Abstract
RNA methyltransferase is responsible for transferring methyl and resulting in methylation on the bases or ribose ring of RNA, which existed widely but mostly remains an open question. A recombinant protein PH1948 predicting RNA methyltransferase from Pyrococcus horikoshii OT3 has been crystallized. The crystals of selenomethionyl PH1948 belong to space group C2, with unit-cell parameters a=207.0 Å, b=43.1 Å, c=118.2 Å, β=92.1°, and diffract X-rays to 2.2 Å resolution. The V M value was determined to be 2.8 Å3/Da, indicating the presence of four protein molecules in the asymmetric unit.
Keywords: Pyrococcus horikoshii, Methyltransferase, X-ray diffraction, Protein crystallography
INTRODUCTION
Methyl transfers are alkylation reactions central to cellular biochemistry, by far, S-adenosyl-L-methionine (AdoMet) is the most commonly used methyl donor. AdoMet-dependent methyltransferases (MTases) involved in biosynthesis, detoxification, signal transduction, protein sorting and repair, chromatin regulation and gene silencing (Armengaud et al., 2004; Schubert et al., 2003). Including DNA MTase, RNA MTase, protein MTase, phospholipids and small molecule MTase, there are more than fifty MTases structures revealed by X-ray crystallography and NMR (Forouhar et al., 2003; Cheng and Roberts, 2001). Among these MTases, RNA MTase high capability to act on RNA directly relates to gene transcription and expression. Such as methylation of the 5′-terminal cap of mRNA involved its transportation, efficient translation, and stability (Chiang et al., 1996).
For MTases, the lack of appropriate substrates, instability, or low activity of the enzymes, and possible redundancy of their functions in vivo, hampered their identification and functional characterization. There are 14 MTases concerning methylations of 23S rRNA in E. coli, but few of the methylating activities have been described to the level of the partial purification, and only one had been well characterized (BÜgl et al., 2002). According to Clusters of Orthologous Groups (COGs) concerning the functional category of translation, ribosomal structure and biogenesis, the number of proteins known or predicted to be RNA MTases is 503. Among them, 15 MTases come from Pyrococcus horikoshii OT3. It is almost impossible to characterize all of them by classical biochemical approach. Solution and analysis of MTases or the complex structures, combining sophisticated sequences comparisons and MTases with known functions, will assist in identifying the biochemical and cellular function of the unknown MTases (Martin and McMillan, 2002).
Protein PH1948 was predicted as a kind of RNA MTase (also called RNA methylase) in Pyrococcus horikoshii OT3, although it has not been fully identified. Here we present preparation, crystallization and preliminary X-ray diffraction analysis of PH1948 for structural analysis.
EXPERIMENTAL DETAILS
Protein expression and purification
The PH1948 gene amplified by PCR from genomic DNA of Pyrococcus horikoshii OT3 was cloned into vector pET26b (Novagen), then the recombinant vector was transformed into Escherichia coli strain B834 (DE3) for protein expression.
Twenty ml aliquots of an overnight culture were inoculated into 2 L LB medium containing 15 μg/ml kanamycin, and grown at 310 K. While optical density at 600 nm was about 0.6, this strain’s cells were induced by the addition of 1 mmol/L isopropyl-β-D-thiogalactopyranoside (IPTG), and grown continuously for 5 h. The harvested cells were resuspended in buffer A (50 mmol/L sodium phosphate pH 6.0) and disrupted with a French press. The lysate was incubated at 343 K for 30 min, then kept on ice for 10 min, finally centrifuged (40000×g for 30 min, 277 K). The supernatant was filtered (0.45 μm, Millipore) and applied onto a HiTrap SP HP column (Amersham Bioscience) equilibrated with buffer A. The bounded protein was eluted with a linear gradient of NaCl (0–1 mol/L, 20 column volumes). The target fraction was loaded onto a column HiLoad 26/60 superdex75pg (Amersham Bioscience), balanced with buffer B (50 mmol/L sodium phosphate pH 6.0, 0.5 mol/L NaCl). The peak fraction was dialyzed overnight against buffer, then applied onto a Resource S column (Amersham Bioscience). With a linear gradient of 0 to 1.0 mol/L NaCl (30 column volumes), PH1948 was eluted at 0.54 mol/L NaCl. The protein buffer was exchanged by dialysis against 10 mmol/L Tris-HCl pH 9.0, then concentrated to a final concentration of 5 mg/ml. Purification of the SeMet-substituted PH1948 (Se-PH1948) is the same as that of the native protein.
Crystallization and X-ray data collection
Hanging-drop vapour diffusion method was applied to perform initial screening with crystallization screening kits. Each drop consisted of 1 μl sample and 1 μl reservoir solution, equilibrated against 100 μl reservoir solution at 293 K. Initial crystals were obtained with Hampton Research Crystal Screen I (No. 14, 0.1 mol/L HEPES-Na pH 7.5, 28% PEG400, 0.2 mol/L CaCl2). Further optimization was done to improve the quality of crystals using hanging-drop method for Se-PH1948. The drop contained 1.5 μl sample and 1.5 μl reservoir solution, then equilibrated against 1 ml reservoir solution. Large crystals reaching dimensions of 0.35 mm×0.25 mm×0.1 mm grew within 2 weeks, which were achieved at the condition of 0.1 mol/L HEPES-Na, pH 7.0–7.2, 28%–30% PEG400 and 0.1 mol/L CaCl2 (Fig.1).
Fig. 1.

Crystals of Se-PH1948
Diffraction data of Se-PH1948 crystals were obtained out at PF-AR NW-12 beamline (Tsukuba, Japan). The crystals were flash-cooled under a stream of nitrogen gas after soaking in mother liquor containing 10% glycerol. To overcome the mosaicity increased by flash-cooling, an in situ flash-annealing technique was used (Yeh and Hol, 1998). All data were processed with HKL2000 and CCP4 software (Collaborative Computational Project, Number 4, 1994).
RESULTS AND DISCUSSION
The high expression level of PH1948 and the application of heat treatment facilitated the separation and purification process without affinity tag, although recombinant protein Se-PH1948 was very sensitive to the buffer pH during concentration. At the relatively low protein buffer pH used (pH below 8.5) aggregates rapidly appeared during concentration. It was no available for inhibiting the aggregation to add sodium chloride. The experimental trial showed that pH 9.0 Tris buffer could inhibit the aggregation. So before concentration of the protein, the buffer was substituted by 10 mmol/L pH 9.0 Tris-HCl.
Initial screening of the crystallization conditions showed that crystals grew in No. 14 of Crystallization Screening I, but that the crystals were thin and diffracted poorly. While protein concentration decreased from 5 to 3 mg/ml, the crystals grew relatively slowly. However, larger and reproducible crystals were obtained. The data-collection statistics are presented in Table 1. The selenium containing crystals belonged to the monoclinic space group C2, with unit-cell parameters a=207.0 Å, b=43.1 Å, c=118.2 Å, β=92.1°, and diffracted to 2.2 Å, The asymmetric unit contained 4 molecules, and the crystal volume per unit molecular weight (V M) was calculated to be 2.8 Å3/Da, correspondingly giving a solvent of 44%.
Table 1.
X-ray data-collection statistics for a Se-PH1948 crystal. Values in parentheses refer to the highest resolution shell
| Beamline | NW12 (PF) |
| Wavelength | 1.00 Å |
| Space group | C2 |
| Unit cell parameters | a=207.0 Å, b=43.1 Å, c=118.2 Å α=90°, β=92.1°, γ=90° |
| Resolution | 50.00−2.20 Å (2.28−2.20 Å) |
| Unique reflections | 53514 |
| Completeness | 99.3% (94.9%) |
| Redundancy | 5.0 (4.1) |
| I/σ | 20.0 |
| Rmerge* | 4.6% (21.8%) |
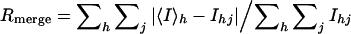 |
Acknowledgments
We thank N. Matsugaki and his colleagues for their kind help during collection of data on beamline NW12 of the Photon Factory, Japan.
Footnotes
Project supported by the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology of Japan
References
- 1.Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J Biol Chem. 2004;279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 2.BÜgl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JCA, Jakob U. RNA methylation under heat shock control. Molecular Cell. 2002;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 3.Cheng XD, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Research. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 5.Forouhar F, Shen J, Xiao R, Acton TB, Montelion G, Liang T. Functional assignment based on structural analysis: Crystal structure of the yggJ protein (HI0303) of Haemophilus influenza reveals an RNA methyltransferase with deep trefoil knot. Proteins: Structure, Function and Genetics. 2003;53:329–332. doi: 10.1002/prot.10510. [DOI] [PubMed] [Google Scholar]
- 6.Martin JL, McMillan FM. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Current Opinion in Structure Biology. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 7.Schubert HL, Blumenthal RM, Cheng XD. Many paths to methyltransfer: A chronicle of convergence. Trends in Biochemical Science. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh JI, Hol WG. A flash-annealing technique to improve diffraction limits and lower mosaicity in crystals of glycerol kinase. Acta Cryst. 1998;D54:479–480. doi: 10.1107/s0907444998004697. [DOI] [PubMed] [Google Scholar]


