Abstract
p21WAF1/CIP1 contributes to positive and negative growth control on multiple levels. We previously mapped phosphorylation sites within the C-terminal domain of p21 that regulate proliferating cell nucear antigen binding. In the current study, a kinase has been fractionated from mammalian cells that stoichiometrically phosphorylates p21 at the Ser146 site, and the enzyme has been identified as an insulin-responsive atypical protein kinase C (aPKC). Expression of PKCζ or activation of the endogenous kinase by 3-phosphoinositide dependent protein kinase-1 (PDK1) decreased the half-life of p21. Conversely, dnPKCζ or dnPDK1 increased p21 protein half-life, and a PDK1-dependent increase in the rate of p21 degradation was mediated by aPKC. Insulin stimulation gave a biphasic response with a rapid transient decrease in p21 protein levels during the initial signalling phase that was dependent on phosphatidylinositol 3- kinase, PKC and proteasome activity. Thus, aPKC provides a physiological signal for the degradation of p21. The rapid degradation of p21 protein during the signalling phase of insulin stimulation identifies a novel link between energy metabolism and a key modulator of cell cycle progression.
Keywords: aPKC/degradation/insulin/p21/phosphorylation
Introduction
p21WAF1/CIP1 is a tumour modifier gene (Jones et al., 1999) and a downstream target for the anti-oncogenic transcription factors p53 (el-Deiry et al., 1993), IRF-1 (Tanaka et al., 1996) and the recently identified prostrate cancer susceptibility gene KLF6 (Narla et al., 2001). The p21 protein contributes to the regulation of cell growth and division on multiple levels, which include: mediation of negative growth signals; activation of cyclin–Cdk4 complexes; positive and negative modulation of cellular differentiation; inhibition of apoptotic cell death; and functions in stem cell renewal (Sherr and Roberts, 1999; Dotto, 2000). The activity of p21 as a cyclin-dependent kinase (CDK) inhibitor has been studied extensively (Ball, 1997; Sherr and Roberts, 1999). However, it is beginning to emerge that CDK-independent functions of p21 also contribute significantly to its biological activity and that these rely on an increasingly complex network of interacting proteins (Dotto, 2000). Although p21 function is dependent on its ability to make protein–protein interactions, little is known about how these interactions are regulated.
We have shown previously that signalling pathways which target the C-terminal regulatory domain of p21 exist in eukaryotic cells, and that phosphorylation of p21 at either Thr145 or Ser146 is sufficient to inhibit the interaction of p21 with proliferating cell nuclear antigen (PCNA; Scott et al., 2000). These data provided the first evidence that phosphorylation of p21 protein may have a regulatory effect on its protein–protein interactions. Subsequent reports have identified protein kinase B (PKB; Akt) as a potential Thr145 kinase and have defined additional functions for modification at this site in terms of both p21 activity and subcellular localization (Rossig et al., 2001; Zhou et al., 2001).
Increasing evidence suggests that the PKC family of kinases are involved in the regulation of critical cell cycle transitions such as cell cycle entry, exit and G1/G2 check points (Black, 2000). PKC-mediated control of these events appears to be dependent on the isoform involved and the precise timing of its activation. Although information on the physiological role(s) of the atypical PKCs (aPKCs; the closely related PKCζ and PKCι) lags behind our knowledge of other family members, they are generally associated with cell proliferation, survival and differentiation (Moscat et al., 2001). The aPKCs play an important role in the regulation of cellular metabolism during the acute response to insulin stimulation. Both PKCζ and ι are activated by insulin through a phosphatidylinositol 3-kinase (PI3K)/3-phosphoinositide dependent protein kinase-1 (PDK1)-dependent mechanism, and insulin stimulation of glucose transport and general protein synthesis depends on the activation of one or both of these enzymes (Mendez et al., 1997; Kotani et al., 1998; Standaert et al., 1999). In addition, the aPKCs may also play a role in an insulin-stimulated negative feedback loop in opposition to PKB (Mao et al., 2000; Liu et al., 2001).
Although the Ser146 site of p21 was shown previously to regulate p21 protein-binding activity (Scott et al., 2000), the signalling pathway that targets this site in vivo is unclear. Here we identify a kinase activity from human cells that phosphorylates p21 at the Ser146 site as an atypical member of the PKC family. Activation of aPKC leads to the degradation of p21 protein, providing a link between insulin signalling and cell cycle control.
Results
Fractionation of mammalian cell lysates to identify potential p21 kinases
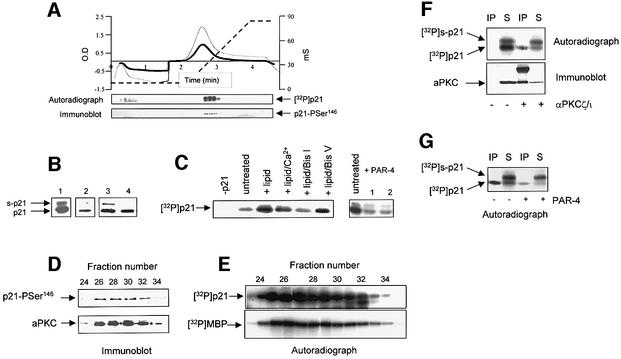
The nature of the signalling pathway(s) that targets the Ser146 phospho-acceptor site of p21 in mammalian cells, or the queues that lead to activation/inactivation of the pathway, remain unclear. HeLa cell extracts were fractionated in an effort to identify a physiologically relevant Ser146 kinase activity. Pooled fractions, containing p21 kinase activity that previously had been subject to chromatography on Q-Sepharose and SP-Poros columns (all the detectable Ser146 kinase activity co-eluted in a single peak from these two columns), were applied to a HS-Poros column and eluted with a linear gradient of 50 mM–1 M NaCl (Figure 1A). The fractions were screened for total p21 kinase activity by incorporation of 32P into full-length p21 and for Ser146 kinase activity using p21-Pser146 IgG by denaturing immunoblot (Scott et al., 2000). Total p21 kinase and Ser146 kinase activity co-eluted in one peak (0.35 M NaCl) on the trailing edge of the total protein peak. The pooled and concentrated active fractions from this column (HS-pool) provided material for characterization of the Ser146 kinase. When the concentrated fractions were assayed for total p21 kinase activity, it was observed, particularly in some preparations, that a second form of p21 with a distinct electrophoretic mobility was generated (labelled s-p21; Figure 1B), whereas the bulk of the phosphorylated protein had the same mobility as unphosphorylated p21 (Figure 1B, lane 4).
Fig. 1. Fractionation of a p21-Ser146 kinase from HeLa cells. (A) The active SP-Sepharose pool was applied to a HS-Poros column and eluted with a linear 0.05–1 M NaCl gradient, and the change in conductivity was measured in milliSiemens (mS; dashed line). Protein was detected at 280 (dark line) and 254 nm (fine line) and is plotted against time. Fractions were collected every 6 s (equivalent to 1 ml) and total p21 kinase activity was measured as the incorporation of 32P into full-length p21 by autoradiography; p21-Ser146 kinase activity was measured by immunoblot using αp21-Pser146 IgG. The active fractions were combined and concentrated to form the HS-pool. (B) Total p21 kinase activity in the HS-pool was determined by the incorporation of 32P into p21 protein by autoradiography (lane 1); phosphorylation at the Ser146 site was detected by immunoblot using p21-Pser146 IgG (lane 2). Lanes 3 and 4 show total p21 protein added to the assay in the presence (lane 3) and absence (lane 4) of the HS-pool and magnesium-ATP. Two forms of p21 were detected and have been labelled p21, which has the same mobility as unphosphorylated protein (lane 4), and s-p21, which has a higher apparent molecular mass. (C) Autoradiographs showing the incorporation of 32P into full-length p21 protein by the HS-pool (left panel) in the presence of lipid mix (100 µg/ml phosphatidylserine and 20 µg/ml diacylglycerol), and lipid mix plus Ca2+ (1 mM), Bis I (10 µM) or Bis V (10 µM) (right panel) in the absence or presence of PAR-4 (1 µg), where PAR-4 had been added immediately prior to the assay (lane 1) or had been pre-incubated with the HS-pool for 10 min (lane 2). (D and E) Fractions from the HS-Poros column were screened for aPKC protein and p21-Ser146 kinase activity by immunoblot using αPKCζ/ι and p21-Pser146 IgG, respectively (D), and for total p21 kinase activity and MBP kinase activity by following the incorporation of 32P using autoradiography (E). (F) aPKC was immunodepleted from the HS-pool using αPKCζ/ι. Incorporation of 32P into p21 was determined by autoradiography (upper panel) and aPKC protein levels by immunoblot using αPKCζ/ι (lower panel) in either the immunoprecipitated material (IP) or the remaining supernatant (S) from a protein G bead alone control (–) or from beads plus αPKCζ/ι (+). (G) The αPKCζ/ι immunoprecipitate (IP) and supernatant (S) from (F) were assayed for incorporation of 32P into p21 protein in the absence (–) or presence (+) of PAR-4 (1 µg). The two forms of p21 are labelled p21 and s-p21.
The Ser146 kinase from proliferating HeLa cells is an aPKC
Characterization of the p21 kinase activity (Figure 1C) showed that it was activated by a lipid mix containing phosphatidylserine but that there was no additional stimulation by Ca2+. The enzyme was inhibited by the general PKC inhibitor bisindolylmaleimide I (Bis I), when used at 10 µM, whereas the inactive derivative, bisindolylmaleimide V (Bis V), had no significant effect. Furthermore, PAR-4, a specific inhibitor of the atypical PKCs, and not other PKC family members (Berra et al., 1997), inhibited the p21 kinase. This preliminary evidence suggested that the Ser146 kinase had the characteristics of an aPKC family member.
Analysis of individual fractions from the HS-Poros column (Figure 1D and E) revealed that aPKC protein co-eluted with the p21-Ser146 kinase activity, total p21 kinase activity and myelin basic protein (MBP) kinase activity (MBP is used commonly as a substrate to assay aPKC activity). Significantly, when aPKC protein was immunodepleted from the HS-pool using αPKCζ/ι, the kinase activity in the precipitate corresponded to p21 and did not give s-p21 (Figure 1F). Conversely, the remaining supernatant had been depleted of the activity giving p21 relative to that giving s-p21. We verified that the immunoprecipitated activity was an aPKC by using PAR-4 to inhibit its activity (Figure 1G). PAR-4 also significantly inhibited the p21 kinase activity remaining in the supernatant, consistent with the fact that not all the αPKCζ/ι reactive material was immunodepleted (Figure 1F). Immunodepletion of the p21 kinase activity using αPKCζ/ι provided strong evidence that the Ser146 kinase from HeLa cells was an aPKC.
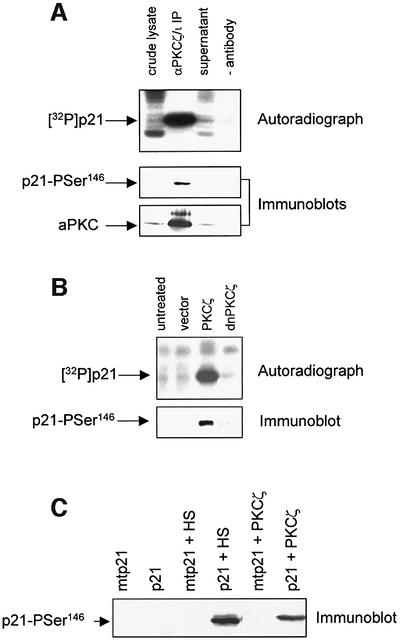
To establish further the link between Ser146 phosphorylation of p21 and the aPKC family, the following experiments were performed. First, endogenous aPKC, immunoprecipitated from crude cell lysates, phosphorylated p21 at the Ser146 site (Figure 2A). Secondly, lysates from HCT116 cells transfected with PKCζ or a kinase-dead PKCζ mutant (dnPKCζ) showed high levels of Ser146 kinase activity only when transfected with wild-type enzyme (Figure 2B). Thirdly, a commercial preparation of recombinant PKCζ phosphorylated p21 at the Ser146 site in vitro, incorporating 0.88 mol of phosphate/mol of p21 protein (Figure 2C), and no signal was detected when the Ser146 site was mutated to alanine. Therefore, endogenous, expressed and purified recombinant aPKC all phosphorylated p21 at the Ser146 site.
Fig. 2. aPKC phosphorylates p21 at the Ser146 site. (A) The incorporation of 32P into p21 protein was detected by autoradiography (upper panel); p21-Ser146 kinase activity and aPKC protein were detected by immunoblot using αp21-Pser146 IgG and αPKCζ/ι, respectively (lower panels), in HCT116 cell lysate (5 µg total protein), αPKCζ/ι immunoprecipitated material (from 250 µg of total cell lysate), the remaining supernatant (5 µg of total protein) or a control in which protein G beads minus αPKCζ/ι were incubated with cell lysate (–antibody). (B) The incorporation of 32P into p21 protein was detected by autoradiography (upper panel) and p21-Ser146 kinase activity by immunoblot (lower blot) using αp21-Pser146 IgG. HCT116 cells were untreated or transfected with 2 µg each of pcDNA3.1, pcDNA3.1/HA-PKCζ or pcDNA3.1/HA-dnPKCζ. (C) Wild-type (p21) or Ser146→Ala (mtp21) p21 protein (1 µg) purified from E.coli was phosphorylated with the HS-pool (5 µl) or 0.5 U/ml recombinant PKCζ (Upstate Biotechnology). Ser146 phosphorylation was detected by immunoblot using αp21-Pser146 IgG.
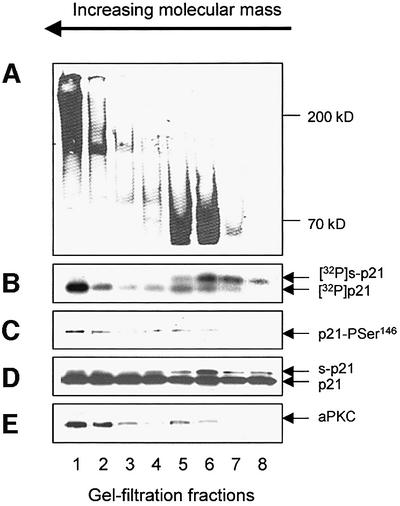
Further characterization of the HS-pool, performed by gel filtration, revealed that aPKC protein was present in a monomeric form (70 ± 5 kDa) and as part of a high molecular weight complex (≥200 kDa). The complex did not represent aggregated protein as it readily entered a 4% native gel and had the mobility of a 200 ± 20 kDa complex when compared with marker proteins (Figure 3A and E). The isolation of a stable aPKC-containing complex is consistent with the aPKCs normally being found in association with regulatory/targeting subunits in vivo (Moscat and Diaz-Meco, 2000). Both the monomeric and complexed form of aPKC had p21-Ser146 kinase activity (Figure 3B and C). Gel filtration provided evidence that the s-p21 species was generated by a distinct enzyme activity that was unrelated to PKCζ/ι (Figure 3). Thus, the s-p21 kinase eluted with an apparent native Mr of 60 ± 5 kDa and did not phosphorylate p21 at the Ser146 site (Figure 3C). Some of the fractions containing s-p21 activity had no aPKC protein (Figure 3E, lanes 7 and 8) and the s-p21 kinase was not inhibited by PAR-4 (data not shown). Furthermore, prior phosphorylation of p21 at the Ser146 site did not inhibit subsequent modification to give s-p21.
Fig. 3. PKCζ can be separated from the s-p21 activity. The HS-pool was subjected to gel filtration on a Superose-12 column, and the included fractions (1–8) were analysed in the following ways. (A) Native gel (4%) electrophoresis/immunoblot developed using αPKCζ/ι. The native molecular mass of the proteins in the gel was determined using a standard curve constructed with marker proteins (B) Incorporation of 32P into p21 protein detected using autoradiography. (C) Phosphorylation of p21 at the Ser146 site detected by immunoblot using αp21-Pser146 IgG. (D) Total p21 protein added to the assay in (B) and (C) was detected by immunoblot using WA-1. (E) aPKC protein determined by immunoblot (50 µg/lane) developed using αPKCζ/ι. s-p21 is protein which runs with a higher apparent molecular mass.
In conclusion, the aPKCs comprise the major p21-Ser146 kinase activity in HeLa cell lysate, and a second enzyme, which causes a shift in the mobility of the p21 substrate, appears to be unrelated to the aPKC family. As aPKCs are associated with cell growth and are inhibited by PAR-4 in response to DNA damage (Berra et al., 1997), they fit well with our criteria for a bona fide Ser146 kinase (Scott et al., 2000).
Phosphorylation of p21 by other AGC kinase superfamily members
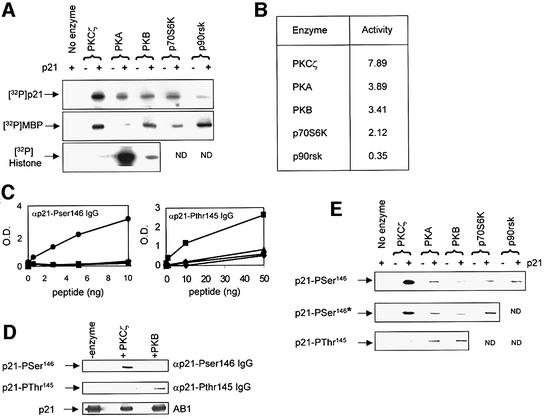
The Ser146 site and the adjoining phospho-acceptor site at Thr145 of p21 (Scott et al., 2000) both lie within con sensus phosphorylation sequences for the AGC family of protein kinases (BXBXXS/T; where B is a basic residue and X can be any amino acid), which includes the aPKCs. Thus, it is possible that other AGC kinases phosphorylate the Ser146 site. We therefore assayed PKA, p70S6K, PKB and p90rsk for p21 kinase activity. The incorporation of 32P into full-length p21 was determined using autoradiography (Figure 4A) and trichloroacetic acid (TCA) precipitation assays (Figure 4B). MBP and histone H1 were used as control substrates for the AGC kinases (Figure 4A). These results show that the AGC kinases are able to phosphorylate p21 to differing degrees; however, the specific activity of PKCζ for p21 was significantly higher than other family members tested. In order to look at the incorporation of phosphate into Ser146 compared with Thr145, we used an αp21-Pthr145 IgG (Figure 4C and D). The αp21-Pthr145 IgG was specific for a Thr145 phosphopeptide, although its affinity for this peptide was significantly lower than that of the αp21-Pser146 IgG for a Ser146 phospho-peptide (Figure 4C). However, the αp21-Pthr145 IgG clearly discriminated between p21 phosphorylated by PKB, which is known to phosphorylate the Thr145 site (Rossig et al., 2001; Zhou et al., 2001), and that phosphorylated at the Ser146 site by PKCζ (Figure 4D). When an equal activity of each AGC kinase (1 U/ml) was added to the assay, PKCζ was the most efficient Ser146 kinase (Figure 4E; upper panel). The same was true when the amount of enzyme added was normalized to the specific activity of each kinase using p21 as the substrate based on the values in Figure 4B (Figure 4E; middle panel). Using the anti-Pthr145 IgG, Thr145 site phosphorylation was readily detected in the presence of PKB and PKA, whereas PKCζ had no detectable activity against this site. Based on our data, and that of other groups (Rossig et al., 2001; Zhou et al., 2001), we would therefore argue against the recent suggestion that PKB is a physiologically relevant Ser146 kinase (Li et al., 2002). This is supported by the fact that a lysine residue at the –5 position (as in the Ser146 site of p21) is a negative determinant for PKB (Alessi et al., 1996) and that efficient phosphorylation of p21 at the Ser146 site by PKB can only be seen when Thr145 is mutated to alanine (Li et al., 2002). In conclusion, although p21 could be phosphorylated by a number of AGC kinases, PKCζ was the most active Ser146 kinase whereas, in agreement with previous studies, PKA (Scott et al, 2000) and PKB (Rossig et al., 2001; Zhou et al., 2001) predominantly phosphorylated p21 at the Thr145 site.
Fig. 4. Phosphorylation of p21 by the AGC family of kinases. (A) PKCζ, PKA, PKB, p70S6K and p90rsk (Upstate Biotechnology) were screened for their ability to incorporate phosphate into full-length p21 protein, MBP and histone H1 by autoradiography when 1 U/ml of each kinase was incubated with magnesium and [32P]ATP. (B) Quantitation of 32P incorporation into p21 protein by the AGC kinases (1 U/ml) determined using TCA precipitation assays. Activity is expressed as pmol of phosphate incorporated per 10 min. (C) Binding of 1 µg/ml αp21-Pser146 IgG (left panel) or αp21-Pthr145 IgG (right panel) to immobilized synthetic peptides was determined by ELISA. The peptide, based on amino acids 141–164 of p21 (KRRQTSMTDFYHSKRRLIFSKRKP), was either unphosphorylated (triangles) or phosphorylated at Thr145 (squares), Ser146 (circles) or Ser153 (diamonds). Peptide amount is plotted against OD at 450 nM. (D) p21 protein was phosphorylated with either PKCζ or PKB (0.5 U/ml) and analysed by immunoblot using αp21-Pser146 IgG or αp21-Pthr145 to detect Ser146 and Thr145 phosphorylated protein, respectively, and AB1 to show total p21 protein added to the assay. (E) p21-Ser146 and p21-Thr145 kinase activities were determined by immunoblots developed using either αp21-Pser146 IgG or αp21-Pthr145 IgG under conditions where 1 U/ml of each kinase were added or when the amount of kinase added was normalized (*) to p21 kinase activity (equivalent to 0.5 U/ml of PKCζ activity) using the values in (B).
Expression of PKCζ in HCT116 cells leads to phosphorylation of p21 at the Ser146 site
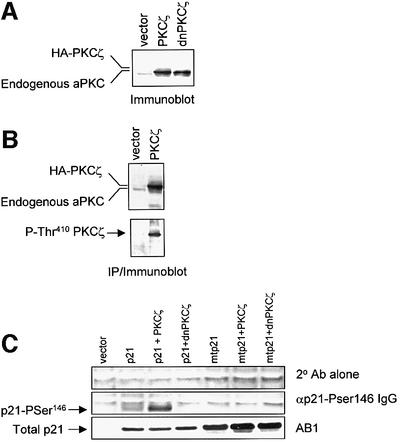
When used to label phosphorylation sites in vivo, radiochemicals have been shown to damage DNA, activating the G1 checkpoint pathway and perturbing kinase/phosphatase enzymes (Bond et al., 1999). As aPKC activity is inhibited by PAR-4 following exposure to DNA-damaging agents (Berra et al., 1997), we chose an alternative approach to determine if PKCζ expression led to the phosphorylation of p21 at Ser146 in cells. First, it was established that PKCζ and dnPKCζ were expressed to a similar extent in HCT116 cells (Figure 5A) and that the expressed PKCζ was activated by phosphorylation at the Thr410 site (Figure 5B). p21 was then expressed in p21–/– HCT116 cells and p21-Pser146 IgG was used to measure phosphorylation at the Ser146 site. Although our p21-Pser146 IgG is highly specific for p21 phosphorylated at the Ser146 site, it has a relatively weak affinity for the phospho-protein (Scott et al., 2000). Conditions were therefore developed under which detection of cellular p21 phosphorylated at Ser146 was optimal. Cell pellets were lysed using denaturing urea buffer and analysed (400 µg of total protein/lane) on 4–10% denaturing mini-gradient gels using a MES buffer system. Ser146 phosphorylated protein was detected at low levels when p21 was expressed in the null cells; when PKCζ was co-expressed, the p21-Pser146 IgG detected a significant increase in phosphorylation, whilst expression of dnPKCζ reduced the amount of detectable protein to background levels (Figure 5C). Although a Ser146→Ala mutant was expressed at higher levels than wild-type p21, no signal was detected with the p21-Pser146 IgG. The data show that expression of PKCζ leads to phosphorylation of p21 at the Ser146 site and that dnPKCζ can inhibit endogenous Ser146 kinase activity efficiently.
Fig. 5. Expression of PKCζ leads to the modification of p21 in cells. (A) Immunoblot developed using αPKCζ/ι for HCT116 cells transfected with 2 µg each of pcDNA3.1 (vector), pcDNA3.1/HA-PKCζ and pcDNA3.1/HA-dnPKCζ (50 μg total protein/lane). (B) aPKC was immunoprecipitated using αPKCζ/ι from HCT116 cell lysates (250 µg total protein) transfected with vector alone or with HA-PKCζ. Total aPKC protein (upper panel) and aPKC phosphorylated at the Thr410 site (lower panel) were detected by immunoblot using αPKCζ/ι and αP-Thr410 serum, respectively. (C) HCT116 p21–/– cells were transfected with 1 µg of wild-type-pcDNA3/p21 (p21) or S146→A-pcDNA3/p21 (mtp21) alone or plus 2 µg each of HA-PKCζ or HA-dnPKCζ. Transfections were normalized for total plasmid DNA using empty vector. Cells were lysed in denaturing urea solution and analysed (400 or 50 µg/lane for Ser146 phospho-p21 and total p21, respectively) on 4–10% gradient mini-gels. The immunoblots were probed with secondary antibody alone (upper panel), αp21-Pser146 IgG (middle panel) or AB1 (lower panel).
PKCζ decreases the half-life of p21 protein in cells
Having shown that PKCζ can phosphorylate p21 at the Ser146 site in HCT116 cells, experiments were designed to study aPKC-dependent signalling to p21 and the effect of phosphorylation on the endogenous protein. This approach was chosen in preference to the use of p21 phosphorylation site mutants as, in agreement with previous studies (Cayrol and Ducommun, 1998), we found that the properties of overexpressed p21 protein were different from those of the endogenous protein (data not shown).
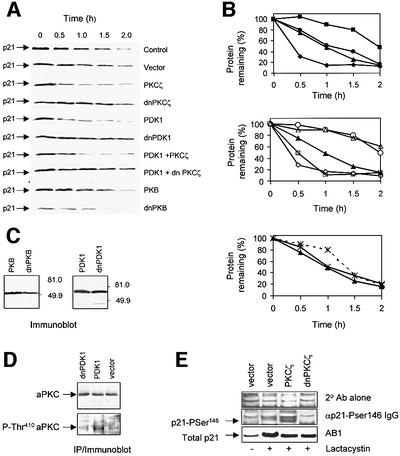
Previous studies have shown that the rate of p21 protein degradation may depend on its ability to interact with certain partner proteins, so that p21 mutants that can no longer bind to PCNA are turned over more rapidly (Cayrol and Ducommun, 1998; Touitou et al., 2001). As we, and others, have shown previously that phosphorylation at the Ser146 site can inhibit PCNA binding (Scott et al., 2000; Rossig et al., 2001), we sought to determine whether PKCζ-dependent phosphorylation of p21 affected the rate at which the protein was degraded. The half-life of endogenous p21 was determined by treating cells with cycloheximide, to inhibit translation, and the decay of existing protein was determined by denaturing immunoblot (Figure 6A). In untreated HCT116 cells or cells treated with vector alone, the half-life of p21 was 55 and 60 min, respectively (Figure 6A and B). In the presence of wild-type PKCζ, this decreased to 27 min. Conversely, dnPKCζ increased the half-life of the p21 protein (>100 min), consistent with its ability to inhibit endogenous Ser146 activity (Figures 6A and B, and 5C). Activation of the aPKCs requires phosphorylation within the T-loop at Thr410 by the upstream kinase PDK1 (Le Good et al., 1998). Expression of PDK1 in HCT116 cells (Figure 6C) enhanced phosphorylation of endogenous aPKC at Thr410 (Figure 6D) and, like PKCζ, PDK1 decreased the half-life of p21 (Figure 6A and B). Co-expression of dnPKCζ with PDK1 prevented the PDK1-dependent decrease in half-life, providing evidence that PDK1 is promoting p21 protein degradation in an aPKC-dependent manner. Similarly, a kinase-inactive PDK1 (dnPDK1), which would be expected to inhibit the phosphorylation and activation of endogenous aPKC, increased the half-life of p21 (Figure 5A and B). In contrast, expression of PKB (Figure 6C) had no significant effect on the half-life of p21 (Figure 6A and B). Consistent with PKB being involved in promoting p21 mRNA expression (Lawlor and Rotwein, 2000), the dnPKB construct significantly decreased the levels of endogenous p21 protein and, although it increased the half-life of the protein, the effect was minor when compared with dnPKCζ or dnPDK1 (Figure 6A and B).
Fig. 6. Expression of PKCζ or modulation of endogenous aPKC activity changes the rate of p21 degradation. (A) HCT116 cells were untransfected (control) or transfected with 2 µg each of pcDNA3.1(vector), pcDNA3.1/HA-PKCζ, pcDNA3.1/HA-dnPKCζ, pCMV5/Myc-PDK1, pCMV5/Myc-dnPDK1, pCMV5/PKB or pCMV5/dnPKB as detailed in the figure. After 24 h, the cells were treated with cycloheximide (30 µg/ml) and harvested at 0, 0.5, 1.0, 1.5 and 2.0 h. Endogenous p21 protein (50 µg total protein/lane) was detected by denaturing immunoblots developed using AB-1. Each time course is representative of at least three separate experiments. (B) p21 protein quantitation was carried out as described in Materials and methods and plotted as percentage protein remaining against time, where the 0 time point is 100%, for control cells (filled triangles) vector alone (filled circles), PKCζ (filled diamonds), dnPKCζ (filled squares), PDK1 (open diamonds), dnPDK1 (open circles), PDK1 and PKCζ (open squares), PDK1 and dnPKCζ (open triangles), PKB (*×+) and dnPKB (…*×+…). (C) HCT116 cells were transfected with 2 µg of pCMV5/myc-PDK1, pCMV5/myc-dnPDK1, pCMV5/PKB or pCMV5/dnPKB and harvested after 24 h; PDK1 and PKB were detected by immunoblot (50 µg total protein/lane) using anti-myc or anti-PKB serum, respectively. (D) aPKC was immunoprecipitated using αPKCζ/ι from HCT116 cell lysate (250 µg of total protein) which had been transfected with myc-dnPDK1, myc-PDK1 or vector alone. Immunoblots were used to detect total aPKC and aPKC phosphorylated at the Thr410 site using αPKCζ/ι and anti-P-Thr410 serum, respectively. (E) HCT116 cells were transfected with 2 µg each of pcDNA3.1 (vector), pcDNA3.1/PKCζ or pcDNA3.1/dnPKCζ, and 24 h later the cells were left untreated (–) or 10 µM lactacystin (+) was added for 6 h prior to harvesting. Cells were lysed in denaturing urea solution and analysed (400 or 50 µg/lane for Ser146 phospho-p21 and total p21, respectively) on 4–10% gradient mini-gels. The immunoblots were probed with secondary antibody alone (upper panel), αp21-Pser146 IgG (middle panel) or AB-1 (lower panel).
In order to confirm that expressed PKCζ was stimulating the phosphorylation of endogenous p21, Ser146 phosphorylation was measured in cells treated with lactacystin to prevent proteasome-mediated degradation (Figure 6E). Whilst phosphorylation was barely detectable in lactacystin-treated control cells, detectable Ser146 phosphorylated p21 protein increased in cells expressing PKCζ, whereas phosphorylation was below the level of detection in cell expressing dnPKCζ.
Insulin stimulation leads to a rapid decrease in p21 protein levels
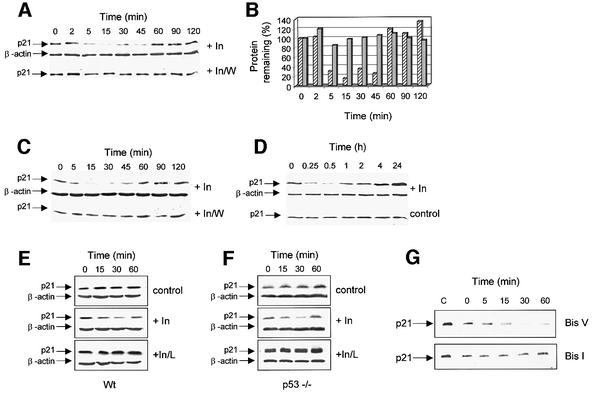
Activation of aPKC occurs via the PI3K pathway in response to insulin stimulation (Standaert et al., 1999). We therefore sought to determine the effect of activating endogenous aPKC using insulin treatment. aPKC activity is maximal within minutes of treatment with 100 nM insulin and returns to basal within 30–60 min (Standaert et al., 1997; Kotani et al., 1998), therefore we looked for effects of low doses of insulin on p21 protein levels during this acute time window. A concentration of 100 nM insulin was chosen, and measurements were taken up to 2 h after treatment. The monoclonal antibody AB-1 was used to detect p21 as it binds to the central domain of the protein (amino acids 61–75) and is therefore not masked by phosphorylation. p21 levels responded rapidly to insulin stimulation so that in cells treated for 5–15 min, the amount of p21 protein had fallen by 70% (Figure 7A and B). Levels remained low up to 45 min post-treatment and returned to basal between 60 and 90 min. If the cells were pre-incubated with 100 nM of the PI3K inhibitor wortmannin, no decrease in p21 protein was seen upon the addition of insulin (Figure 7A and B) demonstrating that insulin is regulating p21 protein levels in a PI3K-dependent manner. The amount of p21 protein in cells treated with insulin carrier alone remained constant, and pre-treatment of cells with dimethylsulfoxide (DMSO; the wortmannin carrier) did not affect the insulin-dependent loss of p21 (not shown). It is important to note that the time given refers to when the cells were removed from the incubator and that subsequent washing and harvesting prior to rapid freezing in liquid nitrogen consistently took 10 min.
Fig. 7. Insulin stimulation leads to the rapid degradation of p21 protein. (A) HCT116 cells at 60% confluence were fasted in serum-free medium for 5 h prior to addition of 100 nM insulin (+In), or the cells were pre-incubated for 15 min with 100 nM wortmannin prior to insulin treatment (+In/W). At the times given, the cells were removed from the incubator, washed twice with PBS, mechanically removed from the plate and harvested by centrifugation before being frozen in liquid nitrogen. The 0 time point was treated with insulin carrier alone. p21 protein levels were determined by denaturing immunoblot (25 µg of total protein/lane) developed using AB-1 and overlaid with AC-15, to detect β-actin. (B) Quantitation of the data in (A) was performed as described in Materials and methods. The percentage protein remaining, where the 0 time point is taken as 100%, is plotted against time and shows insulin alone (hatch) or insulin + wortmannin (grey). (C) HEK293 cells at 60% confluence were treated exactly as described in (A) except that the immunoblot shows 50 µg of total protein/lane. (D) HEK293 cells were fasted as above prior to addition of 1 µM insulin (+In) or insulin carrier alone (control). At the times shown, p21 and β-actin protein levels were detected by denaturing immunoblot (50 µg of total protein/lane). Wild-type (E) or p53–/– cells (F) were fasted as above prior to addition of insulin carrier alone (control) or 100 nM insulin (+In), or pre-incubated with lactacystin (10 µM) for 2 h prior to insulin addition (+In/L). Cells were harvested at the times shown; p21 and β-actin levels were determined as described above. (G) HCT116 cells were fasted as above then treated for 15 min with 10 µM Bis I, or the inactive derivative Bis V, prior to the addition of insulin (100 nM). Cells were collected at the times shown where C is a fasted cell control that was not treated with either insulin or Bis. p21 was detected by immunoblot using AB-1.
In order to determine whether the effect of insulin on p21 protein was seen in other cell types, we used the human embryonic kidney cell line HEK293. Although the kinetics of p21 loss were slightly different in the HEK293 cells, decreases in p21 protein levels were again detected by 15 min and were maximal by 15–30 min, recovering by 60–90 min, and the loss of p21 protein was prevented by wortmannin (Figure 7C). Using a supra-physiological dose of insulin, we confirmed previous reports suggesting that p21 is upregulated by insulin at later time points (Figure 7D). In HEK293 cells treated with 1 µM insulin, p21 levels were elevated by 4 h and continued to increase up to 24 h. The upregulation of p21 protein expression at later time points is associated with the mitogenic activity of insulin and is accompanied by increases in p21 mRNA (Lai et al., 2001).
The half-life of p21 protein in untreated cells is 55 min (Figure 6), suggesting that the rapid loss of the protein in cells 5–15 min after insulin treatment is due to changes in the rate of p21 degradation. When cells were pre-treated with the proteasome-specific inhibitor lactacystin, the loss of p21 in response to insulin was inhibited in both p53 wild-type and p53–/– cells (Figure 7E and F), demonstrating that lactacystin can overcome insulin-stimulated loss of p21 and that this is not due to the accumulation of transcriptionally active p53 (no increase in p21 mRNA level was detected in wild-type cells incubated with lactacystin; data not shown). Finally, we ascertained whether the loss of p21 protein seen in response to insulin could be prevented by the PKC inhibitor Bis I when used at a concentration which inhibits the aPKCs. When HCT116 cells were treated with Bis I, levels of p21 protein were reduced; however, this was not a PKC-specific event as Bis V, an inactive derivative, produced an equivalent decrease in p21 levels. When Bis-treated cells subsequently were incubated with insulin, p21 levels were reduced further in the presence of Bis V (Figure 7G; upper panel), whereas no decrease in p21 protein levels was detected in cells pre-treated with the active PKC inhibitor Bis I (Figure 7G; lower panel). Thus, the signalling pathway linking insulin to p21 most probably contains a PKC signalling component. The data presented here are consistent with a model where aPKC provides a physiological signal for the degradation of the p21 protein.
Discussion
Identification of the aPKCs as enzymes that phosphorylate p21 at the Ser146 site has uncovered a role for p21-regulated processes during the initial response to insulin stimulation. Having previously described Ser146 of p21 as a phospho-acceptor site that can modulate its protein– protein interactions (Scott et al., 2000), our current study shows that the phosphorylation status of this site in cells can influence the rate of its degradation. Thus, the expression of PKCζ or activation of endogenous aPKC by insulin or PDK1 leads to proteasome-dependent degradation of the p21 protein.
p21 degradation is mediated by the proteasomes and can proceed through a ubiquitin pathway (Maki and Howley, 1997) or in a ubiquitin-independent manner through binding to the C8 α-subunit of the 20S proteasome (Sheaff et al., 2000; Touitou et al., 2001). Although there is increasing evidence that changes in the rate of p21 degradation contribute to modulation of its steady-state levels in response to incoming signals (Timchenko et al., 1997; Johannessen et al., 1999; Kobayashi et al., 2000), surprisingly little is known about the regulation of this process. One model is that binding partners determine the rate of p21 degradation. This is based on data showing that the half-life of p21 involved in interactions with C/ERB, PCNA or TSG101 is significantly increased (Cayrol and Ducommun, 1998; Harris et al., 2001; Touitou et al., 2001; Oh et al., 2002), whereas p21 in complex with the cyclin–CDKs is turned over more rapidly (Cayrol and Ducommun, 1998). Thus the inhibition of PCNA binding by Ser146 phosphorylation (Scott et al., 2000; Rossig et al., 2001) is consistent with the increased turnover of p21 in cells expressing PKCζ or PDK1 (Figure 6). However, although, Thr145 phosphorylation has also been shown to inhibit PCNA binding (Scott et al., 2000; Rossig et al., 2001), the expression of PKB/dnPKB (Figure 6A and B) or p21-Thr145 phospho-mimetics (Li et al., 2002) does not significantly affect the rate of p21 turnover. This suggests that the binding of proteins other than PCNA to the C-terminus of p21 may play a critical role in determining its rate of degradation. As a number of C-terminal-binding proteins have been identified (Dotto, 2000), it will be important for future studies to fine map these interactions and to establish whether C-terminal phosphorylation promotes or inhibits the binding of individual factors.
In the current study, although expression of PKCζ correlated with increased phosphorylation of p21 at the Ser146 site, we cannot rule out the possibility that degradation of p21 requires additional PKCζ-stimulated signalling events acting in concert with phosphorylation at the Ser146 site. Phosphorylation of p21 by GSK3 at the Thr57 site has also been linked to the turnover of p21 (Rossig et al., 2002). In this study, the mutation of Thr57 to alanine decreased the rate of p21 degradation, as did inhibition of GSK3 by lithium chloride. Intriguingly, PKCζ can stimulate GSK3 activity, by relieving PKB-imposed inhibition (Doornbos et al., 1999). Thus, activation of aPKC may be involved both directly and indirectly in the regulation of p21 turnover. However, the role of Thr57 in the regulation of p21 is controversial, as Kim et al. (2002) have reported that mutation of Thr57 to alanine promotes, rather than inhibits, degradation. In fact, the Thr57 site illustrates how studies on the mechanism and regulation of p21 protein degradation may be complicated by the use of overexpressed p21 mutants. Consistent with previous observations that high-level expression of p21 can overcome proteasome-dependent regulation of its degradation (Cayrol and Ducommun, 1998), we found that the degradation of overexpressed p21 protein was not increased by either insulin stimulation or PKCζ expression (data not shown). Moreover, whereas Kim et al. (2002) reported that mutation of Ser146 to alanine caused p21 to be expressed at lower levels, in our current study we found that expression of the Ser146→Ala mutant in p21–/– HCT116 cells was significantly higher than that of the wild-type protein (Figure 5C). These discrepancies may reflect the importance of p21-binding proteins for regulation of its degradation and the fact that a portion of the transfected p21 is likely to be unbound and therefore unstructured (Kriwacki et al., 1996). Another potential drawback to the use of phosphorylation site mutants is the complication of affecting the specificity of a kinase for its substrate. For example, we found that mutation of the Ser146 site to alanine inadvertently made the Thr145 site, that was not normally phosphorylated by PKCζ, a target for this enzyme (data not shown).
It has become clear that p21 plays a number of critical functions in the regulation of cell growth (Dotto, 2000). We have demonstrated a new role for p21-regulated processes during the initial response to insulin-stimulated cell signalling which potentially links cell cycle control to the regulation of energy metabolism and biosynthesis. The implications of p21 protein down-regulation by insulin may include changes in CDK activity and modulation of gene expression. p21 stimulates cyclin–Cdk4 complex formation and activity, whilst it inhibits Cdk2-containing complexes (Sherr and Roberts, 1999); thus, decreases in p21 steady-state levels would be expected to activate Cdk2 differentially, compared with Cdk4, suggesting that new Cdk2 targets may include enzymes involved in the control of cellular metabolism. p21 protein levels have also been shown to have a striking effect on gene expression (Chang et al., 2000). Increased levels of p21 lead to selective inhibition of genes involved in mitosis and DNA replication/repair and the upregulation of genes linked to cell ageing and senescence, as well as secreted growth factors. The transient down-regulation of p21 is therefore likely to have consequences for gene expression as well as protein kinase activity.
Materials and methods
Reagents
[γ-32P]ATP was from Amersham Pharmacia. Plasmids were provided as follows: pcDNA3.1/HA-PKCζ and pcDNA3.1/HA-dnPKCζ by J.Moscat (Centro de Biologia Molecular), pGEX-2TK/PAR-4 by S.Roberts (University of Manchester), and pCMV5/myc-PDK-1, pCMV5/myc-dnPDK-1, pCMV5/PKB and pCMV5/dnPKB by D.Alessi (University of Dundee). pcDNA3-p21 has a BamHI fragment encoding human p21 or p21 with a Ser146→Ala mutation. αThr410 IgG and αPthr145 IgG were kind gifts from the MRC Protein Phosphorylation Unit (University of Dundee). HCT116 p21–/– cells were a kind gift from B.Vogelstein (Johns Hopkins). Purified recombinant PKA, PKB, p70S6K, p90rsk and PKCζ were from Upstate Biotechnology. Insulin was from Roche; wortmannin, lactacystin, Bis I and V were from Calbiochem.
Cell culture, transfection and lysis
HCT116 cells were cultured in McCoys medium, and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen); in both cases, the medium was supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin, and cultured at 37°C and 5% CO2. HeLa cells, harvested in exponential phase, were provided by CR UK. The HCT116 cells were transfected with the plasmids indicated in the text using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. HCT116 and HEK293 cells were washed twice in ice-cold phosphate-buffered saline (PBS), pelleted and harvested by low speed centrifugation prior to freezing in liquid nitrogen. The pellets were lysed in an equal volume of buffer A [50 mM HEPES pH 7.4, 150 mM NaCl, 25 mM NaF, 5 mM dithiothreitol (DTT), 1 mM EDTA, 10 mM β-glycerophosphate, 10 µg/ml leupeptin, 4 µg/ml aprotinin, 2 µg/ml pepstatin, 1.2 mM benzamidine, 10 µg/ml soybean trypsin inhibitor and 400 µg/ml pefabloc] plus 1% (v/v) Triton X-100, and the supernatant was collected following centrifugation at 14 000 g for 15 min at 4°C.
Immunoblots
SDS–PAGE/immunoblot and native gel electrophoresis/immunoblot were performed as previously described (Ball et al., 1994; Scott et al., 2000). Total p21 protein was detected using AB-1 (Oncogene Sciences) or WA-1 at 1 and 0.5 µg/ml, respectively. p21 phosphorylated at the Ser146 or Thr145 sites was detected using αp21-Pser146 IgG (1 µg/ml) or αp21-Pthr145 IgG (1 µg/ml). aPKC was detected using αPKCζ/ι serum (C-20; Santa Cruz) at 1 µg/ml; the Thr410 phosphorylated form was detected using αThr410 IgG at 0.5 μg/ml. Myc-tagged PDK1 and dnPDK1 were detected using an anti-c-myc serum provided by S.Lain (University of Dundee) at 1:1000 dilution, PKB was detected using αPKB 1 μg/ml (Upstate Biotechnology) and β-actin levels were determined using AC-15 (Sigma) at a 1:5000 dilution. p21 protein levels determined by immunoblot were quantitated using a Genegenius/Genegnome Bioimaging system and software (Syngene).
Phosphorylation of p21 in cells
Cell pellets were lysed in 2 vols of 50 mM HEPES pH 7.6 containing 8 M urea, 1% (v/v) Triton X-100, 100 mM DTT, 20 mM NaF and 20 mM β-glycerophosphate. Total cell lysate (400 µg/lane) was analysed on 4–10% pre-cast gradient mini-gels using an MES buffer system, and transferred to nitrocellulose following the manufacturer’s instructions (Invitrogen). The membranes were blocked and washed with 3% bovine serum albumin (BSA) in PBS, containing 1% (v/v) Tween-20, 50 mM NaF and 20 mM β-glycerophosphate; the p21-Pser146 IgG was used at a concentration of 2.5 µg/ml in the same solution and was incubated with the membrane for 18 h. Total p21 protein was determined as described in the previous section.
p21 kinase assays
Column fractions (6 µl/assay), purified recombinant proteins (1 U/ml or as detailed in the figure legends) or immunoprecipitates were assayed for p21 kinase activity using recombinant wild type or Ser146→Ala p21 protein purified from Escherichia coli essentially as previously described (Scott et al., 2000). Following incubation, the reactions were stopped either by: (i) removing a 10 µl sample of the reaction mixture, adding to Laemmli sample buffer and subjecting to SDS–PAGE/autoradiography; or (ii) by adding 20 µl to 0.5 ml of ice-cold 25% (w/v) TCA for 1 h prior to harvesting, washing and Cerenkov counting. Alternatively, p21 phosphorylated at Ser146 or Thr145 in the absence of radiochemical was detected by denaturing immunoblot.
Purification of p21 kinase
HeLa pellets (1 × 109 cells) were lysed in buffer A (50 ml) containing 0.01% (v/v) Triton X-100. Insoluble material was removed by centrifugation, the supernatant was diluted to 20 mg/ml in buffer B [25 mM HEPES pH 7.4, 5% (v/v) glycerol, 0.01% (v/v) Triton X-100, 25 mM NaF, 1 mM EDTA, 5 mM DTT, 1 mM benzamidine and 40 µg/ml pefabloc plus 50 mM NaCl] and applied to a Q-Sepharose column (60 mg of total protein/ml of resin) equilibrated in buffer B. The column was washed with buffer B and developed with a linear salt gradient (0.05–1 M NaCl) over five column volumes at a flow rate of 1 ml/min. The active fractions were pooled, concentrated and diluted to 50 mM NaCl, then applied to an SP-Sepharose column equilibrated in buffer B. The column was washed and developed as above. Active fractions were pooled and concentrated. Once diluted to a final salt concentration of 50 mM NaCl, the pooled SP-Sepharose fractions were applied to a HS-Poros column (1.66 ml) equilibrated in buffer B. The column was washed with 10 column volumes of buffer B and developed with a linear salt gradient (0.05–1 M NaCl) over 14 column volumes at a flow rate of 10 ml/min, with 1 ml fractions being collected. Active fractions were pooled and concentrated to >1 mg/ml to form the HS-pool. The HS-pool was concentrated to 100 µl and applied to a Superose-12 column (Pharmacia) equilibrated in buffer B plus 100 mM NaCl; the column was developed with buffer B plus 100 mM NaCl at 0.5 ml/min and 1 ml fractions were collected.
Immunoprecipitation and ELISA
Cell lysate or active fractions were incubated with 1 µg of αPKCζ/ι in buffer C: 50 mM HEPES pH 7.4, 150 mM NaCl, 5 mM NaF, 1 mM DTT, 1% (v/v) Triton X-100, 10 µg/ml leupeptin, 4 µg/ml aprotinin, 2 µg/ml pepstatin, 1.2 mM benzamidine, 10 µg/ml soybean trypsin inhibitor and 400 µg/ml pefabloc, for 2 h at 4°C on a rotating wheel. Protein G beads (20 µl packed volume) were added and the mixture was rotated for 1 h at 4°C. The beads were pelleted and washed three times in buffer C. The beads were either washed twice in kinase buffer (50 mM HEPES pH 7.4, 10 mM MgCl2, 0.8 mM EDTA, 0.8 mM DTT) and subject to a p21 kinase assay or added to Laemmli sample buffer and analysed by SDS–PAGE. Peptide enzyme-linked immunosorbent assays (ELISAs) were performed as described previously (Scott et al., 2000).
Acknowledgments
Acknowledgements
We are very grateful to Dario Alessi for all his kind help with reagents. K.L.B. is a Cancer Research UK Senior Cancer Research Fellow, and the research was funded by CR UK and AICR.
References
- Alessi D.R., Caudwell,F.B., Andjelkovic,M., Hemmings,B.A. and Cohen,P. (1996) Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett., 399, 333–338. [DOI] [PubMed] [Google Scholar]
- Ball K.L. (1997) p21: structure and functions associated with cyclin–CDK binding. Prog. Cell Cycle Res., 3, 125–134. [DOI] [PubMed] [Google Scholar]
- Ball K.L., Dale,S., Weekes,J. and Hardie,D.G. (1994) Biochemical characterization of two forms of 3-hydroxy-3-methylglutaryl-CoA reductase kinase from cauliflower (Brassica oleracia). Eur. J. Biochem., 219, 743–750. [DOI] [PubMed] [Google Scholar]
- Berra E., Municio,M.M., Sanz,L., Frutos,S., Diaz-Meco,M.T. and Moscat,J. (1997) Positioning atypical protein kinase C isoforms in the UV-induced apoptotic signaling cascade. Mol. Cell. Biol., 17, 4346–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.D. (2000) Protein kinase C-mediated regulation of the cell cycle. Front. Biosci., 5, D406–D423. [DOI] [PubMed] [Google Scholar]
- Bond J.A., Webley,K., Wyllie,F.S., Jones,C.J., Craig,A., Hupp,T. and Wynford-Thomas,D. (1999) p53-dependent growth arrest and altered p53-immunoreactivity following metabolic labelling with 32P ortho-phosphate in human fibroblasts. Oncogene, 18, 3788–3792. [DOI] [PubMed] [Google Scholar]
- Cayrol C. and Ducommun,B. (1998) Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene, 17, 2437–2444. [DOI] [PubMed] [Google Scholar]
- Chang B.D., Watanabe,K., Broude,E.V., Fang,J., Poole,J.C., Kalinichenko,T.V. and Roninson,I.B. (2000) Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence and age-related diseases. Proc. Natl Acad. Sci. USA, 97, 4291–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos R.P., Theelen,M., van der Hoeven,P.C., van Blitterswijk,W.J., Verkleij,A.J. and van Bergen en Henegouwen,P.M. (1999) Protein kinase Cζ is a negative regulator of protein kinase B activity. J. Biol. Chem., 274, 8589–8596. [DOI] [PubMed] [Google Scholar]
- Dotto G.P. (2000) p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta, 1471, M43–M56. [DOI] [PubMed] [Google Scholar]
- el-Deiry W.S. et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- Harris T.E., Albrecht,J.H., Nakanishi,M. and Darlington,G.J. (2001) C/EBPα cooperates with p21 to inhibit cdk2 kinase activity and induces growth arrest independent of DNA binding. J. Biol. Chem., 276, 29200–29209. [DOI] [PubMed] [Google Scholar]
- Johannessen L.E., Knardal,S.L. and Madshus,I.H. (1999) Epidermal growth factor increases the level of the cyclin-dependent kinase (CDK) inhibitor p21/CIP1 in A431 cells by increasing the half-lives of the p21/CIP1 transcript and the p21/CIP1 protein. Biochem. J., 337, 599–606. [PMC free article] [PubMed] [Google Scholar]
- Jones J.M., Cui,X.S., Medina,D. and Donehower,L.A. (1999) Heterozygosity of p21WAF1/CIP1 enhances tumor cell proliferation and cyclin D1-associated kinase activity in a murine mammary cancer model. Cell Growth Differ., 10, 213–222. [PubMed] [Google Scholar]
- Kim G.Y., Mercer,S.E., Ewton,D.Z., Yan,Z., Jin,K. and Friedman,E. (2002) The stress-activated protein kinases p38α and JNK1 stabilize p21Cip1 by phosphorylation. J. Biol. Chem., 277, 29792–29802. [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Takada,Y., Hachiya,M. ando,K., Nakajima,N. and Akashi,M. (2000) TNF-α induced p21(WAF1) but not Bax in colon cancer cells with mutated p53: important role of protein stabilization. Cytokine, 12, 1745–1754. [DOI] [PubMed] [Google Scholar]
- Kotani K. et al. (1998) Requirement of atypical protein kinase Cλ for insulin stimulation of glucose uptake but not for Akt activation in 3T3-L1 adipocytes. Mol. Cell. Biol., 18, 6971–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriwacki R.W., Hengst,L., Tennant,L., Reed,S.I. and Wright,P.E. (1996) Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl Acad. Sci. USA, 93, 11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A., Sarcevic,B., Prall,O.W. and Sutherland,R.L. (2001) Insulin/insulin-like growth factor-I and estrogen cooperate to stimulate cyclin E–Cdk2 activation and cell cycle progression in MCF-7 breast cancer cells through differential regulation of cyclin E and p21(WAF1/Cip1). J. Biol. Chem., 276, 25823–25833. [DOI] [PubMed] [Google Scholar]
- Lawlor M.A. and Rotwein,P. (2000) Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol. Cell. Biol., 20, 8983–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Good J.A., Ziegler,W.H., Parekh,D.B., Alessi,D.R., Cohen,P. and Parker,P.J. (1998) Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science, 281, 2042–2045. [DOI] [PubMed] [Google Scholar]
- Li Y., Dowbenko,D. and Lasky,L.A. (2002) AKT/PKB phosphorylation of p21Cip1/WAF1 enhances protein stability of p21Cip1/WAF1 and promotes cell survival. J. Biol. Chem., 276, 52. [DOI] [PubMed] [Google Scholar]
- Liu Y.F. et al. (2001) Insulin stimulates PKCζ-mediated phosphorylation of insulin receptor substrate-1 (IRS-1). J. Biol. Chem., 276, 14459–14465. [DOI] [PubMed] [Google Scholar]
- Maki C.G. and Howley,P.M. (1997) Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol., 17, 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M., Fang,X., Lu,Y., Lapushin,R., Bast,R.C.,Jr and Mills,G.B. (2000) Inhibition of growth-factor-induced phosphorylation and activation of protein kinase B/Akt by atypical protein kinase C in breast cancer cells. Biochem. J., 352, 475–482. [PMC free article] [PubMed] [Google Scholar]
- Mendez R., Kollmorgen,G., White,M.F. and Rhoads,R.E. (1997) Requirement of protein kinase Cζ for stimulation of protein synthesis by insulin. Mol. Cell. Biol., 17, 5184–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J. and Diaz-Meco,M.T. (2000) The atypical protein kinase Cs. Functional specificity mediated by specific protein adapters. EMBO rep., 1, 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Sanz,L., Sanchez,P. and Diaz-Meco,M.T. (2001) Regulation and role of the atypical PKC isoforms in cell survival during tumor transformation. Adv. Enzyme Regul., 41, 99–120. [DOI] [PubMed] [Google Scholar]
- Narla G. et al. (2001) KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science, 294, 2563–2566. [DOI] [PubMed] [Google Scholar]
- Oh H., Mammucari,C., Nenci,A., Cabodi,S., Cohen,S.N. and Dotto,G.P. (2002) Negative regulation of cell growth and differentiation by TSG101 through association with p21(Cip1/WAF1). Proc. Natl Acad. Sci. USA, 99, 5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossig L., Jadidi,A.S., Urbich,C., Badorff,C., Zeiher,A.M. and Dimmeler,S. (2001) Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol. Cell. Biol., 21, 5644–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossig L., Badorff,C., Holzmann,Y., Zeiher,A.M. and Dimmeler,S. (2002) Glycogen synthase kinase-3 couples AKT-dependent signaling to the regulation of p21Cip1 degradation. J. Biol. Chem., 277, 9684–9689. [DOI] [PubMed] [Google Scholar]
- Scott M.T., Morrice,N. and Ball,K.L. (2000) Reversible phosphorylation at the C-terminal regulatory domain of p21(Waf1/Cip1) modulates proliferating cell nuclear antigen binding. J. Biol. Chem., 275, 11529–11537. [DOI] [PubMed] [Google Scholar]
- Sheaff R.J., Singer,J.D., Swanger,J., Smitherman,M., Roberts,J.M. and Clurman,B.E. (2000) Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell, 5, 403–410. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts,J.M. (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev., 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Standaert M.L., Galloway,L., Karnam,P., Bandyopadhyay,G., Moscat,J. and Farese,R.V. (1997) Protein kinase C-ζ as a downstream effector of phosphatidylinositol 3-kinase during insulin stimulation in rat adipocytes. J. Biol. Chem., 272, 30075–30082. [DOI] [PubMed] [Google Scholar]
- Standaert M.L. et al. (1999) Insulin activates protein kinases C-ζ and -λ by an autophosphorylation-dependent mechanism and stimulates their translocation to GLUT4 vesicles and other membrane fractions in rat adipocytes. J. Biol. Chem., 274, 25308–25316. [DOI] [PubMed] [Google Scholar]
- Tanaka N. et al. (1996) Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature, 382, 816–818. [DOI] [PubMed] [Google Scholar]
- Timchenko N.A., Harris,T.E., Wilde,M., Bilyeu,T.A., Burgess-Beusse,B.L., Finegold,M.J. and Darlington,G.J. (1997) CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol., 17, 7353–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou R., Richardson,J., Bose,S., Nakanishi,M., Rivett,J. and Allday,M.J. (2001) A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 α-subunit of the 20S proteasome. EMBO J., 20, 2367–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.P., Liao,Y., Xia,W., Spohn,B., Lee,M.H. and Hung,M.C. (2001) Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol., 3, 245–252. [DOI] [PubMed] [Google Scholar]