Abstract
Innate immunity, an ancient form of defense against microbial infection, is well described for animals and is also suggested to be important for plants. Discrimination from self is achieved through receptors that recognize pathogen-associated molecular patterns (PAMPs) not found in the host. PAMPs are evolutionarily conserved structures which are functionally important and, thus, not subject to frequent mutation. Here we report that the previously described peptide elicitor of defense responses in parsley, Pep-13, constitutes a surface-exposed fragment within a novel calcium-dependent cell wall transglutaminase (TGase) from Phytophthora sojae. TGase transcripts and TGase activity are detectable in all Phytophthora species analyzed, among which are some of the most destructive plant pathogens. Mutational analysis within Pep-13 identified the same amino acids indispensable for both TGase and defense-eliciting activity. Pep-13, conserved among Phytophthora TGases, activates defense in parsley and potato, suggesting its function as a genus-specific recognition determinant for the activation of plant defense in host and non-host plants. In summary, plants may recognize PAMPs with characteristics resembling those known to trigger innate immune responses in animals.
Keywords: elicitor/innate immunity/pathogen-associated molecular pattern/Phytophthora/transglutaminase
Introduction
The innate immune response is a well-studied phenomenon in human, mice and insects, and its molecular basis shows remarkable evolutionary conservation across kingdom borders (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000). Activation of inflammatory responses or production of antimicrobial compounds relies on the recognition through Toll-like receptors (TLRs) of pathogen-associated molecular patterns (PAMPs) (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000; Imler and Hoffmann, 2001; Underhill and Ozinsky, 2002). Common features of such immune modulators are their highly conserved structures, their functional importance for the microorganism and their presence in a broad range of microbial species. Recognized PAMPs that trigger innate immune responses include bacterial lipopolysaccharide (LPS), lipoproteins and flagellin, in addition to fungal cell wall-derived carbohydrates and proteins (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000; Imler and Hoffmann, 2001; Underhill and Ozinsky, 2002). Plants also possess non-self recognition systems (receptors) for numerous microbe-derived molecules which mediate activation of plant defense responses in a non-cultivar-specific manner and have been described as ‘general elicitors’ (Heath, 2000; Cohn et al., 2001; Dangl and Jones, 2001). These include β-heptaglucan structures from oomycete cell walls, fungal cell wall chitin fragments and an N-terminal fragment of bacterial flagellin, flg22 (Felix et al., 1999; Nürnberger and Scheel, 2001). In particular, flg22, which was found in several but not all bacterial flagellins, triggered defense responses in a range of different plants (Felix et al., 1999). However, in none of the cases to date has any such motif been shown to be indispensable for the host microbe and, hence, to be physiologically equivalent to the PAMPs described for humans and Drosophila.
TGases (R-glutaminyl-peptide:amine-γ-glutamyltransferase, EC 2.3.2.13), which catalyze an acyl transfer reaction between peptide-bound glutamine residues and primary amines including the ε-amino group of peptide-bound lysine residues, form intra- or intermolecular isopeptide bonds resulting in irreversible protein cross-linking (Folk, 1980; Aeschlimann and Paulsson, 1994). TGase activity has been implicated in a variety of physiological activities in animals, including neuronal growth and regeneration, bone development, angiogenesis, wound healing, cellular differentiation and apoptosis (Liu et al., 2002). However, no physiological function has been elucidated for either bacterial, fungal or plant TGases.
We have previously identified a peptide fragment (Pep-13), within an abundant cell wall glycoprotein (GP42) from the phytopathogenic oomycete Phytophthora sojae, that was necessary and sufficient for receptor-mediated defense gene expression and synthesis of antimicrobial phytoalexins in parsley (Nürnberger et al., 1994; Hahlbrock et al., 1995). Now, we provide evidence that GP42 is a P.sojae cell wall-associated Ca2+-dependent TGase, which is the first such enzyme reported from an oomycete species. TGases with a highly conserved Pep-13 motif were found in all Phytophthora species analyzed. Mutational analysis within the Pep-13 motif revealed that the same amino acid residues that were shown to be important for plant defense-eliciting activity in parsley (Nürnberger et al., 1994) and potato were also essential for TGase activity. Our data support the intriguing view that plants may have evolved receptors that recognize genus-specific, ‘epitope-like’ motifs present within, and essential for, the function of pathogen-derived molecules. Thus, Pep-13 exhibits characteristics reminiscent of PAMPs modulating innate immune responses in vertebrate and invertebrate organisms (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000; Underhill and Ozinsky, 2002). Implications of PAMP-mediated pathogen recognition for the activation of defense responses, in both host plants (potato) and non-host plants (parsley), will be discussed.
Results
The P.sojae GP42 is a TGase
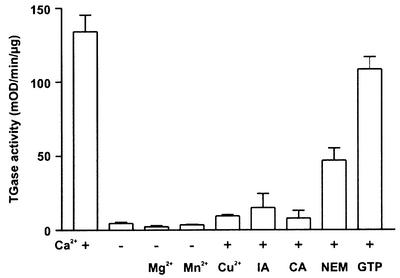
We have previously reported that an oligopeptide fragment (Pep-13) identified within a 42 kDa glycoprotein elicitor from P.sojae (GP42) is necessary and sufficient to trigger a multifaceted defense response in parsley (Nürnberger et al., 1994; Hahlbrock et al., 1995; Jabs et al., 1997; Ligterink et al., 1997; Zimmermann et al., 1997; Blume et al., 2000). Since the GP42 amino acid sequence exhibited significant homology to a recently purified Phytophthora cactorum TGase (our unpublished data), we tested whether the P.sojae GP42 possessed TGase activity. Therefore, a solid-phase microtiter plate assay based on the incorporation of 5-(biotinamido) pentylamine into N,N-dimethylcasein was employed. TGase activity was shown to be associated with both recombinant (Figure 1) and purified GP42 (not shown). The activity of both purified and recombinant P.sojae TGase was strictly dependent on Ca2+ [KM(pentylamine) = 0.249 mM at 5 mM Ca2+], which could not be substituted by 5 mM Mg2+ or Mn2+. The TGase inhibitors Cu2+, iodoacetamide, cystamine and N-ethylmaleimide blocked this Ca2+-dependent activity efficiently. GTP, an inhibitor of human tissue TGase (Melino and Piacentini, 1998), did not significantly affect TGase activity. In addition, as reported for guinea pig liver TGase (Folk, 1980), the oomycete enzyme catalyzed Ca2+-dependent auto- oligomerization (data not shown). In summary, the Phytophthora TGase shares biochemical characteristics of mammalian Ca2+-dependent TGases (Folk, 1980; Aeschlimann and Paulsson, 1994; Melino and Piacentini, 1998). The lack of sequence homology to any TGases present in databases (Sacks et al., 1995) suggests, however, that GP42 belongs to a novel class of these enzymes.
Fig. 1. TGase activity of the P.sojae GP42 elicitor protein. Recombinant P.sojae GP42 was expressed in A.oryzae and assayed for TGase activity in the presence (+) or absence (–) of 5 mM Ca2+. The specificity for Ca2+ was determined by replacement with 5 mM Mg2+ or 5 mM Mn2+, respectively. TGase inhibitors, including 1 mM Cu2+, 10 µM iodoacetamide (IA), 10 mM cystamine (CA) or 10 mM N-ethylmaleimide (NEM), inhibited this Ca2+-dependent activity. No significant inhibition was observed in the presence of 5 mM GTP. Bars represent the mean values ± SD of three independent experiments.
The Pep-13 motif is highly conserved among Phytophthora TGases
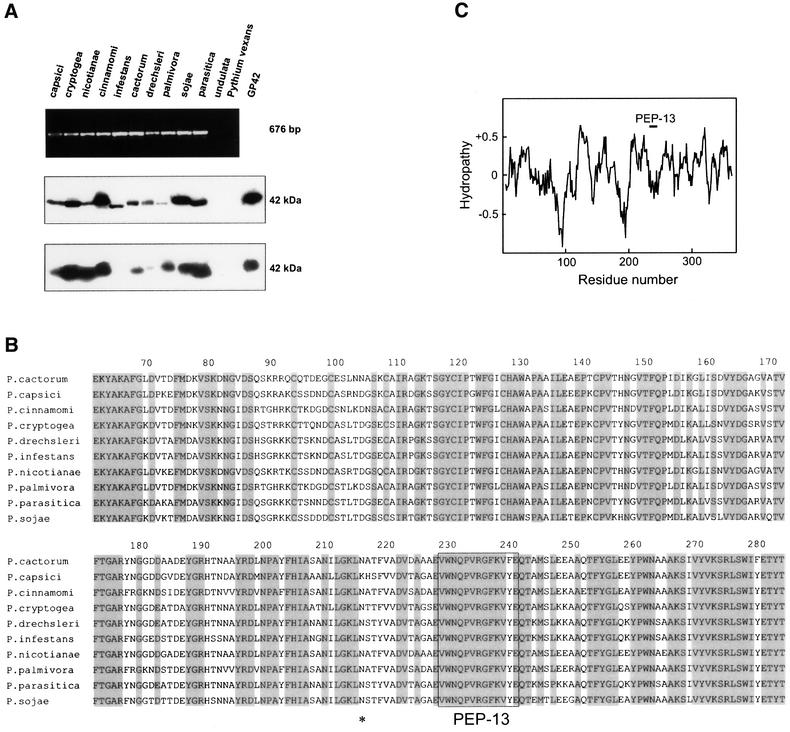
Non-self recognition through phytopathogen-derived PAMPs requires that the motifs selected as recognition determinants are not present within the recipient organism, but are widely found among various microbial species (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000; Imler and Hoffmann, 2001; Underhill and Ozinsky, 2002). Database analysis of all plant sequences, including the fully sequenced Arabidopsis thaliana genome (The Arabidopsis Genome Initiative, 2000), against the complete TGase sequence (Sacks et al., 1995), or against the Pep-13 sequence (NCBI Blast Search for short nearly exact matches), suggested that plants possess neither orthologs of the Phytophthora TGase nor proteins containing peptide motifs with Pep-13 elicitor activity (not shown). In contrast, genomic DNA blot analysis demonstrated that several species of the oomycete genus Phytophthora, but not of the related genus Pythium, possess a gene family encoding GP42 TGase-related proteins (Sacks et al., 1995). A RT–PCR analysis performed on poly(A)+ RNA from 10 Phytophthora species revealed the presence of GP42 TGase homologs in all species tested (Figure 2A). However, no GP42 TGase-related transcripts were obtained from Pythium vexans, Phytophthora undulata, the latter being taxonomically more closely related to Pythium species (Erwin and Ribeiro, 1996), or from the obligate biotroph Peronospora parasitica (not shown). Consistently, a peptide antiserum raised against the Pep-13 motif of P.sojae GP42 recognized a protein of ∼42 kDa in the culture filtrate of all Phytophthora species tested that was not detected in P.undulata or P.vexans (Figure 2A). A zymogram of TGase activity associated with these 42 kDa proteins confirmed that homologs of GP42 possessing TGase activity were present in all Phytophthora species (Figure 2A). Interestingly, enzyme activity prepared from Phytophthora infestans was reproducibly found to be associated with an 85 kDa protein, which cross-reacted with the anti-Pep-13 antiserum and most likely represents a TGase dimer (not shown).
Fig. 2. GP42 TGase homologs containing the surface-exposed Pep-13 motif are highly conserved among the genus Phytophthora. (A) RT–PCR demonstrated the presence of GP42 TGase-related transcripts in Phytophthora species (upper panel). Immunoblot analysis of culture filtrates (50 µg protein/lane) using anti-Pep13 antibodies revealed that each species possessed a GP42-like protein containing the Pep-13 motif (middle panel). In-gel TGase assays demonstrated that the GP42-related proteins possessed TGase activity (lower panel). Purified P.sojae GP42 (100 ng) was used as a positive control for both immunodetection and TGase activity (lane GP42). (B) The alignment of the deduced amino acid sequences of the RT–PCR products highlights the conservation of the Pep-13 motif (boxed). The asterisk marks the position of the sole N-glycosylation site of the P.sojae GP42. (C) The hydropathy plot based on the Eisenberg algorithm (Eisenberg et al., 1984) predicts the Pep-13 motif to reside in a hydrophilic region of the protein.
Analysis of the partial TGase sequences at the deduced amino acid level revealed >60% identity between all sequences (Figure 2B). Remarkably, the sequence comprising Pep-13, the peptide fragment essential for activation of defense responses, was highly conserved among all species analyzed (Figure 2B). The only exception involved a tyrosine residue (corresponding to Y241 in the P.sojae protein), which in two species was replaced by another aromatic amino acid, phenylalanine. A synthetic peptide containing this amino acid substitution was found to bind to the Pep-13 receptor and activate defense responses in parsley in a manner indistinguishable from Pep-13 (data not shown). Hydrophobicity analysis predicted Pep-13 to be present within a hydrophilic region of the enzyme (Figure 2C). In addition, secondary structure prediction analysis (Rost, 1996) suggested that Pep-13 resides in a surface-exposed loop structure containing the sole N-glycosylation site (Parker et al., 1991; Sacks et al., 1995). Thus, the strong sequence conservation and surface exposure of the Pep-13 motif are consistent with its role as a recognition determinant for the activation of plant defense responses during the interaction with Phytophthora species.
W231 and P234 are important for both elicitor activity and TGase activity
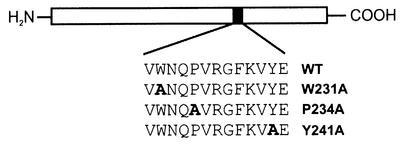
The strict conservation of the Pep-13 motif within the Phytophthora TGases prompted us to investigate whether this sequence was important for enzyme activity. We had previously shown that Pep-13 was sufficient for the activation of plant defense responses by intact GP42 (Nürnberger et al., 1994). In addition, replacement within Pep-13 of each individual amino acid revealed W231 and P234 to be important for elicitor activity (Nürnberger et al., 1994). Moreover, conservative mutations W231F (phenylalanine) and P234Hyp (hydroxyproline) retained the ability of the Pep-13 mutants to trigger phytoalexin production in parsley, whereas non-conservative mutations W231T (threonine) and P234I (isoleucine) abolished the elicitor activity of these mutant peptides (not shown). Thus, non-conservative mutations introduced into the codons encoding either W231 or P234 would be likely to impair TGase-mediated recognition of P.sojae by the plant. To test whether such mutations would affect the TGase activity, P.sojae wild-type GP42 TGase and mutant proteins containing single alanine exchanges within the Pep-13 sequence (Figure 3) were expressed and purified from Aspergillus oryzae. In control experiments, neither TGase activity nor TGase protein was detected in culture supernatants of A.oryzae transformed with an empty expression vector (not shown). Both elicitor and TGase activities of the mutant proteins were determined and compared with the activity of the wild-type protein. Significantly, mutations that compromised the ability of the protein to elicit defense responses in parsley protoplasts also markedly reduced TGase activity (Table I). Substitution of Trp231 by alanine (W231A) resulted in a 98% reduction in TGase activity. This substitution also abolished the elicitor activity of the protein and thus we were unable to determine an EC50 value. Replacement of Pro234 by alanine (P234A) resulted in a reduction in TGase activity to ∼6% of wild-type activity and a concurrent 20-fold higher EC50 value for elicitor activity. In contrast, substitution of Tyr241 by alanine (Y241A) had only a modest effect on both TGase and elicitor activities. These data demonstrate that amino acid residues important for the TGase activity of the GP42 protein are identical to those necessary to elicit defense reactions in parsley protoplasts. Thus, it appears that the evolutionary stability of this functionally indispensable epitope may have favored its selection as a PAMP that is recognized by the plant in order to detect and respond to attack by Phytophthora species.
Fig. 3. Schematic representation indicating the position of mutations introduced into the Pep-13 sequence of the P.sojae GP42 TGase. WT represents the wild-type GP42 sequence. W231A, P234A and Y241A correspond to single amino acid exchanges of Trp231, Pro234 and Tyr241 with alanine, respectively. All proteins were heterologously expressed in A.oryzae and prepared from culture filtrate as described in Materials and methods.
Table I. Mutational analysis of GP42 reveals amino acid residues indispensable for both TGase and elicitor activitiesa.
| Protein | TGase activity (mOD/min/µg) | Elicitor activity [EC50 (nM)] |
|---|---|---|
| WT TGase | 104.0 ± 22.8 | 0.5 |
| W231A | 1.5 ± 1.3 | –b |
| P234A | 6.5 ± 2.6 | 11.2 |
| Y241A | 98.9 ± 2.2 | 1.1 |
aElicitor activity of TGase is expressed as the EC50 value, which corresponds to the protein concentration required to half-maximally stimulate phytoalexin production in parsley protoplasts (Parker et al., 1991).
bNo detectable activity (tested up to 50 nM).
Pep-13-mediated defense responses in potato
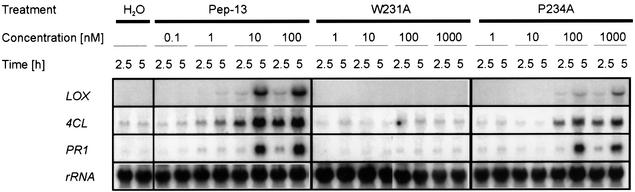
The interaction between potato and P.infestans, the causal agent of late blight disease, can result in devastating crop losses, as illustrated by the Irish potato famine of the 19th century (Govers, 2001; Kamoun, 2001). Both infection of potato plants and treatment of potato cells with P.infestans culture filtrate triggered defense gene expression and the synthesis of antimicrobial compounds (Rohwer et al., 1987; Göbel et al., 2001). We therefore tested whether potato has the ability to recognize and respond to the conserved Pep-13 motif. Treatment of potato cells with Pep-13 led to the accumulation of defense-related transcripts encoding lipoxygenase, 4-coumarate:CoA ligase and pathogenesis-related protein 1 (Figure 4). Likewise, increased transcript levels of the same genes were detected in intact potato leaves upon infiltration of Pep-13 (not shown). Dose–response experiments revealed that 10 nM Pep-13 was sufficient to induce accumulation of defense gene transcripts in potato cells (Figure 4). We next tested whether activation of defense in potato through recognition of Pep-13 resembled that described for parsley, using the Pep-13 mutant derivatives. No defense-related transcript accumulation was detectable in potato cells treated with Pep-13 containing the W231A mutation (Figure 4), even at higher concentrations (1 µM), demonstrating that this mutation abolishes the ability of Pep-13 to elicit defense responses in potato. This compares favorably with our failure to measure phytoalexin production in parsley protoplasts treated with GP42 TGase containing the W231A mutation. However, treatment of potato cells with Pep-13 containing the P234A mutation induced defense gene activation (Figure 4) only at concentrations significantly higher than wild-type Pep-13 (≥100 nM). This is in agreement with a significantly higher EC50 value for initiation of defense responses in parsley following treatment with the corresponding GP42 TGase mutant. Thus, the ability of Pep-13 to elicit defense responses in potato correlates quantitatively and qualitatively with the elicitor activity described for parsley. These data suggest that this recognition capacity for Phytophthora species is a more widespread feature of plants, and that the Pep-13 motif could function as a PAMP for the activation of innate defense reactions during these interactions.
Fig. 4. Functionally indispensable residues selected as recognition determinants for the activation of defense in parsley also serve as defense-inducing determinants in potato. RNA isolated from potato cells 2.5 and 5 h following treatment with Pep-13 (0.1–100 nM), or with Pep-13 mutant derivatives W231A or P234A (1–1000 nM), was analyzed for the accumulation of defense-related transcripts. The RNA blots were probed with radiolabeled cDNA fragments encoding lipoxygenase (LOX), 4-coumarate:CoA ligase (4CL), pathogenesis-related protein 1 (PR1) and 25S rRNA (rRNA) as loading control.
Discussion
Here we describe for the first time a microbial TGase that is Ca2+ dependent. Microbial TGases known to date are of bacterial or plasmodial origin, but these enzymes are very sequence divergent from the animal and Phytophthora TGases, and their enzyme activities were found to be independent of Ca2+ (Kanaji et al., 1993; Adini et al., 2001). Ca2+-dependent TGases have been isolated from a variety of animal species (Aeschlimann and Paulsson, 1994; Noguchi et al., 2001; Ahvazi et al., 2002), but not from fungi (including yeast) or plants. Interestingly, Phytophthora TGases also lack sequence similarity to all of the known Ca2+-dependent TGases, but share with these enzymes (in those cases where tested) a similar co-factor requirement and inhibitor sensitivity. Limited, but significant, sequence similarity between Phytophthora and mammalian TGases, as well as cysteine proteases, was observed in the regions adjacent to the catalytic site cysteine residue of these enzymes (our unpublished data). Thus, it is intriguing to speculate whether the apparent sequence dissimilarity between the TGases is the result of divergent evolution or, alternatively, whether similarities in the active site domain are indicative of convergent evolution.
Seminal reviews (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000; Imler and Hoffmann, 2001; Underhill and Ozinsky, 2002) have highlighted striking similarities between the molecular organization of innate immunity in vertebrates and insects. The authors referred to pathogen-derived signals as PAMPs, which enable potential host cells to discriminate between potential microbial pathogens and self, no matter whether these microbes are pathogenic or not. Receptor-mediated PAMP recognition results in the production of antimicrobial compounds. PAMPs are not only shared by particular pathogen races, but are broader signatures of a given class of microorganisms. They constitute evolutionarily conserved structures that are unique to microorganisms, have important roles in microbial physiology and may therefore not be subject to frequent mutation. Typical PAMPs include LPS of Gram-negative bacteria, bacterial flagellin, and fungal cell wall-derived carbohydrates or proteins, some of which were also shown to trigger plant defense in a non-cultivar-specific manner (Boller, 1995; Nürnberger and Brunner, 2002). The concept of PAMP-mediated non-self recognition has found renewed interest among plant biologists, as it may provide an explanation for why plants may recognize and respond to non-race-specific elicitors of plant defense.
In this study, we attempted to show whether Pep-13 exhibits characteristics of PAMPs. Such studies are part of our general attempt to elucidate why microbe-specific surface structures do induce plant defense reactions, and if such perception systems resemble those described in vertebrates and insects. We show that Pep-13 is a surface-exposed motif present within a cell wall TGase, which is apparently unique to Phytophthora species and is not found in potential host plants. In addition, mutational analyses within the Pep-13 motif revealed amino acid residues to be important for both TGase activity and elicitor activity of the parent protein. Thus, mutations within this conserved and functionally important region may not allow the microbe to evade Pep-13-mediated recognition by plants. The genus-wide presence of the protein (including the highly conserved Pep-13 region) is indicative of an important function of this protein for the life cycle of Phytophthora. Unfortunately, our attempts to inactivate P.infestans TGase gene expression failed. Among the 14 stable transformant cell lines produced, none was found to exhibit significantly reduced TGase levels (F.Brunner, I.Vijn, F.Glovers and T.Nürnberger, unpublished data). However, as gene silencing in Phytophthora (and oomycetes in general) is not yet a routine application, we were unable to produce significantly larger numbers of transgenic lines. Neverthe less, it is reasonable to assume that the genus-wide presence of the TGase (including the highly conserved Pep-13 region) is supportive of an essential function for this protein. Taken together, the elicitor Pep-13 shows hallmarks of PAMPs known to evoke innate immune responses in vertebrates and insects, and may thus serve a similar role in the interaction between plants and phytopathogenic Phytophthora.
Elicitors, such as Pep-13, may act (often as one of many) as non-self recognition determinants for the activation of plant defense responses in a non-cultivar-specific manner, but may not necessarily mediate resistance. This (in addition to the broad distribution among pathogen races) clearly distinguishes PAMPs from avirulence gene products conferring plant cultivar-specific pathogen recognition and disease resistance (Nürnberger et al., 1994; Cohn et al., 2001; Dangl and Jones, 2001). PAMP-based alert systems seem to function with different efficiencies in both host and non-host plants. In the case of the non-host plant, parsley, receptor-mediated recognition of this PAMP may trigger defense reactions that contribute to, or are sufficient for, resistance against Phytophthora infection (Nürnberger et al., 1994). However, in the potato–P.infestans disease-causing interaction, pathogen recognition through the Pep-13 motif is clearly insufficient to provide resistance. It is assumed that during evolution plant species resistance was overcome by phytopathogens through the acquisition of virulence factors, which enabled them to either evade or (partially) suppress host plant defense mechanisms. Such newly evolved pathogen race-specific virulence factors have driven the co-evolution of plant resistance genes and thus the development of phylogenetically more recent pathogen race/plant cultivar-specific disease resistance (Heath, 2000; Dangl and Jones, 2001; Kamoun, 2001). Importantly, susceptibility of host cells in spite of PAMP-mediated pathogen recognition (probably through repression of host defense by the pathogen) is found in animals as well. Stimulation of the innate immune system in human or Drosophila by, for example, bacterial LPS or flagellin, may not in all cases sufficiently protect the host from infection by Gram-negative bacteria displaying either one or both PAMPs. Nevertheless, PAMP-mediated activation of innate immune responses was shown to contribute to successful defense against microbial invasion in both Drosophila (Lemaitre et al., 1996) and mammals (Medzhitov and Janeway, 1997; Aderem and Ulevitch, 2000; Underhill and Ozinsky, 2002). Importantly, two natural mutations (lps) that render mice insensitive to Gram-negative bacterial LPS, yet highly susceptible to Gram-negative infection, were shown to be defective in the TLR4 receptor (Poltorak et al., 1998; Hoshino et al., 1999), a homolog of which was first cloned from human and shown to be essential for activation of adaptive immune responses as well (Medzhitov et al., 1997). In total, 10 vertebrate TLR receptors sensing different microbial molecules have been identified, covering the whole array of pathogens with which a potential host must cope. Gene knockout work confirmed that various TLR–/– mice showed decreased resistance to a variety of Gram-positive as well as Gram-negative bacteria (Underhill and Ozinsky, 2002). Intriguingly, the repertoire for the recognition of TLR stimuli can be significantly enlarged through hetero-oligomerization of different TLRs (Ozinsky et al., 2000).
It will now be important to determine whether and to what extent PAMP-mediated pathogen recognition may contribute to the activation of innate immune responses in both host and non-host plants. For example, plant varieties susceptible under certain test conditions may not necessarily be susceptible under less favorable infection conditions determined by humidity, temperature or reduced inoculum densities (Tyler, 2002). Moreover, PAMP-mediated alert systems for the activation of plant defense in non-host plants may contribute to non-host resistance, which is the predominant form of plant disease resistance (Heath, 2000; Kamoun, 2001; Nürnberger and Brunner, 2002). Instrumental to the assessment of individual PAMP recognition events for overall disease resistance will be the genetic inactivation of PAMP-encoding pathogen genes (such as the TGase gene in Phytophthora species) as well as those encoding PAMP receptors in plants. Once technically feasible, inactivation of PAMP-encoding genes in Phytophthora will enable assessment of the effects of such mutations on overall fitness of the pathogen as well as on its virulence. Likewise, we have previously devised a strategy to isolate the Pep-13 receptor from parsley (Nennstiel et al., 1998), which upon completion will allow us to isolate the encoding receptor genes from parsley and potato. Subsequently, upon inactivation of this gene in potato, it will be interesting to assess whether such a mutation would result in an enhanced susceptibility phenotype. However, it should also be taken into consideration that another, yet unexplored role of PAMP-mediated ‘non-self’ recognition might be to establish/maintain a beneficial homeostasis between plants and commensal or potentially mutualistic symbionts.
The finding that an elicitor of non-cultivar-specific plant defense exhibits characteristics of PAMPs known to trigger innate immune responses in animals adds to the growing list of parallels in the molecular organization of innate immunity in various kingdoms. The intriguing view of an evolutionary conservation of innate defense mechanisms across kingdom borders (Cohn et al., 2001; Dangl and Jones, 2001; Nürnberger and Brunner, 2002) is further supported by structural similarities found between the flagellin receptor in human (TLR5) (Hayashi et al., 2001) and the Arabidopsis flg22 receptor FLS2 (Gomez-Gomez and Boller, 2000), as well as by the identification of MAP kinase cascades implicated in the activation of innate immune responses in both plants and animals (Asai et al., 2002; Dong et al., 2002).
Materials and methods
Cultivation of oomycetes and plant cell cultures
Oomycetes were grown on vegetable juice agar (Rohwer et al., 1987). Liquid cultures were harvested after 3–4 weeks of growth and filtered through a 200 µm nylon mesh. The filtrate was cleared by centrifugation at 4100 g for 20 min. The culture filtrate material, stored as freeze-dried preparations, was dissolved in water and concentrated in Centriprep or Centricon YM10 filters (Millipore).
Dark-grown, 5-day-old suspension-cultured potato cells (cv. Désirée) or parsley cells were used for elicitor treatment (Göbel et al., 2001) or protoplast preparation (Parker et al., 1991). Quantification of furanocoumarin phytoalexin production in parsley protoplasts was performed as described previously (Parker et al., 1991). Elicited potato cells were harvested by filtration and stored at –80°C.
Protein biochemistry
Proteins were separated on 12.5% SDS–polyacrylamide gels and blotted according to standard protocols. Both primary (anti-Pep-13) and secondary (goat anti-rabbit IgG–horseradish peroxidase conjugate; Bio-Rad) antibodies were used at 1:5000 dilution. Immunodetection was performed using the ECL Plus detection system (Amersham Pharmacia Biotech). For in-gel determination of TGase activity, a PVDF membrane was first incubated overnight in a 10–20 mg/ml N,N-dimethylcasein solution in 0.1 M Tris–HCl pH 8.5. The membrane was blocked with non-fat dry milk (1% in 0.1 M Tris–HCl pH 8.5) for 1 h, followed by two washes in 0.1 M Tris–HCl pH 8.5 and two washes with 0.1 M sodium acetate pH 5.2.
In-gel denaturation and renaturation of TGases were performed as described previously (Usami et al., 1995). After two washes in 50 mM Tris–HCl pH 8.5 and 0.1 M sodium acetate pH 5.2 for 5 min, the gel was overlaid upon the N,N-dimethylcasein-coated PVDF membrane and maintained immersed in TGase buffer [0.1 M sodium acetate pH 5.2, 0.5 mM 5-(biotinamido) pentylamine, 10 mM DTT, 5 mM CaCl2] for 12 h at 20°C. The membrane was washed twice with 0.2 M EDTA pH 8.0 and twice with PBS buffer prior to 1 h incubation with avidin–horseradish peroxidase (Bio-Rad), diluted 1:5000 in PBS buffer, 1% non-fat dry milk, 0.05% Tween-20. After four additional washes with PBS buffer, TGase activity was detected by chemiluminescence.
The TGase solid-phase microtiter plate assays were carried out as described previously (Slaughter et al., 1992) with the following modifications: 100 ng/ml recombinant wild-type or mutant GP42 was used in each reaction. The reaction was performed in 0.1 M sodium acetate pH 5.2 and the streptavidin–alkaline phosphatase (1000 U/ml; Roche) was diluted 1:1000. A kinetic measurement of absorbance at 405 nm was determined at 15 min intervals for a period of 1–4 h using an MRX microplate reader (Dynatech Laboratories).
Recombinant protein expression in A.oryzae
The P.sojae GP42 encoding cDNA (U10471) was introduced into the pBluescript II KS(-) vector (Stratagene) and subjected to site-directed mutagenesis using the GeneEditor in vitro site-directed mutagenesis system (Promega). Trp231, Pro234 and Tyr241 were each substituted by alanine. The mutant cDNA constructs were sequenced and confirmed to contain only the expected mutations. The constructs were subcloned into the pHD464 expression vector (Dalbøge and Heldt-Hansen, 1994) and transformation of A.oryzae JaL228 was carried out as described previously (Fuglsang et al., 2000). Transformants were grown for 3 days at 30°C before harvesting the culture supernatant. Aliquots of this material were desalted by gel filtration on PD-10 columns (Amersham Pharmacia Biotech) and stored at –80°C until use in assays for TGase and elicitor activity.
RNA blot and RT–PCR analysis
Twenty micrograms of total RNA isolated from elicitor-treated potato cells were subjected to RNA blot analysis as described previously (Göbel et al., 2001). As probes, the following potato cDNA fragments were used: a 0.4 kb PCR fragment from LOX (POTLX-3; Kolomiets et al., 2000), a 2.0 kb EcoRI fragment from 4CL and a 0.3 kb EcoRI fragment from PR1 (Göbel et al., 2001). For standardization, blots were probed with a 1.3 kb BamHI fragment from potato 25S rRNA.
Oomycete poly(A)+ RNA was purified from 100 µg of total RNA (Dunsmuir et al., 1989) using oligo(dT) cellulose (Amersham Pharmacia Biotech). First-strand cDNA was synthesized from 50 ng of poly(A)+ RNA using the First-Strand cDNA Synthesis Kit (Amersham Pharmacia Biotech). PCR amplification was performed using degenerate primers 5′-gatgaattcGA(A/G)AA(A/G)TA(C/T)GC(N)AA(A/G)GC(N)TT(C/T)GG-3′ (sense) and 5′-cccgggtcgaCGT(A/G)TA(N)GT(C/T)TC(A/G)TA(A/G/T)ATCCA-3′ (antisense) encoding, respectively, amino acids (one-letter code) EKYAKAF and WIYETYT in the sequence of the P.sojae GP42. The following PCR conditions were used: 30–32 cycles (1 min, 94°C; 1 min, 54°C; 1.5 min, 72°C). Subsequently, the PCR fragments were cloned into the pGEM-T vector (Promega) and sequenced.
Acknowledgments
Acknowledgements
We thank H.Keller and P.Venard (INRA, Unité Santé Végétale et Environnement, Antibes) for providing oomycete isolates, W.Wirtz for cloning of the TGase wild-type construct and M.-A.Allerslev for skillful technical assistance. We are grateful to Jane Parker (MPIZ Cologne) for kindly providing P.parasitica-infected Arabidopsis leaves. F.B. received support from KWS Einbeck, Germany. J.J.R., J.L., D.S. and T.N. were funded by the EC, DFG and Fonds der Chemischen Industrie. B.Blume and A.Ozinsky are gratefully acknowledged for critical reading of the manuscript.
References
- Aderem A. and Ulevitch,R. (2000) Toll-like receptors in the induction of the innate immune response. Nature, 406, 782–787. [DOI] [PubMed] [Google Scholar]
- Adini A., Krugliak,M., Ginsburg,H., Li,L., Lavie,L. and Warburg,A. (2001) Transglutaminase in Plasmodium parasites: activity and putative role in oocysts and blood stages. Mol. Biochem. Parasitol., 117, 161–168. [DOI] [PubMed] [Google Scholar]
- Aeschlimann D. and Paulsson,M. (1994) Transglutaminases: protein cross-linking enzymes in tissue and body fluids. Thromb. Haemost., 71, 402–415. [PubMed] [Google Scholar]
- Ahvazi B., Kim,H.C., Kee,S.H., Nemes,Z. and Steinert,P.M. (2002) Three-dimensional structure of the human transglutaminase 3 enzyme: binding of calcium ions changes structure for activation. EMBO J., 21, 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena,G., Plotnikova,J., Willmann,M.R., Chiu,W.L., Gomez-Gomez,L., Boller,T., Ausubel,F.M. and Sheen,J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Blume B., Nürnberger,T., Nass,N. and Scheel,D. (2000) Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell, 12, 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. (1995) Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol., 46, 189–214. [Google Scholar]
- Cohn J., Sessa,G. and Martin,G.B. (2001) Innate immunity in plants. Curr. Opin. Immunol., 13, 55–62. [DOI] [PubMed] [Google Scholar]
- Dalbøge H. and Heldt-Hansen,H.P. (1994) A novel method for efficient expression cloning of fungal enzyme genes. Mol. Gen. Genet., 243, 253–260. [DOI] [PubMed] [Google Scholar]
- Dangl J.L. and Jones,J.D.G. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dong C., Davis,R.J. and Flavell,R.A. (2002) MAP kinases in the immune response. Annu. Rev. Immunol., 20, 55–72. [DOI] [PubMed] [Google Scholar]
- Dunsmuir P., Bond,D., Lee,K.J., Gidoni,D. and Townsend,J. (1989) Stability of introduced genes and stability in expression. In Gelvin,S.B. and Schilperoort,R.A. (eds), Plant Molecular Biology Manual, Vol. 1. Kluwer Academic, Dordrecht, The Netherlands, pp. 1–17.
- Eisenberg D., Schwarz,E., Komarony,M. and Wall,R. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol., 179, 125–142. [DOI] [PubMed] [Google Scholar]
- Erwin D.C. and Ribeiro,O.K. (1996) Phytophthora Diseases Worldwide. The American Phytopathological Society Press, St Paul, MN.
- Felix G., Duran,J.D., Volko,S. and Boller,T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J., 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Folk J.E. (1980) Transglutaminases. Annu. Rev. Biochem., 49, 517–531. [DOI] [PubMed] [Google Scholar]
- Fuglsang C.C., Berka,R.M., Wahleithner,J.A., Kauppinen,S., Shuster,J.R., Rasmussen,G., Halkier,T., Dalbøge,H. and Henrissat,B. (2000) Biochemical analysis of recombinant fungal mutanases. A new family of α1,3-glucanases with novel carbohydrate-binding domains. J. Biol. Chem., 275, 2009–2018. [DOI] [PubMed] [Google Scholar]
- Göbel C., Feussner,I., Schmidt,A., Scheel,D., Sanchez-Serrano,J.J., Hamberg,M. and Rosahl,S. (2001) Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor-treated potato cells. J. Biol. Chem., 276, 6267–6273. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L. and Boller,T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Govers F. (2001) Misclassification of pest as ‘fungus’ puts vital research on wrong track. Nature, 411, 633. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K., Scheel,D., Logemann,E., Nürnberger,T., Parniske,M., Reinhold,S., Sacks,W.R. and Schmelzer,E. (1995) Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc. Natl Acad. Sci. USA, 92, 4150–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F. et al. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature, 410, 1099–1103. [DOI] [PubMed] [Google Scholar]
- Heath M. (2000) Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol., 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Hoshino K., Takeuchi,O., Kawai,T., Sanjo,H., Ogawa,T., Takeda,Y., Takeda,K. and Akira,S. (1999) Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol., 162, 3749–3752. [PubMed] [Google Scholar]
- Imler J.-L. and Hoffmann,J.A. (2001) Toll receptors in innate immunity. Trends Cell Biol., 11, 304–311. [DOI] [PubMed] [Google Scholar]
- Jabs T., Tschöpe,M., Colling,C., Hahlbrock,K. and Scheel,D. (1997) Elicitor-stimulated ion fluxes and O2– from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl Acad. Sci. USA, 94, 4800–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S. (2001) Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr. Opin. Plant Biol., 4, 295–300. [DOI] [PubMed] [Google Scholar]
- Kanaji T., Ozaki,H., Takao,T., Kawajiri,H., Ide,H., Motoki,M. and Shimonishi,Y. (1993) Primary structure of microbial transglut aminase from Streptoverticillium sp. strain s-8112. J. Biol. Chem., 268, 11565–11572. [PubMed] [Google Scholar]
- Kolomiets M.V., Chen,H., Gladon,R.J., Brown,E.J. and Hannapel,D.J. (2000) A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiol., 124, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Nicolas,E., Michaut,L., Reichhart,J. and Hoffmann,J. (1996) The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell, 86, 973–983. [DOI] [PubMed] [Google Scholar]
- Ligterink W., Kroj,T., zur Nieden,U., Hirt,H. and Scheel,D. (1997) Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science, 276, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Liu S., Cerione,R.A. and Clardy,J. (2002) Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl Acad. Sci. USA, 99, 2743–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. and Janeway,C.A.,Jr (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell, 91, 295–298. [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt,P. and Janeway,C.A.,Jr (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature, 388, 323–324. [DOI] [PubMed] [Google Scholar]
- Melino G. and Piacentini,M. (1998) Tissue transglutaminase in cell death: a downstream or multifunctional upstream effector? FEBS Lett., 430, 59–63. [DOI] [PubMed] [Google Scholar]
- Nennstiel D., Scheel,D. and Nürnberger,T. (1998) Characterization and partial purification of an oligopeptide elicitor receptor from parsley (Petroselinum crispum). FEBS Lett., 431, 405–410. [DOI] [PubMed] [Google Scholar]
- Noguchi K., Ishikawa,K., Yokoyama,K., Ohtsuka,T., Nio,N. and Suzuki,E. (2001) Crystal structure of red sea bream transglut aminase. J. Biol. Chem., 276, 12055–12059. [DOI] [PubMed] [Google Scholar]
- Nürnberger T. and Brunner,F. (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol., 5, 318–324. [DOI] [PubMed] [Google Scholar]
- Nürnberger T. and Scheel,D. (2001) Signal transmission in the plant immune response. Trends Plant Sci., 6, 372–379. [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Nennstiel,D., Jabs,T., Sacks,W.R., Hahlbrock,K. and Scheel,D. (1994) High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell, 78, 449–460. [DOI] [PubMed] [Google Scholar]
- Ozinsky A., Underhill,D.M., Fontenot,J.D., Hajjar,A.M., Smith,K.D., Wilson,C.B., Schroeder,L. and Aderem,A. (2000) The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl Acad. Sci. USA, 97, 13766–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.E., Schulte,W., Hahlbrock,K. and Scheel,D. (1991) An extracellular glycoprotein from Phytophthora megasperma f. sp. glycinea elicits phytoalexin synthesis in cultured parsley cells and protoplasts. Mol. Plant-Microbe Interact., 4, 19–27. [Google Scholar]
- Poltorak A. et al. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science, 282, 2085–2088. [DOI] [PubMed] [Google Scholar]
- Rohwer F., Fritzemeier,K.H., Scheel,D. and Hahlbrock,K. (1987) Biochemical reactions of different tissue of potato (Solanum tuberosum) to zoospores or elicitors from Phytophthora infestans. Planta, 170, 556–561. [DOI] [PubMed] [Google Scholar]
- Rost B. (1996) PredicProtein. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Sacks W.R., Nürnberger,T., Hahlbrock,K. and Scheel,D. (1995) Molecular characterization of nucleotide sequences encoding the extracellular glycoprotein elicitor from Phytophthora megasperma. Mol. Gen. Genet., 246, 45–55. [DOI] [PubMed] [Google Scholar]
- Slaughter T.F., Achyuthan,K.E., Lai,T.-S. and Greenberg,C.S. (1992) A microtiter plate transglutaminase assay utilizing 5-(biotinamido) pentylamine as substrate. Anal. Biochem., 205, 166–171. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Tyler B.M. (2002) Molecular basis of recognition between Phytophthora pathogens and their hosts. Annu. Rev. Phytopathol., 40, 137–167. [DOI] [PubMed] [Google Scholar]
- Underhill D.M. and Ozinsky,A. (2002) Toll-like receptors: key mediators of microbe detection. Curr. Opin. Immunol., 14, 103–110. [DOI] [PubMed] [Google Scholar]
- Usami S., Banno,H., Ito,Y., Nishimama,R. and Machida,Y. (1995) Cutting activates a 46-kilodalton protein kinase in plants. Proc. Natl Acad. Sci. USA, 92, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S., Nürnberger,T., Frachisse,J.-M., Wirtz,W., Guern,J., Hedrich,R. and Scheel,D. (1997) Receptor-mediated activation of a plant Ca2+-permeable ion channel involved in pathogen defense. Proc. Natl Acad. Sci. USA, 94, 2751–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]