and ras2 mutants, the reduced vigor with which conjugation tubes were formed from wild-type cells indicates that ras2 mutants are capable of pheromone secretion, although pheromone production may be reduced or delayed. It is possible that another G protein may either play a minor role in pheromone signaling or be able to substitute, albeit inefficiently, for the loss of Ras2, since pheromone production and conjugation tube formation were observed at reduced levels in ras2 mutants. The detection of several bands with sequences from the ras2 locus after hybridization under low-stringency conditions indicates that additional Ras-like proteins may exist in U. maydis (data not shown).
Certainly, functional overlap between Ras proteins has been documented in Cryptococcus neoformans and S. cerevisiae. For example, overexpression of the C. neoformans RAS2 gene fully suppresses the mating defect of a ras1 mutant and partially suppresses the morphological and high-temperature-growth defects of the ras1 mutant (55). In a similar manner, overexpression of the RAS1 gene of S. cerevisiae restores invasive growth to ras2 mutants (38, 42). Functional redundancy with Ras2 may also be due to an alternative pathway that activates the transcription of genes at the mating type locus. In support of this, the cAMP pathway has been shown to influence pheromone signaling, as ubc1 mutants express elevated levels of mfa1 transcript (29). Taken together, these results show that Ras2 is required for the basal expression of mating pheromone and that a Ras2-independent pathway exists for the amplification of pheromone expression in response to pheromone from compatible cells.
Mutants deficient in components of the putative pheromone response pathway exhibit phenotypes that are similar to that of the ras2 mutant. Much like the ras2 mutant, ubc3 mutants fail to produce aerial hyphae when cospotted on mating medium (36). Further analysis by drop mating and RNA blot assays confirmed that ubc3 mutants produce less pheromone and are incapable of responding to pheromone produced by compatible mating partners (36, 40). In addition, haploid fuz7 mutants show reduced filament formation during mating interactions, and diploid fuz7 mutants are yeastlike after 24 h of growth on charcoal agar (4). Furthermore, pheromone signaling through the MAP kinase cascade leads to the activation of the pheromone response factor encoded by the prf1 gene (21, 36, 40). Thus, it seems likely that Ras2 signals through Fuz7, Ubc3, and Prf1 to regulate pheromone response.
Ras2 is a pathogenicity factor.
The correlation between mating and morphogenesis can be further extended to include pathogenesis because all three processes are intricately connected in U. maydis. Transformation of wild-type strains with the activated ras2Val16 allele resulted in increased pheromone gene expression and an elongated cell morphology. Given that P6D cells expressing the activated ras2Val16 allele were apparently more virulent than the untransformed control, activation of the Ras1 pathway may also serve to enhance filamentous growth in planta, host penetration, or tumor formation. In Cryptococcus neoformans, activation of the cAMP pathway by deletion of the PKR1 gene, encoding the regulatory subunit of PKA, increases virulence in both rabbit and mouse models of cryptococcosis (11). However, there were no observable differences between maize seedlings infected with U. maydis wild-type cells carrying a plasmid containing the activated ras2Val16 allele and cells carrying an empty vector as a control (data not shown). These results indicate that the increased virulence brought about by expression of the ras2Val16 allele may correct a problem specific for the P6D strain. For example, the activation of Ras2 may aid the P6D strain in pathogenicity simply by promoting the filamentous cell morphology. The presence of branched filaments and branch primordia in wild-type dikaryotic filaments may facilitate host tissue invasion (3). Although P6D filaments in planta have yet to be characterized, the development and morphological features of hyphae from dikaryons and diploids are indistinguishable (3).
It is a common finding that diploid strains heterozygous at the mating type loci are less pathogenic on maize (similar to the P6D strain) than dikaryons. It may be that the P6D and diploid strains do not efficiently produce filaments or other virulence traits that are necessary for aggressive proliferation in the host environment. In fact, we observed larger tumors and more obvious disease symptoms in maize seedlings infected with the d132 diploid strain carrying a plasmid containing the activated ras2Val16 allele than in seedlings infected with d132 carrying an empty vector control (data not shown). Thus, Ras2 may function in a pheromone-independent pathway that regulates pathogenicity. Perhaps host signals are less well perceived by diploid and P6D strains than by wild-type dikaryons, and these signals trigger the activation of fungal factors promoting filament proliferation, tumor induction, and teliospore development through a pathway controlled by Ras2.
In U. maydis, all of the factors thought to be associated with the pheromone response MAP kinase cascade, including pheromones, pheromone receptors, Ras2, the scaffold protein Ubc2, the MAP kinase kinase Fuz7, and the MAP kinase Ubc3, have been implicated in both filamentous growth and pheromone response. However, the transcription factor Prf1 appears to be solely responsible for pheromone response, and only Ras2 and Ubc2 are absolutely required for pathogenesis. It seems likely that Ras2 responds to multiple signals and controls different pathways that lead to the activation of diverse targets. The ability of Ras2 to discriminate between different signals and the elucidation of the downstream effectors of Ras2 will be interesting challenges for future research.
Acknowledgments
We gratefully thank Yume Kohno and Adriana De Maria for technical help, Cletus D'Souza for comments on the manuscript, Scott Gold and Flora Banuett for strains, and Regine Kahmann for the prf1 disruption construct.
This work was supported by a grant (to J.W.K.) from the Canadian Institutes of Health Research. N. Lee acknowledges support from a University Graduate Fellowship from UBC. J.W.K. is a Burroughs Wellcome Fund Scholar in pathogenic mycology.
REFERENCES
- 1.Alspaugh, J. A., L. M. Cavallo, J. R. Perfect, and J. Heitman. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352-365. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, D. L., J. D. Egan, M. E. Mayorga, and S. E. Gold. 2000. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 13:781-786. [DOI] [PubMed] [Google Scholar]
- 3.Banuett, F., and I. Herskowitz. 1996. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122:2965-2976. [DOI] [PubMed] [Google Scholar]
- 4.Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAP kinase kinase homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, K. J., S. E. Gold, and J. W. Kronstad. 1993. Identification and complementation of a mutation to constitutive filamentous growth in Ustilago maydis. Mol. Plant-Microbe Interact. 6:274-283. [DOI] [PubMed] [Google Scholar]
- 6.Bölker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 7.Broach, J. R. 1991. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7:28-33. [DOI] [PubMed] [Google Scholar]
- 8.Broach, J. R., and R. J. Deschenes. 1990. The function of ras genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54:79-139. [DOI] [PubMed] [Google Scholar]
- 9.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53:555-566. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, J. F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dürrenberger, F., and J. Kronstad. 1999. The ukc1 gene encodes a protein kinase involved in morphogenesis, pathogenicity and pigment formation in Ustilago maydis. Mol. Gen. Genet. 261:281-289. [DOI] [PubMed] [Google Scholar]
- 13.Dürrenberger, F., R. D. Laidlaw, and J. W. Kronstad. 2001. The hgl1 gene is required for dimorphism and teliospore formation in the fungal pathogen Ustilago maydis. Mol. Microbiol. 41:337-348. [DOI] [PubMed] [Google Scholar]
- 14.Dürrenberger, F., K. Wong, and J. W. Kronstad. 1998. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc. Natl. Acad. Sci. USA 95:5684-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, Q., E. Summers, B. Guo, and G. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukui, Y., T. Kozasa, Y. Kaziro, T. Takeda, and M. Yamamoto. 1986. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell 44:329-336. [DOI] [PubMed] [Google Scholar]
- 17.Giasson, L., and J. W. Kronstad. 1995. Mutations in the myp1 gene of Ustilago maydis attenuate mycelial growth and virulence. Genetics 141:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 19.Gold, S., G. Duncan, K. Barrett, and J. Kronstad. 1994. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8:2805-2816. [DOI] [PubMed] [Google Scholar]
- 20.Gold, S. E., G. Bakkeren, J. E. Davies, and J. W. Kronstad. 1994. Three selectable markers for transformation of Ustilago maydis. Gene 142:225-230. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann, H. A., R. Kahmann, and M. Bölker. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 22.Holliday, R. 1961. The genetics of Ustilago maydis. Genet. Res. Cambridge Soc. 2:204-230. [Google Scholar]
- 23.Holliday, R. 1974. Ustilago maydis, p. 575-595. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum, New York, N.Y.
- 24.Kamper, J., M. Reichmann, T. Romeis, M. Bölker, and R. Kahmann. 1995. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73-83. [DOI] [PubMed] [Google Scholar]
- 25.Kana-uchi, A., C. T. Yamashiro, S. Tanabe, and T. Murayama. 1997. A ras homologue of Neurospora crassa regulates morphology. Mol. Gen. Genet. 254:427-432. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka, T., S. Powers, C. McGill, O. Fasano, J. Strathern, J. Broach, and M. Wigler. 1984. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 37:437-445. [DOI] [PubMed] [Google Scholar]
- 27.Kernkamp, M. F. 1941. The relative effect of environmental and genetic factors on growth types of Ustilago zeae. Phytopathology 32:554-567. [Google Scholar]
- 28.Kronstad, J. W., and S. A. Leong. 1989. Isolation of two alleles of the b locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA 86:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 30.Kusch, G., and K. Schauz. 1989. Light and electron microscopic studies of chlamydospore development in Ustilago maydis. Cryptogamic Bot. 1:230-235. [Google Scholar]
- 31.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schroppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 33.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 34.Maresca, B., and G. S. Kobayashi. 2000. Dimorphism in Histoplasma capsulatum and Blastomyces dermatitidis. Contrib. Microbiol. 5:201-216. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto, K., I. Uno, Y. Oshima, and T. Ishikawa. 1982. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 79:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 37.Mayorga, M. E., and S. E. Gold. 2001. The ubc2 gene of Ustilago maydis encodes a putative novel adaptor protein required for filamentous growth, pheromone response and virulence. Mol. Microbiol. 41:1365-1379. [DOI] [PubMed] [Google Scholar]
- 38.Mosch, H. U., E. Kubler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, P., C. Aichinger, M. Feldbrugge, and R. Kahmann. 1999. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 41.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powers, S., T. Kataoka, O. Fasano, M. Goldfarb, J. Strathern, J. Broach, and M. Wigler. 1984. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell 36:607-612. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, R. L., H. U. Mosch, and G. R. Fink. 1997. 14-3-3 proteins are essential for RAS/MAP kinase cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89:1055-1065. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Herrera, J., C. G. Leon, L. Guevara-Olvera, and A. Carabez-Trejo. 1995. Yeast-mycelial dimorphism of haploid and diploid strains of Ustilago maydis. Microbiology 141:695-703. [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schafer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka, K., H. Nambu, Y. Katoh, M. Kai, and Y. Hidaka. 1999. Molecular cloning of homologs of RAS and RHO1 genes from Cryptococcus neoformans. Yeast 15:1133-1139. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 52.Urban, M., R. Kahmann, and M. Bölker. 1996. The biallelic a mating type locus of Ustilago maydis: remnants of an additional pheromone gene indicate evolution from a multiallelic ancestor. Mol. Gen. Genet. 250:414-420. [DOI] [PubMed] [Google Scholar]
- 53.Wang, J., D. W. Holden, and S. A. Leong. 1988. Gene transfer system for the phytopathogenic fungus Ustilago maydis. Proc. Natl. Acad. Sci. USA 85:865-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20:352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 56.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, H. P., M. White, S. Marcus, and M. Wigler. 1994. Concerted action of RAS and G proteins in the sexual response pathways of Schizosaccharomyces pombe. Mol. Cell. Biol. 14:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yarden, O., M. Plamann, D. J. Ebbole, and C. Yanofsky. 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11:2159-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]