Abstract
Genetic tractability and easy manipulation make Caenorhabditis elegans a good model to study host–pathogen interactions. Dozens of different bacterial species can pathogenically infect C. elegans under laboratory conditions, and all of these microbes are extracellular pathogens to nematodes. Viruses, on the other hand, are obligate intracellular parasites, and yet no viral infections have been reported for C. elegans. We established a procedure allowing vaccinia virus to enter and subsequently replicate in C. elegans. Virus replication was significantly enhanced in ced-3, ced-4, ced-9(gf), and egl-1(lf) mutants, demonstrating that the core programmed cell death (PCD) genes ced-3, ced-4, ced-9, and egl-1 control vaccinia virus replication in C. elegans. The ability of ced-3 and ced-4 alleles to restrict virus replication is correlated with their cell-killing activities. Moreover, the increase in vaccinia virus replication levels in the PCD-defective mutants was not likely to be caused by the extra live cells, as neither the inhibition of PCD by icd-1 overexpression nor the presence of extra cells after extra cell divisions in cul-1 or lin-23 mutants had any significant effect on vaccinia virus replication. Therefore, the core PCD genes possess a unique function in controlling vaccinia virus replication in C. elegans.
Keywords: programmed cell death, virus–host interaction
The issue of virus–host interactions remains a challenging frontier in virology, especially concerning the identity of cellular factors involved in viral replication. The systematic study of such interactions between an animal and a viral pathogen has been hindered largely by the lack of an appropriate model system that is both genetically tractable and amenable to facile low-cost, high-throughput genetic screens. Because of its genetic tractability and easy manipulation, Caenorhabditis elegans has been a particularly successful system to study host–bacterium interactions (1–8).
A number of bacterial species can pathogenically infect C. elegans, and all these microbes multiply themselves as extracellular pathogens to nematodes. Despite the enormous evolutionary distance between nematode and humans, several bacterial virulence factors have been shown to play a role in both C. elegans and mammalian pathogenesis (9). More importantly, C. elegans mutants, which are either more susceptible or resistant to bacterial killing, can be readily identified. Together, these facts make C. elegans a powerful system for studying the molecular basis of bacterial pathogenesis and host innate defense responses (8–13).
Although C. elegans affords sophisticated genetic analyses, no viruses have been reported to infect C. elegans. Considering that >40% of the 19,000 C. elegans predicted proteins have matches in humans, this also makes C. elegans a compelling model for studies of numerous viral diseases. Thus, in this study, we addressed whether C. elegans can be further extended as an artificial host for animal virus infections. We found that, in the presence of polyethylene glycol (PEG), vaccinia virus (VV) can enter and subsequently initiate viral genome replication, and as a result, newly reproduced virions could be generated in certain strains of C. elegans. One of the important features about mammalian–viral interactions is the programmed cell death (PCD) responses of infected targets. PCD in mammals has been implicated in several human diseases, including cancer, autoimmune disease, and neurodegenerative disease (14). Because many of the key components of the mammalian cell death machinery were first identified in C. elegans by genetic studies (15, 16), we began with determining the role of PCD when nematodes are attacked by a viral pathogen. We used this model system to explore how viral genome replication could be influenced by a given mutation of the PCD pathway in C. elegans. The feasibility of this approach on vaccinia–nematode interaction may be expanded to apply to other animal viruses.
Results
VV Can Replicate in PEG-Treated C. elegans.
The notion that common microbial virulence and host defense mechanisms exist has been reinforced by the use of C. elegans as a model system in the studies of bacterial pathogenesis (1–7). We investigated whether C. elegans could serve as a host for virus infection. Using several conventional infection protocols, we found that the wild-type N2 strain of C. elegans could not be infected with VV, Sindbis virus, baculovirus, or adenovirus. The observed insusceptibility of C. elegans to virus infection could be due to the lack of extracellular virus receptors or intracellular permissiveness for viral replication in C. elegans. To further examine whether C. elegans might be devoid of the machinery for virus entry but could remain permissive intracellularly to virus genome replication, we incorporated PEG in our infection protocol to facilitate viral entry. PEG is an uncharged, hydrophilic, linear polymer that improves the efficiency of transduction of a nonenveloped adenovirus to cells and tissues (17) and enhances the infectivity of an enveloped pestivirus to normally insusceptible Madin–Darby bovine kidney cells (18). We first investigated whether VV could enter and subsequently replicate in C. elegans in the presence of PEG. Although an approach of this kind may not be suitable for exploring the initial steps of viral infection, such as attachment, penetration, and uncoating, it is applicable for studies of viral genome replication and virus–host interactions at the intracellular level.
VV is an enveloped DNA virus that belongs to the family Poxviridae, which comprises a number of the largest (200–400 nm) poxviruses. It has a broad spectrum of host ranges and, like other members in the family, is capable of replicating exclusively in the cytoplasm of the infected cells (19). VV contains a double-stranded DNA genome, transcriptional enzymes, and factors. After viral uncoating, VV genomic DNA is replicated to form concatemers, and the intermediate genes, which are located in the newly synthesized progeny DNA, are in turn transcribed and translated to form the late transcriptional factors that guide the viral factory to commence the late phase of VV life cycle in the infected cells (19–22). To monitor viral genome replication in C. elegans, we used the recombinant vaccinia virus, vREβCAT, which contains the lacZ reporter gene encoding β-galactosidase under control by the late promoter of VV F17R gene. The expression of VV late genes appears to reach a steady-state level 4 h after infection (21). Previous studies showed that β-galactosidase is actively expressed only if this recombinant virus is undergoing a productive genome replication that in turn switches on the F17R promoter (19, 21, 23). Thus, the DNA replication level of recombinant VV can be inferred from the β-galactosidase expression level in the infected host.
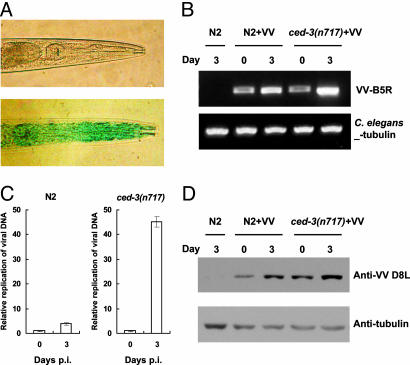
To establish the optimal conditions for PEG-mediated VV infection, we used a spectrophotometric measure of β-galactosidase activity and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) in situ staining as indicators for virus genomic replication inside the worms. Various parameters in PEG-mediated infection were tested, including viral titers, PEG size (molecular weights ranging from 1000 to 4000), concentration (2–10%), incubation time (1–15 min), temperature (4°C or 25°C), and the developmental stage (L1-L4) of the worms. Treatment with PEG 1000 or 4000 alone for 5 min killed ≈25% and 50% of worms, respectively. Therefore, we used PEG 1000 to establish the optimal condition for virus infection. Using the PEG-mediated infection protocol, we could detect the positive X-Gal staining signal in virus-treated worms but not in mock-treated control worms (Fig. 1A), and virus particles, likely in assembling process, could also be seen in C. elegans by electron microscopy (Figs. 5 and 6, which are published as supporting information on the PNAS web site), suggesting that VV could undergo an active replication process in certain PEG-treated worms. As shown in Table 1, which is published as supporting information on the PNAS web site, we found that L3 and L4 worms exposed to 1 × 107 plaque forming units (pfu) per milliliter VV in the presence of 2% PEG 1000 for 5 min at room temperature showed the highest infection rate of 5%. This infection rate could be further increased to ≈18% when the remaining live worms were subsequently soaked in M9 medium containing 1 × 107 pfu/ml of VV for another 6 h at room temperature. This secondary virus exposure in the absence of PEG appears to be crucial for boosting the infection rate from 5% to 18%. Although the mechanism remains unknown, this infection enhancement did not occur when M9 medium containing virus titers of 106 or lower. The 6-h incubation time of the secondary exposure also seems to be important, because a shorter secondary exposure time (e.g., 3 h) did not enhance the infection rate. We performed PCR and real-time PCR analysis to further confirm the accumulation of VV genomic DNA occurring in the PEG-treated worms with time. PCR assay was used to detect the change of viral DNA amounts in the worms between days 0 and 3 postinfection (p.i.) using the primers targeting to the VV B5R gene, and amplification of C. elegans β-tubulin was used as the internal control. As shown in Fig. 1B, increased intensity of PCR products could be readily seen in the DNA samples derived from day 3 as compared to those from day 0. To more accurately measure the fold increases of VV DNA accumulations in the worms between the indicated time points, we carried out the real-time PCR analysis. As shown in Fig. 1C, an ≈4-fold increase of VV replication levels could be seen for the wild-type N2 worms at day 3 as compared to that at day 0, and more dramatically, an ≈45-fold increase of VV replication could also be detected for ced-3(n717) mutant worms (see below). Moreover, as detected by immunoblotting, we found that the amount of VV envelope protein D8L was accumulated significantly to higher levels in both N2 and ced-3(n717) mutant worms at day 3 when compared to that at day 0 (Fig. 1D). These results together clearly illustrate that, with a brief PEG treatment, VV can undergo active amplification process through genomic DNA replication in C. elegans.
Fig. 1.
Replication of VV in PEG-treated C. elegans. Groups of about 1000 worms at L3-L4 stage were subjected to PEG-mediated infection by 1 × 107 pfu/ml of vaccinia virus (VV). (A) Wild-type worms exposed to 2% PEG alone (Upper) or to 2% PEG plus 1 × 107 pfu/ml VV (Lower) on the second day postinfection (p.i.) were fixed and stained with X-Gal. (B) PCR analysis. Accumulation of VV genomic DNA was analyzed by PCR in wild-type N2 and ced-3(n717) mutant worms at day 0 or day 3 p.i. (C) Real-time PCR analysis. The relative replication level of VV genomic DNA in wild-type N2 or ced-3(n717) mutant worms was determined by real-time PCR using primers specific for VV B5R gene. The result represents the abundance of replicative DNA in the sample collected at day 3 relative to that at day 0 p.i. The Ct values are shown separately in Table 2, which is published as supporting information on the PNAS web site. All experiments were repeated three times independently and data are means ± SD. (D) Immunoblot. The amounts of VV proteins expressed in N2 and ced-3(n717) mutant at the indicated time points were analyzed by immunoblotting using antibody specific for VV structural D8L protein. For simplicity, we defined day 0 as the time when the 5-min virus exposure with PEG followed by the 6-h virus treatment without PEG had been completed.
As controls, neither plaque-forming nor β-galactosidase activities were detected in infection assays using UV-killed VV or dead worms. VV also failed to replicate in worms that were exposed to virus-containing PEG solution for <1 min. In addition, when the virus-containing stock solution was filtered through 0.1-μm pores, the resulting filtrates could no longer infect worms. By contrast, the virions purified by sucrose cushion could readily infect worms in the presence of PEG. We have also ruled out the possibility that, without vREβCAT replication, F17R-drived lacZ reporter in VV, which is regulated by a viral late gene promoter, may spuriously express by itself due to the viral DNA introduced into the PEG-permeablized animals (see Fig. 7, which is published as supporting information on the PNAS web site).
Blue X-Gal staining was observed in many tissues and organs, including pharynx, intestine, muscle, hypodermis, and occasionally germ line. All of the worms stained positive for X-Gal showed the blue signal in the intestine. About 80% of these worms exhibited an additional signal in the pharynx (Fig. 1A), and 30–40% of infected worms exhibited a broad signal distribution in intestine, pharynx, muscle, and hypodermis. These observations suggest a different susceptibility of cells to VV entry or replication in our infection procedure to the wild-type worms.
To examine possible vertical transmission of VV to progeny, virus-treated worms were stained with X-Gal by using a protocol that killed the mothers but spared the offspring (24). Positively X-Gal-stained worms and eggs were collected and plated on fresh 60-mm plates containing of E. coli OP50 at confluence. After hatching, worms at different stages from L2 to L4 were collected, fixed, and stained with X-Gal for signs of VV replication. No vertical transmission of VV was observed in 100 progeny of 30 vaccinia-infected worms. Moreover, we found all eggs from infected hermaphrodites could hatch normally as negative control. However, we did not detect any VV from progenies of these hatched eggs. Thus, it is possible that these embryos were able to clear the viruses during development.
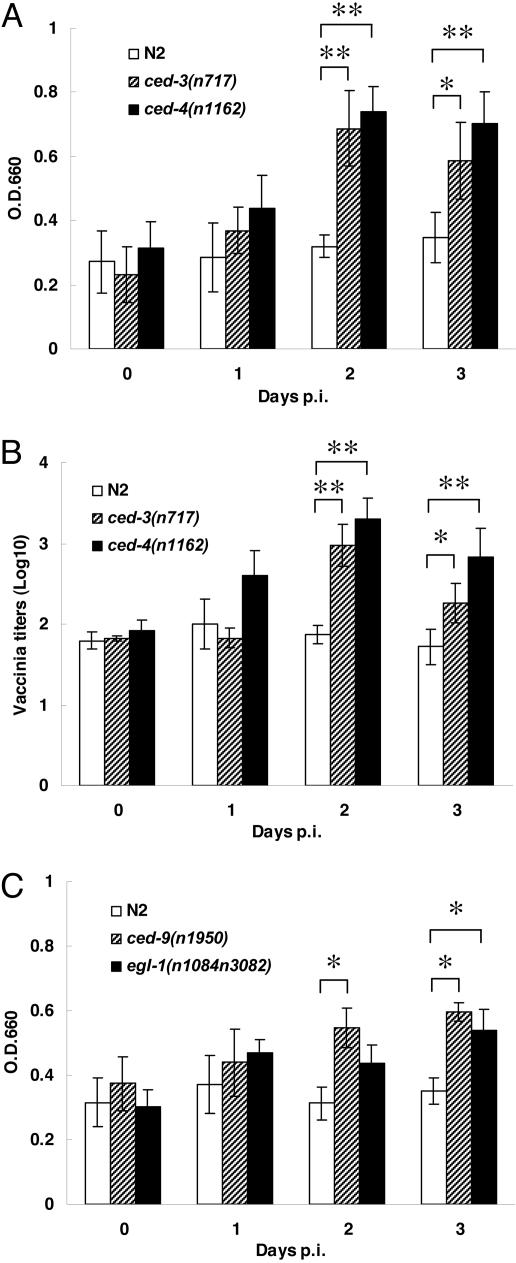
Consistent with the X-Gal staining results, β-galactosidase activity was detected in VV-treated worms during the 3 days p.i. (Fig. 2A). To quantify virus production in the treated worms, we performed a standard plaque-forming assay on BHK-21 cells using infected worm lysates. Viable VV can be detected up to three days p.i. (Fig. 2B). Interestingly, when compared with other mutant strains in Fig. 2, the VV replication levels in wild-type N2 worms did not increase with time after PEG-mediated infection, as judged by β-galactosidase activities (Fig. 2A) and virus titers (Fig. 2B). This observation suggests that intracellular VV replication might somehow be restricted in wild-type worms (see below).
Fig. 2.
The PCD mutations enhance VV replication in PEG-treated C. elegans. About 1,000 worms (stages L3-L4) of each strain were infected with vaccinia virus in the presence of PEG. (A) VV titers in lysates of ced-3(n717) and ced-4(n1162) mutant strains were measured for β-galactosidase activity at OD660 at different days p.i. (B) After infection, the titers of VV from each C. elegans strain were determined at the indicated days by the standard plaque-forming assay. (C) Virus replication levels in the ced-9(n1950) and egl-1(n1084n3082) mutant strains were determined from the spectrophotometric β-galactosidase activity at OD660 at the indicated times. All experiments were repeated three times independently (∗, P < 0.05; ∗∗, P < 0.01 by Student’s t test). Data are means ± SD.
The same protocol was also successfully used for a recombinant Sindbis virus, an enveloped RNA virus that carries a reporter gene of enhanced GFP (EGFP) under the control of the virus’ own subgenomic promoter. Using the EGFP signal as an indicator of replication, we found that this recombinant Sindbis virus could enter and then undergo genome replication process in ≈10% of PEG-treated worms (Fig. 8, which is published as supporting information on the PNAS web site). Therefore, this PEG-mediated infection protocol is not specific to VV but rather is general to some viruses for their entrance and replication in C. elegans.
Increases of VV Replication in Core PCD-Defective C. elegans.
We next addressed whether the genes in the PCD pathway may play a role in VV replication in C. elegans. PCD, or apoptosis, is an evolutionarily conserved cellular process that is important for development and tissue homeostasis of multicellular organisms. During the development of C. elegans hermaphrodites, 131 of the 1,090 somatic cells generated undergo PCD (25–27). Genetic studies have identified four genes, ced-3, ced-4, ced-9, and egl-1, which govern the core machinery of PCD in C. elegans (28–30). The ced-3, ced-4, and ced-9 genes encode proteins similar to the human caspases, Apaf-1, and Bcl-2, respectively (31–34). EGL-1 is a small BH3-containing protein (30). The strong loss-of-function (lf) mutations ced-3(n717), ced-4(n1162), and egl-1(n1084n3082), as well as the gain-of-function (gf) mutation ced-9(n1950), block most, if not all, PCD during C. elegans development. In these mutants, very few or no cell corpses are observed, and >80% of cells that normally undergo PCD in the pharynx survive (28–30). Although VV infection rates mediated by PEG were indistinguishable among ced-3(n717), ced-4(n1162), and wild-type worms, the ced-3(n717) and ced-4(n1162) mutants had significantly increased both β-galactosidase activities and virus titers at days 2 and 3 p.i. compared with wild-type controls (Fig. 2 A and B). This finding suggests that mutations in ced-3 and ced-4 may facilitate virus replication. Because viral titers measured by plaque-forming assay correlated well with spectrophotometric β-galactosidase activities, we used the latter as our indicator for VV replication level in the experiments described below. VV in ced-3 and ced-4 mutants exhibited much stronger intensity as well as a more extended pattern of X-Gal staining than those of wild-type worms on days 2 and 3 p.i., in agreement with the results of β-galactosidase activity measurement (Fig. 2A) and virus titration assay (Fig. 2B). Similar to the elevated viral titers seen in ced-3(n717) and ced-4(n1162) worms, ced-9(n1950), and egl-1(n1084n3082) mutants showed higher VV replication levels than the wild-type control (Fig. 2C).
Restriction of Vaccinia Replication in C. elegans Depends on the Core PCD Genes.
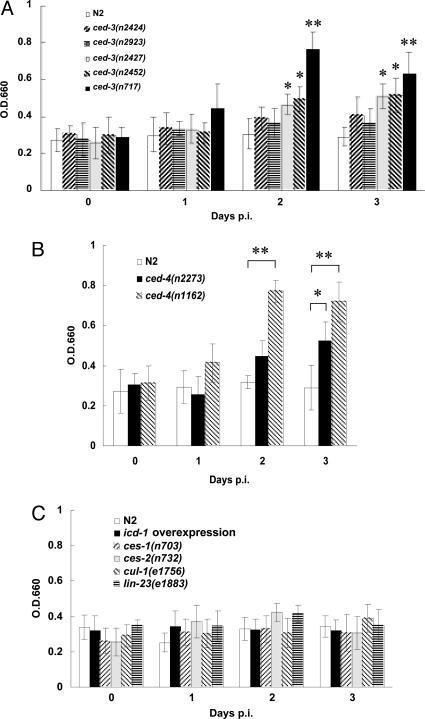
To investigate the relationship between virus replication and the cell-killing activities of ced-3 and ced-4 genes, we examined the replication levels of VV in ced-3 or ced-4 mutants with different allelic strengths. The PCD defect of ced-3 mutants can be quantified by the number of extra surviving cells present in the anterior region of the pharynx (35). Animals homozygous for the alleles n2424, n2923, n2427, n2452, and n717 have on average 0, 0, 1.2, 9.5, and 11.3 extra cells in the anterior pharynx, respectively, indicating that these ced-3 lesions debilitate CED-3 cell-killing activities in a graded manner. In Fig. 3A, the weak mutations n2424 and n2923 did not increase the viral replication level, whereas the stronger mutations n2427 and n2452 modestly supported VV replication and the strongest ced-3(n717) mutation greatly enhanced the VV replication starting from day 2 p.i. Consistently, VV replicated more efficiently in the strong ced-4(n1162) mutants than in the weak ced-4(n2273) mutants (P < 0.05 by Student’s t test) (Fig. 3B). Thus, the cell-killing activities of ced-3 and ced-4 genes were positively associated with VV replication levels in C. elegans.
Fig. 3.
The effects of different ced-3 and ced-4 alleles on vaccinia virus replication in C. elegans. About 1,000 worms (stages L3–L4) of the indicated strains were infected with VV in the presence of PEG. The level of virus replication in each strain was determined from the β-galactosidase activity at OD660. (A) Viral replication levels in most of the ced-3 mutants tested were higher than those in wild type. (B) VV replication levels in ced-4 mutants were significantly higher than those in wild-type worms at the second day p.i. (C) Overexpression of icd-1 or mutations in ces-1, ces-2, cul-1, and lin-23 did not affect VV titers. All experiments were repeated three times. (∗, P < 0.05; ∗∗, P < 0.01 by Student’s t test). Data are means ± SD.
In contrast to genes of the core PCD machinery, we found that the recently identified cell-death inhibitor icd-1 was not involved in the control of VV replication (Fig. 3C). The icd-1 gene encodes the β-subunit of the nascent polypeptide-associated complex (β-NAC) and can function to protect cells from PCD in a ced-4-dependent but ced-3-independent way (36). To study the involvement of icd-1 in virus replication, we used the strain carrying the transgene Phsp::icd-1, which contains icd-1 cDNA under the control of the C. elegans heat shock promoter (36). Overexpression of icd-1 appears to inhibit apoptosis of the cells that are normally programmed to die. Even though the heat-shock-induced expression of icd-1 blocked the PCD, it did not enhance VV replication in C. elegans (Fig. 3C). Similarly, VV replication was not affected by ces-1(n703) and ces-2(n732) mutations, which prevent the deaths of a specific set of cells (37) (Fig. 3C). Thus, not all gene components of the PCD pathway control VV replication in worms, only those of the core PCD machinery, ced-3, ced-4, ced-9, and egl-1.
On the other hand, although ced-3(lf), ced-4(lf), ced-9(gf), and egl-1(lf) mutations result in extra cells, several of our observations suggest that these extra cells cannot account for the increase of VV replication levels in these mutants. For example, compared with wild type, the strong ced-3(n717) and ced-4(n1162) mutants displayed a 60–150% increase in VV titers and β-galactosidase activities on the day 2 p.i. (Fig. 2 A and B), whereas these mutants have only ≈12% (131 of 1,090 at most) increase in somatic cell numbers. In addition, overexpression of icd-1, which results in the survival of an averaged of 3.8 extra cells in the anterior pharynx (36), did not enhance VV replication (Fig. 3C), whereas the ced-3(n2427) mutation, which causes fewer extra cells (≈1.2 cells) to survive in the pharynx, did (Fig. 3A). We further investigated whether the presence of surplus cells in cul-1 and lin-23 mutants could enhance VV replication. The cul-1 and lin-23 mutations fail to restrain cell proliferation in various cell types, thereby resulting in supernumerary cell production in C. elegans (38, 39). Compared with wild type, cul-1, and lin-23 mutants have, on average, 115 and 70 extra somatic cells, respectively (38, 39). However, we found that β-galactosidase activities in cul-1(e1756) and lin-23(e1883) mutants were indistinguishable from those of wild type (Fig. 3C). Thus, the increase of VV replication levels in the PCD-defective mutants was unlikely to be caused by the extra live cells. Together, these results suggest that the restriction in VV replication is mediated through the core PCD genes including ced-3, ced-4, ced-9, and egl-1.
Killing of C. elegans by VV Infection Is PCD-Independent.
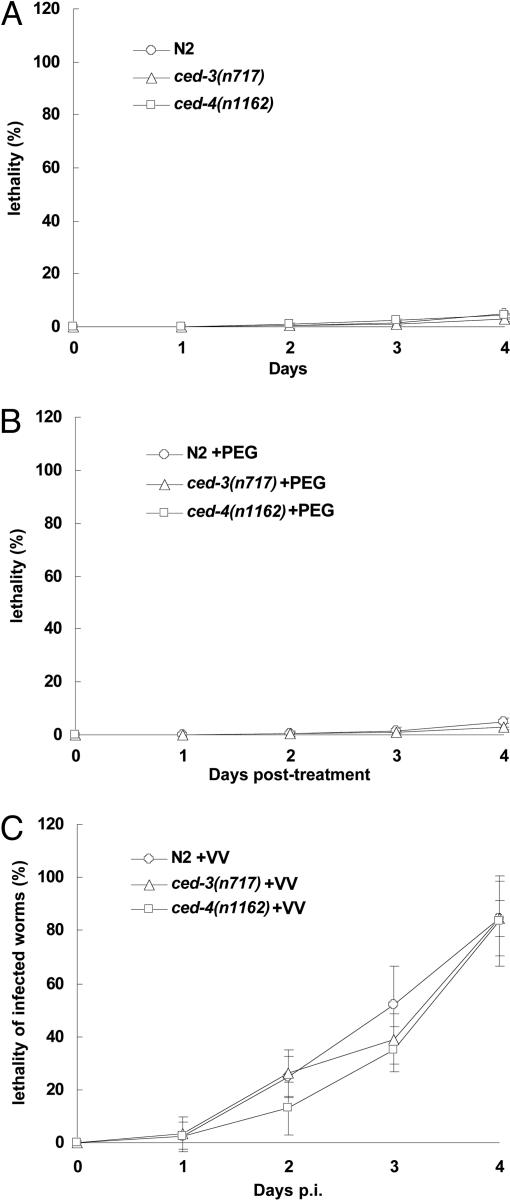
While analyzing vaccinia virus-containing worms, we noticed that many worms moved slowly and died after the PEG-mediated virus treatment. We observed no obvious abnormality at the cellular level under a Nomarski microscope in worms stained positive with X-Gal. We then investigated whether the death of worms was a consequence of VV infection. Although PEG treatment alone caused the rapid death of ≈25% of worms, this treatment did not appear to cause any further death in worms during the 4 days p.i. (Fig. 4A and B). In contrast, PEG-mediated VV infection rapidly killed >80% of the treated worms within 4 days (Fig. 4C), and as a control, the UV-inactivated VV failed to kill the worms, indicating that the VV-mediated killing depended on intracellular viral genome replication. It has been previously demonstrated that both replication and bacteria-mediated killing of Pseudomonas aeruginosa is independent of PCD in C. elegans (3). Although ced-3(717) and ced-4(n1162) mutations caused a marked enhancement for VV replication, they had virtually the same killing effect as wild type on the associated lethality (Fig. 4C), suggesting that, in wild-type worms, even at the basal levels, the process of viral genome replication is cytotoxic enough to kill the infected hosts. Thus, PEG-mediated VV infection appears to kill C. elegans within 4 days through an unknown PCD-independent mechanism.
Fig. 4.
Vaccinia-mediated killing of C. elegans is independent of ced-3 and ced-4. About 100 worms (stages L3–L4) of each C. elegans strain were treated under the indicated conditions. Dead worms were picked on indicated days and tested by using X-Gal staining. The percentage of the dead infected worms of each strain was calculated by the following formula: (the number of total X-Gal-positive worms that died by the indicated days/the number of total infected worms) × 100%. The number of total infected worms was calculated by the number of total animals times the infection rate. (A) No PEG or virus treatment. (B) PEG treatment without virus. (C) PEG plus virus treatment. All data (means ± SD) were obtained from four independent experiments.
Discussion
VV, like many animal viruses, can readily trigger infected cells to undergo apoptotic or necrotic cell death. However, no cellular components involved in either of these death processes have been identified to modulate VV replication in infected mammalian cells (40). Given the coevolutionary stresses between virus and host, it is actually not unexpected that there are no mammalian ced-related genes that can influence VV replication in vitro and in vivo. To successfully harness mammalian cells as efficient viral factories, vaccinia virus must have gained various effective strategies to antagonize the unfavorable effects caused by its mammalian ced counterparts. VV could replicate substantially only in ced mutants of C. elegans, demonstrating that these ced genes possess a previously unidentified function for VV restriction. Conceivably, only the ced genes from C. elegans, but not their mammalian homologs, can play such a role in the control of VV replication. Alternatively, VV restriction mediated by ced-related genes may be evolutionally and functionally conserved both in C. elegans and mammals, and yet, because of the discrepancy in cellular milieus between C. elegans and mammals, this restriction capability can only be revealed in C. elegans rather than mammalian system. Thus, it is of interest to investigate whether human ced-related genes can functionally substitute the worm ced genes to restrict VV replication and, reciprocally, whether the worm ced genes can inhibit VV replication in the mammalian system.
We show here that the ced-3, ced-4, ced-9, and egl-1 genes, composing the core PCD machinery, control VV replication inside C. elegans. In addition, the effects of ced-3 or ced-4 mutations on the cell-killing activity and VV replication levels in worms appear to be closely associated. However, our data also suggest that the increase in viral replication levels in ced-3 and ced-4 mutants is not a consequence of extra surviving cells, as icd-1 overexpression and cul-1 and lin-23 mutations, despite having more cells in the worms, all failed to augment VV replication (Fig. 3C). A robust expression of icd-1 induced by heat-shock treatment has previously been shown to inhibit PCD during the development of wild-type C. elegans (36). Intriguingly, we observed that such an icd-1 overexpression failed to boost VV replication in our experimental setting, suggesting that vaccinia restriction in wild-type C. elegans cannot be lifted by the icd-1-controlled PCD pathway. Thus, in wild-type worms, the components of the core PCD machinery seem to possess another unknown PCD-independent mechanism that suppresses intracellular VV replication. Because no somatic cell death can be seen in soma of wild-type hermaphroditic worms after the L2 stage, the ced-3/ced-4 pathway may conceivably play a role in activation of defense responses to block VV replication in our study.
The genes ced-3, ced-4, and egl-1, but not the cell-death-specification genes ces-1 or ces-2, protect C. elegans from Salmonella typhimurium-induced killing (3). It has been hypothesized that colonization of S. typhimurium to the worm’s intestine might activate a ced-3/ced-4-dependent signaling pathway that triggers cell death in the germ line but activates defense responses in somatic cells (3). How extracellular bacteria induce PCD-associated signaling in adult C. elegans remains elusive. On the other hand, cytoplasmic VV replication has been shown to readily trigger the apoptotic signaling pathway (40); moreover, we found the core PCD-defective mutants of C. elegans appeared to loosen the control over VV replication. Our observations and a previous study on S. typhimurium (3) together suggest that the core PCD machinery participates in an innate defense mechanism for C. elegans in response to attacks by either intracellular or extracellular pathogens.
Although the ced-3(n2452) allele could increase VV replication, the enhanced replication level was significantly lower than that caused by the ced-3(n717) allele on the second day p.i. (Fig. 3A). The ced-3(n2452) is the allele that deletes the protease domain of CED-3/caspase (35). It is possible that the CED-3 protein in the ced-3(n2452) mutant may contain a nonprotease killing activity that is not disrupted by the deletion mutation. This possibility supports the notion that ced-3 may restrict VV replication in a protease-independent pathway. However, it is also possible that the CED-3 protein of the n717 mutant may have an abnormal activity that interferes with the components that regulate programmed cell death as well as virus replication. In fact, VV encodes its own caspase inhibitor, SPI-2, to modulate the apoptotic cascade occurring in the infected mammalian cells. SPI belongs to the serpin family, which specifically inhibits interleukin-1β-converting enzyme (ICE), caspase 8, and granzyme B (41). It is as yet unclear whether SPI-2 can suppress the CED-3 caspase activity, and might therefore be involved in VV replication in C. elegans.
In summary, we have established a protocol allowing VV to enter and then replicate in C. elegans. VV replication could be significantly enhanced in the core PCD-defective mutants of ced-3, ced-4, ced-9, and egl-1, but not in other PCD mutants. Moreover, the increase of VV replication levels in the PCD-defective mutants was unlikely to be caused by the extra live cells, because both icd-1 overexpression and cul-1 and lin-23 mutants, although retaining more viable cells, failed to boost VV replication. Therefore, the core PCD genes possess a unique function to restrict VV replication in C. elegans. In addition to VV, our infection protocol was also successfully used for Sindbis virus to replicate in wild-type worms. Therefore, the PEG-mediated infection protocol may be a practical approach to induce other viruses to enter and replicate in C. elegans. Virus-infected C. elegans mediated by PEG might thus serve as a valuable model to study virus–host interactions from a new perspective.
Materials and Methods
Worm and Virus Strains.
All C. elegans strains were maintained on NGM agar with E. coli OP50 as a food source using standard methods (see Supporting Text, which is published as supporting information on the PNAS web site). The wild-type strain of Bristol N2 C. elegans and its derivative mutants were used throughout this study. The recombinant VV vREβCAT has dual reporters in which the late F17R/F18R promoter drives the lacZ gene and the early promoter of the VV growth factor gene controls the chloramphenicol acetyltransferase (CAT) gene (23). vREβCAT viruses were routinely amplified in BSC40 cells by collecting both intracellular mature virus and extracellular enveloped virus particles (23).
PEG-Mediated Viral Infection in C. elegans.
About 1,000 worms at L3 and/or L4 stages were washed off plates by using M9 buffer and collected by low-speed centrifugation. Worms were washed twice with M9 buffer and then mixed with 200 μl of M9 medium containing 2% PEG 1000 (Sigma) plus 1 × 107 pfu/ml VV, or 2 × 108 pfu/ml Sindbis virus, for 5 min at room temperature. PEG caused ≈25% sudden death during this treatment. The survivors were washed and further soaked for 6 h at room temperature in M9 solution containing 1 × 107 pfu/ml VV or 2 × 108 pfu/ml Sindbis virus. The resulting worms were washed with M9 buffer twice to remove excess virus particles and placed on 60-mm plates containing confluent bacteria. The treated worms were harvested at different times for phenotypic analysis, plaque-forming assay, β-galactosidase assay, and X-Gal staining.
C. elegans Fixation and X-Gal Staining.
The X-Gal staining protocol was modified from a described method (45). Briefly, the infected worms were washed off plates by using M9 buffer, and further washed twice in M9 solution to remove bacteria. Worms were resuspended in 0.5 ml of M9 buffer and mixed with 0.5 ml of ice-cold staining solution (160 mM KCl/40 mM NaCl/20 mM Na2EGTA/10 mM spermidine-HCl/30 mM Na Pipes, pH 7.4/50% methanol) containing 4% formaldehyde. The mixture was frozen at −80°C and then quickly thawed in a 70°C water bath. Worms were washed in a Tris-Triton buffer (100 mM Tris·HCl, pH 7.4/1% Triton X-100/1 mM EDTA) and incubated in 1 ml of 1% 2-mercaptoethanol/Tris-Triton buffer for 1–2 h at 37°C. Worms were further washed in 1 ml of borate buffer (25 mM H3BO3/12.5 mM NaOH) and incubated in 1 ml of 10 mM DTT/borate buffer for 15 min at room temperature. Finally, worms were washed again in 1 ml of borate buffer and stained with 120 μg/ml X-Gal at room temperature for 8–10 h.
VV-Mediated Killing Assays.
About 100 L4 worms were washed off plates and infected with VV as described. The infected animals were transferred to fresh plates containing E. coli OP50. Plates were scored every 24 h, and four replicates were performed. A worm was considered dead when it no longer responded to being touched with a platinum wire. The dead worms were collected and examined for viral infection by using the X-Gal staining technique. The percentage of the dead infected worms of each strain was calculated by the following formula: (the number of total X-Gal positive worms that died by the indicated days/the number of total infected worms) × 100%. The number of total infected worms is calculated by the number of total animals times the infection rate. The infection rate was determined at the second or third days p.i. The values of the VV infection rates were similar for wild-type, ced-3, and ced-4 worms.
Supplementary Material
Acknowledgments
We thank R. H. Horvitz, J. Rothman, and the Caenorhabditis Genetics Center for C. elegans strains and Wen Chang (Academic Sinica) for the recombinant vREβCAT virus and anti-D8L antibody. This work was supported in part by National Health Research Institutes Grant NHRI-EX94-9330SI, National Science Council Grants NSC94-3112-B-016-001 and NSC91-2320-B-182-034, National Research Program for Genomic Medicine Grant 93GM040, and C. Y. Foundation for advancement of Sciences (Taipei, Taiwan, Republic of China).
Glossary
Abbreviations:
- PEG
polyethylene glycol
- VV
vaccinia virus
- PCD
programmed cell death
- pfu
plaque-forming unit
- p.i.
postinfection
Note.
After our submission of this manuscript to the PNAS Editorial Office, two papers describing RNA virus infection to C. elegans were published. Wilkins et al. (43) showed that vesicular stomatitis virus was able to infect the primary cells derived from C. elegans (43), and Lu et al. (44) used a reverse genetic approach to show that Flock house virus (FHV) could replicate its genomic RNA in the FHV-DNA transgenic worms. The results of both studies suggest that RNA interference (RNAi) is an antiviral immune defense mechanism to RNA viruses in the worm. On the other hand, our study reveals that VV, one of the medically important DNA viruses, is able to replicate in the body of PEG-treated C. elegans, whose components in PCD pathway play a role in restriction of VV replication. Thus, C. elegans appears a good model organism for the studies of virus–host interactions.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darby C., Cosma C. L., Thomas J. H., Manoil C. Proc. Natl. Acad. Sci. USA. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodgkin J., Kuwabara P. E., Corneliussen B. Curr. Biol. 2000;10:1615–1618. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- 3.Aballay A., Ausubel F. M. Proc. Natl. Acad. Sci. USA. 2001;98:2735–2739. doi: 10.1073/pnas.041613098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurz C. L., Ewbank J. J. Nat. Rev. Genet. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 5.Schulenburg H., Kurz C. L., Ewbank J. J. Immunol. Rev. 2004;198:36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- 6.Alegado R. A., Campbell M. C., Chen W. C., Slutz S. S., Tan M. W. Cell. Microbiol. 2003;5:435–444. doi: 10.1046/j.1462-5822.2003.00287.x. [DOI] [PubMed] [Google Scholar]
- 7.Gravato-Nobre M. J., Hodgkin J. Cell. Microbiol. 2005;7:741–751. doi: 10.1111/j.1462-5822.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 8.Sifri C. D., Begun J., Ausubel F. M. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Kurz C. L., Ewbank J. J. Trends Microbiol. 2000;8:142–144. doi: 10.1016/s0966-842x(99)01691-1. [DOI] [PubMed] [Google Scholar]
- 10.Mylonakis E., Ausubel F. M., Perfect J. R., Heitman J., Calderwood S. B. Proc. Natl. Acad. Sci. USA. 2002;99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., Murray B. E., Calderwood S. B., Ausubel F. M. Proc. Natl. Acad. Sci. USA. 2001;98:10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garsin D. A., Villanueva J. M., Begun J., Kim D. H., Sifri C. D., Calderwood S. B., Ruvkun G., Ausubel F. M. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 13.Kim D. H., Ausubel F. M. Curr. Opin. Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Thompson C. B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 15.Hedgecock E. M., Sulston J. E., Thomson J. N. Science. 1983;220:1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- 16.Horvitz H. R., Shaham S., Hengartner M. O. Cold Spring Harbor Symp. Quant. Biol. 1994;59:377–385. doi: 10.1101/sqb.1994.059.01.042. [DOI] [PubMed] [Google Scholar]
- 17.Croyle M. A., Yu Q. C., Wilson J. M. Hum. Gene Ther. 2000;11:1713–1722. doi: 10.1089/10430340050111368. [DOI] [PubMed] [Google Scholar]
- 18.Flores E. F., Kreutz L. C., Donis R. O. J. Gen. Virol. 1996;77:1295–1303. doi: 10.1099/0022-1317-77-6-1295. [DOI] [PubMed] [Google Scholar]
- 19.Buller R. M., Palumbo G. J. Microbiol. Rev. 1991;55:80–122. doi: 10.1128/mr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldick C. J., Jr., Cassetti M. C., Harris N., Moss B. J. Virol. 1994;68:6052–6056. doi: 10.1128/jvi.68.9.6052-6056.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldick C. J., Jr., Moss B. J. Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright C. F., Moss B. Proc. Natl. Acad. Sci. USA. 1987;84:8883–8887. doi: 10.1073/pnas.84.24.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsey-Ewing A., Moss B. Virology. 1995;206:984–993. doi: 10.1006/viro.1995.1021. [DOI] [PubMed] [Google Scholar]
- 24.Xie G., Jia Y., Aamodt E. Genet. Anal. 1995;12:95–100. doi: 10.1016/1050-3862(95)00112-3. [DOI] [PubMed] [Google Scholar]
- 25.Sulston J. E., Horvitz H. R. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 26.Sulston J. E., Schierenberg E., White J. G., Thomson J. N. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 27.Kimble J., Hirsh D. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 28.Ellis H. M., Horvitz H. R. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 29.Hengartner M. O., Ellis R. E., Horvitz H. R. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 30.Conradt B., Horvitz H. R. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 31.Yuan J., Shaham S., Ledoux S., Ellis H. M., Horvitz H. R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 32.Hengartner M. O., Horvitz H. R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 33.Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 34.Yuan J., Horvitz H. R. Development (Cambridge, U.K.) 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 35.Shaham S., Reddien P. W., Davies B., Horvitz H. R. Genetics. 1999;153:1655–1671. doi: 10.1093/genetics/153.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bloss T. A., Witze E. S., Rothman J. H. Nature. 2003;424:1066–1071. doi: 10.1038/nature01920. [DOI] [PubMed] [Google Scholar]
- 37.Ellis R. E., Horvitz H. R. Development (Cambridge, U.K.) 1991;112:591–603. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]
- 38.Kipreos E. T., Lander L. E., Wing J. P., He W. W., Hedgecock E. M. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 39.Kipreos E. T., Gohel S. P., Hedgecock E. M. Development (Cambridge, U.K.) 2000;127:5071–5082. doi: 10.1242/dev.127.23.5071. [DOI] [PubMed] [Google Scholar]
- 40.Kettle S., Alcami A., Khanna A., Ehret R., Jassoy C., Smith G. L. J. Gen. Virol. 1997;78:677–685. doi: 10.1099/0022-1317-78-3-677. [DOI] [PubMed] [Google Scholar]
- 41.Turner P. C., Moyer R. W. Semin. Virology. 1998;8:453–469. [Google Scholar]
- 42.Blasco R., Moss B. J. Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkins C., Dishongh R., Moore S. C., Whitt M. A., Chow M., Machaca K. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 44.Lu R., Maduro M., Li F., Li H. W., Broitman-Maduro G., Li W. X., Ding S. W. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fire A., Harrison S. W., Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.