Abstract
Shoot architecture and flowering time in angiosperms depend on the balanced expression of a large number of flowering time and flower meristem identity genes. Loss-of-function mutations in the Arabidopsis EMBRYONIC FLOWER (EMF) genes cause Arabidopsis to eliminate rosette shoot growth and transform the apical meristem from indeterminate to determinate growth by producing a single terminal flower on all nodes. We have identified the EMF1 gene by positional cloning. The deduced polypeptide has no homology with any protein of known function except a putative protein in the rice genome with which EMF1 shares common motifs that include nuclear localization signals, P-loop, and LXXLL elements. Alteration of EMF1 expression in transgenic plants caused progressive changes in flowering time, shoot determinacy, and inflorescence architecture. EMF1 and its related sequence may belong to a new class of proteins that function as transcriptional regulators of phase transition during shoot development.
INTRODUCTION
Higher plants undergo successive developmental phase changes that are regulated by factors extrinsic and intrinsic to the shoot apical meristem (Poethig, 1990). In Arabidopsis, the main shoot development begins with the rosette/vegetative phase, that is, production of a rosette shoot comprising leaves with long petioles but no internode elongation. The transition from the vegetative to the reproductive phase results in the production of the inflorescence shoot, which displays elongated internodes with nodes that produce coflorescence shoots in the axils of cauline leaves and flowers without subtending cauline leaves. Eventually, the main shoot apical meristem ceases growth but does not form a flower.
Environmental and endogenous cues control the length of each developmental phase (Levy and Dean, 1998). In Arabidopsis, ∼40 genes are involved in regulating the transition to flowering (for a recent review, see Blazquez, 2000). Impairments in early- or late-flowering genes, such as CONSTANS (CO), EARLY FLOWERING3 (ELF3), and FLOWERING LOCUS T (FT), hasten or delay the transition from rosette to inflorescence development (Koornneef et al., 1991; Zagotta et al., 1992). Mutations in floral meristem identity genes, such as APETALA1 (AP1) and LEAFY (LFY), lengthen the inflorescence and delay the initiation of flower development (Schultz and Haughn, 1991; Huala and Sussex, 1992; Weigel et al., 1992).
The TERMINAL FLOWER1 (TFL1) gene and the two EMBRYONIC FLOWER (EMF) genes, EMF1 and EMF2, are involved in delaying both the vegetative to reproductive transition and flower initiation in Arabidopsis (Shannon and Meeks-Wagner, 1991; Sung et al., 1992; Schultz and Haughn, 1993; Yang et al., 1995). Loss-of-function mutations in TFL1 shorten both rosette and inflorescence development (Alvarez et al., 1992; Bradley et al., 1997). Loss-of-function emf mutants display more dramatic phase-reduction phenotypes: there is no rosette shoot development; only a reduced inflorescence with several flowers lacking petals is produced.
Mutants of the TFL1 and EMF1 genes display another similar phenotype, the conversion of the inflorescence apex from indeterminate to determinate growth by production of a terminal flower (Alvarez et al., 1992; Chen et al., 1997). The TFL1 ortholog in Antirrhinum, CENTRORADIALIS (CEN), also specifies shoot indeterminacy, which led Bradley et al. (1996) and Amaya et al. (1999) to propose that a common mechanism underlies indeterminacy in Antirrhinum and Arabidopsis. Shoot determinacy affects many aspects of inflorescence architecture. For example, development of a terminal flower could cause a reduction in apical dominance, activating the emergence of a secondary inflorescence and drastically changing the shoot architecture. The EMF1 and TFL1 genes may play an important role in specifying the two major inflorescence types in angiosperms, determinate (cymose) and indeterminate (racemose) (Cronquist, 1988).
To enable investigation of the molecular function of EMF1, we cloned the EMF1 gene by positional cloning. It encodes a new class of regulatory proteins also found in rice, a monocot. By manipulating EMF1 activity in transgenic plants, we confirmed that EMF1 activity controls shoot indeterminacy and flowering time. The possible function of EMF1 and its role in regulating rosette and inflorescence development are discussed.
RESULTS
Positional Cloning of the EMF1 Locus
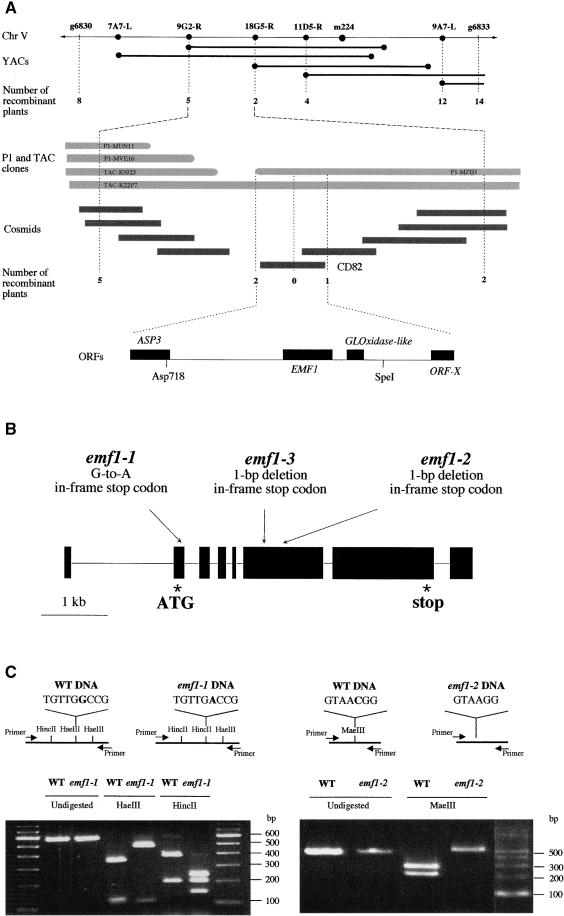
The EMF1 locus has been mapped to the upper part of chromosome V (Yang et al., 1995). Figure 1A diagrams the positional cloning experiments. By using restriction fragment length polymorphism (RFLP) markers and a segregating population representing ∼1200 meiotic events, EMF1 was fine mapped between markers 9G2-R and 18G5-R, which were derived from the ends of yeast artificial chromosome (YAC) clones included in a large contig covering that region. P1 and transformation-competent bacterial artificial chromosome (TAC) clones were anchored to these markers, and a contig was assembled based on information available at www.kazusa.or.jp/kaos/kazusa/chr5/pmap/P1_map_3.html. The 9G2-R and 18G5-R ends were used as probes to initiate a chromosome walk. Cosmid clones were isolated from binary cosmid libraries. Internal fragments of cosmid or P1 clones were converted into RFLP markers and used to delimit the EMF1 locus on cosmid clone CD82.
Figure 1.
Map-Based Cloning and Gene Structure of EMF1.
(A) Summary of the physical and genetic positions of EMF1 on chromosome (Chr) V. The top horizontal line represents the markers used to fine map the EMF1 locus between 9G2-R and 18G5-R on chromosome V. Values indicate the number of recombinant plants identified between the EMF1 locus and a given RFLP marker (circles). The bottom horizontal line shows the organization of the four genes deduced from the sequence of the CD82 clone: ORF-X (a putative ORF), the GLOxidase-like gene, EMF1, and ASP3 (aspartate aminotransferase3).
(B) Structure of the EMF1 gene and positions of the three mutations in the mutant alleles, emf1-1, emf1-2, and emf1-3. Black boxes indicate exons, and lines between the boxes indicate introns.
(C) Allele-specific RFLPs created by the emf1-1 and emf1-2 mutations. The positions of primers and enzyme restriction sites for each genotype, wild type (WT) and mutants (emf1-1 and emf1-2), are depicted in the schemes. Below, the left and right panels show the allele-specific RFLP analysis associated with the emf1-1 and emf1-2 alleles, respectively. Molecular mass markers are identified to the right of each gel.
Sequence analysis of the 17,341-bp CD82 clone and sequence data from bacterial artificial chromosome clone F15N18 revealed the presence of four open reading frames (ORFs) on cosmid clone CD82 (Figure 1A). RFLP mapping localized the EMF1 locus to a fragment common to P1 clone MZH1, TAC K22P7, and cosmid CD82 (Figure 1A). We subcloned a SpeI–Asp718 fragment from the genomic DNA of TAC K22P7 and transformed emf1-1 and emf1-2 segregating plants. T2 seed from T1 lines carrying the emf1 alleles were sown on kanamycin (Km) medium, and Km resistance and the emf mutant phenotype were scored in progeny of four emf1-1 T1 lines totaling 231 plants and six emf1-2 T1 lines totaling 884 plants (Table 1). The absence of any Km-resistant emf mutants showed that this region contained a functional EMF1 gene that complements the emf1 phenotypes. The complementing fragment has two ORFs (Figure 1A). One ORF has sequence homology with l-gulono-lactone oxidase (GLOxidase) and encodes a GLOxidase-like protein (Koshizaka et al., 1988). The second ORF has homology with two expressed sequence tags (ESTs) from Arabidopsis encoding putative polypeptides of unknown function and a hypothetical protein from the rice genomic sequencing project.
Table 1.
Complementation of emf1 Mutant Alleles by the SpeI–Asp718 Genomic DNA
| Number of T2 plantsb
|
||||||
|---|---|---|---|---|---|---|
| T1 Line | Mutant Allelea |
KmR WT | KmS WT | KmR Mutant |
KmS Mutant |
Total |
| 1-4 | emf1-1 | 44 | 10 | 0 | 2 | 56 |
| 3-12 | emf1-1 | 45 | 15 | 0 | 5 | 65 |
| 3-11 | emf1-1 | 47 | 0 | 0 | 9 | 56 |
| 3-10 | emf1-1 | 51 | 3 | 0 | 0 | 54 |
| Total | 187 | 28 | 0 | 16 | 231 | |
| 1-2 | emf1-2 | 107 | 32 | 0 | 11 | 150 |
| 2-2 | emf1-2 | 134 | 0 | 0 | 8 | 142 |
| 7-2 | emf1-2 | 110 | 0 | 0 | 36 | 146 |
| 8-2 | emf1-2 | 128 | 0 | 0 | 18 | 146 |
| 9-1 | emf1-2 | 81 | 11 | 0 | 8 | 100 |
| 9-2 | emf1-2 | 141 | 45 | 0 | 14 | 200 |
| Total | 701 | 88 | 0 | 95 | 884 | |
a Mutant alleles of T1 transgenic plants were confirmed by the allele-specific RFLP or by the occurrence of KmS emf1 mutants in T2 populations.
b KmR, kanamycin-resistant; KmS, kanamycin-sensitive; WT, wild type.
We sequenced these two candidate genes from genomic DNA isolated from the emf1 mutants and corresponding wild-type plants. All three emf1 mutant alleles (emf1-1, emf1-2, and emf1-3) have a single base pair mutation in the second ORF: each mutation creates a stop codon interrupting the predicted ORF (Figure 1B). The emf1-1 and emf1-2 mutations generate allele-specific RFLPs that were detected using a polymerase chain reaction (PCR)-based method derived from the cleaved amplified polymorphic sequence marker (Konieczny and Ausubel, 1993; Michaels and Amasino, 1998). A G-to-A change in emf1-1 and a deleted C in emf1-2 changed a HaeIII restriction site to HincII in emf1-1 and deleted a MaeIII site in emf1-2. The PCR products obtained from wild type, emf1-1, and emf1-2 and digested with the relevant restriction enzymes had the predicted fragment length polymorphisms (Figure 1C). Based on the complementation experiment and the presence of mutations in all three mutant alleles, we concluded that we had identified the EMF1 gene.
EMF1 and OsEMF1: A Novel Class of Putative Transcriptional Regulators
Using reverse transcription (RT)-PCR, we generated a 3.8-kb cDNA (see Methods) that is likely the full-length EMF1 cDNA because the first ATG initiating a 1096–amino acid polypeptide is preceded by one or more stop codons in all frames. This cDNA detected a single low-abundance transcript of ∼4 kb on poly(A)+ RNA gel blots (see below). Sequence comparison between Arabidopsis genomic DNA and this cDNA product revealed seven introns (Figure 1B), all of which display the consensus border sequence GT/AG (Hanley and Schuler, 1988). The first and last introns are located in the untranslated transcribed regions; the other five introns are located within the coding region (Figure 1B). The EMF1 gene encodes a predicted 121.7-kD protein (Figure 2A) with similarity to two ESTs from Arabidopsis and a rice sequence, as mentioned above. The two EST clones are identical to the cDNA clone amplified by RT-PCR. Analysis of the size of the two ESTs showed that they are partial cDNA clones. Furthermore, analysis of the Arabidopsis genome sequence did not reveal any other sequence closely related to the EMF1 gene. Thus, we conclude that the EMF1 gene is a single- copy gene in the Arabidopsis genome.
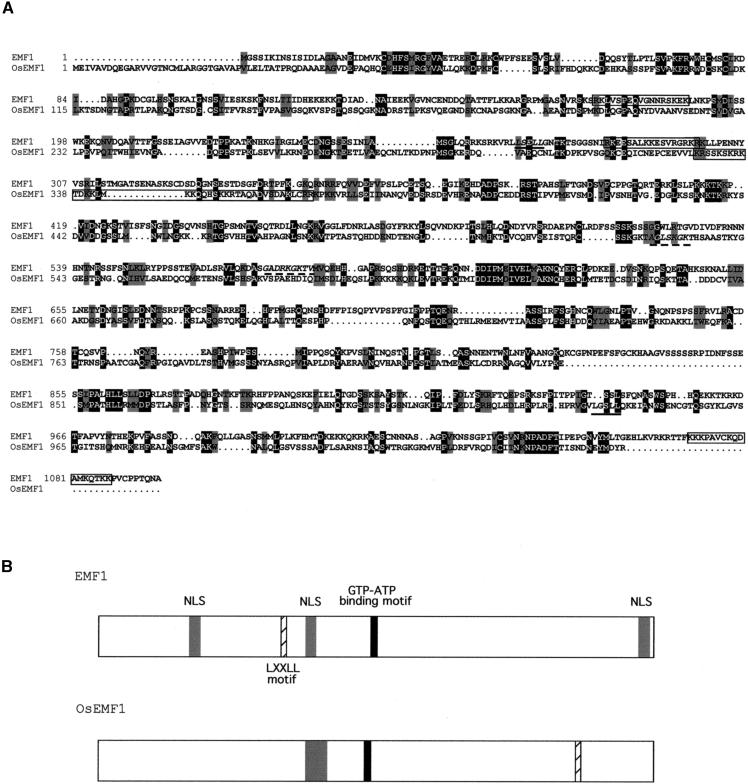
Figure 2.
Alignment between Predicted EMF1 and OsEMF1 Amino Acid Sequences and Protein Structures.
(A) Alignment between predicted EMF1 and OsEMF1 amino acid sequences. Identical amino acid residues are shaded in black, and similar amino acid residues are shaded in gray. Dots denote gaps introduced by the alignment program. Boxed sequences indicate putative nuclear localization signals (NLSs). In Arabidopsis, each box includes one possible NLS, whereas in rice, the box includes four possible overlapping NLSs. The solid underlines mark the LXXLL motif. The dashed underlines mark the GTP-ATP binding motif (P-loop motif).
(B) Predicted EMF1 and OsEMF1 protein structures. Gray boxes, NLSs; black boxes, GTP-ATP binding motif (P-loop motif); hatched boxes, LXXLL motif.
To better characterize the rice EMF1 homolog (OsEMF1), we isolated the corresponding cDNA clone by the rapid amplification of cDNA ends technique (see Methods). The OsEMF1 cDNA of 3896 nucleotides predicts a 1057–amino acid polypeptide (estimated molecular mass, 116.4 kD) that is 328 amino acids shorter than the predicted protein in BAA94774.1. The organization of introns and exons predicted at the 5′ end in BAA94774.1 was not confirmed by the sequence of the OsEMF1 cDNA (Figure 2A). The OsEMF1 cDNA likely includes a complete ORF because several stop codons are found in all three possible reading frames upstream of the first ATG initiating the 1057–amino acid polypeptide. The Arabidopsis and rice predicted protein sequences display 37% similarity and 20% identity over their entire lengths.
Neither EMF1 nor OsEMF1 displays significant homology with proteins of known function from any organism. Nevertheless, several domains could be identified in the predicted EMF1 and OsEMF1 polypeptides (Figure 2B), including nuclear localization signals (Raikhel, 1992), phosphorylation sites, an ATP/GTP binding motif (P-loop) (Walker et al., 1982), and an LXXLL motif. The LXXLL motif has been demonstrated to mediate the binding of steroid receptor coactivator complexes to a nuclear receptor (Heery et al., 1997; Torchia et al., 1997). In plants, it has been identified in the RGA and GAI proteins, two putative transcriptional regulators in the gibberellic acid signal transduction pathway (Peng et al., 1997; Silverstone et al., 1998). A PSI-BLAST homology search (Altschul et al., 1997) revealed a region of the EMF1 protein between amino acids 901 and 1034 that displays similarity (identity, 23%; positive, 37%) with two members of a nuclear receptor gene family. This gene family comprises one of the most abundant groups of transcriptional regulators in mammals, with members involved in various developmental processes (Sluder et al., 1999). Furthermore, the EMF1 protein displays homopolymeric stretches of serine residues, as do the two putative transcriptional regulators RGA and GAI (Silverstone et al., 1998). The identification of these motifs indicates that EMF1 and OsEMF1 could represent a new class of molecules that could function as transcriptional regulators during shoot development in higher plants.
Ubiquitous Expression of EMF1
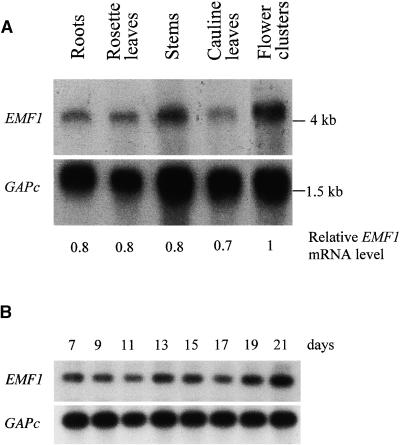
To investigate the molecular mechanism of EMF1-regulated shoot development, we studied the spatial and temporal expression of EMF1. RNA gel blot analysis found EMF1 mRNA in all organs examined: roots, rosette leaves, stems, cauline leaves, and flower clusters (Figure 3A). Using the glyceraldehyde 3-phosphate dehydrogenase c (GAPc) gene as a loading control, we found that EMF1 was expressed in all vegetative organs and was more abundant (∼20 to 30%) in flower clusters, which contain the inflorescence meristem, many flower meristems, and flowers of all stages. Thus, EMF1 RNA appeared to be expressed constitutively. Figure 3B shows that EMF1 RNA levels remained constant throughout the development of wild-type Arabidopsis plants grown under short-day conditions. Although EMF1 RNA expression did not decrease during Arabidopsis development, as proposed previously (Chen et al., 1997), EMF1 protein activity may be modulated during development by protein modification via phosphorylation, nuclear localization, or other means.
Figure 3.
Expression of EMF1 RNA in Wild-Type Arabidopsis.
(A) Autoradiographic determination of the relative amount of EMF1 RNA from a blot containing 1 μg of poly(A)+ RNA isolated from different tissues. The RNA gel blot was hybridized with an EMF1 radioactive probe and, after stripping, with a GAPc probe as a loading control. The numbers below the gels indicate the relative amounts of EMF1 RNA after standardization using the GAPc signal as a reference. Tissue samples were from roots of 2-week-old plants on agar plates; rosette leaves of 3- to 4-week-old plants in soil under short-day conditions; stems, cauline leaves, and flower clusters from an inflorescence shoot apex with developing buds and open flowers.
(B) An autoradiograph from semiquantitative RT-PCR analysis of the EMF1 level in wild-type plants grown under short-day conditions. Total RNAs were isolated from seedlings at the times indicated. RT-PCR products were amplified with EMF1 primers and GAPc primers and hybridized with an EMF1 probe (top) or a GAPc probe (bottom).
Modulation of the EMF1 Level Alters Flowering Time and Shoot Determinacy
To study the function of EMF1, we attempted to decrease EMF1 expression in wild-type plants. Three constructs containing the EMF1 coding sequence extending 0.6, 2.4, and 3.3 kb from the translation initiation codon in the antisense orientation under the control of the 35S cauliflower mosaic virus promoter (35S) (see Methods) were introduced into wild-type Arabidopsis (Bechtold et al., 1993). The 2226 T1 transgenic plants carrying the three different antisense constructs displayed a spectrum of emf1-like, early-flowering, and wild-type–like phenotypes (Figures 4A and 4B). The emf1-like plants were sterile, whereas the early-flowering plants were fertile and could grow in soil. The proportion of the three phenotypic categories observed varied among the constructs (Table 2). The two longer antisense constructs (2.4 and 3.3 kb) gave higher proportions of emf1-like transgenic plants and lower proportions of early-flowering plants than the shortest construct. The emf1-like transgenic plants, like emf1 mutants (Figure 4C), lacked rosette leaves and flowered at 14 to 16 days after sowing. Early-flowering transgenic plants produced two to eight normal petiolated rosette leaves and flowered at 16 to 20 days after sowing (Figure 4A); in the same growth conditions, wild-type–like plants produced 10 to 13 rosette leaves and flowered at ∼25 days after sowing. The endogenous EMF1 transcript levels of the early-flowering and emf1-like antisense plants were decreased greatly relative to those of wild-type–like antisense plants and wild-type plants (Figure 4E).
Figure 4.
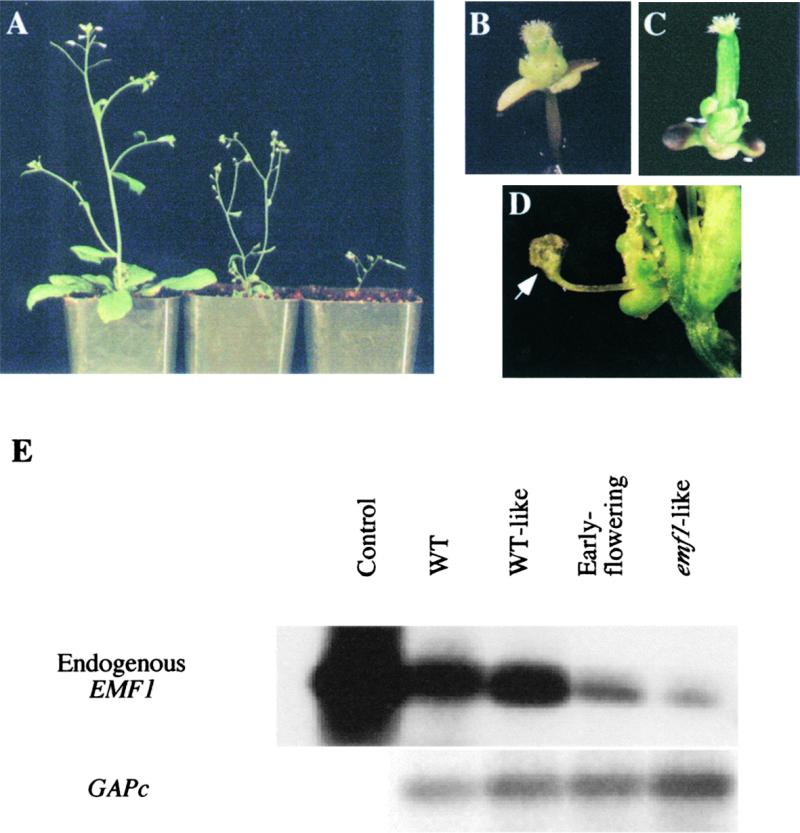
Phenotypes and EMF1 mRNA Levels of 35S::Antisense EMF1 Transgenic Plants.
Phenotypes of antisense transgenic plants and emf1-1 mutants are shown in (A) to (D).
(A) Thirty-four-day-old plants grown under long-day conditions. A wild-type–like plant is shown on the left, and two early-flowering antisense transgenic plants are shown on the right.
(B) A 25-day-old emf1-1–like transgenic plant grown under short-day conditions.
(C) A 25-day-old emf1-1 mutant grown under short-day conditions.
(D) A flower of a 32-day-old early-flowering transgenic plant with ovule-like structures on stamens (arrow) or sepals.
(E) Semiquantitative RT-PCR analysis of endogenous EMF1 levels in 25-day-old Columbia wild-type (WT) plants and antisense transgenic plants grown under short-day conditions. Total RNA in the wild-type plants, wild-type–like transgenic plants, and early-flowering transgenic plants was isolated from rosette/cauline leaves, and total RNA in emf1-like transgenic plants was isolated from seedlings. Control EMF1 fragments were amplified using EMF1 cDNA as a template. Shown are autoradiographs of RT-PCR products of endogenous EMF1 mRNA (top) and GAPc mRNA (bottom).
Table 2.
Phenotypes of 35S::Antisense EMF1 T1 Transgenic Plants
| Length of Antisense cDNA (kb) | Percent of emf1-Like Plants | Percent of Early-Flowering Plants | Percent of WTa-Like Plants |
No. of Transgenic Plants Analyzedb |
|---|---|---|---|---|
| 0.6 | 49.3 | 28.7 | 22.0 | 608 |
| 2.4 | 64.6 | 4.2 | 31.2 | 1025 |
| 3.3 | 63.9 | 6.8 | 29.3 | 593 |
a WT, wild-type.
Analyzed at 25 to 30 days after germination.
All of the emf1-like and early-flowering antisense plants made the shift from indeterminate to determinate growth by producing terminal flowers (Figures 4A and 4B). Additionally, some early-flowering plants showed a sympodial branching phenotype during shoot development, a phenotype that is seen in nature (Foster and Gifford, 1974) but that is never observed in wild-type Arabidopsis. We also found evidence of the sensitivity of flower organ differentiation to EMF1 level. Some emf1-like and early-flowering antisense plants with three or four rosette leaves produced stigmatic papillae and ovule-like structures on stamens or sepals (Figure 4D).
To study the effect of ectopic EMF1 expression on shoot development, we generated 35S::sense EMF1 transgenic plants (see Methods). Approximately 400 T1 plants and 3000 T2 plants were analyzed. The sense transgenic plants displayed the same flowering time phenotypes as the antisense transgenic plants: emf1-like, early-flowering, and wild-type–like plants. None of the 35S::sense EMF1 transgenic plants were late flowering. In the emf1-like sense transgenic plants, no EMF1 RNA was detected by RT-PCR and RNA gel blot analyses (data not shown). Thus, the emf1-like sense transgenic plant phenotypes are best explained by the occurrence of post-transcriptional gene silencing in response to the level of overexpression of EMF1 RNA (Hamilton and Baulcombe, 1999). Transgenic plants that were verified to overexpress EMF1 RNA by RNA gel blot analysis had wild-type–like phenotypes (data not shown).
DISCUSSION
Cloning of EMF1 revealed that it encodes a novel protein that might function as a transcriptional regulator. The ubiquitous presence of EMF1 RNA in Arabidopsis tissues indicates limited transcriptional regulation; however, the activity of the EMF1 protein may be modulated during development by protein phosphorylation, nuclear localization, or other means. EMF1 affects diverse developmental processes. The phenotypes of the transgenic plants indicate that EMF1 regulates flowering time, shoot determinacy, and floral organ identity. These various EMF1 functions could be explained by involvement in multiple pathways via interaction with different partners or by varying EMF1 activities in different developmental stages. Below, we analyze the phenotypes of EMF1 transgenic plants in an attempt to elucidate the function of the EMF1 gene.
The Role of the EMF1 Gene in Regulating Flowering (Bolting) Time
Flowering time is known to be regulated by photoperiod, vernalization, nutrients, and hormones (Blazquez, 2000). The relationship between the proteins involved in these flowering pathways and the EMF1 protein remains to be characterized. EMF1 loss-of-function mutants form no petiolated or rosette leaves; rather, they develop an inflorescence shoot directly from the embryo. This observation raised the possibility that EMF1 specifies rosette leaf development and influences flowering time indirectly. The early-flowering phenotype would be a pleiotropic effect of defective leaf development. However, transgenic plants with an early-flowering phenotype caused by suppression of EMF1 activity formed normal rosette leaves with long petioles before flowering, although there were fewer such leaves (Figure 4). This observation indicates that EMF1 activity affects the number of rosette leaves produced before flowering but not leaf size or morphology.
Antisense EMF1 transgenes caused wild-type plants to flower early, supporting the notion that EMF1 suppresses flowering. Furthermore, the reduction of EMF1 expression in the antisense transgenic plants correlated with flowering time (Figure 4E). This result suggests that a decrease in EMF1 expression is required for flowering in wild-type plants. However, results from RT-PCR analysis showed that the level of EMF1 expression did not decrease during development (Figure 3), suggesting that post-translational regulation of EMF1 may be required in wild-type plant development.
Ectopic expression of EMF1 in sense transgenic plants produced no late-flowering plants. On the contrary, we observed a gradient of early-flowering phenotypes, from extremely early, as in an emf1 mutant, to moderately early, as in a tfl1 mutant. Because these phenotypes were also observed in antisense transgenic plants, we postulate that they may result from gene silencing (Hamilton and Baulcombe, 1999). The normal phenotype of the transgenic plants that actually overexpressed EMF1 suggests that overexpression of EMF1 alone cannot delay flowering in wild-type plants. If other factors are required for a functional EMF1 complex, overexpression of these genes as well as EMF1 may be required for a delayed-flowering phenotype.
Role of EMF1 in Inflorescence Development
Based on mutant analysis, a role of EMF1 in controlling Arabidopsis shoot meristem identity was proposed (Chen et al., 1997). The early-flowering transgenic plants with a determinate inflorescence and normal leaves demonstrated that this role was played by EMF1. The inflorescence development might be regulated by the level of EMF1 activity, as shown by the various levels of EMF1 RNA found in the antisense transgenic plants with terminal flowers or determinate inflorescences.
Previous studies have suggested that both the EMF1 and TFL1 genes interact with the LFY and AP1 genes in a reciprocal or mutual negative regulation manner (Chen et al., 1997; Liljegren et al., 1999). The production of a terminal flower was correlated with the ectopic expression of AP1 and LFY on the tfl1 inflorescence meristem (Weigel et al., 1992; Bowman et al., 1993; Gustafson-Brown et al., 1994; Bradley et al., 1997). In emf1 mutants, the AP1 promoter was activated prematurely in the shoot apical meristem (Chen et al., 1997). Plants constitutively expressing LFY and AP1 produced a phenotype similar to that of the tfl1 mutant (Mandel and Yanofsky, 1995; Weigel and Nilsson, 1995). Interestingly, alteration in EMF1 expression also produced the tfl1 phenotype, as seen in antisense transgenic plants; this finding suggests that, in contrast to AP1 and LFY, which are negative regulators of TFL1, EMF1 activities may be required by TFL1. Furthermore, modification in the overexpression of AP1 or LFY generated new phenotypes, such as an umbel-like shoot in 35S::AP1 lfy plants; this phenotype could be enhanced greatly under short-day conditions (Liljegren et al., 1999).
Modification of EMF1 activities in transgenic plants also produced such a phenotype under short-day conditions (data not shown). In a 35S::LFY 35S::TFL1 population, some carpelloid structures that developed secondary flowers terminated a shoot (Ratcliffe et al., 1999). This feature also was seen in some of our transgenic plants (data not shown). It is unclear whether the similar inflorescence structures produced by alteration of AP1, LFY, TFL1, and EMF1 activities ultimately were mediated by EMF1 activities or by a common downstream gene regulated by a combination of these genes' activities. The molecular basis of these phenotypes awaits further study. Nevertheless, these results show that EMF1 mediates the same process regulated by TFL1, AP1, and LFY during inflorescence development and that EMF1 is one of the key regulators of a network of genes that regulate global shoot architecture.
METHODS
Plant Material and Growth Conditions
Surface-sterilized Arabidopsis thaliana seed were stratified for 3 days in the cold and germinated on agar plates containing half-strength Murashige and Skoog (1962) salts and 15 g/L sucrose. Seedlings were grown under short-day conditions (8 hr of light/16 hr of dark) for 10 days and then transferred to soil and grown under long-day conditions (16 hr of light/8 hr of dark) except as indicated. The emf1 mutants or emf1-like transgenic plants were grown on agar plates continuously under short-day conditions.
Identification of the Arabidopsis and Rice EMF1 cDNAs
The longest Arabidopsis EMF1 cDNA was identified from cDNA prepared by reverse transcription–polymerase chain reaction (RT-PCR) using 1 μg of total RNA from 4-day-old seedlings (AMLV-RT; Promega, Madison, WI). One-fifth of the reaction was amplified using a pfu Taq polymerase (Stratagene, La Jolla, CA). The PCR products were purified by agarose gel electrophoresis. DNA was isolated from the gel using a gel extraction kit (Qiagen, Valencia, CA) and cloned into a blunt end vector (pCR-Blunt; Invitrogen, Carlsbad, CA). Comparison of amplified sequences with genomic sequences revealed the intron positions. The sequence of the 3′ end primer was 5′-CCCTCTCTTTGTATCCCTC-3′. Several primers were designed from the genomic sequence at the putative 5′ end of the EMF1 transcript. The primer 5′-ATCGAGCTCGAATCTCGC-3′, situated 1033 bp upstream of a putative start codon that initiates the longest open reading frame (ORF), gave the longest RT-PCR product (∼3.8 kb).
To clone the OsEMF1 cDNA, we isolated total RNA from 7-day-old rice (Oryza sativa cv Nipponbare) seedlings using the RNeasy plant mini kit (Qiagen). Using 1 μg of total RNA and the SMART RACE (rapid amplification of cDNA ends) cDNA amplification kit (Clontech, Palo Alto, CA), we synthesized first-strand cDNAs and amplified cDNAs corresponding to the OsEMF1 transcript. The amplified PCR fragments were cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced.
Constructs and Plant Transformation
For the complementation experiment, an SpeI–Asp718 genomic fragment of the transformation-competent bacterial artificial chromosome K22P7 was cloned into the binary vector pPZP211 (Hajdukiewicz et al., 1994) and transformed into emf1-1 and emf1-2 segregating plants. Seed of T1 transgenic plants were sown on kanamycin (Km) Murashige and Skoog medium, and Km resistance and the emf1 mutant phenotypes were scored. T1 lines with emf1 mutant alleles were identified by allele-specific restriction fragment length polymorphism (RFLP) or by the occurrence of Km-sensitive emf1 mutants in T2 populations.
The antisense constructs were made by inserting ORF fragments extending 0.6 and 2.4 kb from the initiation codon into the Asp718 site of pGA1535 and inserting a 3.3-kb fragment into the XbaI–Asp718 site of pGA1535 in the antisense orientation. For the sense construct, a uidA gene in pBI121 was replaced with the fragment containing the longest cDNA of EMF1 in the sense orientation.
Constructs were transformed into Agrobacterium tumefaciens strain GV3101 (pMP90) and then into Arabidopsis plants as recommended by Bechtold et al. (1993). Seed from the transformed plants were selected with 50 mg/L Km.
RT-PCR Experiments
The amount of EMF1 transcript was determined by semiquantitative RT-PCR. The RT conditions were as described for the cloning of EMF1 cDNA. The PCR amplification was performed in a final volume of 12.5 μL using a Promega Taq polymerase according to the recommendations of the supplier. Amplification was verified as being in the exponential phase: 15 to 20 cycles for the EMF1 gene, 10 to 15 cycles for the GAPc gene. The data shown are representative of the tissues or time points in at least three independent RT-PCR experiments.
Poly(A)+ RNA Isolation and Hybridization
Total RNA was isolated according to the protocol established by Logemann et al. (1987). Poly(A)+ RNA was then purified using the Oligotex mRNA kit (Qiagen). RNA and DNA gel blots were analyzed as described (Sambrook et al., 1989). The hybridization was performed with Church buffer (Church and Gilbert, 1984). After hybridization, membranes were washed at 65°C in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS, then in 1 × SSC and 0.1% SDS, and finally in 0.1 × SSC and 0.1% SDS for 20 min each.
GenBank Accession Numbers
The GenBank accession numbers are as follows: bacterial artificial chromosome clone F15N18, AL163815; ESTs from Arabidopsis encoding putative polypeptides, N96450 and Z46543; hypothetical protein from the rice genomic sequencing project, BAA94774-1; EMF1 gene, AF319968; OSEMF1 cDNA of 3896 nucleotides, AF326768.
Acknowledgments
We are grateful to T. Ito and K. Shinozaki (Institute of Physical and Chemical Research, RIKEN, Tsukuba, Japan) for sharing unpublished information and for their efforts in the search for the EMF1 cDNA clones, to S. Tabata and S. Sato (Kazusa DNA Research Institute, Kisarazu, Chiba, Japan) for sharing their data before publication, and to Denise Schichnes and Steven Ruzin (Biological Imaging Facility, College of Natural Resources, University of California) for assistance with image processing. We also thank W. Lukowitz (Carnegie Institution of Washington, Department of Plant Biology, Stanford, CA), G. An (POSTECH, Pohang, Korea), and P. Maliga (Waksman Institute, Rutgers University, New Brunswick, NJ) for providing aliquots of binary cosmid library and binary vectors, and the Arabidopsis Biological Resource Center at the Ohio State University (Columbus) for providing the cDNA library and clones. This work was supported by U.S. Department of Agriculture Grant 99-35301-7984, by funds from the Norvatis Agricultural Discovery Institute to Z.R.S., and by a Korean Government fellowship (KOSEF) to Y.-H.M.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, J., Guli, C.L., Yu, X.-H., and Smyth, D.R. (1992). terminal flower: A gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2, 103–116. [Google Scholar]
- Amaya, I., Ratcliffe, O.J., and Bradley, D.J. (1999). Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell 11, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Blazquez, M. (2000). Flower development pathways. J. Cell Sci. 113, 3547–3548. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Alvarez, J., Weigel, D., Meyerowitz, E.M., and Smyth, D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743. [Google Scholar]
- Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S., and Coen, E. (1996). Control of inflorescence architecture in Antirrhinum. Nature 379, 791–797. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- Chen, L., Cheng, J.C., Castle, L., and Sung, Z.R. (1997). EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9, 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronquist, A. (1988). The Evolution and Classification of Flowering Plants, 2nd ed. (Bronx, NY: New York Botanical Garden).
- Foster, A.S., and Gifford, E.M., Jr. (1974). Comparative Morphology of Vascular Plants. (San Francisco: W.H. Freeman).
- Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1994). Regulation of the floral homeotic gene APETALA1. Cell 76, 131–143. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hanley, B.A., and Schuler, M.A. (1988). Plant intron sequences: Evidence for distinct groups of introns. Nucleic Acids Res. 16, 7159–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery, D.M., Kalkhoven, E., Hoare, S., and Parker, M.G. (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Huala, E., and Sussex, I.M. (1992). LEAFY interacts with floral homeotic genes to regulate floral development. Plant Cell 4, 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and Van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Koshizaka, T., Nishikimi, M., Ozawa, T., and Yagi, K. (1988). Isolation and sequence analysis of a complementary DNA encoding rat liver l-gulono-gamma-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis. J. Biol. Chem. 263, 1619–1621. [PubMed] [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10, 1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G., and Yanofsky, M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1998). A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14, 381–385. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923–930. [DOI] [PubMed] [Google Scholar]
- Raikhel, N. (1992). Nuclear targeting in plants. Plant Physiol. 100, 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Bradley, D.J., and Coen, E.S. (1999). Separation of shoot and floral identity in Arabidopsis. Development 126, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schultz, E.A., and Haughn, G.W. (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, E.A., and Haughn, G.W. (1993). Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119, 745–765. [Google Scholar]
- Shannon, S., and Meeks-Wagner, D.R. (1991). Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 3, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder, A.E., Mathews, S.W., Hough, D., Yin, V.P., and Maina, C.V. (1999). The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 9, 103–120. [PubMed] [Google Scholar]
- Sung, Z.R., Belachew, A., Shunong, B., and Bertrand-Garcia, R. (1992). EMF, an Arabidopsis gene required for vegetative shoot development. Science 258, 1645–1647. [DOI] [PubMed] [Google Scholar]
- Torchia, J., Rose, D.W., Inostroza, J., Kamei, Y., Westin, S., Glass, C.K., and Rosenfeld, M.G. (1997). The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Walker, J.E., Saraste, M., Runswick, M.J., and Gay, N.J. (1982). Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Yang, C.H., Chen, L.J., and Sung, Z.R. (1995). Genetic regulation of shoot development in Arabidopsis: Role of the EMF genes. Dev. Biol. 169, 421–435. [DOI] [PubMed] [Google Scholar]
- Zagotta, M.T., Shannon, S., Jacobs, C., and Meeks-Wagner, R. (1992). Early flowering mutants of Arabidopsis thaliana. Aust. J. Plant Physiol. 19, 411–418. [Google Scholar]