Abstract
A cytosine DNA methyltransferase containing a chromodomain, Zea methyltransferase2 (Zmet2), was cloned from maize. The sequence of ZMET2 is similar to that of the Arabidopsis chromomethylases CMT1 and CMT3, with C-terminal motifs characteristic of eukaryotic and prokaryotic DNA methyltransferases. We used a reverse genetics approach to determine the function of the Zmet2 gene. Plants homozygous for a Mutator transposable element insertion into motif IX had a 13% reduction in methylated cytosines. DNA gel blot analysis of these plants with methylation-sensitive restriction enzymes and bisulfite sequencing of a 180-bp knob sequence showed reduced methylation only at CpNpG sites. No reductions in methylation were observed at CpG or asymmetric sites in heterozygous or homozygous mutant plants. Our research shows that chromomethylase Zmet2 is required for in vivo methylation of CpNpG sequences.
INTRODUCTION
Cytosine methylation is a DNA modification that is correlated with gene expression in eukaryotes. In plants, hypermethylation is associated with inactivation of transposable elements (Chandler and Walbot, 1986; Schwartz and Dennis, 1986; Banks et al., 1988), instances of imprinting and paramutation (Kermicle, 1996; Walker, 1998), and transgene silencing (Linn et al., 1990; Ingelbrecht et al., 1994). Control of gene expression mediated by DNA methylation also may play a role in plant development (Kakutani et al., 1995; Finnegan et al., 1996).
DNA methylation patterns are established and maintained by DNA methyltransferases, which catalyze the transfer of the methyl group from S-adenosylmethionine (SAM) to the C-5 position in the pyrimidine ring of cytosine. In plants, 5-methylcytosine residues are found predominantly at symmetric CpG and CpNpG sequences (Gruenbaum et al., 1981). Methylated cytosines are found at a lower frequency at asymmetric positions such as CpTpT and CpApT. At least three types of methyltransferase activity are likely to exist in plants. De novo methyltransferase activity is required to establish methylation at unmethylated sites and to propagate methylation at asymmetric sites. Separate CpG and CpNpG methyltransferase activities maintain symmetric methylation patterns during the process of DNA replication and cell division. Although the CpG and CpNpG methyltransferase activities likely are distinct, evidence in pea suggests the possibility that these activities may be conferred by products of a single gene (Pradhan and Adams, 1995; Pradhan et al., 1998). CpNpG methylation is common in plants but infrequent or absent in mammals.
The focus of this study was the analysis of a putative maize methyltransferase gene homologous with the CMT family of chromomethylase genes in Arabidopsis. Chromomethylases were first identified in a database search for genes containing chromodomains (Henikoff and Comai, 1998; Rose et al., 1998). Chromodomains have been found in several proteins involved in the chromatin-based regulation of gene expression and may be critical for the targeting of these proteins within the nucleus (Cavalli and Paro, 1998). Chromomethylases contain conserved methyltransferase domains at the C-terminal end, whereas the N-terminal portion of chromomethylases differs substantially from that of other methyltransferases. Chromomethylases lack ∼750 amino acids in their N-terminal domain relative to the Dnmt1 class of methyltransferases, which is exemplified by the MET1 (Finnegan and Dennis 1993) and Zmet1 genes in plants. Chromomethylases are similar in size to the Dnmt3 class of de novo methyltransferases, which is represented by the Zmet3 and Drm2 genes in plants (Cao et al., 2000), but they differ substantially on the basis of the organization of the conserved methyltransferase motifs and the structure of the N-terminal portion of the gene. Chromomethylases have not been identified in the genomic sequences of any organism other than plants. To date, no function has been assigned to the chromomethylase class of methyltransferases. The CMT1 chromomethylase of Arabidopsis has been deemed nonessential because several Arabidopsis ecotypes contain genes with a retroelement insertion that disrupts the coding region or a frameshift mutation that results in truncated proteins (Henikoff and Comai, 1998). No obvious differences in DNA methylation have been correlated with these alleles.
We have characterized one member of a small gene family of chromomethylases in maize, Zea methyltransferase2 (Zmet2). In this study, we show that this methyltransferase is required for the methylation of CpNpG sequences.
RESULTS
Zmet2 Is a Chromomethylase
Zmet2 was discovered in an expressed sequence tag search of the Pioneer Hi-Bred International databases using conserved methyltransferase domains as the query sequence. The full-length cDNA sequence of Zmet2 was obtained by 3′ and 5′ rapid amplification of cDNA ends, and the sequence was analyzed against the GenBank and SWISS-PROT databases. The full-length genomic sequence was obtained by screening a genomic library.
Screening of the genomic library also resulted in the recovery of a class of clones with significant similarity to Zmet2. Genomic sequences of the homologous clones were obtained, and the cDNA sequences were confirmed by sequencing rapid amplification of cDNA ends products. This second gene is designated Zmet5. ZMET2 and ZMET5 belong to a class of DNA methyltransferases termed chromomethylases (Henikoff and Comai, 1998). The Zmet2 and Zmet5 nucleotide sequences are 90% identical over the coding regions.
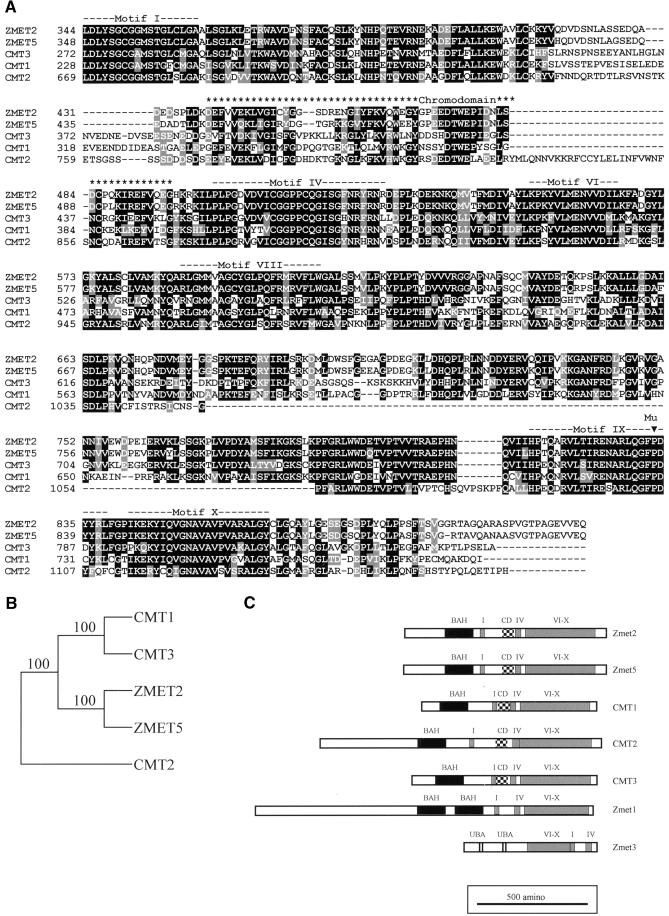
Amino acid sequence alignments of ZMET2 and ZMET5 with the Arabidopsis chromomethylases CMT1, CMT2, and CMT3 (Figure 1A) showed conservation in motifs I, IV, VI, VIII, IX, and X (motifs defined by Posfai et al., 1989; Kumar et al., 1994). Phylogenetic analysis indicated that the ZMET2 and ZMET5 proteins are more closely related to CMT1 and CMT3 (GenBank accession numbers provided in Methods) than to CMT2 (Figure 1B). Alignments of ZMET2 and CMT1 revealed 44% amino acid identity and 57% similarity. CMT1 and ZMET2 have 87% amino acid similarity across the six conserved functional domains.
Figure 1.
Maize Zmet2 Encodes a Chromomethylase.
(A) The conserved methyltransferase motifs of the ZMET2 and ZMET5 inferred amino acid sequences are aligned with the Arabidopsis chromomethylases CMT1, CMT2, and CMT3. Black shading indicates identical residues, and gray shading indicates similarity. Dashes in the sequences represent gaps introduced by CLUSTAL W to optimize the alignments. Alignments were processed by BOXSHADE. The locations of the six conserved methylase motifs are indicated above the sequences. The chromodomain is located upstream of and adjacent to motif IV. The location of the Mu insertion in the zmet2-m1::Mu allele is indicated by an arrowhead above the sequences.
(B) Relationships of maize and Arabidopsis chromomethylases. The aligned conserved methyltransferase motifs of ZMET2, ZMET5, CMT1, CMT2, and CMT3 (from the beginning of motif I to the end of motif X) were analyzed using PHYLIP. The resulting tree is shown with bootstrap values indicated at the nodes.
(C) Diagrams of methyltransferase proteins. The chromomethylases of maize and Arabidopsis are shown with Zmet1 and Zmet3. The location of the chromodomains (CD), the conserved methyltransferase motifs (I, IV, and VI to X), the BAH domains, and the ubiquitin-associated domains (UBA) are indicated by different shading.
The N-terminal domain of ZMET2 is smaller than those found in the dnmt1 class of maintenance methyltransferases, but it does contain putative nuclear localization signals, as defined by Raikhel (1992). A chromodomain is present between motifs I and IV, and the amino acid sequence and position are conserved between ZMET2 and CMT1. The inferred ZMET2 protein, using the first predicted translation start site located within a consensus Kozak sequence (Kozak, 1991), is 912 amino acids in length with a predicted mass of 101 kD.
The ZMET5 and CMT protein sequences were tested for the presence of recognizable domains by using both the PFAM and SMART protein prediction World Wide Web servers (Schultz et al., 2000). In addition to containing a chromodomain and a conserved methyltransferase domain, all chromomethylases contain a bromo adjacent homology (BAH) domain. The location of this domain is indicated in Figure 1C. BAH domains have been implicated in linking DNA methylation, replication, and transcriptional regulation in mammals (Callebaut et al., 1999). ZMET1 and MET1 both contain two BAH domains in the N-terminal regulatory region. This may indicate common mechanisms controlling these two classes of methyltransferases or targeting them in the nucleus. This finding also supports the phylogenetic evidence suggesting that the chromomethylases and MET1-type enzymes have a common ancestor (Cao et al., 2000).
Mutant Analysis Reveals That Zmet2 Is Required For CpNpG Methylation
A reverse genetics approach was used to determine the function of Zmet2. An F2 family segregating for a Mutator transposable element (Mu) insertion in Zmet2 was identified from Pioneer Hi-Bred International's TUSC (Trait Utility System for Corn) populations by using a polymerase chain reaction primer for Mu and a gene-specific primer for Zmet2. This allele is called zmet2-m1::Mu. The Mu element is inserted into exon 18, which encodes motif IX. To determine the likely effect of the Mu insertion, the aberrant transcript resulting from this insertion event was sequenced. The aberrant transcript contains a stop codon after amino acid 833 (Figure 1A). The resulting protein lacks motif X, which is required for SAM binding (Cheng et al., 1993), and is expected to lack enzymatic function.
Twelve individual plants from an F4-derived F5 family, composed of three wild-type plants, seven plants heterozygous for the zmet2-m1::Mu allele, and five plants homozygous for the zmet2-m1::Mu allele, were analyzed by HPLC to assess the effect of the mutation on global methylation levels. A 12.6% decrease in 5-methylcytosine was observed in plants homozygous for zmet2-m1::Mu relative to siblings homozygous for wild-type Zmet2 (Table 1). Heterozygous plants had significantly (α < 0.001) less methylation than did the homozygous normal plants. The reduction in methylation of the heterozygous class relative to the homozygous normal class was ∼27% of the reduction in methylation observed in the homozygous zmet2-m1::Mu class.
Table 1.
Cytosine Methylation Levels in an F4-Derived F5 Family Segregating for zmet2-m1::Mua
| Genotype | Plants | Total 5-me Cytosine (%)b | Wild-Type Levels (%) | Decrease (%) |
|---|---|---|---|---|
| Wild type | 3 | 24.80 a | 100.0 | 0.0 |
| Heterozygous zmet2-m1::Mu | 7 | 23.96 b | 96.6 | 3.4 |
| Homozygous zmet2-m1::Mu | 5 | 21.68 c | 87.4 | 12.6 |
a The 5-methylcytosine content of DNA extracted from tissue of immature leaves was determined by reverse phase HPLC. Percentages of 5mC content [5mC/(5mC + C)] were calculated from concentrations determined from integration of peak areas and comparisons with known standards.
b Letters following means indicate groupings that were significantly different (α < 0.001).
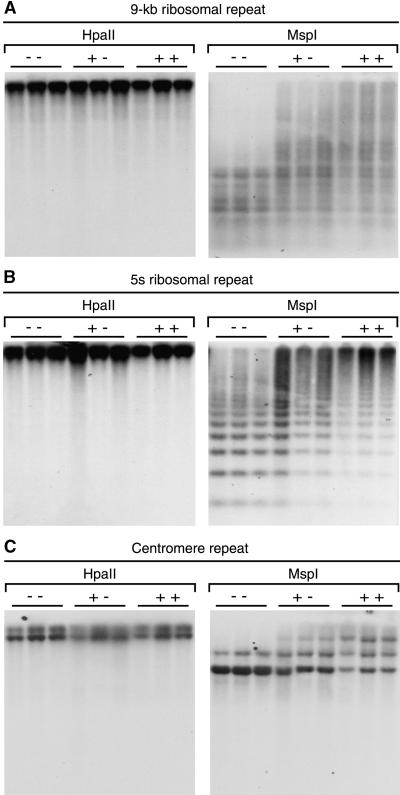
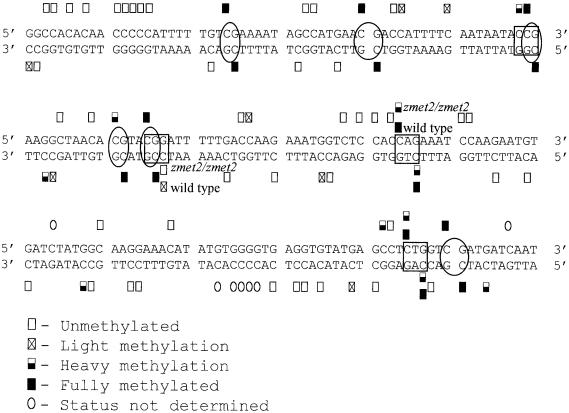
Restriction enzyme analysis of the same DNA samples was used to test for site-specific changes in DNA methylation. DNA gel blots of DNA cut with methylation-sensitive restriction enzymes and probed with repetitive sequences revealed significant reductions in cytosine methylation at mCpCpG sites (Figure 2) and mCpA/TpG sites (data not shown). Plants homozygous for the zmet2-m1::Mu allele had the largest reduction in methylation, whereas plants heterozygous for zmet2-m1::Mu were intermediate in their digestion patterns. No changes were observed at mCpG sites, indicated by unchanged HpaII (Figure 2) and HhaI (data not shown) restriction profiles. Genomic bisulfite sequencing of the 180-bp knob sequence (Figure 3) confirmed that only CpNpG methylation was reduced in the zmet2-m1::Mu mutant. The 180-bp knob sequence was methylated at both symmetric and asymmetric cytosines. The bisulfite sequencing analysis revealed that asymmetric methylation was not reduced by this mutation.
Figure 2.
Gel Blot Analysis of Repetitive DNA Methylation Patterns.
Decreased methylation is observed in mutant plants (− −) relative to nonmutant plants (+ +) digested with MspI, which is sensitive to methylation at mCpCpG sequences. No changes in methylation patterns at mCpG sites are observed in mutant plants, as indicated by the lack of digestion with HpaII. Plants heterozygous for zmet2-m1::Mu (+ −) also show decreases at mCpCpG sites. DNA gel blots were hybridized with probes for repetitive DNA: the 9-kb 26s-5.8s-17s ribosomal repeat (A), the 5s ribosomal repeat (B), and the centromeric repeat pSau3A9 (C).
Figure 3.
The DNA Methylation Patterns of Both Strands of the 180-bp Knob Repeat Were Determined by Direct Genomic Bisulfite Sequencing.
CpG, CpNpG, and asymmetric methylation all were detected in the knob sequence of plants wild type for Zmet2. CpG dinucleotides are indicated by ovals, and CpNpG trinucleotides are indicated by rectangles. The symbols above and below the alignment indicate the amount of DNA methylation observed in wild-type and homozygous zmet2-m1::Mu plants. When only one symbol is shown, the methylation was the same in wild-type and zmet2-m1::Mu plants. When the methylation status differed, two symbols are shown, with the top symbol showing the methylation status of that base in zmet2-m1::Mu plants and the bottom symbol showing the methylation status of that base in wild-type siblings. The only differences in methylation were found at CpNpG sequences. No changes in CpG or asymmetric methylation were observed.
Our analysis of the zmet2-m1::Mu allele indicates that reductions in methylation are restricted to CpNpG sites. The HPLC data indicate an overall reduction in global methylation of 12.6%. Calculations based on work by Gruenbaum et al. (1981) indicate that CpNpG methylation in wheat seedlings accounts for ∼30 to 40% of total cytosine methylation. The 12.6% reduction in total cytosine methylation in the homozygous zmet2-m1::Mu plants explains a 30 to 50% reduction in methylation at CpNpG sites.
Complete reduction in methylation at a given methylated site was observed rarely, but nearly every site analyzed showed some reduction. This indicates that the partial reduction in CpNpG methylation is caused by random reduction of methylation at most or all CpNpG sites, rather than complete reduction at some sites with no reduction at others. Our analysis did not allow us to determine if there are sectors of methylation types within the leaf tissue analyzed. That is, we do not know at present whether DNA from certain cells is completely devoid of CpNpG methylation whereas others have normal amounts of methylation, or conversely, if CpNpG methylation is lost randomly at sites along a DNA strand.
There are several potential explanations for an incomplete reduction in CpNpG methylation in homozygous zmet2-m1::Mu plants. The most likely explanation is that Zmet5, a homolog of Zmet2, has at least partial overlapping function and expression. This notion is supported by overlapping expression profiles of Zmet2 and Zmet5 (data not shown) and a high degree of conservation among the proteins. A second possibility is that an enzyme unrelated to Zmet2 is capable of maintaining CpNpG methylation at a reduced frequency. The cloned MET1 gene from pea displayed both CpG and CpA/TpG methyltransferase activities in vitro (Pradhan and Adams, 1995; Pradhan et al., 1998). No CpCpG activity was found for this protein, however. In addition, MET1 antisense plants showed some reduction at CpNpG sites (Finnegan et al., 1996), supporting the possibility that this class of methyltransferases may have CpNpG activity in vivo. Finally, it is possible that the zmet2-m1::Mu insertion does not reduce the activity of the enzyme completely, although our sequence analysis of the altered transcript indicates that the protein will lack a critical domain and provides little support for this possibility.
Restriction analysis and HPLC quantitation support an intermediate level of methylation reduction in heterozygous zmet2-m1::Mu plants relative to homozygous normal plants (Tables 1 and 2). This intermediate level of methylation in the heterozygote class was unexpected for the zmet2-m1::Mu allele, which should have lost enzymatic function. One possible explanation for the intermediate methylation of the heterozygote is that the amount of CpNpG methylation in the genome is stoichiometrically determined by the amount of ZMET2 protein. To test whether the amount of ZMET2 stoichiometrically determines the level of methylation in cells, hypoploids, euploids, and hyperploids, with one, two, and three copies of chromosome 10L, respectively, were generated using a stock with a maize A chromosome arm translocated to a B chromosome fragment containing the B chromosome centromere. Zmet2 maps distal to R on chromosome 10L (data not shown) and should be contained on the translocation chromosomes. Methylation levels of all three classes were not significantly different from each other (data not shown). Assuming that altered copy number of chromosome arms containing Zmet2 translates into varying levels of ZMET2 enzyme, this result indicates, but does not prove conclusively, that enzyme amount likely does not limit methylation levels.
Table 2.
Remethylation of Backcross Progeny from a Heterozygous zmet2-m1::Mu Plant Backcrossed to the Nonmutant Mo17 Parental Linea
| Genotype | nb | Total 5-me Cytosine (%)c |
|---|---|---|
| Mo17 wild-type parent | 3 | 24.63 a |
| zmet2-m1::Mu homozygous mutant parent | 3 | 20.50 d |
| Mo17 × zmet2-m1::Mu F1 | 3 | 22.96 c |
| Homozygous normal Zmet2 BC1 progeny | 3 | 24.05 b |
| Heterozygous zmet2-m1::Mu BC1 progeny | 3 | 23.24 c |
a Mean cytosine methylation levels are given for each family type. Percentages of 5mC content [5mC/(5mC + C)] were calculated from concentrations determined from integration of peak areas and comparisons with known standards.
n, number of individual plants tested. Two duplicates of each sample were analyzed by HPLC.
c Letters following means indicate groupings that were significantly different (α < 0.01).
A second possible explanation for a partial reduction of methylation in heterozygous zmet2-m1::Mu plants is that the mutant protein acts as a competitive inhibitor of the normal ZMET2 protein, and possibly the ZMET5 protein, producing a dominant negative effect. Because methyltrans-ferases are thought to function enzymatically as monomers, a dominant negative effect would have to be caused by target site competition rather than production of nonfunctional enzyme complexes. The target site could be either a protein complex associated with DNA or a direct association with the DNA itself. zmet2-m1::Mu is likely to produce a protein that contains the domains involved in targeting and DNA binding, including the chromodomain and the BAH domain, but it lacks the SAM binding domain, thereby abolishing enzyme function. Under the dominant negative hypothesis, reduction in methylation would result from the localization of the mutant protein to hemimethylated sites without completing the methylation reaction. The mutant enzyme would remain associated with the target site, precluding an interaction of the hemimethylated site with a functional methyltransferase. This would result in a reduction in methylation even if some amount of functional protein were present in the cell.
Inheritance of Methylation Status
To test the stability of methylation levels over generations, homozygous zmet2-m1::Mu mutant plants derived from heterozygous, F3-derived plants were compared with homozygous zmet2-m1::Mu mutant plants derived from several generations of self-pollination of homozygous mutant plants. The percentage of methylated cytosines was consistent among all homozygous mutant progeny regardless of pedigree and did not decrease upon self-pollination of homozygous mutants (data not shown). Unfortunately, because there is a methylation reduction in heterozygotes, it is impossible to assess a “first generation” effect because the nature of the reverse genetics approach used does not produce sufficient DNA or sibling controls for a valid analysis of the founder heterozygous mutant plant. Therefore, we cannot conclude unambiguously that reduction in methylation does not increase in the generation subsequent to the primary mutation event. It is only possible to conclude that the level of methylation is relatively consistent thereafter.
To determine the extent of remethylation when the zmet2-m1::Mu mutation is removed by segregation, we produced backcross plants with a homozygous zmet2-m1::Mu plant as a grandparent. The inbred line, Mo17, was crossed to a homozygous zmet2-m1::Mu plant, and the resulting F1 plant was then backcrossed to the Mo17 parent line. Restriction enzyme analysis of backcross progeny indicated that all individuals without the Mu insertion displayed substantial remethylation of repetitive centromeric sequences. Furthermore, analysis of the same DNA samples by HPLC indicated that genomic levels of cytosine methylation in homozygous normal backcross progeny were less than those of Mo17, the nonmutant parent, but significantly greater than those of heterozygous backcross siblings (Table 2). These data are consistent with the hypothesis that methylation levels are partially, but not completely, restored in the first generation of homozygote wild-type progeny obtained from a homozygous mutant parent.
The observation that remethylation occurs in normal progeny of zmet2-m1::Mu plants indicates either that ZMET2 has in vivo, de novo activity and is responsible for the establishment of CpNpG methylation patterns or that a separate de novo methyltransferase functions only early in development and that Zmet2 is responsible for maintaining these patterns. Our current data do not allow us to determine which of these possibilities is correct.
DISCUSSION
Our data indicate that chromomethylases function in vivo to maintain CpNpG symmetrical methylation patterns. This is consistent with two observations. First, CpNpG is found in most angiosperm and gymnosperm genomes but is limited in frequency in organisms other than plants. Second, chromomethylases have been found in plant species ranging from monocots to dicots but have not been found in the genomes of any other organisms. Therefore, chromomethylases, which apparently evolved after the divergence of plants from other organisms, offer plant genomes a second means to propagate methylated cytosines. The conservation of chromomethylase function across species as diverse as Arabidopsis and maize suggests that these genes provide a function that offers an evolutionary advantage to the organism.
The conserved domains of chromomethylases may provide insight into the purpose of these enzymes. In addition to the conserved methyltransferase domains, chromomethylases contain a chromodomain and a BAH domain. Chromodomains are found in several proteins involved in chromatin-level repression of transcription (Cavalli and Paro, 1998). For two of these proteins, Polycomb and HP1, the chromodomain is critical for the proper targeting. The chromodomains of the Drosophila melanogaster dosage compensation proteins MOF and MSL-3 are involved in binding to noncoding RNA molecules (Akhtar et al., 2000). The interaction of chromodomains with RNA may be the mechanism for targeting proteins containing chromodomains to specific regions of chromosomes. In plants, some RNA molecules have been shown to induce DNA methylation of homologous sequences (Wassenegger et al., 1994; Jones et al., 1999; Mette et al., 2000; Wassenegger, 2000). Chromomethylases may be the enzymes responsible for RNA-directed DNA methylation in plants. Recently, Swi6, an HP1 homolog, was shown to interact directly with methylated Lys-9 of histone H3 (Rea et al., 2000; Nakayama et al., 2001). This finding suggests the alternative possibility that chromomethylases are targeted by their chromodomain to heterochromatic regions marked by H3 Lys-9 methylation.
A BAH domain also is found in the N-terminal portion of chromomethylases. It is interesting that all Dnmt1/Met1 DNA methyltransferases and chromomethylases contain BAH domains. The Dnmt1/Met1 proteins contain two BAH domains, whereas chromomethylases contain only one BAH domain. This common feature may suggest a similar function or targeting for these two groups of methyltransferases. One function proposed for the BAH domain is to link DNA methylation to replication. These two classes of methyltransferases both may be involved in maintaining symmetric methylation patterns, with the Dnmt1/Met1 class acting on hemimethylated CpG sites and the chromomethylases methylating hemimethylated CpNpG sites soon after replication.
The distribution of CpNpG methylation in the genome also may provide insight into the function of chromomethylases. In general, methylation is found at lower levels in expressed genes and single-copy sequences than in repetitive sequences. However, CpG methylation is found in some regions of promoters and genes, depending on the gene and the stage of development. CpNpG methylation is almost never found in genic regions and appears to be restricted to repetitive DNA, with the highest abundance in heterochromatic regions. Therefore, the purpose of CpNpG methylation may be to reinforce the heterochromatic state and assist in the silencing of transposable and retrotransposable elements. The function of the chromodomain may be to target CpNpG methylation to these repeats. The increase in methylation across these repeats likely accelerates primary sequence degeneration attributable to mutation via spontaneous deamination of methylated cytosines. The addition of CpNpG methylation is expected to nearly double the number of methylated cytosines in regions susceptible to this type of methylation, thereby nearly doubling the frequency of C-to-T transition mutations caused by deamination of cytosines.
In summary, we have determined that the maize gene Zmet2 is required for the methylation of CpCpG and CpA/TpG sites in vivo. Plants containing a Mu transposable element insertion that disrupts Zmet2 have an ∼13% reduction in methylation. The observations that the chromomethylase family of genes is unique to plants, that chromomethylase genes are required for CpNpG methylation, and that CpNpG methylation is much more abundant in plants than in other organisms all support the notion that the chromomethylase family of genes evolved to provide and maintain CpNpG methylation in plants. Future research will determine whether the abundance of CpNpG methylation in plants plays a unique role in the control of gene expression and genome stability or if this family of methyltransferases evolved as a redundant mechanism to CpG methylation and other silencing pathways.
METHODS
Cloning and Sequencing of zmet2
A partial cDNA clone (CGET064) from an immature tassel cDNA library was identified in the Pioneer Hi-Bred International expressed sequence tag collection. The sequence of this clone, which represents the 3′ end of the transcript, was used to design forward and reverse primers for 5′ and 3′ rapid amplification of cDNA ends (RACE). RACE was conducted using the Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA) according to the manufacturer's protocols on cDNA prepared from Mo17 10-day-old seedling mRNA. Total RNA was extracted using Trizol (Gibco BRL) according to the manufacturer's protocol, and mRNA was isolated using oligo(dT)-cellulose columns (Pharmacia). RACE products were isolated, and ends were sequenced using Marathon primers and gene-specific primers (zmet2-RT, 5′-CTACAACATCATAGTTGGGCAGAGG-3′; and zmet2-5F, 5′-TAAAGG-GCGTGAGGGTTGGA-3′). The remaining sequence was obtained from polymerase chain reaction (PCR) products by primer walking (zmet2-1R, 5′-CCAGCTCAGCTCAGATCTGTCATCCTTT-3′; zmet2-1F, 5′-TGGTTGCTATGGTCTGCCACAGTTCAG-3′; and zmet2-3R, 5′-TCTCTA-ATTTTCTGCGGGCAG-3′; zmet2-8R, 5′-GCAATCAAGCACATTGTCGTTCTTTTCCTC-3′; zmet2-9R, 5′-TTCTTTGCGGCAGTGCTG-CG-3′; and zmet2-9F, 5′-GAAGAAGAGGGTGGGGAGAAGGAACG-3′). Two sequencing passes were made on the cDNA ends, and four sequencing passes were made on the intervening regions. PCR products were sequenced using Big Dye terminator cycle sequencing on an ABI sequencer (Perkin-Elmer Applied Biosystems) at the University of Wisconsin Biotechnology Center Sequencing Facility.
Identification of the zmet2-m1::Mu Mutant Allele
A mutant containing a Mu transposable element insertion was identified in a collection of indexed mutagenized F2 families derived from several Mu active stocks (Bensen et al., 1995). The mutant was identified using a Mu-specific primer (5′-AGAGAAGCCAACGCCA(A/T) CGCCTC(C/T)ATTTCGTC-3′) and zmet2 gene-specific primers (zmet2-1F, 5′-TGGTTGCTATGGTCTGCCACAGTTCAG-3′; and zmet2-1R, 5′-CCAGCTCAGCTCAGATCTGTCATCCTTT-3′). Because the Mutator population is quite variable, heterozygous zmet2-m1::Mu F2 seed was advanced by selfing to produce the F4-derived F5 segregating family used in this analysis.
DNA Extraction and Gel Blot Analysis for Genotyping and Methylation Analysis
The fifth to seventh immature leaf tips were collected from 15 plants of the F4-derived F5 segregating family and frozen immediately in dry ice. Tissue was ground in liquid nitrogen, and DNA was extracted as described (Saghai Maroof et al., 1984). The genotype at Zmet2 was determined by digesting DNA (10 μg) with BamHI and EcoRI, which cut on each side of the Mu insertion. The digested DNA was electrophoresed through a 0.8% agarose–0.5 × Tris-borate-EDTA gel. Gels were treated with 0.25 N HCl for 15 min, denatured in 0.2 N NaOH and 0.6 M NaCl for 30 min, and then neutralized in 0.5 M Tris and 1.5 M NaCl for 30 min. DNA was transferred to an Immobilon nylon membrane (Millipore, Bedford, MA) with 5 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate). Blots were dried at 80°C for 1.5 hr. Prehybridization was performed in 5 × SSC, 50 mM Tris, pH 8.0, 0.2% SDS, 10 mM EDTA, 1 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), and 0.1 mg/mL single-stranded sheared herring DNA overnight (8 to16 hr) at 65°C.
Hybridization conditions were similar to prehybridization conditions except for the addition of 5% dextran sulfate to the hybridization solution. Probes (25 to 50 ng) (clone CGET064 for genotyping) were labeled with 32P-dCTP (50 μCi) by using random priming. After overnight hybridization at 65°C, blots were washed two times (0.15 × SSC and 0.1% SDS) for 30 to 45 min at 65°C. Hybridized blots were exposed to Kodak BioMax film. DNA gel blot analysis with methylation-sensitive restriction enzymes was conducted on a subset of the plants using the same protocols used for genotyping except that 5 μg of DNA was digested. Enzymes included in the study were the differentially methylation-sensitive isoschizomers HpaII-MspI and EcoRII-BstNI as well as other methylation-sensitive enzymes (BamHI, BglII, HhaI, PstI, PvuII, SacI, and ScrFI). Blots were hybridized with probes for repetitive-sequence regions of the maize genome, a 9-kb clone for the maize 26s-5.8s-17s repeat (McMullen et al., 1991), the 5s ribosomal subunit clone (Mascia et al., 1981), and centromere probe pSau3A9 (Jiang et al., 1996).
HPLC Analysis
HPLC was conducted according to protocols described previously (Gehrke et al., 1984). Duplicate preparations for each of 15 plants were analyzed. Five homozygous zmet2-m1::Mu plants, seven heterozygous zmet2-m1::Mu plants, and three homozygous normal plants were analyzed. For the remethylation study, duplicate preparations from three plants of each class were analyzed. HPLC analysis was conducted at the University of Wisconsin Biotechnology Center. A volume of 50 μL was injected into a Brownlee Laboratories Spheri-5 RP-8 column (Alltech, Deerfield, IL). Nucleosides were separated at a flow rate of 0.75 mL/min. All samples were analyzed on a Beckman Instruments System Gold chromatograph, and nucleosides were detected at A254 and A280. Nucleoside and nucleotide standards (Sigma) were used to determine nucleoside peak positions and to create standard curves to determine nucleoside concentration. The ratio of 5-methylcytosine to total cytosine was calculated relative to known standards as the mean value from the two wavelength readings, and statistical analysis was conducted using SAS (SAS Institute, Cary, NC). The significance of the variance among genotypic classes was tested using the variance from plants nested within genotypes as the error term. Pairwise differences among classes were assessed using t tests, with the plants nested within genotypes also used as the error term.
Genomic Bisulfite Sequencing of the 180-bp Knob Repeat
DNA was denatured initially by adding 1.5 μL of 10 N NaOH to 8 μg of DNA in 50 μL of distilled, deionized water followed by incubation for 15 min at 37°C. After denaturation, 2.5 μL of 8-hydroxyquinoline and 450 μL of a 3.24 M urea–2 M sodium metabisulfite solution were added to the DNA solution and mixed gently. The solution was then divided into 100-μL aliquots and overlaid with mineral oil. The bisulfite modification was conducted using 20 alternating cycles of denaturation (95°C for 1 min) and modification (55°C for 15 min). After bisulfite modification, the DNA solutions were pooled and precipitated. The DNA was desalted with a Qiagen (Valencia, CA) PCR prep kit.
The 180-bp repeat was sequenced directly from the bisulfite-modified genomic DNA using Big Dye terminator cycle sequencing on an ABI sequencer according to the manufacturer's instructions (Perkin-Elmer Applied Biosystems). Sequencing reactions were performed using the buffer and enzyme supplied by the manufacturer in a 10-μL volume with 320 ng of DNA and 10 pg of primer. Sequencing reactions were conducted separately on each strand using the primers 180-bp Forward (5′-CCACACAACCCCCATTTTT-3′) to sequence one strand and 180-bp Reverse (5′-TCATACACCTCACCCCACAT-3′) to sequence the complementary strand.
Sequence Analysis
Sequence data were processed using tools available through the World Wide Web at http://dot.imgen.bcm.tmc.edu. CLUSTAL W was used for multiple sequence alignments. Sequence alignments for presentation were processed using BOXSHADE, which was available through http://www.ch.embnet.org. Sequence comparisons and database searches using BLAST 2.0 were made through http://www.ncbi.nlm.nih.gov. Phylogenetic analysis was conducted using the PHYLIP programs available through http://bioweb.pastuer.fr. Phylogenetic trees were generated using the parsimony method.
GenBank Accession Numbers
The GenBank accession numbers are as follows; Arabidopsis chromomethylases CMT1 (AF039367), CMT2 (CAA16759), and CMT3 (AAG52543); Zmet1 (AF063403).
Acknowledgments
We appreciate the assistance of Thomas Wright and his crew at the West Madison Research Station, where the nursery is located. We are grateful for the assistance of Lynn Hummel and Laura van Slyke, who helped to maintain plants in the greenhouse. We thank Steve Jacobsen for ongoing discussions and review of the manuscript and for sharing his data on plant chromomethylases. This work was supported by Pioneer Hi-Bred International, Inc., and the University of Wisconsin Graduate School.
References
- Akhtar, A., Zink, D., and Becker, P.B. (2000). Chromodomains are protein–RNA interaction modules. Nature 407, 405–409. [DOI] [PubMed] [Google Scholar]
- Banks J.A., Masson, P., and Fedoroff, N. (1988). Molecular mechanisms in the developmental regulation of the Suppressor-mutator transposable element. Genes Dev. 2, 1364–1380. [DOI] [PubMed] [Google Scholar]
- Bensen, R.J., Johal, G.S., Crane, V.C., Tossberg, J.T., Schnable, P.S., Meeley, R.B., and Briggs, S.B. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut, I., Courvalin, J.C., and Mornon, J.P. (1999). The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193. [DOI] [PubMed] [Google Scholar]
- Cao, X., Springer, N.S., Muszynski, M.G., Phillips, R.L., Kaeppler, S., and Jacobsen, S.E. (2000). Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. USA 97, 4979–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli, G., and Paro, R. (1998). Chromo-domain proteins: Linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol. 10, 354–360. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., and Walbot, V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X., Kumar, S., Posfai, J., Pflugrath, J.W., and Roberts, R.J. (1993). Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell 74, 299–307. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., and Dennis, E.S. (1993). Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 10, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke, C.W., McCune, R.A., Gama-Sosa, M.A., Ehrlich, M., and Kuo, K.C. (1984). Quantitative reverse-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J. Chromatogr. 301, 199–219. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. (1981). Sequence specificity of methylation in higher plant DNA. Nature 292, 860–862. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Comai, L. (1998). A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 148, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelbrecht, I., van Houdt, H., van Montagu, M., and Depicker, A. (1994). Posttranslational silencing of reporter transgenes in tobacco correlates with DNA modification. Proc. Natl. Acad. Sci. USA 91, 10502–10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Nasuda, S., Dong, F., Scherrer, C.W., Woo, S.S., Wing, R.A., Gill, B.S., and Ward, D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., and Richards, E.J. (1995). Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L. (1996). Epigenetic silencing and activation of a maize r gene. In Epigenetic Mechanisms of Gene Regulation, V. Russo, R. Martienssen, and A. Riggs, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 267–287.
- Kozak, M. (1991). An analysis of vertebrate mRNA sequences: Intimations of translational control. J. Cell Biol. 115, 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Cheng, X., Klimasauskas, S., Mi, S., Posfai, J., Roberts, R.J., and Wilson, G.G. (1994). The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn, F., Heidmann, I., Saedler, H., and Meyer, P. (1990). Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: Role of numbers of integrated gene copies and state of methylation. Mol. Gen. Genet. 222, 329–336. [DOI] [PubMed] [Google Scholar]
- Mascia, P.N., Rubenstein, I., Phillips, R.L., Wang, A.S., and Xiang, L.Z. (1981). Localization of the 5S rRNA genes and evidence for diversity in the 5S rDNA region of maize. Gene 15, 7–20. [DOI] [PubMed] [Google Scholar]
- McMullen, M.D., Phillips, R.L., and Rubenstein, I. (1991). Molecular analysis of the nucleolus organizer region in maize. In Chromosome Engineering in Plants: Genetics, Breeding, and Evolution, P.K. Gupta and T. Tsuchiya, eds (New York: Elsevier), pp. 561–576.
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Rice, J.C., Strahl, B.D., Allis, D.C., and Grewal, S.I.S. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Posfai, J., Bhagwat, A.A., Posfai, G., and Roberts, R.J. (1989). Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan, S., and Adams, R.L.P. (1995). Distinct CG and CNG DNA methyltransferases in Pisum sativum. Plant J. 7, 471–481. [DOI] [PubMed] [Google Scholar]
- Pradhan, S., Cummings, M., Roberts, R.J., and Adams, R.L.P. (1998). Isolation, characterization and baculovirus-mediated expression of the cDNA encoding cytosine DNA methyltransferase from Pisum sativum. Nucleic Acids Res. 26, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel, N. (1992). Nuclear targeting in plants. Plant Physiol. 100, 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B.D., Sun, Z.W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C.P., Allis, C.D., and Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Rose, T.M., Schultz, E.R., Henikoff, J.G., Pietrokovski, S., McCallum, C.M., and Henikoff, S. (1998). Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai Maroof, M.A., Soliman, K.M., Jorgensen, R.A., and Allard, R.W. (1984). Ribosomal spacer-length polymorphisms in barley: Mendelian inheritance, chromosome location, and population dynamics. Proc. Natl. Acad. Sci. USA 81, 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. (2000). SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D., and Dennis, E. (1986). Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol. Gen. Genet. 205, 476–482. [Google Scholar]
- Walker, E.L. (1998). Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148, 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M. (2000). RNA-directed methylation. Plant Mol. Biol. 43, 203–220. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., Riedel, L., and Sanger, H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76, 567–576. [DOI] [PubMed] [Google Scholar]