Abstract
We present a comprehensive structural, evolutionary and molecular dynamics (MD) study of the G/U wobble basepairs in the ribosome based on high-resolution crystal structures, including the recent Escherichia coli structure. These basepairs are classified according to their tertiary interactions, and sequence conservation at their positions is determined. G/U basepairs participating in tertiary interactions are more conserved than those lacking any interactions. Specific interactions occurring in the G/U shallow groove pocket—like packing interactions (P-interactions) and some phosphate backbone interactions (phosphate-in-pocket interactions)—lead to higher G/U conservation than others. Two salient cases of unique phylogenetic compensation are discovered. First, a P-interaction is conserved through a series of compensatory mutations involving all four participating nucleotides to preserve or restore the G/U in the optimal orientation. Second, a G/U basepair forming a P-interaction and another one forming a phosphate-in-pocket interaction are replaced by GNRA loops that maintain similar tertiary contacts. MD simulations were carried out on eight P-interactions. The specific GU/CG signature of this interaction observed in structure and sequence analysis was rationalized, and can now be used for improving sequence alignments.
INTRODUCTION
Comparisons of basepair frequencies and positions in conserved RNA sequences, such as rRNA, can lead to important predictive models for other RNAs. Since the sequencing of tRNA in the 1960's, comparative sequence analysis has been used extensively to infer the common secondary structures of homologous RNAs. More recently it has been applied to infer the locations of tertiary interactions that stabilize folded RNA 3D structures. The inferred tertiary interactions in several cases have been applied to construct 3D models for biologically active RNAs, which subsequently were verified by X-ray crystallography. Recent examples include RNase P and the Group I intron (1–4). In this work we explore the potentials of tertiary interactions made by G/U wobbles to be used in similar ways.
The cis Watson–Crick (WC) G/U basepair is the most common non-classical basepair present in RNA. It was first recognized by Crick in 1966 in the context of the tRNA—mRNA anticodon–codon interaction (5). Subsequently, G/U's at specific positions have been shown to be essential for the function of various RNAs (6). Critical functional roles also have been inferred for highly conserved G/U basepairs found near the active sites of certain ribozymes, such as the Group I introns (4).
The first systematic study of the cis WC G/U basepairs was done by Gutell et al. (7) who investigated G/U variations in rRNA among broadly divergent phylogenetic taxa and classified them into several types according to their sequence conservation. In a similar way, Gautheret et al. (8) classified the G/U basepairs according to their phylogenetic patterns combined with their positions in the secondary structures. These authors recognized that at least 50% of WC-paired positions in 16S and 23S rRNAs contain >1% G/U in sequence alignments, and ∼10% of all paired positions display 50% or more G/U substitutions. They also noted that most of these positions are often substituted by various classical WC pairs (A/U, U/A, G/C or C/G) but some highly conserved G/U's have specific variation patterns—such as G/U to U/G or G/U to A/C (8).
Earlier studies describing G/U basepairs were not based on knowledge of X-ray structures (7,8). Thus the role of tertiary interactions in constraining the G/U pairs could not be considered. Here, we analyze phylogenetic substitution patterns of the G/U basepairs in different structural contexts. Our work has several aims. First, to provide complete structural and phylogenetic characterization of all G/U basepairs in rRNA and classify their tertiary interactions. Second, to identify G/U-specific interactions and describe their sequence signatures. Third, to improve sequence alignments based on the tertiary interactions.
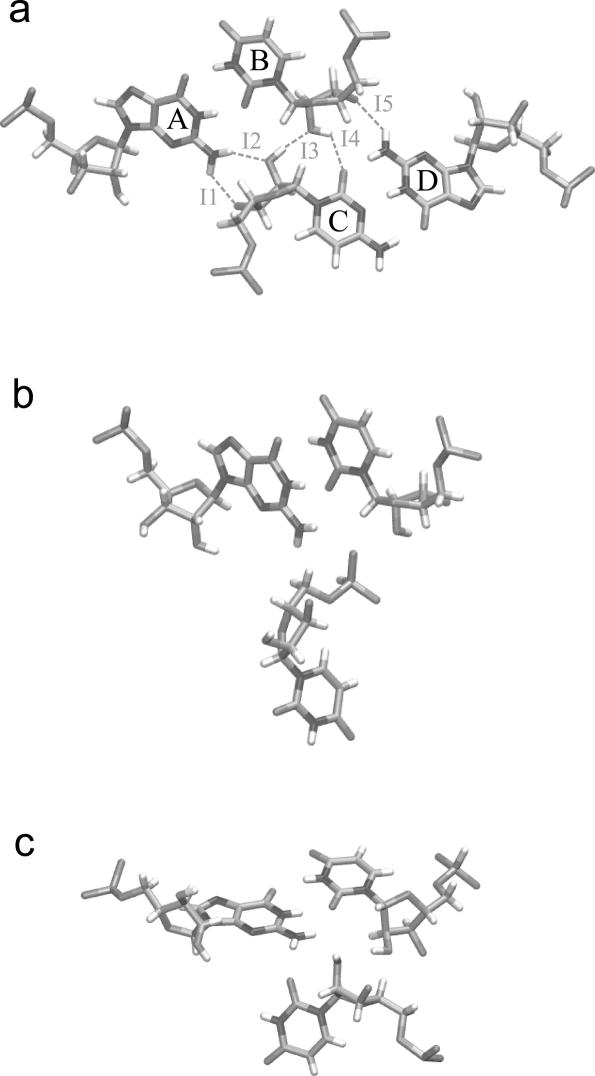
The packing interaction (P-interaction) (9) involves two basepairs, usually cis WC G/U and cis WC C/G basepairs. It somewhat resembles a ‘type 0’ A-minor motif (10), but with better and deeper packing of the interacting helices along the G/U shallow (minor) groove pocket (Figure 1a). Both the P-interaction and ‘type 0’ A-minor motif are variants of ribose zippers (4). We show that P-interactions have the highest G/U conservation among all G/U interactions. We identify a novel quadruple compensatory mutation (involving 4 nt at once) between a G/U and another basepair, reinstating the interaction in a reversed orientation. We suggest that the P-interaction can be used for improving sequence alignments because of its specific GU/CG signature and, consequently, we refine the sequence alignments at several positions where a P-interaction takes place.
Figure 1.
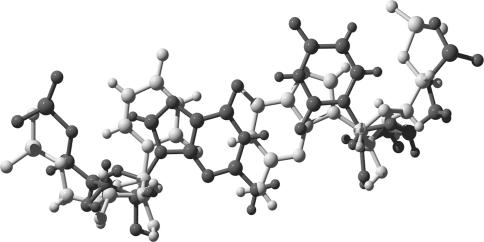
SGP interactions made by G/U basepairs. (a) P-interaction between the 4 nt A, B, C and D. This interaction is optimal when one of the basepairs is cis WC G/U, resulting in the five H-bonds indicated (I1-I5). (b) Phosphate-in-pocket interaction. (c) O2′-in-pocket interaction. (a) and (b) are from PDB file 1S72, (c) is from PDB file 1J5E. Produced by DeepView (14).
We identify two new types of highly conserved tertiary interactions involving the G/U shallow groove pocket (SGP). Phosphate backbone interaction (Phosphate-in-pocket interaction) has a phosphate embedded deep in the G/U SGP (Figure 1b), in analogy with the sulfate ion interaction in the same pocket (11). O2′-in-pocket interaction (Figure 1c) involves the sugar O2′ coming from outside the plane of the G/U and binding at the G/U SGP.
We further show that tertiary contacts of P- and phosphate-in-pocket interactions can be conserved even upon substantial change of the local motifs involved. Thus, we found G/U's forming a P-interaction or phosphate-in-pocket interaction in some ribosomal X-ray structures to be replaced by GNRA loops in others while keeping similar tertiary contacts. These unique motif swaps underline the precedence of tertiary over secondary structure in their particular contexts.
MATERIALS AND METHODS
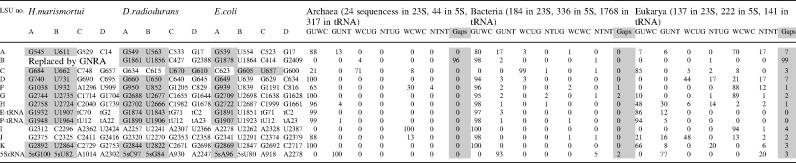
X-ray crystal structures were obtained from the Nucleic Acid Database and the Protein Data Bank (PDB) (12,13) and were visualized in 3D with DeepView (14). We studied all cis WC G/U basepairs in the 16S rRNA of the bacterium Thermus thermophilus (Tt)—PDB files 1IBM, 3.31Å resolution, and 1J5E, 3.05Å (15), and in the 23S/5S rRNA structures of the archaeon Haloarcula marismortui (Hm)—PDB files 1JJ2 and 1S72, both at 2.4 Å (16) and the bacterium Deinococcus radiodurans (Dr)—PDB files 1KPJ and 1LNR, both at 3.10 Å (17). In addition, all Escherichia coli (Ec) cis WC G/U basepairs were studied in recent 70S structures—PDB files 2AVY, 2AW4, 2AW7 and 2AWB, solved at 3.5 Å (18). Structures with bound mRNA and tRNA or substrate analogues also were examined (19–26).
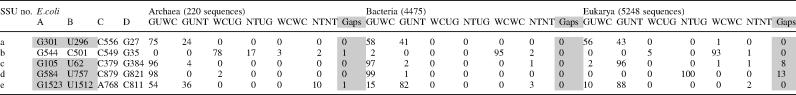
The alignments for the 16S-like and the 23S-like rRNA were obtained from the European ribosomal RNA database (27,28). The 16S-like rRNA alignments included 220 archaeal, 4475 bacterial and 5248 eukaryal unique sequences (sequences with only one representative from each species; thus the alignments are made of broadly divergent taxa, and their sequence analysis is less biased). The 23S-like rRNA alignments included 24 archaeal, 184 bacterial and 137 eukaryal unique sequences. The 5S-like seed RFAM rRNA alignment was used in our study (29,30). It contains 37 archaeal, 336 bacterial and 222 eukaryal unique sequences. Genomic tRNA alignments were obtained from the compilation of tRNA sequences and sequences of tRNA genes (31), and they are composed of 317 archaeal, 1768 bacterial and 141 eukaryal unique sequences. The sequence analysis was carried out with ‘ribostral’, a specially designed MATLAB program (A. Mokdad and N.B. Leontis, in preparation). This is a user-friendly program that includes a sequence viewer able to display substitution patterns of basepairs in an RNA alignment colored according to their isostericity with the actual basepair observed in the crystal structure. Thus, the 3D structure information is directly related to the sequence alignment, and substitutions that belong to the same or different isosteric subfamilies for each basepair are easily recognized. Ribostral also measures substitution patterns of multiple nucleotides at once, such as base triples or quadruples, which was useful for simultaneous analysis of all 4 nt participating in P-interactions. Further details and a preliminary stand-alone version of ribostral can be found at http://rna.bgsu.edu/mokdad/ribostral.
Our study includes all cis WC G/U basepairs seen in the high-resolution crystal structures of the ribosome and its subunits. Further, other basepairs are considered if they have >50% G/U substitutions in sequence alignments of archaeal or bacterial sequences (as these prove to be more trustworthy than eukaryal alignments). Secondary structures from the comparative RNA website (32) were first used to identify all such pairs. However, X-ray structure examination revealed that some of them were either not cis WC-paired or not paired at all. These basepairs were excluded from this study (for a list of basepairs omitted in this way see Supplementary Table S2). This process produced a final list of 80 bp from 16S, 186 bp from 23S and 7 bp from 5S rRNAs.
Molecular dynamics (MD) analysis was carried out on eight systems each composed of two 24 nt-long helices coming together and forming a P-interaction in their center. The first of these systems, extracted from PDB file 1S72, contains a G/U packed with C/G (P-interaction, abbreviated as GU![]() CG). This central P-interaction was then modified using InsightII (from Accelrys) to obtain the following eight different combinations: GU
CG). This central P-interaction was then modified using InsightII (from Accelrys) to obtain the following eight different combinations: GU![]() CG (original), AC
CG (original), AC![]() CG, GU
CG, GU![]() GC, AC
GC, AC![]() GC, GC
GC, GC![]() CG, GC
CG, GC![]() GC, AU
GC, AU![]() CG and AU
CG and AU![]() GC. The explicit solvent MD simulations were carried out using the AMBER7.0 package (33) with the parm99 Cornell et al. force field (34–36). The RNA was solvated in a rectangular box of TIP3P waters (37) and neutralized by minimal number of sodium cations (38) initially placed by the LeaP module at points of favorable electrostatic potential close to the RNA. The standard protocols (39) were used for the equilibration and production simulations, performed by the Sander module of AMBER7.0. The production runs were carried out at 300 K with constant-pressure periodic boundary conditions and the particle mesh Ewald method (40) applied. The MD trajectories were then analyzed using the ptraj and carnal modules of the AMBER7.0 package and our own scripts and visualized by the program VMD (41).
GC. The explicit solvent MD simulations were carried out using the AMBER7.0 package (33) with the parm99 Cornell et al. force field (34–36). The RNA was solvated in a rectangular box of TIP3P waters (37) and neutralized by minimal number of sodium cations (38) initially placed by the LeaP module at points of favorable electrostatic potential close to the RNA. The standard protocols (39) were used for the equilibration and production simulations, performed by the Sander module of AMBER7.0. The production runs were carried out at 300 K with constant-pressure periodic boundary conditions and the particle mesh Ewald method (40) applied. The MD trajectories were then analyzed using the ptraj and carnal modules of the AMBER7.0 package and our own scripts and visualized by the program VMD (41).
RESULTS AND DISCUSSION
Specific features of the cis WC G/U basepair
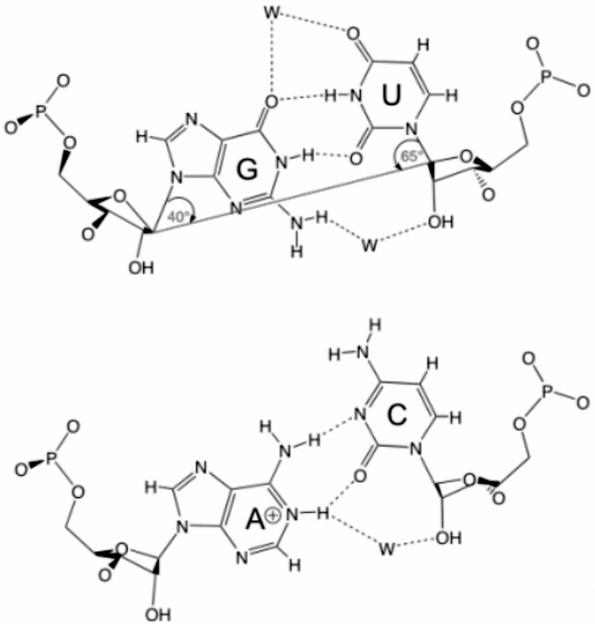
The C1′–C1′ distance of the cis WC G/U basepair is ∼10.2 Å, which is very close to the distances in classical WC basepairs (10.3 Å). The U is shifted towards the deep (major) groove to allow for hydrogen bonding between G(O6) … U(N3) and G(N1) … U(O2). The angle between the C1′–C1′ axis and the glycosidic bonds is ∼40° for G and 65° for U, instead of the symmetric 54° angle in the canonical basepairs (Figure 2). This causes helices containing G/U's to locally overtwist or undertwist depending on the orientation of the wobble basepair and its neighbors (6). Therefore, the only other basepair that is completely isosteric to the cis WC G/U basepair is the cis WC A+/C basepair, which is rare due to the A(N1) protonation requirement (42,43). The canonical WC basepairs are nearly isosteric to G/U or U/G (6) while, due to their asymmetry, G/U and A+/C are not isosteric upon reversal to U/G and C/A+. The pocket created by G(N2), U(O2), and U(O2′) in the shallow groove often coordinates an integral water molecule by forming two H-bonds. Another feature of the cis WC G/U basepair is the unique H-bond donor and acceptor distribution around its grooves. The hydrogens of the amino group G(N2) are free to interact in the shallow groove, and the electronegative G(N7), G(O6) and U(O4) atoms in the deep groove create a strong electronegative surface (44,45) that binds metal ions. This may help in RNA folding or ribozymatic activity (46,47). In addition, the cis WC G/U basepair is approximately as thermodynamically stable as the classical cis WC A/U basepair (45,48–53). Finally, the cis WC G/U basepair possesses a unique conformational flexibility (6) that allows it to respond to sequence contexts and crystal packing much more easily than classical basepairs (54), hence allowing for recognition of interacting proteins or other RNAs by induced fit (55,56). These characteristics of G/U basepairs play major roles in determining the types of RNA–RNA, RNA–protein or RNA–metal ion interactions in which they participate, and in their preferred substitution patterns throughout evolution.
Figure 2.
Top, cis Watson–Crick G/U (wobble) basepair with water molecules (W) in its SGP and DGP. The angles formed between the C1′–C1′ axis and the glycosidic bonds show the asymmetry of this basepair compared with the classical WC basepairs. Bottom, the isosteric A+/C basepair. Produced by ChemDraw (CambridgeSoft Corporation).
Survey of G/U basepairs
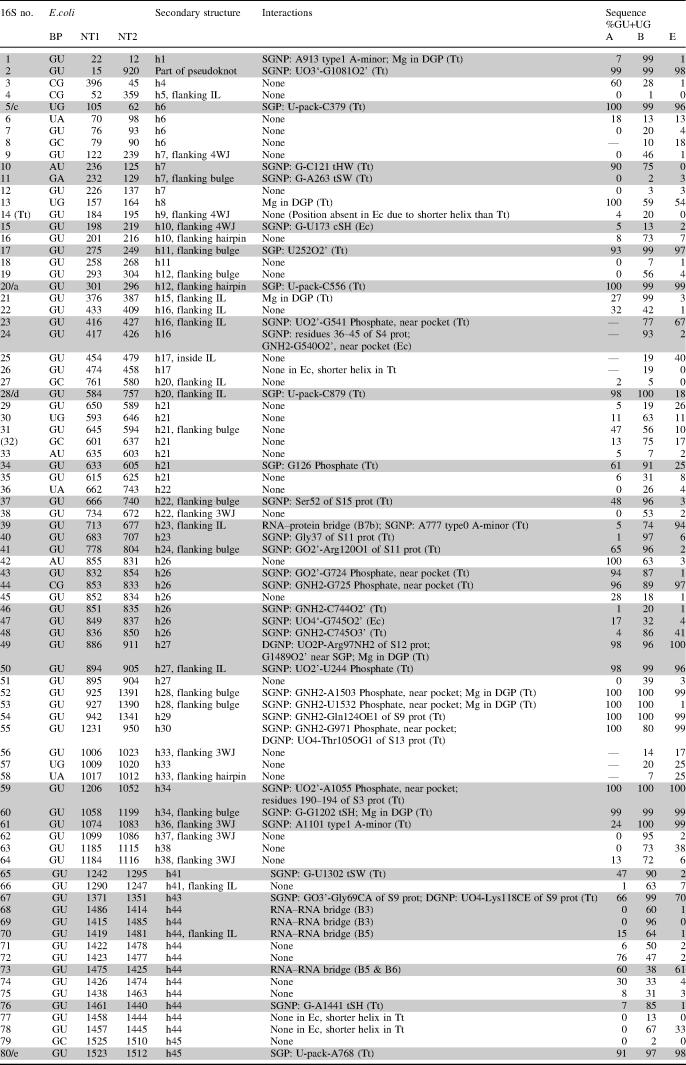
Table 1 is a compilation of the secondary structure features and tertiary interactions of all the G/U basepairs that occur in crystal structures of 16S rRNA (a similar compilation for tRNA and 23S/5S rRNA interactions can be found in Supplementary Table S3). Interactions are classified according to their location, i.e. in shallow groove or in deep groove. Shallow groove interactions are subdivided further into those that involve the G/U pocket (SGP), forming H-bonds with at least two groups among GNH2, UO2 and UO2′, and those that do not (Shallow Groove not in Pocket, SGNP). Deep groove interactions are subdivided into those that involve the deep groove pocket (DGP), forming H-bonds with GO6 and UO4 simultaneously, and those that do not (Deep Groove not in Pocket, DGNP). In most cases, the Tt and Hm structures prove to be the best for observing 16S and 23S/5S interactions, respectively. The 23S Dr and 70S Ec structures are at slightly lower resolutions and are mainly used for comparison and for inferring motions or motif swaps, as well as observing basepairs that are not G/U's in Tt or Hm. Table 1 also displays the conservation of the G/U in sequence alignments at each of the positions. The main goal for our classification was to test whether the interactions in the G/U SGP are more specific for G/U than others occurring elsewhere around this basepair.
Table 1.
Interactions of the cis WC G/U basepairs in 16S rRNA, based on the crystal structures of T.thermophilus and E.coli
Initials of the structure best for observing each interaction appear in parentheses in the interactions column. Basepairs with interactions are shaded. The letters (a, b, …) in the first column after the basepair number mark the individual basepairs forming P-interactions (indicated by -pack- in the fourth column) and are also used in Table 4 and Figure 6 and referenced in the text. Basepair number (32) is not G/U in any crystal structure, but is included in the study for having >50% GU content in bacterial sequences. The last three columns indicate GU+UG (%) content at those positions in sequence alignments of archaea (A), bacteria (B) and eukarya (E). A dash (—) in these columns means that sequence alignments are all gaps (insertions) at the corresponding location. Abbreviations: h, helix; IL, internal loop; WJ, way-junction; SGP, shallow groove pocket; SGNP, shallow groove not in pocket; DGP, eep groove pocket; and DGNP, deep groove not in pocket. For the 23S/5S rRNA analysis see Supplementary Table S3.
Among the 80 G/U basepairs that occur in the 16S rRNA, 48 (60%) are inside helices, and the remaining 32 (40%) are at the ends of helices or within other motifs. Among the 193 G/U basepairs of 23S and 5S rRNA, 122 (63%) are inside helices, and the remaining 71 (37%) are elsewhere. This agrees with the previous counts of 56% intra-helical and 44% at the ends of helices compiled by Gautheret et al. (8), although our criteria for finding the G/U pairs are slightly different. The cis WC G/U basepairs have greater probability to be found within helices compared with cis WC A/G basepairs, which have significantly larger C1′–C1′ distance and are thus found almost exclusively at the ends of helices (57).
Of the 80 16S basepairs, 28 (35%) form SGNP interactions, 6 (8%) form interactions in SGP and 2 (3%) form DGNP interactions. Five basepairs form intermolecular bridges with ribosomal proteins or the large ribosomal subunit (58). No interactions are observed in the DGP. Instead, this area is occupied by a cation in several cases, consistent with the high electronegativity of this site (44,45). About one-half of the 16S G/U basepairs (42 out of 80) show no evidence of any tertiary interaction in the available crystal structures (interactions with ions are not considered as tertiary interactions). Similarly, of the 193 23S/5S G/U basepairs, 62 (32%) form SGNP contacts, 21 (11%) form interactions in SGP, 4 (2%) form DGNP interactions and 3 form interactions in DGP. Seven basepairs form intermolecular bridges with tRNA (58). A cation binds simultaneously to GO6 and UO4 in several cases, and a water molecule occupies the G/U SGP in about one-third of the cases (only Hm crystal structure contains water molecules). Again, about one-half of the 23S/5S G/U basepairs do not form any tertiary interactions (103 out of 193).
These data demonstrate almost total agreement between the statistics of the two ribosomal subunits, showing that most tertiary interactions involving G/U occur in the shallow groove. Almost all G/U tertiary and quaternary interactions in 16S Tt and Ec and in 23S/5S Hm, Dr and Ec are seen at equivalent positions. Exceptions are those interactions that occur in a variable region that is either absent in one or two of the structures, or significantly different between them. Another possible reason for seeing interactions in some of the crystal structures and not all of them is the presence of a proximal kink-turn (59). When the intrinsically flexible kink-turns change between open and closed conformations, different sets of tertiary contacts become possible. Kink-turns are like hinges that allow the motion of attached helices with respect to the body of the molecule. Upon opening or closing of the kink, the motion propagates from the fulcrum of the kink to the attached helical arms (39). Since each crystal structure shows only a static snapshot, different X-ray structures can capture the flexible K-turns in different substates. An example of this is the position corresponding to Hm G798/U815 in H34 (H and h symbols followed by a number denote helices in the large and small subunit, respectively). Here, a tertiary interaction is seen in the Hm structure but not in the Dr or Ec. Another example is the G/U basepair corresponding to Hm G1646/U1539 in H56. A tertiary interaction is seen in the Hm structure between G1646(NH2) and A1597(NH2). This interaction is possible because GNH2 can assume a pyramidal configuration due to the NH2 partial sp3 hybridization (57). No equivalent interactions are seen in Dr or Ec. Note that the limited resolution of the X-ray structures may affect the appearance of some of the interactions.
Classification of observed tertiary interactions and their sequence patterns
Three distinct types of shallow groove interactions involve the G/U pocket (SGP): P-interactions, phosphate-in-pocket interactions and ribose O2′-in-pocket interactions (Figure 1). Aside from the G/U pocket, several other types of interactions occur in the shallow groove. These include phosphate single H-bond interactions, ‘type 0’ A-minor motifs, ‘type I’ A-minor motifs (also known as trans sugar edge/sugar edge or tSS basepair), H-bonding to GNH2 or ribose sugars of a G/U, and other edge-to-edge interactions, the most common of which are cSS (cis sugar edge/sugar edge, also known as ‘type II’ A-minor motif) and tWS (trans Watson–Crick/sugar edge) interactions. There also are >20 non-specific interactions with proteins. A few G/U basepairs participate in more than one type of tertiary/quaternary interactions at once. Some others participate in interactions that look like SGNP interactions in the crystal structure, but could easily fall into the SGP category after a subtle geometrical rearrangement (‘potential’ SGP interactions).
Tables 2 and 3 represent sequence analysis of G/U basepairs classified into four major categories: basepairs with SGP interactions, potential SGP interactions (see above), other types of interactions and no interactions. The percentage of basepairs at each position is measured after ignoring all sequences with gaps (deletions) (i.e. they are not percentages over all sequences). For each position, however, we also report the percentage of sequences with gaps over all sequences (shaded). If no motif swaps take place between domains or classes at these locations, then this percentage of gaps can be considered a measure of the quality of alignments at the corresponding positions.
Table 2.
Sequence analysis of cis WC G/U basepairs in 16S 5S rRNA
Percentages are measured after ignoring sequences with gaps (deletions). The percent of gaps is separately given in the last column (shaded) as a measure of the quality of alignments at the corresponding positions, assuming that no motif swaps take place. Abbreviations: SGP, shallow groove pocket; WC, any combination of the four classical Watson–Crick basepairs (G/C, C/G, A/U and U/A); and NTs, any basepair other than GU, AC, UG, CA or WC.
Table 3.
Sequence analysis of cis WC G/U basepairs in 23S and 5S rRNA
Percentages are measured after ignoring sequences with gaps (deletions). The percent of gaps is separately given in the last column (shaded) as a measure of the quality of alignments at the corresponding positions, assuming that no motif swaps take place. Abbreviations: SGP, shallow groove pocket; WC, any combination of the four classical Watson–Crick basepairs (G/C, C/G, A/U and U/A); and NTs, any basepair other than GU, AC, UG, CA or WC.
In general, we observe that when G/U basepairs are not conserved they are replaced primarily by A/C and by classical WC basepairs (Tables 2 and 3). Much less common are substitutions by U/G or C/A or other non-classical basepairs marked as ‘NTs’ (other nucleotides). Although A/C and C/A basepairs are relatively scarce, and thus a trend is difficult to measure, G/U's seem to get more commonly substituted by A/C's than C/A's. As mentioned above, this can be explained by isostericity (42). This type of substitution pattern is more apparent in the category of non-interacting G/U's, whereas G/U's that make SGP interactions, and in some cases those that make potential SGP interactions are much more conserved (Tables 2 and 3). This indicates that these interactions are specific for G/U basepairs. G/U mutations at such locations can disturb 3D structure which might affect the fitness of the ribosome. One exception occurs in 16S rRNA, where the G/U's with SGP interactions have 84% GU and 7% UG content in archaeal sequence alignments. Although it seems to violate the isostericity principle, it has a simple explanation. There are only 6 bp in this category in 16S, and the 7% UG observed is due to a single position, G633/U605, which is occupied by G/U in 22% and by U/G in 39% of the sequences. This basepair makes a phosphate-in-pocket interaction, which can still take place if G/U flips to U/G, since position of GNH2 that makes one of the two H-bonds in this interaction remains virtually unchanged (Figure 3). This means that specific orientation of G/U is not required for the phosphate-in-pocket interaction.
Figure 3.
G/U superimposed on U/G along the C1′–C1′ axis without distorting the backbone, showing that position of the GNH2 is invariant with basepair reversal.
The high gap count in some highly conserved regions, like those with specific SGP interactions, may suggest either mistakes in the available alignments, or motif swaps. This seems to be more apparent in the case of eukaryal alignments in both subunits. For this reason, we consider eukaryal alignments of lower quality than archaeal and bacterial alignments.
Contemporary sequence alignment methods depend almost exclusively on the local basepairing and covariation patterns of classical WC basepairs, and some G/U basepairs, and do not take into consideration tertiary interactions that impose unique constraints. Including the tertiary interaction data in sequence alignment methods would greatly enhance the quality of those alignments, and is simple to accomplish for some particular interactions with strong signatures. An example of this is 23S Hm U731/G740 which in crystal structures superposes with Dr U650/G660, both making the same P-interaction. Current sequence alignment protocols, however, do not properly align these positions. We were able in this case to use the structure data to manually adjust the alignments accordingly.
P-interaction
The most conserved interaction that G/U basepairs make is the P-interaction. It takes place between the ribose O2′ belonging to a nucleotide from a basepair in one helix packing deep into the G/U SGP of a second helix, forming up to five H-bonds along the interface. One nucleotide from the first basepair makes most of the contact with one nucleotide from the second basepair. These 2 nt are known together as the internal pair. We designate this interaction as AB![]() CD, where AB and CD are the basepairs from the two helices, with B and C forming the internal pair (as seen in Figure 1a). A previous study noted that the P-interaction plays important roles in tRNA binding to the 50S subunit and in translocation, and that it is conserved in all three phylogenetic domains of the large subunit (9). No prior sequence analysis work on the small subunit has been reported, and no attempt was made to divide the results according to the three domains. We exhaustively searched the 3D structures for this type of interaction using several methods including FR3D, a structural motif search program of our design (M. Sarver, C.L. Zirbel, J. Stombaugh, A. Mokdad and N.B. Leontis, manuscript in preparation). Five such interactions are found in the small ribosomal subunit and thirteen in the large subunit, as reported in Tables 4 and 5. The tables include locations and substitution patterns at all four positions participating in this interaction. Among the thirteen P-interactions of the large subunit there is one between 23S and 5S, one between 23S and E-site tRNA, and one between 23S and P-site tRNA. U
CD, where AB and CD are the basepairs from the two helices, with B and C forming the internal pair (as seen in Figure 1a). A previous study noted that the P-interaction plays important roles in tRNA binding to the 50S subunit and in translocation, and that it is conserved in all three phylogenetic domains of the large subunit (9). No prior sequence analysis work on the small subunit has been reported, and no attempt was made to divide the results according to the three domains. We exhaustively searched the 3D structures for this type of interaction using several methods including FR3D, a structural motif search program of our design (M. Sarver, C.L. Zirbel, J. Stombaugh, A. Mokdad and N.B. Leontis, manuscript in preparation). Five such interactions are found in the small ribosomal subunit and thirteen in the large subunit, as reported in Tables 4 and 5. The tables include locations and substitution patterns at all four positions participating in this interaction. Among the thirteen P-interactions of the large subunit there is one between 23S and 5S, one between 23S and E-site tRNA, and one between 23S and P-site tRNA. U![]() C is the most commonly seen internal pair, but there is no apparent preference for a Y
C is the most commonly seen internal pair, but there is no apparent preference for a Y![]() Y over Y
Y over Y![]() R interactions, since U
R interactions, since U![]() G, U
G, U![]() A, and other Y
A, and other Y![]() R also are seen. There is, however, clear preference for these over R
R also are seen. There is, however, clear preference for these over R![]() R interactions (none of these are observed). G/U on the other hand is the most commonly seen basepair making this interaction, as only 3 of the 18 P-interactions do not involve a G/U (P-motifs denoted as b, I and J in Tables 4 and 5). In addition, two such interactions involve a non-WC basepair (e and 5S). A total of four P-interactions were not reported before (e, B, I and 5S).
R interactions (none of these are observed). G/U on the other hand is the most commonly seen basepair making this interaction, as only 3 of the 18 P-interactions do not involve a G/U (P-motifs denoted as b, I and J in Tables 4 and 5). In addition, two such interactions involve a non-WC basepair (e and 5S). A total of four P-interactions were not reported before (e, B, I and 5S).
Table 4.
Sequence analysis of all P-interactions in the small ribosomal subunit (SSU)
The letter code in the first column is used to abbreviate the individual P-interactions and is also applied in Table 1 and Figure 6 as well as referenced in the text. All 4 nt participating in each interaction are compared. These interactions occur mostly between one helical element with a G/U and another helical element with a regular WC basepair (the G/U element is shaded). However, some occur between two helical elements without any G/U (b), and some even occur between one helical element and one non-helical element (16S A768/G811 is trans Hoogsteen/sugar edge basepair). Abbreviations: WC, any combination of the four classical Watson–Crick basepairs (G/C, C/G, A/U and U/A); and NT, any basepair other than GU, AC, UG, CA or WC.
Table 5.
Sequence analysis of all P-interactions in the large ribosomal subunit (LSU)
The letter code in the first column is used to abbreviate the individual P-interactions and is also applied in Table S3 and Figure S3 as well as referenced in the text. All 4 nt participating in each interaction are compared. These interactions occur mostly between one helical element with a G/U and another helical element with a regular WC basepair (the G/U element is shaded). However, some occur between two helical elements without any G/U (I and J), and some even occur between one helical element and one non-helical element (23S A1014/A2302 is trans Hoogsteen/Hoogsteen basepair). Abbreviations: WC, any combination of the four classical Watson–Crick basepairs (G/C, C/G, A/U and U/A); and NT, any basepair other than GU, AC, UG, CA or WC.
The high conservation of G/U's participating in P-interactions is remarkable (Tables 4 and 5). Even in the cases where the crystal structure did not contain a G/U basepair (b, I and J), alignments of some domains displayed a very high G/U content in the first helix or U/G in the second helix (95% U/G in the second helix in archaeal alignments for b, 100% G/U in the first helix in bacterial sequences for I, and 88 and 98% G/U in the first helix in archaeal and bacterial sequences for J). Even more striking is that in the rare cases when the G/U in a P-interaction is lost, there is great tendency for all 4 nt forming the interaction to mutate in a way to finally recreate a G/U in the right orientation in either one of the two involved helices. This is best demonstrated by the P-interaction corresponding to Hm G684/U662![]() C748/G657 (C in Table 5, between H28 and H27). It seems that a quadruple compensatory mutation between archaea and bacteria has occurred. The GU
C748/G657 (C in Table 5, between H28 and H27). It seems that a quadruple compensatory mutation between archaea and bacteria has occurred. The GU![]() CG in Hm is replaced by a GC
CG in Hm is replaced by a GC![]() UG in Dr, and by CG
UG in Dr, and by CG![]() UG in Ec. In archaeal sequence alignments, these positions are 21% GU–WC, 71% WC–UG and 8% WC–WC. The latter number could correspond to organisms that survived with the intermediary mutation. Almost all (99%) of bacterial sequences are WC–UG, and most eukaryal sequences (85%) are GU–WC. The most parsimonious explanation is that originally there was a WC–UG variant, which is still present in bacteria today. Sometime after the archaeal/eukaryal line diverged from bacteria a mutation occurred, causing some archaeal organisms to have WC–WC at those positions. Some of these organisms live till today. However, most archaea had other compensatory mutations following the first one causing either their return to the original WC–UG bacterial variant, or their ‘flipping’ into the opposite GU–WC variant. Some archaea also may have retained the original bacterial variant without ever mutating because they may have originated before the first mutation ever occurred. Eukarya most probably separated from archaea before the first mutation. These observed patterns of substitution strongly suggest that the P-interaction was conserved in most, if not all organisms, even when the identity of the individual nucleotides making it had changed. It also is clear that the G/U basepair with U at the internal position was favored. This is the first time that such a covariation involving 2 bp, rather than just 2 nt, has been reported.
UG in Ec. In archaeal sequence alignments, these positions are 21% GU–WC, 71% WC–UG and 8% WC–WC. The latter number could correspond to organisms that survived with the intermediary mutation. Almost all (99%) of bacterial sequences are WC–UG, and most eukaryal sequences (85%) are GU–WC. The most parsimonious explanation is that originally there was a WC–UG variant, which is still present in bacteria today. Sometime after the archaeal/eukaryal line diverged from bacteria a mutation occurred, causing some archaeal organisms to have WC–WC at those positions. Some of these organisms live till today. However, most archaea had other compensatory mutations following the first one causing either their return to the original WC–UG bacterial variant, or their ‘flipping’ into the opposite GU–WC variant. Some archaea also may have retained the original bacterial variant without ever mutating because they may have originated before the first mutation ever occurred. Eukarya most probably separated from archaea before the first mutation. These observed patterns of substitution strongly suggest that the P-interaction was conserved in most, if not all organisms, even when the identity of the individual nucleotides making it had changed. It also is clear that the G/U basepair with U at the internal position was favored. This is the first time that such a covariation involving 2 bp, rather than just 2 nt, has been reported.
There is, on the other hand, one case where a P-interaction in bacteria (corresponding to Ec numbers G1878/U1864![]() C414/G2409) is completely lost and replaced by a GNRA loop interaction in archaea (and probably in eukarya as well, as suggested by their sequence alignments). The GNRA loop substitutes for one of the two helices (H68 flanked by the G/U) without much distortion of structure, because it makes a similar 3D contact as the P-interaction (Supplementary Figure S1). This proves that the presence and orientation of an interaction (which defines the 3D folding) is far more important than the actual identity or type of that interaction.
C414/G2409) is completely lost and replaced by a GNRA loop interaction in archaea (and probably in eukarya as well, as suggested by their sequence alignments). The GNRA loop substitutes for one of the two helices (H68 flanked by the G/U) without much distortion of structure, because it makes a similar 3D contact as the P-interaction (Supplementary Figure S1). This proves that the presence and orientation of an interaction (which defines the 3D folding) is far more important than the actual identity or type of that interaction.
Tables 4 and 5 also shows that no G/U was substituted by U/G (in the same helix), which can be explained first by the fact that cis WC U/G is not isosteric to G/U, and second by the fact that the P-interaction, unlike the phosphate-in-pocket interaction, is structurally directional. It cannot occur as UG![]() CG because of the specific asymmetric orientation of the G/U basepair that is more open for SGP interactions coming from the side of the U rather than the side of the G.
CG because of the specific asymmetric orientation of the G/U basepair that is more open for SGP interactions coming from the side of the U rather than the side of the G.
Molecular dynamics analysis of the P-interaction
Most of the P-interactions observed in crystal structures are between a G/U pair and a C/G pair, with U and C being at the internal positions (GU![]() CG). The purpose of MD analysis was to explain this behavior, as well as reveal why the G/U's forming P-interactions almost never are substituted by the isosteric A/C basepairs throughout evolution.
CG). The purpose of MD analysis was to explain this behavior, as well as reveal why the G/U's forming P-interactions almost never are substituted by the isosteric A/C basepairs throughout evolution.
MD simulations were carried out for eight different combinations of P-interactions, namely GU![]() CG, AC
CG, AC![]() CG, GU
CG, GU![]() GC, AC
GC, AC![]() GC, GC
GC, GC![]() CG, GC
CG, GC![]() GC, AU
GC, AU![]() CG and AU
CG and AU![]() GC. The studied motifs were embedded in two A-type helices, each with 24 residues (Figure 4). The helices were identical in all simulations, except for their centrally positioned P-interactions. The above combinations were chosen to represent Y
GC. The studied motifs were embedded in two A-type helices, each with 24 residues (Figure 4). The helices were identical in all simulations, except for their centrally positioned P-interactions. The above combinations were chosen to represent Y![]() Y and R
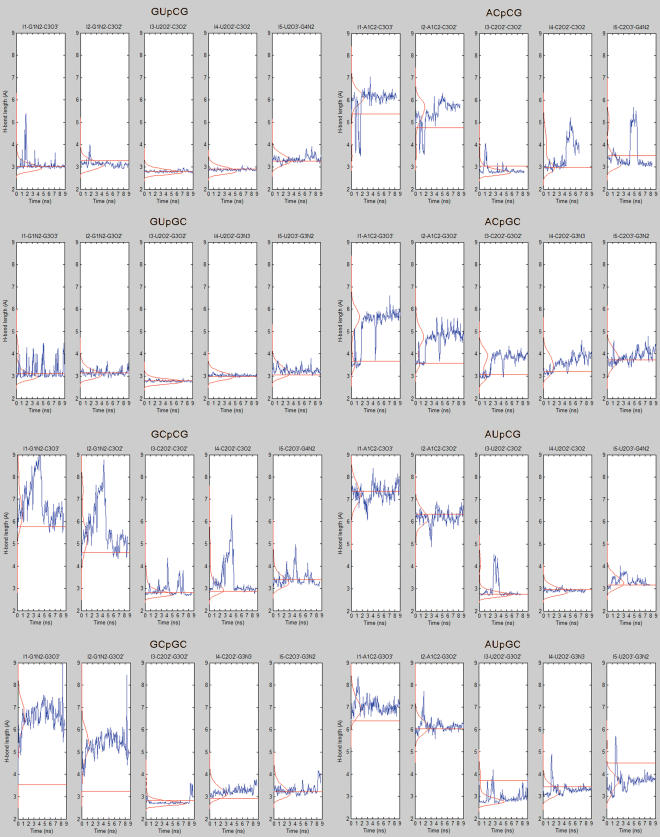
Y and R![]() Y types of interaction, as well as G/U and other isosteric (A/C) or nearly isosteric (A/U and G/C) basepairs embedded in one helix, combined with G/C and C/G basepairs embedded in the other helix. The simulations were extended to ∼10 ns each, which appears to be sufficient for the purpose of this study. Figure 5 shows time development of the five H-bonds constituting the P-interaction in each system (as defined in Figure 1a, except that H-bond lengths are measured via the heavy atoms in the simulations). The eight structures were ranked in order of decreasing stabilities which was indirectly estimated based on the number of H-bonds and their dynamic behavior in the simulations. (Note that such classification is more relevant to judge the stability of RNA interactions than, for example, evaluation of direct base–base interaction energies. This would neglect the interplay between that base–base interaction and all the other effectors such as adjacent basepairs, solvent screening and the like). The suggested stability order is as follows: GU
Y types of interaction, as well as G/U and other isosteric (A/C) or nearly isosteric (A/U and G/C) basepairs embedded in one helix, combined with G/C and C/G basepairs embedded in the other helix. The simulations were extended to ∼10 ns each, which appears to be sufficient for the purpose of this study. Figure 5 shows time development of the five H-bonds constituting the P-interaction in each system (as defined in Figure 1a, except that H-bond lengths are measured via the heavy atoms in the simulations). The eight structures were ranked in order of decreasing stabilities which was indirectly estimated based on the number of H-bonds and their dynamic behavior in the simulations. (Note that such classification is more relevant to judge the stability of RNA interactions than, for example, evaluation of direct base–base interaction energies. This would neglect the interplay between that base–base interaction and all the other effectors such as adjacent basepairs, solvent screening and the like). The suggested stability order is as follows: GU![]() CG =GU
CG =GU![]() GC (these complexes maintained all five H-bonds throughout the simulations) >> GC
GC (these complexes maintained all five H-bonds throughout the simulations) >> GC![]() GC = AU
GC = AU![]() GC =AU
GC =AU![]() CG = GC
CG = GC![]() CG (these lost the first two H-bonds, but generally maintained the others) > AC CG = AC
CG (these lost the first two H-bonds, but generally maintained the others) > AC CG = AC![]() GC (most H-bonds lost early in the simulations). Thus, a P-interaction involving A/C is the weakest among the tested systems. This is related to the fact that the A+/C SGP is less electronegative than the G/U pocket, and it has a much lower H-bonding potential due to the loss of NH2 group. Hence, it cannot accommodate the ribose O2′ in the same way as G/U does (for a comparison of electrostatic potential between G/U and A+/C see Supplementary Figure S2). Supplementary Data further present free energy calculations (MM-GBSA and MM-PBSA methods, Supplementary Table S1) of relative stabilities of the studied P-interactions. The calculations were also performed for UG
GC (most H-bonds lost early in the simulations). Thus, a P-interaction involving A/C is the weakest among the tested systems. This is related to the fact that the A+/C SGP is less electronegative than the G/U pocket, and it has a much lower H-bonding potential due to the loss of NH2 group. Hence, it cannot accommodate the ribose O2′ in the same way as G/U does (for a comparison of electrostatic potential between G/U and A+/C see Supplementary Figure S2). Supplementary Data further present free energy calculations (MM-GBSA and MM-PBSA methods, Supplementary Table S1) of relative stabilities of the studied P-interactions. The calculations were also performed for UG![]() GC combination, which can be considered as unsuitable for P-interaction. These calculations confirm superior stability of the GU
GC combination, which can be considered as unsuitable for P-interaction. These calculations confirm superior stability of the GU![]() CG = GU
CG = GU![]() GC motifs and provide some additional insights. One has to point out, however, that such thermodynamics calculations are presently based on substantial approximations and that is why we prefer to assess the simulations primarily based on the structural dynamics seen in Figure 5.
GC motifs and provide some additional insights. One has to point out, however, that such thermodynamics calculations are presently based on substantial approximations and that is why we prefer to assess the simulations primarily based on the structural dynamics seen in Figure 5.
Figure 4.
P-interaction between two helices showing the whole system used for MD analysis; the nucleotides forming the P-interaction are highlighted in red. Produced by VMD (41).
Figure 5.
MD of the eight P-interaction systems studied. All five H-bonds involved in the generic P-interaction are monitored (as defined in Figure 1a, but here are measured from the heavy atoms). The red horizontal lines mark H-bond lengths in starting structures (average over first 50 ps); the red curves describe the probability distribution of H-bond lengths in the simulation and blue curves displays the actual time development of the H-bond lengths along the trajectory.
Thus, since GU![]() CG is the most commonly seen P-interaction in crystal structures and sequence alignments, and also among the two most stable systems in our simulations, it can be considered as the signature of this interaction.
CG is the most commonly seen P-interaction in crystal structures and sequence alignments, and also among the two most stable systems in our simulations, it can be considered as the signature of this interaction.
Other shallow groove pocket interactions
There are two additional SGP interactions in the ribosome, phosphate-in-pocket interaction, and O2′-in-pocket interaction (Figure 1). The latter means that O2′ is inserted in the SGP of G/U, but it can not be classified as P-interaction or ‘type 0’ A-minor interaction. Both of these SGP interactions lead to high conservation of the G/U's. The phosphate-in-pocket interaction is less directional than the P-interaction, having no preference for a specific G/U orientation. This G/U to U/G covariation is also the case of some other interactions in the shallow groove, especially some single H-bond interactions. Thus, it is not unique to phosphate-in-pocket interactions but at least it is one of its indicators. Similar to P-interaction, we detected one case where a G/U forming a phosphate-in-pocket interaction is replaced by a GNRA loop, with no substantial distortion of the 3D folding. This occurs at the positions corresponding to Ec G1740/U1720–C1550P, bringing together H63 and H56. Here, both Hm and Dr have a shorter H63 than Ec, so the GNRA loop that normally is at the end of H63 is brought to a position homologous to the G/U position in Ec (Supplementary Figure S1).
Figure 6 and Supplementary Figure S3 summarize positions of all SGP interactions on the secondary structures of the small and large ribosomal subunits, respectively.
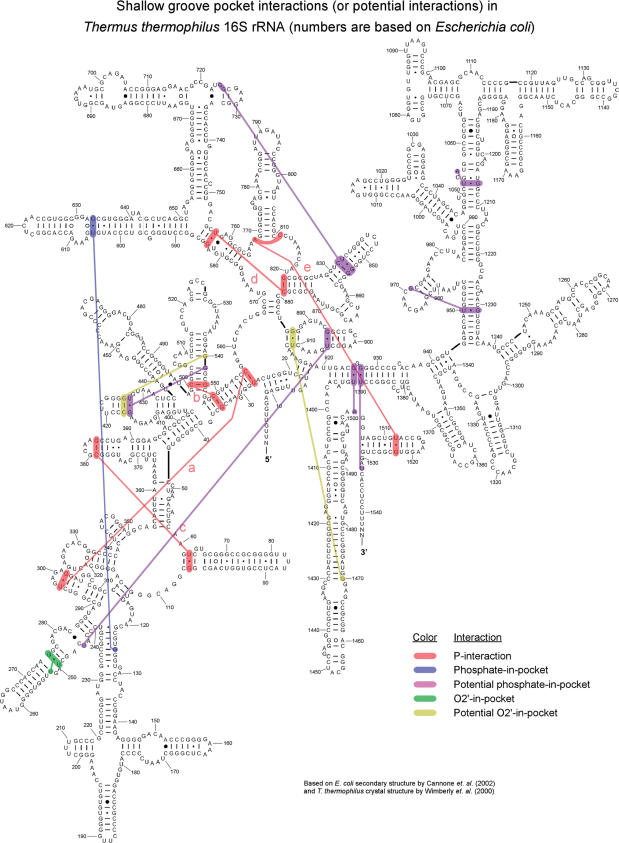
Figure 6.
SGP interactions marked on the 16S rRNA secondary structure. Labels of P-interactions correspond to Tables 1 and 4. Color code displayed at the bottom right. For the tRNA and 23S/5S rRNA analysis see Figure S3.
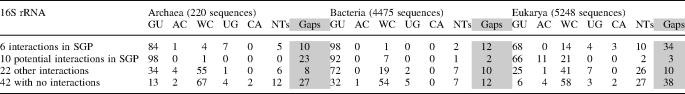
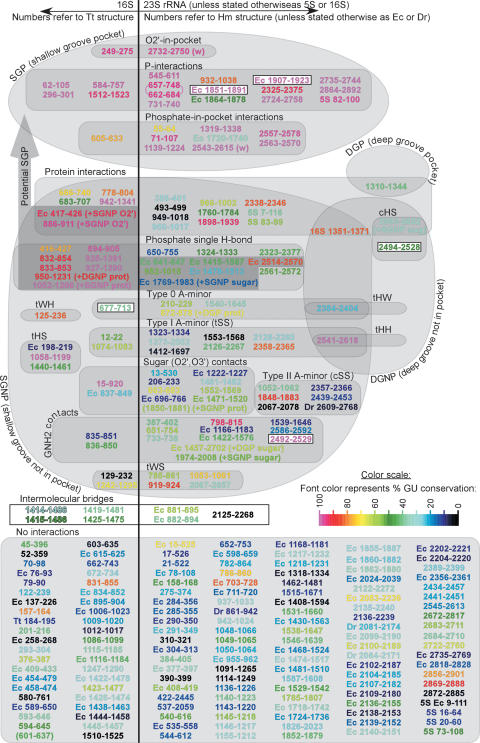
There are 273 positions in the ribosome occupied by cis WC G/U basepairs in one or more of the available crystal structures. This corresponds to roughly 15% of all ribosomal basepairs. About half of these are involved in tertiary or quaternary interactions. Therefore, they have important roles in the assembly and 3D folding of the ribosome and its subunits. Figure 7 is a Venn diagram representing the sequence data from archaeal and bacterial alignments mapped to the structural data of each of the positions. This provides valuable insights into the possible functions and mechanisms for some specific families of interactions. The diagram clearly shows that G/U's forming specific SGP interactions are most highly conserved. These are the P-interactions, phosphate-in-pocket interactions, and ribose O2′-in-pocket interactions. Other tertiary interactions also are included in the figure, showing less G/U conservation in most cases. Protein interactions were discussed in detail in (60) and are not fully addressed here.
Figure 7.
Venn diagram representing locations and types of interactions of ribosomal cis WC G/U basepairs and their sequence conservations. Nucleotide numbers are taken from the main crystal structure in each category (Tt in 16S, Hm in 23S and 5S), unless otherwise specified (by Dr or Ec preceding the numbers). Each basepair is colored according to its average % GU+UG content in sequence alignments of archaea and bacteria. Water mediated interactions are indicated by (w); open boxes indicate intermolecular bridges between ribosomal subunits. The following abbreviations are used: c, cis; t, trans; W, Watson–Crick; H, Hoogsteen; S, sugar edge (64); SGP, shallow groove pocket; SGNP, shallow groove not in pocket; DGP, deep groove pocket and DGNP, deep groove not in pocket; additional interactions, if present, are noticed in parentheses after plus (e.g. ‘+SGNP sugar’ and ‘+DGP prot’ mean additional SGNP involving G/U sugar and DGP involving protein, respectively). The two positions connected by curved lines in P-interactions (657–748 and 662–684) represent the P-interaction with quadruple compensatory mutation, discussed in detail in the text. The two basepairs in parentheses (16S 601–637 and 23S 1850–1881) are not GU in any crystal structure, but are included in the study for having >50% GU content in bacterial and archaeal sequences, respectively.
Conservation patterns of potential SGP interactions
As noted above, potential SGP interactions are those that could form SGP interactions after a modest structural rearrangement (Tables 2 and 3). The substitution patterns in Tables 2 and 3 suggest that most potential SGP interactions in 16S actually form SGP interactions. They have high G/U conservation and phylogenetically they behave as actual SGP interactions. In contrast, most such 23S basepairs are not expected to form SGP interactions because of their low G/U conservation, except for Ec 2514-2570.
G/U interactions can mediate flexible and transient contacts in the ribosome
The ribosome is a large dynamical molecular machine. Movements of its parts must be well coordinated for correct and efficient functioning. Transient interactions, like those with tRNA or between the ribosomal subunits, have to be formed and broken easily, and therefore must be relatively weak while still stereochemically precise. This may be possible through the association and dissociation of multiple weak tertiary and quaternary interactions or via consecutive transformation between different types of similar interactions. The similarity between P-interactions, ‘type 0’ A-minor motifs, O2′-in-pocket interactions, and some potential SGP interactions (all having an O2′ buried at different depths in the G/U SGP) enables smooth switches from one interaction to another. A similar scenario was suggested for tertiary interactions comprising two or more A-minor motifs (61). Such dynamic behavior could explain why some G/U basepairs without apparent interactions are conserved. A possible example of this kind is 16S G886/U911 interacting with G1489/C1411, which is observed as a potential O2′-in-pocket interaction. However, sequence analysis shows that this position is >98% GU–WC (mainly GU–GC and GU–CG) in all domains, making it very similar in its evolutionary conservation to a P-interaction. We suggest that as a P-interaction relaxes or starts to dissociate, it may convert to an O2′-in-pocket interaction or a ‘type 0’ A-minor interaction because of the orientational similarity between them.
Other conserved G/U basepairs
A few other basepairs are highly conserved in archaeal and bacterial alignments, but are not taking part in SGP or potential SGP interactions, or are not interacting at all (Figure 7). Some of these may be involved in transient interactions that are in their dissociated states in crystal structures. Others, especially those flanking internal loops or junctions which make up more than one-third of all G/U's, may be highly conserved because of the important roles they play in providing specific stacking interactions that stabilize these nearby motifs. Examples of these are 23S 1848–1883 flanking a four-way-junction at the base of H66, 23S 1898–1939 flanking the internal loop of H68, which is a conserved motif similar to C-loop, and 23S 2869–2888 flanking the internal loop of H101. Other intriguing cases include 23S 2541–2618, which is a specific basepair forming ‘type I’ A-minor motif with the terminal A76 of the tRNA CCA acceptor stem. The G/U here permits the interaction to become more compact by allowing the unstacked A76 to insert deeper in its pocket, making the G/U the preferred basepair at this location. The 23S 798–815 bp flanking the internal loop of H34 is another interesting case where GNH2 forms an H-bond with A1598O2′, which belongs to the hairpin of H58. This helix is joined to the rest of 23S subunit by a kink-turn, and the G/U here may be involved in stabilizing conformational changes at this flexible region located at the interface with the small subunit. Another conserved G/U is 23S 2492–2529 which is a part of a complex motif forming a receptor for a GNRA loop.
CONCLUSIONS
Large functional RNA molecules, like the ribosomal RNAs, are compactly folded and form complex tertiary and quaternary RNA–RNA and RNA–protein contacts mediated by several types of recurrent motifs and interactions. We have carried out a complete structural and sequence analysis of the G/U basepairs in rRNA and classified their tertiary interactions.
The SGP interactions of G/U basepairs reaching deep in shallow grooves of helices are clearly the most prominent G/U interaction patterns. They include the P-interaction identified earlier (9) and phosphate-in-pocket interactions and O2′-in-pocket interactions reported here for the first time (Figure 1). We also detected several P-interactions not noticed before. We show that the P-interaction G/U's are the most conserved ones, closely followed by the remaining SGP interactions.
We identify a novel quadruple compensatory mutation (involving 4 nt at once) between a G/U and another basepair, reinstating the P-interaction in a reversed orientation. Further, we show that tertiary contacts of P- and phosphate-in-pocket interactions can be conserved upon considerable change of the local motifs involved. Thus, one G/U forming a P-interaction in E.coli and D.radiodurans is replaced by a GNRA loop in H.marismortui. Still, similar tertiary contacts are present between the equivalent areas of the structures. Another G/U making a phosphate-in-pocket interaction in E.coli is replaced by a GNRA loop in both H.marismortui and D.radiodurans while again keeping similar tertiary contacts, preventing any major differences in the overall folds. These unique motif swaps underline the precedence of tertiary over secondary structure in their particular contexts. All these findings also clearly demonstrate the importance of G/U SGP interactions in folding and function of ribosomes.
We also identify interactions that could fall into the SGP category after a subtle geometrical rearrangement and we call them potential SGP interactions. This could indicate a transient formation of SGP interactions in these positions. Indeed, most potential SGP interactions in 16S rRNA actually appear to form SGP interactions because phylogenetically they behave as SGP interactions. In contrast, most such 23S rRNA basepairs are not expected to form SGP interactions because of their low G/U conservation. The similarity between P-interactions, ‘type 0’ A-minor motifs, and O2′-in-pocket interactions, and some potential SGP interactions indicates that they could be involved in smooth structural switches utilizing a set of consecutive similar interaction patterns.
Most of these G/U interactions are long-range contacts bringing together different RNA molecules or distant helices in the same RNA molecule. Some of them, however, are short range interactions that are nonetheless extremely conserved, such as the P-interactions b, C, D and J, the phosphate-in-pocket interactions occurring in h34, between H6 and H7, and in H91, and the O2′-in-pocket interaction occurring in h11 (refer to Figure 6 and Supplementary Figure S3). Interestingly, none of these interactions takes place between the ‘head’ of the 30S subunit corresponding to domain III in 16S rRNA and the rest of the small subunit. This agrees with previous studies showing that only one A-minor motif interaction occurs between domain III and the rest of the small subunit (61). It also supports the observation in the 70S Ec that the ‘head’ is subject to large-scale rigid body motions relative to the rest of the ribosome during the protein synthesis cycle, especially the ratchet-like motion that accompanies translocation (18,62,63).
In contrast to SGP interactions, G/U DGP interactions are very rare. There are just three occurrences of these in both ribosomal subunits. These occurrences do not resemble each other structurally, preventing their classification and characterization. They also do not show any specific sequence signatures, and the G/U's forming them are not highly conserved in general (Figure 7).
As noted above, few G/U's participating in P-interactions and phosphate-in-pocket interactions, which are some of the most conserved interactions in RNA, can be replaced by other elements (like GNRA loops) that are able to form similar tertiary contacts. This proves that these tertiary contacts are essential for the survival of the organisms. This, however, does not challenge the importance of the G/U basepair itself since it still remains the preferred moderator of such interactions. This knowledge can be helpful in refining sequence alignments by looking for the signatures of some of these interactions, such as the GU![]() CG signature of the P-interaction, and the G/U covariation with U/G which is one of the indications of a phosphate-in-pocket interaction.
CG signature of the P-interaction, and the G/U covariation with U/G which is one of the indications of a phosphate-in-pocket interaction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Scott Rogers for helpful discussions and critical reading of this work, and Kamila Reblova and Filip Razga for valuable additions. Funding for this work was provided by NIH Grant (2 R15 GM055898-03) and PRF Grant (PRF# 42357–AC 4) to N.B.L. J.S. was supported by Wellcome Trust International Senior Research fellowship in Biomedical Science GR067507, grants GA203/05/0388 and GA203/05/0009 by Grant Agency of the Czech Republic, and research project AVO Z5 004 0507 (IBP) and grant MSM0021622413 by Ministry of Education of the Czech Republic (J.S.). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Massire C., Jaeger L., Westhof E. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA. 1997;3:553–556. [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai H.Y., Masquida B., Biswas R., Westhof E., Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 3.Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 4.Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 5.Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 6.Varani G., McClain W.H. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutell R.R., Larsen N., Woese C.R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautheret D., Konings D., Gutell R.R. G.U base pairing motifs in ribosomal RNA. RNA. 1995;1:807–814. [PMC free article] [PubMed] [Google Scholar]
- 9.Gagnon M.G., Steinberg S.V. GU receptors of double helices mediate tRNA movement in the ribosome. RNA. 2002;8:873–877. doi: 10.1017/s135583820202602x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masquida B., Sauter C., Westhof E. A sulfate pocket formed by three GoU pairs in the 0.97 A resolution X-ray structure of a nonameric RNA. RNA. 1999;5:1384–1395. doi: 10.1017/s1355838299991173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman H.M., Zardecki C., Westbrook J. The Nucleic Acid Database: a resource for nucleic acid science. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1095–1104. doi: 10.1107/s0907444998007926. [DOI] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 15.Wimberly B.T., Brodersen D.E., Clemons W.M., Jr, Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 16.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 17.Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 18.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 19.Schmeing T.M., Moore P.B., Steitz T.A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen J.L., Schmeing T.M., Moore P.B., Steitz T.A. Structural insights into peptide bond formation. Proc. Natl Acad. Sci. USA. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy F.V.T., Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nature Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 22.Murphy F.V.t., Ramakrishnan V., Malkiewicz A., Agris P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nature Struct. Mol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 23.Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 24.Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 25.Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 26.Schmeing T.M., Seila A.C., Hansen J.L., Freeborn B., Soukup J.K., Scaringe S.A., Strobel S.A., Moore P.B., Steitz T.A. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nature Struct. Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 27.Wuyts J., De Rijk P., Van de Peer Y., Winkelmans T., De Wachter R. The European large subunit ribosomal RNA database. Nucleic Acids Res. 2001;29:175–177. doi: 10.1093/nar/29.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuyts J., Van de Peer Y., Winkelmans T., De Wachter R. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 2002;30:183–185. doi: 10.1093/nar/30.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case D.A., Pearlman D.A., Caldwell J.W., Cheathan T.E., III, Wang J., Ross W.S., Simmerling C.L., Darden T.A., Merz K.M., Stanton R.V., et al. AMBER 7. San Francisco, CA: University of California San Francisco; 2002. [Google Scholar]
- 34.Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Jr, Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 35.Cheatham T.E., Cieplak P., Kollman P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 36.Wang J.M., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 37.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phy. 1983;79:926–935. [Google Scholar]
- 38.Aqvist J. Ion water interaction potentials derived from free-energy perturbation simulations. J. Phy. Chem. 1990;94:8021–8024. [Google Scholar]
- 39.Razga F., Koca J., Sponer J., Leontis N.B. Hinge-like motions in RNA kink-turns: the role of the second a-minor motif and nominally unpaired bases. Biophys. J. 2005;88:3466–3485. doi: 10.1529/biophysj.104.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle Mesh Ewald method. J. Chem. Phy. 1995;103:8577–8593. [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 42.Leontis N.B., Stombaugh J., Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doudna J.A., Cormack B.P., Szostak J.W. RNA structure, not sequence, determines the 5' splice-site specificity of a group I intron. Proc. Natl Acad. Sci. USA. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allain F.H., Varani G. Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res. 1995;23:341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDowell J.A., Turner D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 46.Draper D.E., Grilley D., Soto A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 47.Konforti B.B., Abramovitz D.L., Duarte C.M., Karpeisky A., Beigelman L., Pyle A.M. Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell. 1998;1:433–441. doi: 10.1016/s1097-2765(00)80043-x. [DOI] [PubMed] [Google Scholar]
- 48.Jaeger J.A., Turner D.H., Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 50.McDowell J.A., He L., Chen X., Turner D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2. Biochemistry. 1997;36:8030–8038. doi: 10.1021/bi970122c. [DOI] [PubMed] [Google Scholar]
- 51.Schroeder S.J., Turner D.H. Thermodynamic stabilities of internal loops with GU closing pairs in RNA. Biochemistry. 2001;40:11509–11517. doi: 10.1021/bi010489o. [DOI] [PubMed] [Google Scholar]
- 52.Strazewski P., Biala E., Gabriel K., McClain W.H. The relationship of thermodynamic stability at a G × U recognition site to tRNA aminoacylation specificity. RNA. 1999;5:1490–1494. doi: 10.1017/s1355838299991586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugimoto N., Kierzek R., Freier S.M., Turner D.H. Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry. 1986;25:5755–5759. doi: 10.1021/bi00367a061. [DOI] [PubMed] [Google Scholar]
- 54.Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 55.Chang K.Y., Varani G., Bhattacharya S., Choi H., McClain W.H. Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc. Natl Acad. Sci. USA. 1999;96:11764–11769. doi: 10.1073/pnas.96.21.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramos A., Varani G. Structure of the acceptor stem of Escherichia coli tRNA Ala: role of the G3.U70 base pair in synthetase recognition. Nucleic Acids Res. 1997;25:2083–2090. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sponer J., Mokdad A., Sponer J.E., Spackova N., Leszczynski J., Leontis N.B. Unique tertiary and neighbor interactions determine conservation patterns of cis Watson–Crick A/G base-pairs. J. Mol. Biol. 2003;330:967–978. doi: 10.1016/s0022-2836(03)00667-3. [DOI] [PubMed] [Google Scholar]
- 58.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 59.Klein D.J., Schmeing T.M., Moore P.B., Steitz T.A. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 61.Noller H.F. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 62.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 63.Moore P.B. Structural biology. A ribosomal coup: E.coli at last! Science. 2005;310:793–795. doi: 10.1126/science.1120539. [DOI] [PubMed] [Google Scholar]
- 64.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.