Abstract
This report describes a new genotyping method capable of detecting low-abundant point mutations in a homogeneous, separation-free format. The method is based on integration of oligonucleotide ligation with a semiconductor quantum dot (QD)-mediated two-color fluorescence coincidence detection scheme. Surface-functionalized QDs are used to capture fluorophore-labeled ligation products, forming QD-oligonucleotide nanoassemblies. The presence of such nanoassemblies and thereby the genotype of the sample is determined by detecting the simultaneous emissions of QDs and fluorophores that occurs whenever a single nanoassembly flows through the femtoliter measurement volume of a confocal fluorescence detection system. The ability of this method to detect single events enables analysis of target signals with a multiple-parameter (intensities and count rates of the digitized target signals) approach to enhance assay sensitivity and specificity. We demonstrate that this new method is capable of detecting zeptomoles of targets and achieve an allele discrimination selectivity factor >105.
INTRODUCTION
The analysis of DNA sequence variants in human genome plays an increasingly important role in diagnosis of genetic and malignant diseases. Since the majority of sequence variants in genetic disorders are associated with variations such as single nucleotide substitutions, deletions and insertions, a rapid, sensitive and cost-effective DNA diagnostic method capable of differentiating single nucleotide polymorphisms (SNPs) is highly desired. A variety of methods have been used to detect SNPs, such as allele-specific hybridization, restriction-enzyme digestion, nuclease-based cleavage, primer extension, mass spectroscopy and oligonucleotide ligation (1–4). Most of these methods require cumbersome gel electrophoresis or a solid support phase as a means for sample separation prior to detection. Development of separation-free methods for SNP detection has been receiving increasing attention because they allow simple assay protocols, instrument automation and high analysis rate (5,6). In addition, performing molecular reactions and detection in a homogeneous, separation-free format facilitates more effective binding kinetics (7,8), thereby improving sensitivity and throughput. A number of separation-free methods for SNP detection have been developed based on microbead-based flow cytometry (5,6,9–11) and fluorescence resonance energy transfer, such as molecular beacons (12–18) and Taqman probes (19,20). Recently, functional nanomaterials such as gold nanoparticles have been used to facilitate separation-free SNP detection by taking advantages of the change of materials properties upon hybridization (21–24).
Ultrasensitive methods that can detect low-abundant DNA sequences in a separation-free format (25–27) have also been developed based on single-molecule photon burst detection (28–32). These methods commonly incorporate two distinctly dye-labeled probes [peptide nucleic acids (26) or oligonucleotides (25)] that can hybridize with a specific target sequence and a confocal laser-induced fluorescence (LIF) spectroscopic system that can detect single fluorophore. Coincident photon bursts detected by two separate photodiodes at different wavelengths indicate the presence of specific targets, while noncoincident signals indicate the absence of targets. This two-color fluorescence coincidence detection approach allows detection of rare targets with reduced or even without PCR amplification. However, this detection platform does not have the capability for identifying point variations within nucleotide sequences.
In this report, we aim to develop a separation-free detection method capable of detecting low-abundant point mutations by incorporating oligonucleotide ligation assay (OLA) (33,34) and semiconductor quantum dots (QDs) (35,36) into the two-color fluorescence coincidence detection platform. The presence of ligation products and thus the genotype of the sample can be determined by detecting the coincident fluorescent signals upon formation of QD-oligonucleotide nanoassemblies. QDs have several unique photophysical properties such as large Stokes shift, broad absorption and narrow emission spectra (35–38). The use of QDs as fluorescent tags therefore avoid emission cross-talk that otherwise complicates two-color fluorescence analysis. QD also functions as a nanoscaffold that confines multiple fluorescent ligation products (FLPs) within a nanoscale domain, amplifying the target signal. These features enable unambiguous detection of coincident fluorescent signals, leading to highly sensitive point mutation detection. The feasibility of this QD-based ligation assay has been exemplified using Kras point mutation detection as a model.
MATERIALS AND METHODS
Oligonucleotides
All the oligonucleotides (Table 1) used in the experiments were purchased from Integrated DNA Technologies (Coralville, IA). The β1 and β2 target sequences were derived from human β-globin gene (33). The two allele-specific ligation probes, βDP1 and βDP2, were biotinylated at the 5′ ends. The common probe, βRP, was labeled with Oregon Green 488 (OG488). For Kras oncogene mutation analysis, two allele-specific probes, K-WDP and K-MDP, and a common probe, K-Com, were designed to detect the point mutation (G→T) on codon 12 of Kras gene. All probes were HPLC purified.
Table 1.
Nucleotide sequences used in QD-mediated point mutation detection
| Code | Description | Sequence (5′ to 3′) |
|---|---|---|
| β1 | A segment in human β-globin gene | ACG GCA GAC TTC TCC TCA GGA GTC AGG TGC AC |
| β2 | Identical to β1 except one nucleotide (C→G) | ACG GCA GAC TTC TCC TGA GGA GTC AGG TGC AC |
| βDP1 | Biotinylated discrimination probe (perfect match to β1) | Biotin-GTG CAC CTG ACT CCT G |
| βDP2 | Biotinylated discrimination probe (perfect match to β2) | Biotin-GTG CAC CTG ACT CCT C |
| βRP | Oregon Green 488 labeled report probe | p AGG AGA AGT CTG CCG T-OG488 |
| K-WDP | Wild-type discrimination probe for Kras gene | OG488-CTC TTG CCT ACG CCA C |
| K-MDP | Mutant type discrimination probe for Kras gene | OG488-CTC TTG CCT ACG CCA A |
| K-Com | Biotinylated common probe for Kras gene | p CAG CTC CAA CTA CCA C-Biotin |
| SLP | Synthesized ligation product | Biotin-GTG CAC CTG ACT CCT GAG GAG AAG TCT GCC GT-OG488 |
Genomic DNA extraction from cell lines and PCR amplification
Genomic DNA samples were extracted from ovarian borderline tumors with known Kras mutation status (39). A palm laser capture microdissection microscope (Zeiss) was used to enrich the tumor epithelium and a PicoPure DNA extract kit (Arcturus, Mountain View, CA) was applied to prepare genomic DNA. The primers for PCR were as follows: TAA GGC CTG CTG AAA ATG ACT G (forward) and TGG TCC TGC ACC AGT AAT ATG C (reverse). After a 2 min denaturation step, 1.5 U Amplitaq DNA polymerase (Perkin-Elmer, Norwalk, CT) was added under hot-start conditions, and amplification was achieved by thermal cycling for 40 cycles at 95°C for 30 s, 60°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 3 min. PCR products were stored at −20°C until required for the ligation assays.
Oligonucleotide ligation assay
T4 and Taq DNA ligase were purchased from New England BioLabs, Inc. (Beverly, MA) with an initial concentration of 400 and 40 U/µl, respectively. For human β-globin gene point mutation detection, a 20 µl reaction mixture comprised of 2.4 pmol reporter probes (βRP), 2.4 pmol discrimination probes (βDP1 or βDP2), 2.4 pmol templates (β1 or β2), 1 U of T4 DNA ligase and 1× company supplied T4 ligation buffer [50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM adenosine triphosphate, and 25 µg/ml BSA], was prepared. After carrying out ligation at 22°C for half an hour, 1 µl aliquots were removed from each ligation reaction solution and were diluted with 98 µl phosphate-buffered saline (PBS) buffer (10 mM sodium phosphate, 100 mM NaCl, pH 7.0) in a test tube. The test tube was placed in an 85°C hot water bath for 5 min to stop the ligation and to completely denature probe/template duplexes, followed by cooling in an ice water bath for 5 min. The tube was then stored in a 4°C refrigerator until required for QD coupling reaction.
For Kras gene point mutation detection, a 20 µl reaction mixture comprised of 0.24 pmol K-Com probes (K-Com), 0.24 pmol discrimination probes (K-WDP or K-MDP), 0.5 µl PCR products (Kras homogeneous wild-type or heterozygous type), 2 U of Taq thermal stable DNA ligase, and 1× company supplied Taq ligation buffer [20 mM Tris–HCl (pH 7.6), 25 mM potassium acetate, 10 mM magnesium acetate, 10 mM DTT, 1 mM NAD+, 0.1% Triton X-100] was prepared. A standard procedure for ligation reaction detection (LDR) was then performed on this mixture (17,40). Thermal cycling was performed in a MultiGene II thermal cycler (Denville, Metuchen, NJ) for 5 cycles at 95°C for 2 min, 55°C for 4 min and a final extension at 40°C for 3 min. After LDR thermal cycling, 10 µl aliquots were removed from each ligation reaction solution and were diluted with 89 µl PBS buffer inside a test tube. The test tube was placed in an 85°C hot water bath for 5 min to denature probe/template duplexes, followed by cooling in an ice water bath for 5 min.
Coupling fluorescent ligation products to quantum dots
CdSe-ZnS QDs (peak emission wavelength at 605 nm) were purchased from Quantum Dot Corporation (Hayward, CA). The QDs have been coupled to streptavidins directly through a carbodiimide-mediated coupling reaction (Streptavidin conjugates user manual, PN 90-0003 Rev 5, Quantum Dot Corp.). QDs were first diluted with PBS buffer to a concentration of 4 nM. One microliter of 4 nM QD solution was then added to each test tube which contained 99 µl diluted and heat denatured ligation products, resulting in a molar ratio of 30 between probes and QDs. Since each of the QDs used in the current assay has about 45–75 available biotin binding sites, it is important to keep the molar ratio between biotinylated probes and QDs used in the assay lower than 75 so that the biotin binding sites will not be depleted by the excess unligated probes (see Supplementary Data). The incubation was carried out at room temperature for 20 min, with mild agitation only at the beginning. To avoid non-specific adsorption of streptavidin-coated QDs on container surfaces, all test tubes where QDs would come into contact were pre-treated with purified water with 0.075% BSA before usage.
Instrumentation for two-color fluorescence coincidence detection
A custom-made single wavelength-excitation, dual-emission confocal LIF spectroscopic system was used for fluorescence detection (Figure 1B). A 488 nm air-cooled argon laser (Melles Griot, Irvine, CA) was used as an excitation light source. A 100× 1.3 N.A. oil immersion apochromatic objective (Olympus, Melville, NY) was used to focus the laser beam inside a 100 µm wide microcapillary (Polymicro Tech., Phoenix, AZ). The emitted fluorescence signal was collected by the same objective. Dichroic mirror 1 (Chroma Technology, Rockingham, VT) was chosen to allow light of wavelengths >505 nm to pass. A 50 µm pin hole (Melles Griot, Irvine, CA) was used to reject the out of focus fluorescence and background noise, thus enhancing the signal-to-noise ratio. The detection volume of the LIF system was estimated, according to the focal diameter of the focused laser beam (∼0.8 µm) and the depth of focus (∼2 µm) defined by the pinhole, to be ∼1.5 fl. Dichroic mirror 2 allowed light of wavelengths >565 nm to pass. Two avalanche photodiodes (APDs) (EG&G, Vandreuil, Canada) were incorporated to register the two filtered emission wavelengths, one from Oregon Green 488 dyes (OG488, emission maxima at 524 nm) and the other from QDs (emission maxima at 605 nm). A digital counter (National Instrument, Austin, TX) and a program written in LabView (National Instrument, Austin, TX) were implemented to perform data acquisition and data analysis. The excitation laser power was kept at 150 µW at all time. In the current experiment, the formed QD-fluorescent ligation products (QD-FLPs) nanoassemblies were further diluted with PBS buffer to reach a final concentration of 4 pM prior to detection. A small volume of sample (∼5 µl) introduced to a microcapillary was driven via hydrodynamic pumping at a flow rate of 1 µl/min during detection.
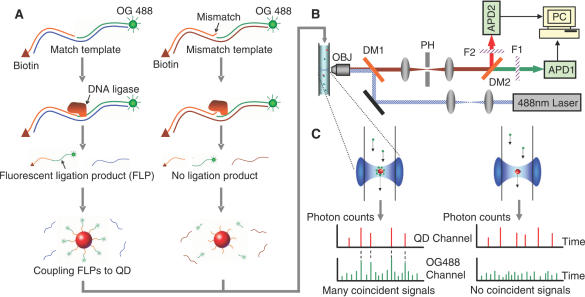
Figure 1.
Schematic concept for homogeneous point mutation detection using QD-mediated two-color coincidence analysis (A) Process flow for OLA: QD-FLPs nanoassemblies are formed in the presence of match templates (left). No QD-FLPs nanoassemblies are formed in the presence of the mismatch templates (right). (B) Detection of ligation products is carried out using a single wavelength-excitation, dual wavelength-emission confocal LIF spectroscopic system. OBJ, objective; DM(1 and 2), dichroic mirrors; PH, pin hole; F(1 and 2), filters; APD(1 and 2), avalanche photodiodes. (C) Left: When a QD-FLPs nanoassembly flows through the detection volume, simultaneous burst signals, or coincident signals (marked by dash lines), are detected in the two detection channels. Right: QDs are bound only with nonfluorescent probes in the mismatch experiments so no coincident signals will be seen. The coincident signals are therefore served as indicators of perfect match targets.
RESULTS
Assay principle
Schematic representation of the assay method is illustrated in Figure 1. First, a biotin-conjugated discrimination probe and an OG488-labeled reporter probe are covalently linked by T4 or Taq DNA ligase in the presence of perfect match targets (Figure 1A, left). After denaturing the duplex by heat, the FLP is separated from the target template. Multiple FLPs can be captured by a single streptavidin-conjugated QD through biotin–streptavidin interaction. Whenever a QD-FLPs nanoassembly flows through the miniscule detection volume of the confocal LIF system (Figure 1B), QD and OG488 are simultaneously excited. The resulting simultaneous emissions are separately detected by two photodiodes and are shown as coincident photon bursts (coincident signals) in Figure 1C. On the other hand, ligation does not occur when mismatch targets are used as templates (Figure 1A, right). Since QDs and organic fluorophores in this mismatch experiment are not associated to each other, they pass the detection volume independently without generating coincident signals. Therefore, perfect match targets can be distinguished from single-base mismatch targets by detecting the coincident signals.
Detection of single-base variations in oligonucleotide targets
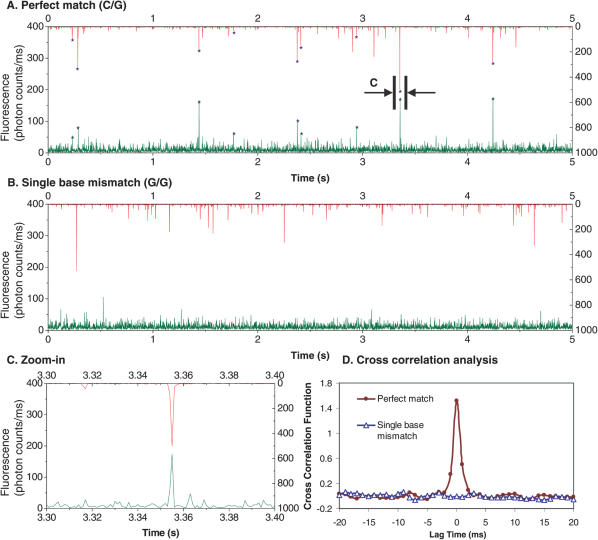
To reduce the effects of variability between DNA samples in evaluation of the assay, we began by analyzing single-base variations in synthetic oligonucleotide targets, β1 and β2, which were derived from variants of human β-globin gene sequences. Figure 2A shows a representative trace of fluorescence signals measured from the experiments with the presence of perfect match targets (β1 versus βDP1). The coincident signals, marked by asterisks, evidenced the formation of QD-FLPs nanoassemblies that brought QDs and OG488 together. In our confocal LIF system, the analyte is measured within a stationary femtoliter laser-focused detection volume, rendering an extremely low fluorescence background. Consequently, photon bursts with high signal-to-noise ratio emitted from single nanoassemblies could be effectively distinguished from the background (Figure 2C). In contrast, coincident signals were barely detected for the experiments with the presence of single-base mismatch targets (β2 versus βDP1, Figure 2B), suggesting that the formation of the nanoassemblies was prevented because of the lack of FLPs in the ligation process using mismatch targets as templates.
Figure 2.
Fluorescence burst signals and cross-correlation analysis. Each burst represents the passage of one entity, which can be a QD (red bursts, upper), a reporter probe (green bursts, lower), or a QD-FLPs assembly (coincident signals, marked by asterisks). The data were binned in 1 ms. (A) As a result of successful ligation on the perfect match templates (T1/DP1), many coincident signals were seen. (B) In the presence of single-base mismatch templates (T2/DP1), no such coincident signals were observed. (C) A close examination of a set of coincident signals. (D) Cross-correlation analysis: strong correlation was observed for perfect match templates at zero lag time (closed circle). However, for mismatch templates (open triangle), no correlation was seen.
The fluorescence signals from both detection channels were also analyzed by performing cross-correlation analysis using the following cross-correlation equation G(τ) (41):
where Ch1 and Ch2 are fluorescence intensities in the QD (red) and OG488 (green) detection channels, respectively, and τ denotes the lag time. As shown in Figure 2D, strong cross-correlation was seen at zero lag time for perfect match experiments but not for single-base mismatch experiments, further validating the QD-based two-color coincidence assay as an effective approach for detecting point mutations.
Target signal amplification
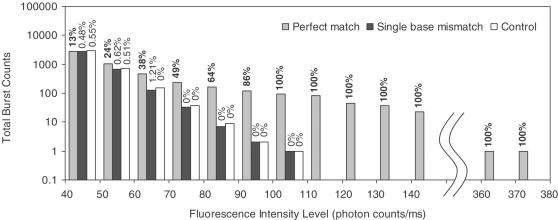
To address the effect of target signal amplification induced by the presence of multiple FLPs associated to a QD, we compared the intensity histograms of OG488 photon bursts detected in the perfect match experiments, the single-base mismatch experiments and additional control experiments (Figure 3). The control experiments were conducted by measuring a sample containing only targets and ligation probes, but without T4 DNA ligase. Since ligation was prevented in control experiments, the fluorescent bursts detected in the green channel should only come from the free reporter probes. As shown in Figure 3, the OG488 photon bursts detected in the control experiment had an intensity level typically <100 photon counts/ms. A similar result was also observed in the mismatch experiments. On the contrary, photon bursts with intensity as high as 300 photon counts/ms or higher were detected in the perfect match experiments. The percentages of the photon bursts that were actually the coincident signals were calculated and were also shown in Figure 3. The majority of the high-intensity photon bursts detected in the perfect match experiments were found associated with the coincident events. In particular, all the bursts were coincident signals at intensities >100 photon counts/ms, suggesting that these high-intensity photon bursts were indeed emitted from the QD-FLPs nanoassemblies. The effect of signal amplification through the formation of the nanoassemblies facilitates effective differentiation between the binding-induced coincident signals and the statistical background coincident signals, which come from free QDs and reporter probes simultaneously present in the detection volume of the confocal LIF system.
Figure 3.
Histogram (in log scale) of total burst counts from perfect match experiment (gray), single-base mismatch experiment (black) and control experiment (white) based on six consecutive 100 s measurements. Clearly the OG488 fluorescence signals are greatly enhanced due to the multiple binding of FLPs to each QD. The number above each bar represents the percentage of bursts that are actually coincident signals.
Selectivity assessment
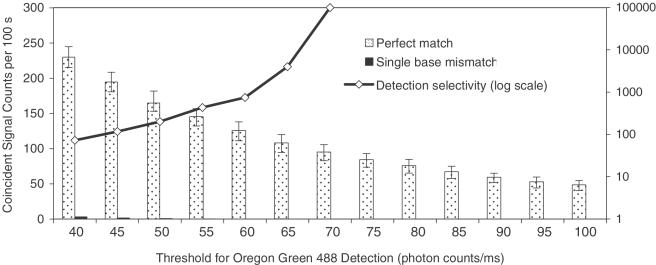
The point mutation selectivity of this method was evaluated by directly comparing the number of coincidence events measured from the perfect match and the single-base mismatch experiments. As shown in Figure 4, at any setting of intensity threshold in OG488 detection channel, the perfect match and mismatch targets were effectively differentiated by the distinct difference in the number of coincidence events. It was noted that the ratio of coincidence events between perfect match and mismatch targets was larger at a higher threshold. A small number of coincident events were also counted at a low intensity threshold when solely mismatch targets were present. This could be attributed to the statistical background coincident signals and the occasional non-specific FLPs produced by T4 DNA ligase (33). One unique feature of this assay is that the amplified coincident signals upon formation of QD-FLPs allow the use of a high-intensity threshold to avoid the complications of these background signals. As a result, an allele discrimination selectivity factor (the ratio of coincidence events detected in the samples comprised of perfect match and mismatch targets) >105 can be achieved when using a burst intensity threshold >70 photon counts/ms in the current experiment.
Figure 4.
Coincident signal counts for perfect match experiments (gray bar) and single-base mismatch experiment (black bar) at different thresholds for OG488 detection. The allele discrimination selectivity factor is plotted in log scale (black line). Error bars represent the standard deviations of six consecutive measurements.
Kras mutation detection
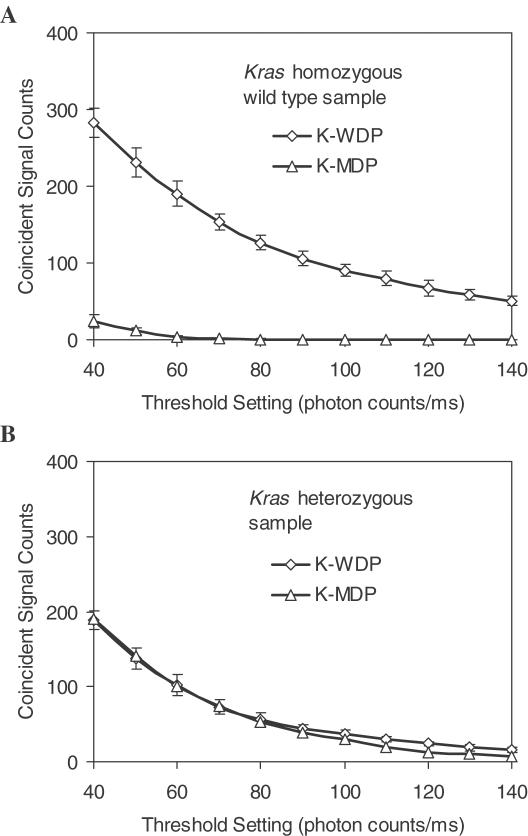
To further validate this QD-based two-color ligation assay, we applied this method to detect Kras point mutations in PCR products from clinical specimens. Kras mutations have been found in a variety of human neoplasms including ovarian serous borderline tumors (39). The Kras mutations in ovarian serous borderline tumors always occur at codon 12 resulting in amino acid alterations that lead to constitutive activation of this oncogene. The mutation status of Kras in six borderline tumors were validated by sequencing analysis using an ABI sequencer (ABI, Foster City, CA) Three of the samples contained GGT→GTT mis-sense mutation at codon 12 and the other three samples contained wild-type Kras sequence. Figure 5A and B show the results from Kras homozygous wild-type sample (G/G) and Kras heterozygous sample (G/T), respectively. Only samples containing the targets perfectly complementary to the discrimination probes gave positive results (i.e. high-intensity coincident signals).
Figure 5.
Kras mutation analysis: (A) Coincident signal counts from a mixture of Kras homozygous wild-type sample and one of the two types of discrimination probes, K-WDP (open diamond) and K-MDP (open triangle) (B) Results from Kras heterozygous sample and one of the two types of discrimination probes, K-WDP (open diamond) and K-MDP (open triangle), based on different thresholds for OG488 detection.
DISCUSSION
The need for genotyping with greater sensitivity, selectivity, simplicity, cost effectiveness and throughput in a wide range of applications (4) has provided the driving force for continuous development of new strategies and technologies for analysis of genetic variations (42–44). In this report, we describe a new method for point mutation detection, which is characterized by simple and separation-free assay protocols as well as high sensitivity and allele discrimination selectivity. This method was developed based on oligonucleotide ligation, semiconductor QDs and a two-color fluorescence coincidence detection and analysis scheme. To our knowledge, this is the first study to incorporate QDs into two-color coincidence detection and apply it to point mutation detection. The incorporation of QDs in two-color coincidence detection has resulted in several advantages. The broad absorption spectra of QDs allow the use of a single laser source to excite both QDs and organic dyes that have distinct emission spectra, thereby eliminating the cumbersome alignment issues associated with dual-laser excitation. The features of narrow and near symmetric emission spectra of QDs also avoid the cross-talk problem in two-color detection. In addition, each QD was coated with 15–25 streptavidins, allowing multiple FLPs associated to a single QD. This multivalency property makes QDs serve as nanoscaffolds to locally concentrate biotinylated ligation products. This facilitates generation of fluorescence burst signals with much enhanced signal-to-noise ratio (>30) from single QD-FLPs nanoassemblies. Consequently, the digitized target signals can be effectively distinguished from the background with a confocal LIF system. Compared with microparticle-based flow cytometry assays, which typically requires capture of more than 105–106 fluorescent targets by each microbead to achieve effective detection of target signal (5,6,9–11), our system can unambiguously detect target signal from ∼30 or less fluorescent targets coupled to a single QD.
Streptavidin–biotin interaction was specifically chosen for this assay. This binding interaction is quick, reliable and strong. It dramatically reduces the sample preparation time from tens of hours, for a thiol-mediated system (23), to just tens of minutes. However, the surface-functionalized QDs showed aggregation and precipitation at elevated temperature and salt concentration. To circumvent this, QDs were added to the reaction solution after all the required temperature cycling was done. The addition of the surface-functionalized QDs after reactions also prevents them from altering probe-target hybridization kinetics and the enzymatic reactions. The non-specific absorption of oligonucleotides to streptavidin-functionalized QDs, which could affect the detection accuracy, was found minimal, mainly because streptavidin (pI = 6.0) was only slightly negatively charged at buffer condition (pH 7) in the experiments (45).
The ability of this method to detect single events, together with the use QDs to amplify target signals, enables a unique approach for analysis of target signals with multiple parameters (fluorescence burst intensities and burst count rates). This allows accurate discrimination of target signals from background signals, thereby enhancing assay sensitivity and specificity. We have, for example, demonstrated that an allele discrimination selectivity factor >105 can be achieved using a burst intensity threshold >70 photon counts/ms (Figure 4). Another feature of the detection method developed in this study is that it could eventually allow the use of a highly reduced assay volume without compromising sensitivity and specificity. Although we introduced 5 µl of QD-FLPs (4 pM) into the two-color confocal LIF system for detection, the volume of sample actually analyzed by the system was only ∼0.5 nl, which was characterized according to the volume of sample flowing through the laser-focused detection region during the 100 s measurement time course. This sub-nanoliter sample contained only minute amount of QD-FLPs nanoassemblies (∼2 zeptomoles) that could still be effectively detected by their easily identified discrete coincident signals (Figure 2A).
Detection requiring a minimal assay volume can allow the sample distribution to a large number of detection chambers, which would enhance assay capacity for detecting multiple SNPs in a single reaction. The reduction in detection volume can also minimize the consumption of reagents and probes, thereby contributing to a much lower running-cost. However, handling of such a small volume of samples and reagents requires a specially designed microfluidic device that allows in situ molecular and enzymatic reactions and effective transport of molecules through the minuscule laser-focused detection volume. On-going work will explore the potential of integrating microfluidic networks with the confocal fluorescence detection system for low-cost, high-throughput screening.
Finally, this QD-based two-color coincidence detection and analysis method can serve as a versatile detection platform, to which other reaction principles for identifying known polymorphisms and mutations such as single base extension (5,6,9) and allele-specific hybridization (46) can also be incorporated. In addition, the superior photophysical properties of QDs allow high flexibility in selection of multiple QD/organic pairs for multiplex analysis in future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
This work was supported by NSF (DBI-0352407) and the Whitaker Foundation. Funding to pay the Open Access publication charges for this article was provided by NSF (DBI-0352407).
Conflict of interest statement. None declared.
REFERENCES
- 1.Twyman R.M., Primrose S.B. Techniques patents for SNP genotyping. Pharmacogenomics. 2003;4:67–79. doi: 10.1517/phgs.4.1.67.22582. [DOI] [PubMed] [Google Scholar]
- 2.Kirk B.W., Feinsod M., Favis R., Kliman R.M., Barany F. Single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res. 2002;30:3295–3311. doi: 10.1093/nar/gkf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwok P.Y. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics. Hum. Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 4.Syvanen A.C. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 5.Vaarno J., Ylikoski E., Meltola N.J., Soini J.T., Hanninen P., Lahesmaa R., Soini A.E. New separation-free assay technique for SNPs using two-photon excitation fluorophore. Nucleic Acids Res. 2004;32:e108. doi: 10.1093/nar/gnh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai H., White P.S., Torney D., Deshpande A., Wang Z.L., Marrone B., Nolan J.P. Flow cytometry-based minisequencing: A new platform for high-throughput single-nucleotide polymorphism scoring. Genomics. 2000;66:135–143. doi: 10.1006/geno.2000.6218. [DOI] [PubMed] [Google Scholar]
- 7.Nam J.M., Thaxton C.S., Mirkin C.A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 8.Storhoff J.J., Lucas A.D., Garimella V., Bao Y.P., Muller U.R. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J.W., Iannone M.A., Li M.S., Taylor J.D., Rivers P., Nelsen A.J., Slentz-Kesler K.A., Roses A., Weiner M.P. A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res. 2000;10:549–557. doi: 10.1101/gr.10.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M.Y., Gao X.H., Su J.Z., Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 11.Xu H.X., Sha M.Y., Wong E.Y., Uphoff J., Xu Y.H., Treadway J.A., Truong A., O'Brien E., Asquith S., Stubbins M., et al. Multiplexed SNP genotyping using the Qbead (TM) system: a quantum dot-encoded microsphere-based assay. Nucleic Acids Res. 2003;31:e43. doi: 10.1093/nar/gng043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B., Kinzler K.W. Digital PCR. Proc. Natl Acad. Sci. USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giesendorf B.A.J., Vet J.A.M., Tyagi S., Mensink E., Trijbels F.J.M., Blom H.J. Molecular beacons: a new approach for semiautomated mutation analysis. Clin. Chem. 1998;44:482–486. [PubMed] [Google Scholar]
- 14.Marras S.A., Kramer F.R., Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 15.Mhlanga M.M., Malmberg L. Using molecular beacons to detect single-nucleotide polymorphisms with real-time PCR. Methods. 2001;25:463–471. doi: 10.1006/meth.2001.1269. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S., Bratu D.P., Kramer F.R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 17.Wabuyele M.B., Farquar H., Stryjewski W., Hammer R.P., Soper S.A., Cheng Y.W., Barany F. Approaching real-time molecular diagnostics: Single-pair fluorescence resonance energy transfer (spFRET) detection for the analysis of low abundant point mutations in K-ras oncogenes. J. Am. Chem. Soc. 2003;125:6937–6945. doi: 10.1021/ja034716g. [DOI] [PubMed] [Google Scholar]
- 18.Knemeyer J.P., Marme N., Sauer M. Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem. 2000;72:3717–3724. doi: 10.1021/ac000024o. [DOI] [PubMed] [Google Scholar]
- 19.Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 20.Livak K.J. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet. Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 21.Elghanian R., Storhoff J.J., Mucic R.C., Letsinger R.L., Mirkin C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 22.Sato K., Hosokawa K., Maeda M. Rapid aggregation of gold nanoparticles induced by non-cross-linking DNA hybridization. J. Am. Chem. Soc. 2003;125:8102–8103. doi: 10.1021/ja034876s. [DOI] [PubMed] [Google Scholar]
- 23.Sato K., Sawayanagi M., Hosokawa K., Maeda M. Single-base mutation detection using neutravidin-modified polystyrene nanoparticle aggregation. Anal. Sci. 2004;20:893–894. doi: 10.2116/analsci.20.893. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell D.J., Taylor J.R., Nie S.M. Self-assembled nanoparticle probes for recognition and detection of biomolecules. J. Am. Chem. Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 25.Eigen M., Rigler R. Sorting single molecules: application to diagnostics and evolutionary biotechnology. Proc. Natl Acad. Sci. USA. 1994;91:5740–5747. doi: 10.1073/pnas.91.13.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castro A., Williams J.G.K. Single-molecule detection of specific nucleic acid sequences in unamplified genomic DNA. Anal. Chem. 1997;69:3915–3920. doi: 10.1021/ac970389h. [DOI] [PubMed] [Google Scholar]
- 27.Li H.T., Ying L.M., Green J.J., Balasubramanian S., Klenerman D. Ultrasensitive coincidence fluorescence detection of single DNA molecules. Anal. Chem. 2003;75:1664–1670. doi: 10.1021/ac026367z. [DOI] [PubMed] [Google Scholar]
- 28.Shera E.B., Seitzinger N.K., Davis L.M., Keller R.A., Soper S.A. Detection of single fluorescent molecules. Chem. Phys. Lett. 1990;174:553–557. [Google Scholar]
- 29.Wang T.H., Peng Y., Zhang C., Wong P., Ho C.M. Single-molecule tracing on a micro fluidic chip for quantitative detection of low-abundance nucleic acids. J. Am. Chem. Soc. 2005;127:5354–5359. doi: 10.1021/ja042642i. [DOI] [PubMed] [Google Scholar]
- 30.Nie S.M., Chiu D.T., Zare R.N. Probing individual molecules with confocal fluorescence microscopy. Science. 1994;266:1018–1021. doi: 10.1126/science.7973650. [DOI] [PubMed] [Google Scholar]
- 31.Barnes M.D., Ng K.C., Whitten W.B., Ramsey J.M. Detection of single rhodamine-6G molecules in levitated microdroplets. Anal. Chem. 1993;65:2360–2365. [Google Scholar]
- 32.Zhang C., Chao S.Y., Wang T.H. Comparative quantification of nucleic acids using single-molecule detection and molecular beacons. Analyst. 2005;130:483–488. doi: 10.1039/b415758c. [DOI] [PubMed] [Google Scholar]
- 33.Landegren U., Kaiser R., Sanders J., Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 34.Nickerson D.A., Kaiser R., Lappin S., Stewart J., Hood L., Landegren U. Automated DNA diagnostics using an Elisa-based oligonucleotide ligation assay. Proc. Natl Acad. Sci. USA. 1990;87:8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruchez M., Moronne M., Gin P., Weiss S., Alivisatos A.P. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 36.Chan W.C.W., Nie S.M. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C.Y., Yeh H.C., Kuroki M.T., Wang T.H. Single quantum dot-based DNA nanosensor. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 38.Medintz I.L., Uyeda H.T., Goldman E.R., Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 39.Ho C.L., Karman R.J., Dehari R., Wang T.L., Shih I.M. Mutations of BRAF and KRAS precede the development of ovarian serous borderline tumors. Cancer Res. 2004;64:6915–6918. doi: 10.1158/0008-5472.CAN-04-2067. [DOI] [PubMed] [Google Scholar]
- 40.Barany F. Genetic-Disease Detection and DNA Amplification Using Cloned Thermostable Ligase. Proc. Natl Acad. Sci. USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwille P., MeyerAlmes F.J., Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., Chu X., Liu Y., Jiang J.H., He Z., Zhang Z., Shen G., Yu R.Q. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005;33:e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takatsu K., Yokomaku T., Kurata S., Kanagawa T. A new approach to SNP genotyping with fluorescently labeled mononucleotides. Nucleic Acids Res. 2004;32:e60. doi: 10.1093/nar/gnh054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou G.H., Gotou M., Kajiyama T., Kambara H. Multiplex SNP typing by bioluminometric assay coupled with terminator incorporation (BATI) Nucleic Acids Res. 2005;33:e133. doi: 10.1093/nar/gni132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hermanson G.T. Bioconjugate techniques. San Diego, CA: Academic Press; 1995. [Google Scholar]
- 46.Prince J.A., Feuk L., Howell W.M., Jobs M., Emahazion T., Blennow K., Brookes A.J. Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Cancer Res. 2001;11:152–162. doi: 10.1101/gr.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.