Abstract
Specific recognition of pathogens is mediated by plant disease resistance (R) genes and translated into a successful defense response. The extent of associated hypersensitive cell death varies from none to an area encompassing cells surrounding an infection site, depending on the R gene activated. We constructed double mutants in Arabidopsis between positive regulators of R function and a negative regulator of cell death, LSD1, to address whether genes required for normal R function also regulate the runaway cell death observed in lsd1 mutants. We report here that EDS1 and PAD4, two signaling genes that mediate some but not all R responses, also are required for runaway cell death in the lsd1 mutant. Importantly, this novel function of EDS1 and PAD4 is operative when runaway cell death in lsd1 is initiated through an R gene that does not require EDS1 or PAD4 for disease resistance. NDR1, another component of R signaling, also contributes to the control of plant cell death. The roles of EDS1 and PAD4 in regulating lsd1 runaway cell death are related to the interpretation of reactive oxygen intermediate–derived signals at infection sites. We further demonstrate that the fate of superoxide at infection sites is different from that observed at the leading margins of runaway cell death lesions in lsd1 mutants.
INTRODUCTION
Plants have evolved mechanisms to detect and respond effectively to most pathogens. Analyses of genetic variation in plant responses to pathogens have identified corresponding gene pairs (resistance or R genes in the plant and avirulence or avr genes in the pathogen) that mediate recognition and cause induction of plant resistance (Staskawicz et al., 1995). These local plant defenses are usually, although not invariably, associated with a form of programmed plant cell death known as the hypersensitive response (HR). The HR can lead to cell death surrounding the infection site (Holub et al., 1994). Localized necrosis also can induce a plant response called systemic acquired resistance, which heightens defenses in uninoculated tissues against a broad spectrum of pathogens (Yang et al., 1997; McDowell and Dangl, 2000).
One of the earliest biochemical changes associated with the HR is an oxidative burst producing reactive oxygen intermediates (ROI), including superoxide anion (O2−·) as a proximal component, which can be dismutated rapidly to hydrogen peroxide (H2O2) (Lamb and Dixon, 1997; Bolwell, 1999; Grant and Loake, 2000). These may serve both as antimicrobial agents and as signaling molecules in local and systemic plant resistance. Nitric oxide (NO), a redox-active molecule with a critical role in the activation of mammalian defense responses (Schmidt and Walter, 1994), also functions as an important signal in plant resistance against pathogens (Delledonne et al., 1998; Durner et al., 1998). Salicylic acid (SA) accumulates in plant tissue responding to pathogen infection and is essential for the induction of systemic acquired resistance as well as being required for some R gene–mediated responses, at least in Arabidopsis and tobacco (Gaffney et al., 1993; Delaney et al., 1994; Mur et al., 1997). Recent results suggest that the balance and cooperation between NO, ROI, and SA produced early in the plant resistance response is required for the full expression of the HR (Delledonne et al., 1998, 2001; Klessig et al., 2000). However, little is known about the sequence of events that determines local plant resistance. Also unclear is whether signals are transduced from an infection focus to first initiate, and then dampen, the HR.
Arabidopsis is the key genetic system with which to unravel disease resistance pathways (Glazebrook, 1999; Feys and Parker, 2000). Arabidopsis R genes have been cloned that confer specific recognition of viral, bacterial, and oomycete pathogens (Parker et al., 2000). Their products belong to the most prevalent R protein class identified in a range of plant species that contains a central nucleotide binding (NB) domain and varying numbers of C-terminal leucine-rich repeats (LRRs) (Jones, 2000). NB-LRR proteins were further categorized into those with a coiled-coil (CC) motif at their N termini and those that have N-terminal (TIR) similarity to the cytoplasmic domains of human and Drosophila Toll-like receptors (Jones, 2000).
Mutational analyses in Arabidopsis uncovered genes required as positive regulators of basal defense (Glazebrook, 1999; Feys and Parker, 2000). EDS1 is a necessary component of RPP1- and RPP4-specified resistance to the oomycete pathogen Peronospora parasitica (Pp) (Parker et al., 1996; Aarts et al., 1998) and is more generally required for resistance mediated by several tested Arabidopsis R genes encoding TIR-NB-LRR proteins (Aarts et al., 1998). However, EDS1 is not required for resistance conferred by any of the tested CC-NB-LRR R genes (Aarts et al., 1998). Many, but not all, CC-NB-LRR R genes examined are dependent on NDR1, a gene identified through mutational analysis of RPM1-mediated resistance to the bacterial pathogen Pseudomonas syringae expressing avrB (Century et al., 1995). Thus, EDS1 and NDR1 differentiate R gene–mediated events that may, at least in several cases, be conditioned by particular R protein structural types (for the current exceptions, see McDowell et al., 2000). Furthermore, ndr1 mutant plants retain an HR initiated by two R genes, RPM1 and RPS5, even though they fail to prevent bacterial growth, suggesting that resistance and HR are separable (Century et al., 1995). EDS1 encodes a 72-kD lipase-like protein that operates upstream of SA-mediated defenses (Falk et al., 1999), whereas NDR1 encodes a 25-kD protein that has two putative membrane attachment domains (Century et al., 1997).
Mutational screens in Arabidopsis identified several other plant defense signaling genes that are components of SA signaling in the plant response against pathogens. For example, PAD4 (Glazebrook et al., 1997; Zhou et al., 1998) and SID1/EDS5 and SID2/EDS16 (Rogers and Ausubel, 1997; Nawrath and Métraux, 1999) function upstream of SA accumulation, whereas NPR1/NIM1 is an important regulator of responses downstream of SA (Cao et al., 1994; Delaney et al., 1995). Significantly, PAD4 encodes a lipase-like protein with catalytic motifs identical to EDS1 (Jirage et al., 1999). EDS1 and PAD4 operate upstream of pathogen-induced SA accumulation, yet their expression can be enhanced by exogenous applications of SA. This finding reinforces evidence of an SA-associated positive feedback loop that may potentiate plant defense (Shirasu et al., 1997; Falk et al., 1999; Jirage et al., 1999). The pad4 mutation affects the same spectrum of R gene functions detailed above for eds1, but the loss of resistance in pad4 is typically not as complete as in eds1 (Glazebrook et al., 1997; Aarts et al., 1998; Feys et al., 2001).
Other Arabidopsis mutations deregulate disease resistance responses and/or HR-like plant cell death responses, suggesting that negative control of plant defense pathways also occurs (Morel and Dangl, 1997). Some of these display a “disease lesion mimic” phenotype that is a feature of several well-characterized crop plant mutants, in which necrotic lesions form spontaneously or can be induced by various biotic or abiotic stresses (Dangl et al., 1996; Büschges et al., 1997; Gray et al., 1997). Importantly, Arabidopsis plants carrying the recessive null lsd1 allele exhibit normal HR after infection by various incompatible pathogens, but runaway cell death (RCD) is initiated subsequently at the margins of these sites (Dietrich et al., 1994). Spreading lesions in lsd1 can be induced by provision of O2−· (Jabs et al., 1996) in uninfected tissues. This, together with observations that O2−· accumulation precedes lesion formation (Jabs et al., 1996), suggests that LSD1 responds to a superoxide-dependent signal(s) emanating from an infection site. SA possibly potentiates this pathway, because lsd1 plants are acutely responsive to treatments with SA or chemically active SA analogs (Dietrich et al., 1994; Jabs et al., 1996). Thus, lsd1 lowers the threshold for both initiation and propagation of plant cell death beyond the HR. lsd1 plants also exhibit enhanced resistance to several normally virulent pathogens in a prelesioned state (Dietrich et al., 1994). We infer from these null phenotypes that LSD1 negatively regulates a signaling pathway(s) for basal defense and cell death and thereby may contribute to establishing a boundary to the plant HR (Dietrich et al., 1994). LSD1 encodes a zinc finger protein with homology with GATA-type transcription factors, and it has been suggested that the LSD1 protein functions either to negatively regulate a pro-death pathway component or to activate a repressor of plant cell death (Dietrich et al., 1997).
We constructed double mutant lines between the eds1, pad4, or ndr1 mutations and lsd1 and assessed their effects on RCD and disease resistance phenotypes after pathogen infection, treatment with benzothiodiazole (BTH), a functional SA mimic (Görlach et al., 1996), or a superoxide generator. We demonstrate that lsd1 does not affect the eds1, pad4, and ndr1 pathogen response phenotypes. However, both EDS1 and PAD4 are necessary for lsd1-conditioned RCD initiated by each tested stimulus. In contrast, NDR1 is required for RCD in response to superoxide and partially reduces lsd1 RCD after pathogen inoculation or BTH treatment. The requirement for EDS1, PAD4, or NDR1 in lsd1 RCD is separable from processes associated with the local HR and disease resistance; therefore, it is likely to operate at the level of defense signal potentiation in cells surrounding an infection site.
RESULTS
EDS1 and PAD4 Are Required for lsd1 RCD Induced by BTH and Pathogens
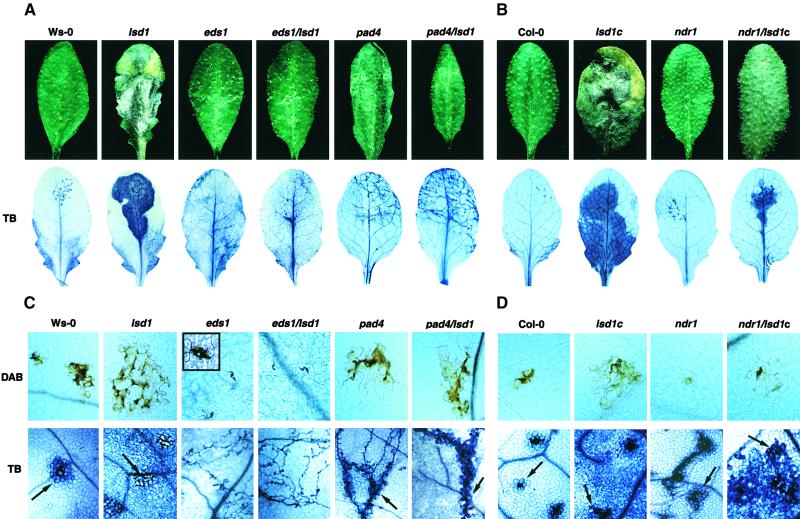
We first examined the responses of short-day-grown eds1/lsd1, pad4/lsd1, and ndr1/lsd1c plants to a known inducer of RCD in lsd1, the SA mimic BTH (all mutants used were null alleles; see Methods). As shown in the top row of Figures 1A and 1B, no phenotype was observed in leaves from either wild-type plants or plants with single mutations in eds1, pad4, or ndr1. Leaves from lsd1 or lsd1c plants, in contrast, formed the expected lesions in response to BTH 3 days after treatment. We did not observe any lesions in leaves of plants with double mutations in eds1/lsd1 and pad4/lsd1, but lesions were observed in leaves of ndr1/lsd1c plants. However, these lesions were not as extensive as those observed in leaves of lsd1 plants (Figure 1B). Thus, mutations in eds1 or pad4 abolish BTH-induced RCD in lsd1 plants. Similar results were observed in these plants after treatment with another inducer of lsd1-mediated RCD, a shift in growing conditions from short- day to long-day conditions (data not shown).
Figure 1.
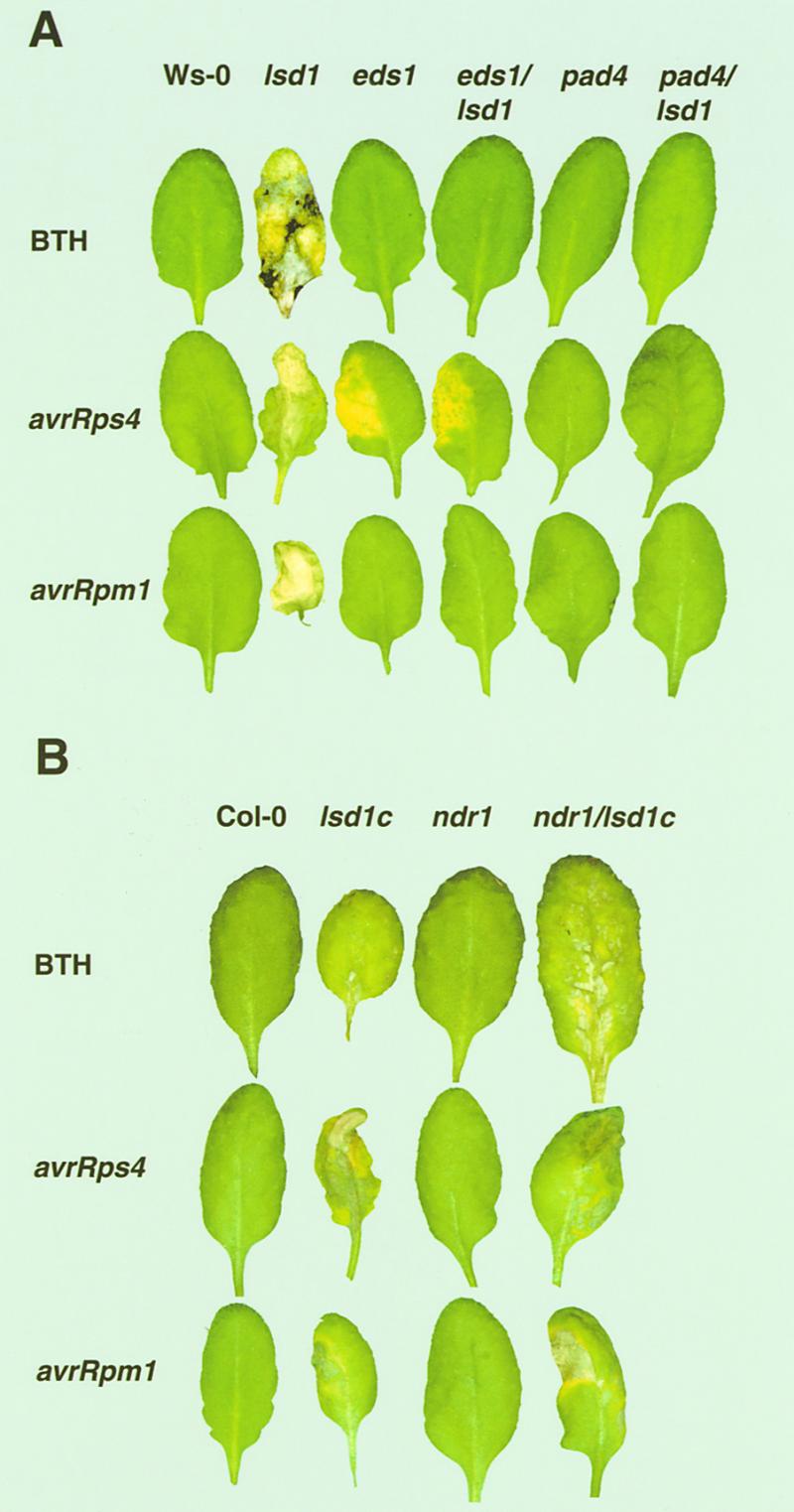
Lesion Phenotypes of Plant Lines after BTH Treatment or Bacterial Pathogen Inoculation.
Leaves of 4-week-old wild-type, single mutant, or double mutant plants were sprayed with 0.35 mM BTH or infiltrated on one side with low titer suspensions (105 colony-forming units/mL) of P. syringae pv DC3000 expressing avrRps4 or avrRpm1. See Methods for further details. Leaves were photographed after 6 days of incubation, and each leaf is representative of 12 to 15 leaves. All treatments were repeated with similar results.
(A) Phenotypes of plant lines in accession Ws-0.
(B) Phenotypes of plant lines in accession Col-0.
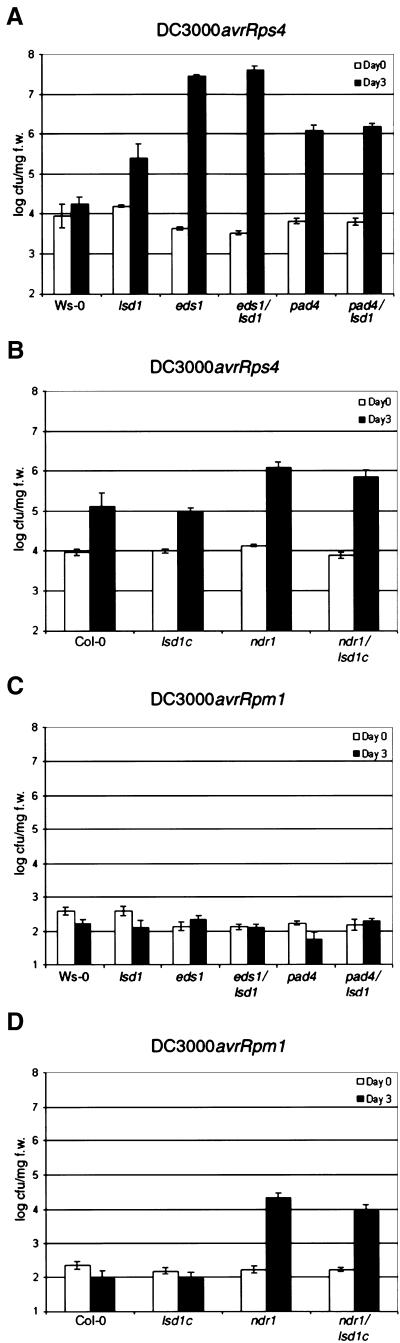
We next assessed the interactions between these double mutant plants and normally avirulent strains of P. syringae pv tomato (DC3000) expressing either avrRpm1 or avrRps4. The different signaling requirements for RPM1 and RPS4 mentioned in the Introduction allowed us to measure the effects of the eds1, pad4, and ndr1 mutations on lsd1-induced phenotypes in the context of both an intact (resistant) and a defective (susceptible) local plant response by using isogenic P. syringae strains differing only in the avr gene they express. Plants were infiltrated with low doses of DC3000/avrRps4 to examine the genetic interactions between eds1 or pad4 in combination with lsd1. As expected, Wassilewskija (Ws-0) and lsd1 plants were resistant (Figure 2A). However, Ws-0 plants exhibited no visible phenotype, whereas lsd1 plants displayed lesions 3 to 4 days postinoculation (DPI) (Figure 1A). In contrast, eds1 and eds1/lsd1 double mutant plants were susceptible (Figure 2A). Additionally, eds1 and eds1/lsd1 double mutant plants developed characteristic chlorotic disease symptoms, but no spreading lesions were observed in the eds1/lsd1 double mutant plants (Figure 1A). Plants with mutations in pad4 or both pad4 and lsd1 were intermediate; bacterial growth was ∼10-fold less than in plants with mutations in eds1 (Figure 2A). However, pad4/lsd1 plants did not exhibit chlorosis associated with disease or pathogen-induced lesioning associated with lsd1 RCD (Figure 1A). Thus, lsd1 does not influence the susceptibility of eds1 or pad4 plants to DC3000/avrRps4.
Figure 2.
Bacterial Growth in Wild-Type, Single Mutant, and Double Mutant Plants.
Growth of P. syringae pv DC3000 expressing avrRps4 or avrRpm1 extracted from leaves at 0 (open bars) and 3 (closed bars) days after inoculation (initial titer, 105 colony-forming units/mL). Data from Ws-0
We then challenged plants with low doses of DC3000/avrRpm1. Wild-type, lsd1, eds1, and pad4 plants responded as expected (see Introduction); all genotypes were resistant, and RCD was visible in lsd1 leaves (Figures 1A and 2C). eds1/lsd1 and pad4/lsd1 double mutants also were resistant, but, surprisingly, they did not exhibit RCD. To confirm this observation, we infiltrated leaves with levels of DC3000/avrRpm1 (107/mL) that induce an HR 6 to 8 hr after inoculation (Grant et al., 1995). Plants from all genotypes (Ws-0, lsd1, eds1, eds1/lsd1, pad4, and pad4/lsd1) exhibited an HR. However, spreading lesions were observed in only lsd1 plants and not in eds1/lsd1 or pad4/lsd1 (Table 1). Therefore, EDS1 and PAD4 are required for lsd1 RCD. Importantly, the requirement for EDS1 and PAD4 in RCD is independent of their signaling functions in RPS4-mediated disease resistance and separate from processes controlling RPM1 resistance.
Table 1.
Response Phenotypes of Wild-Type, Single Mutant, and Double-Mutant Lines to Inoculation with Avirulent Bacteria or Treatment with RB
| Arabidopsis Lines |
DC3000/avrRps4
|
DC3000/avrRpm1
|
RB
|
||||
|---|---|---|---|---|---|---|---|
| HR | ROI | RCD | HR | ROI | RCD | RCD | |
| Ws-0 | + | + | − | + | ++ | − | − |
| eds1 | − | − | − | + | ++ | − | − |
| pad4 | + | + | − | + | ++ | − | − |
| lsd1 | + | ++ | + | + | +++ | + | + |
| eds1/lsd1 | − | − | − | + | +++ | − | − |
| pad4/lsd1 | + | + | − | + | ++ | − | − |
| Col-0 | (+) | (+) | − | + | + | − | − |
| ndr1 | − | − | − | * | ++ | − | − |
| lsd1c | (+) | + | + | + | ++ | + | + |
| ndr1/lsd1c | − | − | (+) | * | ++ | (+) | − |
Leaves were dipped in suspensions (107 colony-forming units/mL) of P. syringae pv DC3000 expressing avrRps4 or avrRpm1 or treated with a 2-μL droplet of 20 mM RB. Development of the plant HR and accumulation of ROI were scored 24 and 48 hr, and RCD was scored 4 days, after bacterial inoculation. The scores (+), +, ++, and +++ reflect the intensity of staining with lactophenol–trypan blue for the HR and DAB for ROI. They are representative of at least six leaves per treatment. Asterisks denote an expanded, diffuse HR observed in ndr1 and ndr1/lsd1c plants. Pathogen inoculations were repeated twice with similar results. Leaves were scored for RCD 5 days after rose bengal application. Similar results were obtained in four independent experiments using 10 leaves per plant line.
We also infiltrated ndr1/lsd1c double mutants with either DC3000/avrRps4 or DC3000/avrRpm1. Columbia (Col-0) and ndr1 plants were resistant to DC3000/avrRps4 (Figures 1B and 2B), whereas ndr1 plants were moderately susceptible to infection by DC3000/avrRpm1, consistent with previous analyses (Century et al., 1995) (Figures 1B and 2D). ndr1/lsd1c double mutants were resistant to DC3000/avrRps4 and susceptible to DC3000/avrRpm1, as expected. However, we observed a partial suppression of RCD in these plants after inoculation with either bacterial strain (Figure 1B). Therefore, although ndr1 reduced lsd1 RCD, the level of reduction did not correlate with the loss of RPM1 function observed in ndr1 mutants.
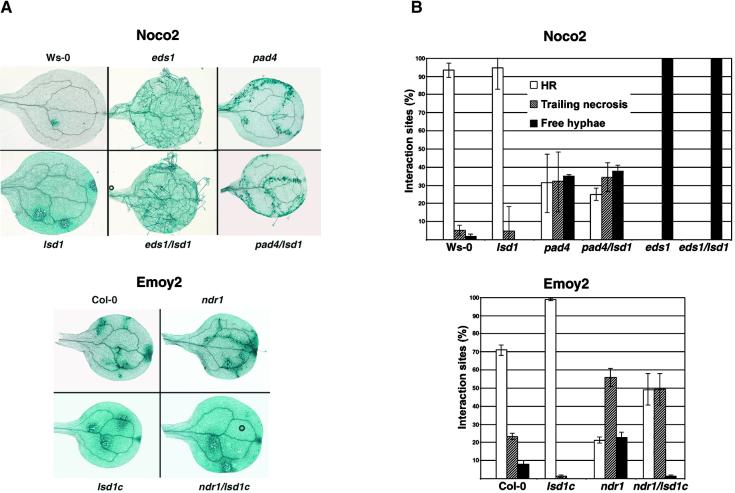
We then examined the responses of the eds1/lsd1, pad4/lsd1, and ndr1/lsd1c mutant lines to normally avirulent isolates of the oomycete pathogen Pp. This extends our analysis to an additional pathogen recognized by R genes that differ in their signaling requirements, as outlined in the Introduction. As shown in Figures 3A and 3B, RPP1-mediated resistance to Noco2 in cotyledons of Ws-0 and lsd1 is manifested as HR at points of attempted pathogen penetration 6 DPI. At this time, developing RCD is visible in lsd1 as an enlargement of the trypan blue–stained zone around an infection site (Figure 3A). In contrast, cotyledons of eds1 and eds1/lsd1 plants were susceptible to Noco2; we observed extensive mycelial growth as well as asexual sporulation (Figures 3A and 3B). Noco2 inoculation failed to elicit an HR in eds1/lsd1 plants. pad4 and pad4/lsd1 plants were partially susceptible to Noco2; we observed trailing necrosis in response to Noco2 (Figure 3A), suggesting that HR was elicited but not sufficient to fully restrict pathogen growth. There was no RCD in either double mutant. Therefore, EDS1 and PAD4 are required for lsd1-mediated RCD, using Pp as an RCD inducer.
Figure 3.
Infection Phenotypes of Plant Cotyledons Inoculated with Pp Isolates Noco2 and Emoy2.
(A) Cotyledons of 10-day-old seedlings were inoculated with Noco2 or Emoy2 (5 × 104 spores/mL) and stained with lactophenol–trypan blue at 6 DPI to reveal Pp mycelium and dead plant cells.
(B) Trypan blue–stained cotyledons were harvested at 6 DPI, and individual plant–pathogen interaction sites were categorized as HR, trailing necrosis, or free mycelium. The percentage of each interaction type was scored from 40 to 80 cotyledons per experiment. Graphs represent the mean and ±se from three independent experiments.
A similar analysis was performed by inoculating Emoy2 onto cotyledons of Col-0, lsd1c, ndr1, and ndr1/lsd1c plants. RPP4-mediated resistance to Emoy2 in Col-0 and lsd1c was associated with HR and the initiation of RCD in lsd1 cotyledons at 6 DPI (Figure 3A). ndr1 partially suppressed RPP4-mediated resistance to Emoy2; we observed an increased frequency of trailing necrosis (Figures 3A and 3B). lsd1c plants expressed strong resistance to Emoy2, as shown by an increase in the proportion of HR sites, compared with Col-0 (Figure 3B). Surprisingly, ndr1/lsd1c double mutants exhibited an intermediate phenotype (Figure 3B). Therefore, the loss of LSD1 function enhanced host resistance to Pp early in the plant–pathogen interaction independent of NDR1 and presumably independent of the recognition conferred by RPP4.
We next examined the responses of leaves from mature (3- to 4-week-old) plants to inoculations of Pp. This analysis allowed us to relate events associated with the plant HR to the initiation and spread of lsd1-conditioned RCD. As shown in Figures 4A and 4B, discrete necrotic flecks formed on leaves of Ws-0 or Col-0 in the area of incompatible Pp inoculation. Leaves of lsd1 produced lesions that spread from the site of the localized resistance response (Figures 4A and 4B). In contrast, asexual sporulation of Pp was observed on infected eds1 and eds1/lsd1 leaves at 6 DPI, and no necrosis was observed. Leaves of pad4 and pad4/lsd1 plants supported some pathogen growth that was accompanied by trailing necrosis (Figure 4A). These results mirror those observed on cotyledons and further support the requirement for EDS1 and PAD4 for lsd1-mediated RCD. Leaves of ndr1 and Col-0 responded in a similar manner to inoculation of Pp Emoy2, although the area of plant tissue undergoing an HR was marginally larger in ndr1 than in Col-0 (Figure 4B; see also Figure 4D). The RCD was severely reduced in ndr1/lsd1c compared with that in lsd1c (Figure 4B). Interestingly, the rate of initial lesion formation at the boundary of the HR was similar in lsd1c and ndr1/lsd1c (data not shown). However, by 3 to 4 DPI, lesions in ndr1/lsd1c ceased to expand, whereas in lsd1c they progressed and consumed the entire leaf by ∼6 DPI (Figures 4B and 4D).
Figure 4.
Disease Resistance and Runaway Plant Cell Death Phenotypes in Adult Leaves after Pp Inoculation.
Leaves of 4-week-old plants from wild-type, single mutant, and double mutant Ws-0 ([A] and [C]) and Col-0 ([B] and [D]) lines were inoculated by placing a 10-μL droplet of Pp spores on the top half of each leaf. Pp isolates Noco2 and Emoy2 (or Emwa1) were used for Ws-0 ([A] and [C]) and Col-0 ([B] and [D]) accession lines, respectively. Macroscopic phenotypes and corresponding trypan blue (TB) staining of plant–pathogen interaction sites are shown for whole leaves ([A] and [B]) and at ×200 magnification ([C] and [D], bottom row) at 6 DPI. Hypersensitive plant cell death in trypan blue–stained leaves is marked with black arrows. Accumulation of H2O2 at interaction sites 32 hr after inoculation of leaves in the dark was measured using DAB and is shown at ×200 magnification ([C] and [D], top row). The inset in the upper left corner of the eds1 image at (C) shows an enlarged view of DAB staining restricted to the pathogen penetration site. Images are representative of four independent experiments using at least five leaves per genotype per experiment.
EDS1 and PAD4 Function in RCD Is Downstream of, or Independent from, the Local HR and Associated ROI Accumulation
An oxidative burst giving rise to local ROI accumulation is an early event associated with the plant HR (Bestwick et al., 1997; Shirasu et al., 1997; Thordal-Christensen et al., 1997). Also, O2−· is necessary and sufficient for lsd1 RCD (Jabs et al., 1996). We examined the production of ROI in wild-type and mutant plants at the point of pathogen penetration to determine whether the effects of eds1, pad4, or ndr1 on lsd1-induced lesion propagation could be related to deficiencies in early ROI accumulation during the HR. Excised leaves were dipped in a solution of 3,3-diaminobenzidine (DAB) to visualize H2O2 and then inoculated with a 10-μL droplet of avirulent Pp conidia, or they were dipped into suspensions of P. syringae. DAB polymerizes as a brown precipitate on contact with H2O2 in the presence of peroxidase (Shirasu et al., 1997; Thordal-Christensen et al., 1997), thus providing a useful marker for total peroxide accumulation.
The results from this analysis are shown in Figures 4C and 4D and are summarized in Table 1. A plant oxidative burst producing detectable local concentrations of H2O2 was observed only in plant genotypes undergoing an HR. Thus, eds1 (and eds1/lsd1) plants challenged with Pp Noco2 or DC3000/avrRps4, in which resistance is suppressed, failed to elicit an oxidative burst or an HR. pad4 (and pad4/lsd1) plants generated high levels of H2O2 and also developed either trailing necrosis or an HR, depending on the pathogen challenge. We conclude that EDS1 activity is required for the oxidative burst in EDS1-dependent R gene–mediated responses, whereas PAD4 functions either downstream or independently of ROI accumulation in the same responses. Both eds1 and pad4 plants produced a wild-type RPM1-mediated oxidative burst and HR after challenge with DC3000/avrRpm1 (Table 1). Thus, neither EDS1 nor PAD4 is required for the HR-associated oxidative burst in this EDS1-independent pathway. Yet, both are required for RCD in any of the tested contexts. Significantly, therefore, the requirements for EDS1 and PAD4 during lsd1-dependent RCD are unrelated to their effects on local R gene–mediated HR. We conclude from these results that EDS1 and PAD4 provide necessary signaling functions leading to lsd1 RCD that are either downstream or independent of the local HR and associated ROI accumulation.
Interestingly, ndr1 exhibited a reduction in the intensity of HR-associated DAB staining compared with that of Col-0 in response to Pp Emoy2, even though more host cells died, as measured by trypan blue (Figure 4D). This is in contrast to the enhanced H2O2 accumulation in lsd1 and lsd1c (Figures 4C and 4D). The response of ndr1/lsd1c plants was intermediate between that of ndr1 and lsd1c alone, and the RCD boundary was less well defined than it was in wild-type plants (Figure 4D). These data suggest that the reduced ROI production in ndr1 may be responsible for the attenuated RCD observed in the ndr1/lsd1c double mutant.
Fate of Superoxide at HR Margins and RCD Sites Is Different
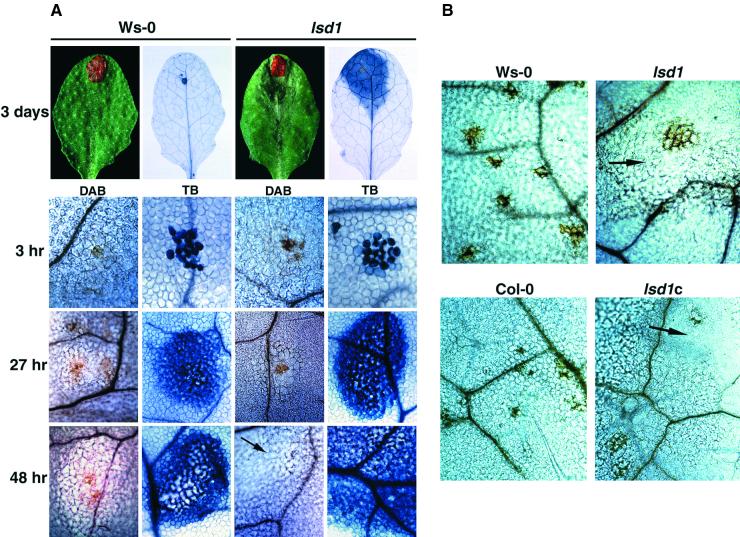
Because provision of O2−· is both necessary and sufficient to induce RCD in lsd1 plants (Jabs et al., 1996), we addressed directly whether EDS1, PAD4, and NDR1 act as signaling intermediates between superoxide and the LSD1-controlled cell death pathway. We elicited superoxide production by applying a discrete spot of rose bengal (RB) to leaves. In the presence of light, RB generates singlet oxygen that reduces to superoxide, which is rapidly dismutated to the more stable H2O2 (Knox and Dodge, 1984; Baker and Orlandi, 1995). RB-induced plant cell death was confined to the application site in wild-type Ws-0 leaves, but it induced RCD in lsd1 leaves, as shown in trypan blue–stained leaves at 3 DPI (Figure 5A). We assessed H2O2 accumulation over a time course (3 to 48 hr) of RB treatment. Within 3 hr, we observed intense DAB and trypan blue staining in the area of RB application (Figure 5A). From 27 hr onward, cell death foci were fixed in Ws-0 but expanded in lsd1 (Figure 5A). In several independent experiments, RB treatments of wild-type, eds1/lsd1, pad4/lsd1, and ndr1/lsd1 plants failed to elicit RCD (Table 1). The same responses were observed in leaves of all genotypes infiltrated with a xanthine/xanthine oxidase superoxide-generating system that was previously shown to induce lesions in lsd1 (Jabs et al., 1996; data not shown).
Figure 5.
Localized H2O2 Accumulation in Wild-Type and lsd1 Leaves after RB Application or Pp Infection.
(A) A single 5-μL spot of 20 mM RB was applied to leaves of 4-week-old plants. H2O2 accumulation was measured over 7 days by staining with DAB. Leaves also were stained with lactophenol–trypan blue (TB) to measure the extent of plant cell death. Localized RB application gives rise to local H2O2 accumulation associated with a discrete patch of dead plant cells. In Ws-0, expansion of plant cell necrosis ceases by 27 hr, whereas in lsd1, the lesions expand. DAB-staining material is detected in the area of RB application but is not associated with the spreading RCD in lsd1 (black arrows).
(B) Leaves of 4-week-old plants were inoculated with a 10-μL droplet of 5 × 104 spores/mL Noco2 (Ws-0 and lsd1) or Emoy2 (Col-0 and lsd1c). Leaves were photographed 5 days after treatment. H2O2 accumulation, measured by DAB staining, was detected at plant–pathogen interaction sites but was not associated with spreading lesions, seen here as clear, unstructured cells (black arrows). Images are representative of three independent experiments using eight leaves per genotype per time point.
In the earlier analysis by Jabs et al. (1996), superoxide accumulation was observed in live plant cells bordering the RCD lesions of lsd1 leaves. We expected to see DAB precipitation at the leading margins of lsd1 lesions that would be generated upon dismutation of O2−· to H2O2. However, there was no detectable H2O2 accumulation associated with lsd1 RCD lesions after either RB-induced cell death (Figure 5A) or Pp inoculation (Figure 5B). Superoxide production, measured by nitroblue tetrazolium (NBT) staining, was not detected at any point associated with the Pp-induced HR or RB-induced cell death (data not shown). Superoxide accumulation, however, was observed at the boundaries of developing lesions in lsd1, confirming previous results (Jabs et al., 1996; data not shown). These results suggest that the fate of superoxide generated as a component of the R gene–dependent HR is different from that produced in association with RCD in lsd1.
DISCUSSION
We demonstrate that EDS1 and PAD4, two positive regulators of plant disease resistance, are essential components of a cell death control pathway regulated by LSD1 in response to pathogen infection, BTH application, or provision of superoxide. Most importantly, the requirement for EDS1 and PAD4 during lsd1 RCD is independent of their roles as mediators of various R gene functions. Additionally, NDR1, a third disease resistance signaling component, contributes to lsd1 RCD during these responses.
EDS1 and PAD4 Potentiate Plant Defense Signaling
Our most important conclusion is that the requirements for EDS1 and PAD4 in lsd1 lesion formation are separable from their roles in localized R gene–mediated plant cell death, as shown in the model in Figure 6. For example, neither EDS1 nor PAD4 function in RPM1 resistance, yet both are required for RCD after RPM1 stimulation in lsd1. EDS1, but not PAD4, is necessary for ROI production and HR after local RPS4- or RPP1-mediated pathogen recognition, yet both EDS1 and PAD4 are required for lsd1 RCD in these responses. We suggest that the activities of EDS1 and PAD4 leading to lesion formation in lsd1 are in defense signal potentiation, downstream or independent of the HR (Figure 6). The finding that eds1 and pad4 suppress lsd1 RCD in response to applications of BTH, a functional mimic of the plant resistance signaling molecule SA, is consistent with this idea. Other studies have shown the involvement of SA in signal potentiation during local and systemic plant defenses (Shirasu et al., 1997; Delledonne et al., 1998; Klessig et al., 2000; Martinez et al., 2000). EDS1 (Falk et al., 1999; Feys et al., 2001) and PAD4 (Zhou et al., 1998; Jirage et al., 1999) operate upstream of SA accumulation during resistance responses in which they are required. In these contexts, their expression levels are enhanced by the application of SA, suggesting that EDS1 and PAD4 are regulated by SA-dependent positive feedback (Falk et al., 1999; Jirage et al., 1999; Feys et al., 2001). We suggest that the flux through this feedback is regulated by LSD1 (Figure 6). Experiments are in progress to determine directly the role of SA and the SA response regulator NPR1/NIM1 (Cao et al., 1997; Ryals et al., 1997) in lsd1-dependent RCD. Notably, ndr1 did not significantly suppress lesioning in lsd1 after BTH treatment. This indicates that, in contrast to EDS1 and PAD4, NDR1 is not essential for BTH signaling (and presumably SA signaling) in relation to LSD1-regulated plant cell death.
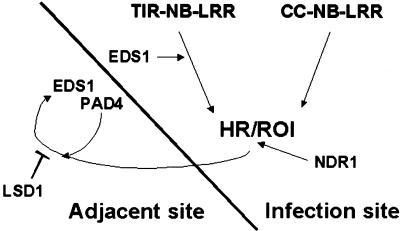
Figure 6.
A Model Positioning EDS1, PAD4, and NDR1 in Relation to LSD1 in Plant Defenses.
As shown in the model, RCD in lsd1 mutants is initiated in tissues adjacent to pathogen infection foci. The roles of EDS1, PAD4, and NDR1 in lsd1 RCD are separable from events controlling the plant HR and its accompanying oxidative burst (ROI) that are elicited upon avirulent pathogen recognition. EDS1, but not PAD4, functions upstream of localized HR and ROI production in resistance conditioned by TIR-NB-LRR–type R genes. In contrast, resistance conditioned by CC-NB-LRR–type R genes operates independently of EDS1 or PAD4 but requires NDR1. Irrespective of the different requirements for EDS1 and PAD4 at initial infection foci, both components are essential for signal relay leading to RCD in lsd1. Because EDS1 and PAD4 are also necessary for lsd1 RCD in response to the artificial provision of ROI or an active SA analog, we propose that EDS1 and PAD4 regulate a ROI/SA-dependent defense signal amplification loop. Flux through this loop is modulated by LSD1. NDR1 also is required for maximal lesion development in lsd1 plants in response to pathogens. The data suggest that NDR1 acts more proximally by regulating ROI balance and transduction of ROI-dependent signals at infection sites (see Discussion for more details).
EDS1, PAD4, and NDR1 Mediate ROI-Dependent Signaling
It was shown previously that RCD in lsd1 plants can be triggered by superoxide furnished by local applications of xanthine and xanthine oxidase (Jabs et al., 1996). Also, superoxide accumulation preceded lesion formation in lsd1 tissue and was detectable in cells bordering the developing lesion by specific NBT staining (Jabs et al., 1996). Thus, LSD1 activity appears to monitor a superoxide-dependent signal. Here, we show that EDS1 and PAD4, and interestingly also NDR1, mediate the ROI-derived signal leading to lsd1 RCD. The most compelling evidence for this is the failure of the eds1/lsd1, pad4/lsd1, and ndr1/lsd1c plants to initiate spreading lesions after local provision of superoxide, supplied either by RB (Table 1) or xanthine/xanthine oxidase applications. These data imply that all three disease resistance regulators express this particular function in unchallenged cells.
The activities of EDS1 and PAD4 in ROI signaling leading to RCD, therefore, are genetically distinct from their roles during the oxidative burst associated with a pathogen-induced HR (Figure 6). This finding strengthens the notion that EDS1 and PAD4 have a second function operating downstream or independently of the HR. We postulate that this second function helps establish the signal normally required to initiate lsd1 RCD. However, three observations suggest a different role for NDR1 in ROI signaling. (1) ndr1 attenuated the oxidative burst during the HR through RPS4 or RPP4, whereas it enhanced the oxidative burst during the HR through RPM1 pathogen recognition. (2) In all of these plant–pathogen combinations, ndr1 diminished lsd1 RCD. (3) NDR1 is not required for lsd1 lesions in response to BTH but is required for lesion development in response to ROI provision. These three points lead us to conclude that NDR1 is important in regulating the local ROI status (Figure 6). Imbalances in this system are likely to affect the efficiency of the HR and consequent local signaling and probably drive RCD in lsd1. Recent studies reveal that the balance of ROI, most particularly O2−·, H2O2, and NO, is crucial for the establishment of the HR (Delledonne et al., 1998, 2001; Klessig et al., 2000).
We propose that EDS1 and PAD4 are regulators of ROI- and SA-dependent signaling in a plant defense potentiation circuit. We suggest that NDR1 is required more proximally for the control of ROI generation and the transduction of a ROI-derived signal at the initial interaction site. In this respect, it is interesting that EDS1 and PAD4, but not NDR1, are components of a basal resistance pathway that limits the growth of virulent pathogens in the absence of plant cell death (Glazebrook et al., 1996; Parker et al., 1996; Reuber et al., 1998). It is conceivable that basal resistance is a reflection of the EDS1 and PAD4 resistance-potentiating activities demonstrated here. Recent analyses revealed a requirement for EDS1 and PAD4 in constitutive SA-dependent resistance pathways induced by the cpr1 and cpr6 mutations (Clarke et al., 2001; Jirage et al., 2001) that is also consistent with resistance-potentiating roles.
ROI Requirements Differ between the HR and LSD1-Controlled Plant Cell Death
Our analysis of ROI accumulation suggests that the nature of ROIs produced by cells undergoing the HR is different from that of ROIs associated with signaling from those cells, and monitored by LSD1. We detected superoxide, but not H2O2, in living cells bordering spreading lsd1 lesions, as shown previously (Jabs et al., 1996). Our failure to observe H2O2 at these margins was surprising, because superoxide would be expected to dismutate to H2O2. LSD1 is required for the SA-dependent induction of antioxidant copper-zinc superoxide dismutase (Cu-Zn SOD) (Kliebenstein et al., 1999) and potentially other antioxidant genes. Thus, a simple explanation is that there is no, or there is delayed, accumulation of Cu-Zn SOD in lsd1 and hence no dismutation. This simple model is weakened by the unlikelihood that Cu-Zn SOD operates in the apoplast, where O2−· is first produced during the oxidative burst (Bolwell, 1999).
Another possibility is that superoxide produced by cells at HR margins, where LSD1 is proposed to function, is converted to something other than H2O2. This could reflect an interplay between O2−· with other ROI molecules, SA, or antioxidant systems. In animal cells, superoxide can react with NO to produce peroxynitrite (ONOO−), a highly reactive redox species that serves as a signal or as a cytotoxic agent, depending on its level and the availability of other redox molecules (Bonfoco et al., 1995; Lin et al., 1995). Delledonne et al. (2001) propose that O2−· production and its dismutation to H2O2 regulate a balance of H2O2/NO that, when disturbed, leads to HR. They argue against a direct role in cell killing for ONOO−. An alternate explanation is that the H2O2 produced is locally unavailable for polymerization with DAB. This could be caused by changes in the cellular pH specific to these mutant backgrounds (DAB staining is effective only at pH values between 5.5 and 6.0; Thordal-Christensen et al., 1997) or by a surge of ROI scavenging enzymes (Vanacker et al., 2000). Cells are permeable to DAB and H2O2 (Thordal-Christensen et al., 1997), ruling out the possibility that H2O2 generated within the cell would be inaccessible for detection. Our data clearly suggest that the fate of superoxide produced in cells undergoing an HR is different from that generated locally during lsd1 lesion development. This implies that signaling extending from infected cells is controlled differently than it is in the infected cells themselves.
Conversely, we observed H2O2, but not superoxide, at infection foci. Superoxide is an unstable redox molecule that rapidly dismutates enzymatically or nonenzymatically to H2O2 (Lamb and Dixon, 1997). Overwhelming evidence suggests that production of superoxide at the cell surface is the proximal event in the plant oxidative burst (Bolwell, 1999). However, other extracellular and intracellular mechanisms may contribute to ROI generation during the oxidative burst (Allan and Fluhr, 1997; Martinez et al., 1998; Bolwell, 1999). The transience of the oxidative burst and the inherent instability of superoxide may account for our failure to observe NBT-reactive material at infection sites or in cells supplied with superoxide by exogenous RB application. RB was applied onto the leaf surface and therefore would release superoxide into the plant apoplast that would be accessible to NBT (Baker and Orlandi, 1995).
Putative Signaling Functions of EDS1, PAD4, and LSD1
Our results draw an important genetic link between the disease resistance–promoting functions of EDS1, PAD4, and NDR1 and the negative regulation of plant cell death exerted by LSD1 (Figure 6), raising questions about the biochemical roles of these proteins in healthy and pathogen-challenged plants. LSD1 encodes a zinc finger protein with similarity to the GATA-type family of transcription factors. EDS1 and PAD4 share homology with the catalytic domains of eukaryotic lipases (Falk et al., 1999; Jirage et al., 1999), although hydrolytic activities have not been demonstrated. It is possible, therefore, that they process ROI-activated signal intermediates spreading from infected to surrounding noninfected cells to perpetuate plant defense responses. In animals (Serhan et al., 1996; Stafforini et al., 1997) and plants (Farmer et al., 1998; Sanz et al., 1998; Rustérucci et al., 1999), activated fatty acids are important signaling molecules produced in response to certain pathogens and after wounding. Thus, EDS1 and PAD4 may potentiate resistance by processing ROI- and SA-activated molecules. The production of such molecules, whether lipid based or otherwise, would normally lead to cell death only if their levels passed a cell death control threshold. Obviously, in an lsd1 null mutant, these levels need not be high to initiate RCD. The biochemical role of NDR1 also remains to be resolved, although its potential membrane association (Century et al., 1997) may be important in regulating cellular communication between external and internal redox systems. Elucidating the activities, cellular localization, and molecular associations of all of these signaling components should provide important insights into their precise functions in plant disease resistance.
METHODS
Plant Material and Cultivation
The origins of eds1-1 (Parker et al., 1996) and lsd1 (Dietrich et al., 1997) in accession Wassilewskija (Ws-0) have been described previously. The pad4-5 T-DNA insertion mutant also was isolated in Ws-0 (Feys et al., 2001). The T-DNA is inserted 35 bp 5′ to the end of the single intron in the PAD4 gene. The ndr1-1 mutant line in accession Columbia (Col-0) (Century et al., 1997) was kindly provided by Dr. Brian Staskawicz (University of California, Berkeley). Seed were sown on low nutrient compost and grown in a chamber under a light period of 8 hr (∼160 μE·m−2·sec−1) at 22°C and 65% relative humidity (RH).
F2 plants derived from selfed F1 plants were genotyped for the lsd1 mutation by polymerase chain reaction (PCR) using a triple primer set (5′-ACCTAACAAAAAGAAAAGTGTGTGAGG-3′, 5′-ATAATAAACCCTACTAGCTCTAACAAG-3′, and 5′-CTGCTACTTTCATCCAAAC-3′). The wild-type LSD1 allele produces a 940-bp product, whereas lsd1 gives a 600-bp product. The Col-0 allele of lsd1 (lsd1c) was constructed by introgressing the Ws-0 allele into a Col-0 line over seven generations and selecting for the mutant allele using the lsd1 PCR described above. The ndr1/lsd1c double mutant was constructed by crossing ndr1-1 plants with lsd1c, selfing the F1 plants, and genotyping the segregating F2 plants for the lsd1 mutation (described above) and the ndr1-1 mutation by using the primer set 5′-GGGACGGTTTCAATTCTGTGATAG-3′ and 5′-CGAGATTGCTCATTGCCATTGG-3′. The eds1-1 mutation was detected in eds1-1 × lsd1 F2 plants using the primer set 5′-GGATAGAAGATGAATACAAGCC-3′ and 5′-ACCTAAGGTTCAGGTATCTGT-3′. PCR products were digested for at least 4 hr with Mse1, and products were resolved on a 2% agarose gel. Cleavage of wild-type EDS1 produces three visible bands of 280, 180, and 150 bp, whereas eds1-1 gives visible products of 240, 180, and 150 bp. PCR-based selection of the pad4-5 mutant allele in pad4-5 × lsd1 F2 plants was as described (Feys et al., 2001). In the initial characterizations of mutant phenotypes, we examined several independent mutant lines. All behaved similarly; hence, more detailed analyses were performed with one representative single and double mutant per genotype.
Pathogen Isolates and Growth Determinations
Peronospora parasitica (Pp) isolates Noco2, Emoy2, and Emwa1 were maintained on the genetically susceptible Arabidopsis accessions Col-0, Oystese, and Ws-0, respectively, as described previously (Dangl et al., 1992). To determine disease symptom development, Pp conidiospores were suspended in water (4 × 104 spores/mL) and sprayed onto 10-day-old (for cotyledon assays) or 4-week-old (for leaf assays) plants. Inoculated plants were kept under a sealed propagator lid to achieve high RH in a growth chamber at 19°C under an 8-hr light period (100 to 160 μE·m−2·sec−1). Alternatively, a 10-μL droplet of Pp conidiospores was placed on the leaf surface, and plants were incubated for up to 7 days under the same conditions as used for Pp growth assays.
Bacterial pathogen induction of runaway cell death (RCD) was measured by infiltrating suspensions (105 colony-forming units/mL) of Pseudomonas syringae pv tomato DC3000 expressing either avrRps4 or avrRpm1 into one side of the leaf using a 1.5-mL needleless syringe. Plants were inspected for disease symptoms and/or spreading lesion formation over 6 days under the same conditions as described for the growth assays. Hypersensitive response (HR) tests were performed using 5 × 107 colony-forming units/mL. Growth of P. syringae pv tomato DC3000 expressing avrRps4 or avrRpm1 in the various lines was determined by dip inoculation and subsequent growth analysis essentially as described (Innes et al., 1993) with modifications (P. Tornero and J.L. Dangl, unpublished data). Briefly, pots containing 2-week-old plants were immersed for 10 to 15 sec in a suspension containing 2.5 × 107 colony-forming units/mL (OD600 = 0.05) and Silwet (200 μL/L). Plants were kept under high humidity for 1 hr, after which time measurement zero was taken. At time 0 and 3 days, bacteria were extracted from the plant tissue and grown on selective agar plates to determine concentration.
Benzothiodiazole Induction of lsd1 RCD
For chemical induction of RCD, leaves of 4-week-old plants were sprayed with 0.35 mM benzothiodiazole (BTH), which was provided as a gift from Syngenta (Research Triangle Park, NC). Plants were maintained under normal growth conditions and inspected for lesion development over 6 days.
Histochemical Analysis of Plant Cell Death and Pp Development
Plant cell necrosis induced by pathogen inoculation or chemical treatment, as well as the development of Pp mycelium inside cotyledon or leaf tissues, was monitored by staining with lactophenol–trypan blue and destaining in saturated chloral hydrate as described (Koch and Slusarenko, 1990). Material was mounted on a slide in 60% glycerol and examined using a light microscope (Axiophot; Zeiss, Jena, Germany). Excised leaves were manipulated in parallel with those used for detection of hydrogen peroxide (H2O2) and maintained under the same conditions (see below).
Histochemical Detection of H2O2 at Interaction Sites
Detection of H2O2 was by endogenous peroxidase-dependent in situ histochemical staining using 3,3-diaminobenzidine (DAB) in a protocol modified from Thordal-Christensen et al. (1997). Leaves of 4-week-old plants were inoculated with a 10-μL droplet of Pp conidiospores placed on the leaf surface. Leaves were then excised and supplied through the cut petiole with a solution of 1 mg/mL DAB for 8 hr in light (100 to 160 μE·m−2·sec−1) or in darkness under the same conditions used to determine Pp growth. Subsequently, the DAB solution was replaced with water, and leaves were maintained under the same conditions as before. For assessment of H2O2 accumulation at P. syringae infection sites, excised leaves were allowed to take up DAB solution for 8 hr and then were dipped in bacterial suspensions and incubated as described for the bacterial growth assays except that leaves were kept in the dark. At different times after pathogen inoculation, leaves were cleared for 5 min in boiling acetic acid/glycerol/ethanol (1:1:3 [v/v/v]) solution. Material was mounted on a slide in 60% glycerol and examined using a light microscope (Axiophot; Zeiss). H2O2 was detectable as reddish brown coloration.
Chemical Provision of ROI in Leaves
Rose bengal (4,5,6,7-tetrachloro-2′,4′,5′,7′-tetraiodofluorescein [RB]; Sigma) is an efficient singlet molecular oxygen (1O2) producer in aqueous solution (Knox and Dodge, 1984). 1O2 gives rise to radical anion superoxide (O2−·) and subsequently to H2O2. RB was applied as a droplet of 10 μL (20 mM solution) onto the surface of excised leaves of 4-week-old plants. These were placed in a growth chamber in the light (160 to 200 μE·m−2·sec−1) for at least 3 hr after RB treatment and maintained for several days under an 8-hr photoperiod at 19°C and 65% RH. Xanthine and xanthine oxidase coinfiltration in leaves of 4- or 5-week-old plants was used to generate superoxide, as described previously (Jabs et al., 1996). Infiltrated plants were maintained under normal plant growth conditions.
Acknowledgments
We thank Jeff Chang for critical comments on the manuscript. Research at the Sainsbury Laboratory is funded by the Gatsby Charitable Foundation. C.R. is a recipient of a Marie Curie postdoctoral research training fellowship from the European Commission. Work at the University of North Carolina, Chapel Hill, was supported by National Institutes of Health Grant 5RO1-GM057171-01 to J.L.D. and by support through the University of North Carolina Curriculum in Genetics National Institutes of Health Training Grant T32 GM07092-26 to D.H.A.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene–mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, A.C., and Fluhr, R. (1997). Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9, 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.J., and Orlandi, E.W. (1995). Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 33, 299–321. [DOI] [PubMed] [Google Scholar]
- Bestwick, C.S., Brown, I.R., Bennett, M.H.R., and Mansfield, J.W. (1997). Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 9, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell, G.P. (1999). Role of active oxygen species and NO in plant defense responses. Curr. Opin. Plant Biol. 2, 287–294. [DOI] [PubMed] [Google Scholar]
- Bonfoco, E., Krainc, D., Ankarcrona, M., Nicotera, P., and Lipton, S.A. (1995). Apoptosis and necrosis: Two distinct events induced, respectively, by mild and intense insults with N-methyl-d-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc. Natl. Acad. Sci. USA 92, 7162–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges, R., et al. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Aarts, N., Feys, B.J., Dong, X., and Parker, J.E. (2001). Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 26, 409–420. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., Holub, E.B., Debener, T., Lehnackers, H., Ritter, C., and Crute, I.R. (1992).Genetic definition of loci involved in Arabidopsis-pathogen interactions. In Methods in Arabidopsis Research, C. Koncz, N.H. Chua, and J. Schell, eds (Singapore: World Scientific Publishers), pp. 393–418.
- Dangl, J.L., Dietrich, R.A., and Richberg, M.H. (1996). Death don't have no mercy: Cell death programs in plant–microbe interactions. Plant Cell 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne, M., Xia, Y.J., Dixon, R.A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Richberg, M.H., Schmidt, R., Dean, C., and Dangl, J.L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. [DOI] [PubMed] [Google Scholar]
- Durner, J., Wendehenne, D., and Klessig, D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95, 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene–mediated disease resistance in Arabidopsis, has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E., Weber, H., and Vollenweider, S. (1998). Fatty acid signaling in Arabidopsis. Planta 206, 167–174. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., and Parker, J.E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidosis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (1999). Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 2, 280–286. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K.-H., Oostendorp, M., Staub, T., Ward, E., Kessmann, H., and Ryals, J. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, J.J., and Loake, G.J. (2000). Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 124, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Gray, J., Close, P.S., Briggs, S.P., and Johal, G.S. (1997). A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89, 25–31. [DOI] [PubMed] [Google Scholar]
- Holub, E.B., Beynon, J.L., and Crute, I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 7, 223–239. [Google Scholar]
- Innes, R.W., Bisgrove, S.R., Smith, N.M., Bent, A.F., Staskawicz, B.J., and Liu, Y.-C. (1993). Identification of a disease resistance locus in Arabidopsis that is functionally homologous to the RPG1 locus of soybean. Plant J. 4, 813–820. [DOI] [PubMed] [Google Scholar]
- Jabs, T., Dietrich, R.A., and Dangl, J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. [DOI] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Zhou, N., Cooper, B., Clarke, J.D., Dong, X., and Glazebrook, J. (2001). Constitutive salicylic acid–dependent signaling in cpr1 and cpr6 mutant requires PAD4. Plant J. 26, 395–407. [DOI] [PubMed] [Google Scholar]
- Jones, D.A. (2000). Resistance genes and resistance protein functions. In Molecular Plant Pathology, Vol. 4, M. Dickinson and J. Beynon, eds (Sheffield, UK: Academic Press), pp. 108–143.
- Klessig, D.F., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Dietrich, R.A., Martin, A.C., Last, R.L., and Dangl, J.L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 12, 1022–1026. [DOI] [PubMed] [Google Scholar]
- Knox, J.P., and Dodge, A.D. (1984). Photodynamic damage to plant tissue by rose bengal. Plant Sci. Lett. 37, 3–7. [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lin, K.-T., Xue, J.-Y., Nomen, M., Spur, B., and Wong, P.Y.-K. (1995). Peroxynitrite-induced apoptosis in HL-60 cells. J. Biol. Chem. 270, 16487–16490. [DOI] [PubMed] [Google Scholar]
- Martinez, C., Montillet, J.L., Bresson, E., Agnel, J.-P., Dai, G.H., Daniel, J.F., Geiger, J.P., and Nicole, M. (1998). Apoplastic peroxidase generates superoxide anions in cells of cotton cotyledons undergoing the hypersensitive reaction to Xanthomonas campestris pv. malvacearum race 18. Mol. Plant-Microbe Interact. 11, 1038–1047. [Google Scholar]
- Martinez, C., Baccou, J.-C., Bresson, E., Baissac, Y., Daniel, J.-F., Jalloul, A., Montillet, J.-L., Geiger, J.-P., Assigbetsé, K., and Nicole, M. (2000). Salicylic acid mediated by the oxidative burst is a key molecule in local and systemic responses of cotton challenged by an avirulent race of Xanthomonas campestris pv malvacearum. Plant Physiol. 122, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., and Dangl, J.L. (2000). Signal transduction in the plant immune response. Trends Biochem. Sci. 25, 79–82. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Cuzick, A., Can, C., Beynon, J., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., and Dangl, J.L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Mur, L., Bi, Y.-M., Darby, R.M., Firek, S., and Draper, J. (1997). Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J. 12, 1113–1126. [DOI] [PubMed] [Google Scholar]
- Nawrath, C., and Métraux, J.-P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Feys, B.J., Van der Biezen, E.A., Noël, N., Aarts, N., Austin, M.J., Botella, M.A., Frost, L.N., Daniels, M.J., and Jones, J.D.G. (2000). Unravelling R gene–mediated disease resistance pathways in Arabidopsis. Mol. Plant Pathol. 1, 17–24. [DOI] [PubMed] [Google Scholar]
- Reuber, T., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E., and Ausubel, F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci, C., Montillet, J.-L., Agnel, J.-P., Battesti, C., Alonso, B., Knoll, A., Bessoule, J.-J., Etienne, P., Suty, L., Blein, J.-P., and Triantaphylides, C. (1999). Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death by cryptogein on tobacco leaves. J. Biol. Chem. 274, 36446–36455. [DOI] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IkB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, A., Moreno, J.I., and Castresana, C. (1998). PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell 10, 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H.H.W., and Walter, U. (1994). NO at work. Cell 78, 919–925. [DOI] [PubMed] [Google Scholar]
- Serhan, C.N., Haeggström, J.Z., and Leslie, C.C. (1996). Lipid mediator networks in cell signaling: Update and impact of cytokines. FASEB J. 10, 1147–1158. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafforini, D.M., McIntyre, T.M., Zimmerman, G.A., and Prescott, S.M. (1997). Platelet-activating factor acetylhydrolases. J. Biol. Chem. 272, 17895–17898. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z.G., Wei, Y.D., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Vanacker, H., Carver, T.L.W., and Foyer, C.H. (2000). Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hypersensitive response in the barley–powdery mildew interaction. Plant Physiol. 123, 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.O., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]