Abstract
The copines are a newly identified class of calcium-dependent, phospholipid binding proteins that are present in a wide range of organisms, including Paramecium, plants, Caenorhabditis elegans, mouse, and human. However, the biological functions of the copines are unknown. Here, we describe a humidity-sensitive copine mutant in Arabidopsis. Under nonpermissive, low-humidity conditions, the cpn1-1 mutant displayed aberrant regulation of cell death that included a lesion mimic phenotype and an accelerated hypersensitive response (HR). However, the HR in cpn1-1 showed no increase in sensitivity to low pathogen titers. Low-humidity-grown cpn1-1 mutants also exhibited morphological abnormalities, increased resistance to virulent strains of Pseudomonas syringae and Peronospora parasitica, and constitutive expression of pathogenesis-related (PR) genes. Growth of cpn1-1 under permissive, high-humidity conditions abolished the increased disease resistance, lesion mimic, and morphological mutant phenotypes but only partially alleviated the accelerated HR and constitutive PR gene expression phenotypes. The disease resistance phenotype of cpn1-1 suggests that the CPN1 gene regulates defense responses. Alternatively, the primary function of CPN1 may be the regulation of plant responses to low humidity, and the effect of the cpn1-1 mutation on disease resistance may be indirect.
INTRODUCTION
The copines are a highly conserved class of proteins found in a wide variety of organisms, including ciliates, plants, Caenorhabditis elegans, mouse, and human (Creutz et al., 1998). The copines are defined as a protein class by their unique combination of two C2 domains in the N-terminal region and an A domain in the C-terminal region. C2 domains are present in a large number of proteins, including protein kinase C (Azzi et al., 1992), phospholipase C (Essen et al., 1996), synaptotagmin (Brose et al., 1995), Doc2 (Duncan et al., 2000), rabphilin (Wang et al., 2000), and Munc13 (Brose et al., 1995). The C2 domain appears to confer Ca2+-dependent phospholipid binding activity to proteins, and mammalian copine I has Ca2+-dependent phospholipid binding activity (Creutz et al., 1998). The A domain may be involved in extracellular protein–protein interactions, as in the case of the von Willebrand factor (Williams et al., 1999). However, available biochemical evidence indicates that copines are intracellular proteins (Creutz et al., 1998; Tomsig and Creutz, 2000). Finally, biochemical analysis of human copine III indicates that it has an intrinsic kinase activity that may be novel, because no homology with known kinase catalytic domains exists in copine III (Caudell et al., 2000). Together, these biochemical data raise the intriguing possibility that copines represent a new group of Ca2+-dependent signal transduction proteins that might be involved in membrane trafficking. However, the biological functions of the copines are unknown at present.
Calcium is an important second messenger involved in a variety of plant responses to stimuli, including pathogens, drought, light, hormones, touch, cold, heat, and oxidative stress (Reddy, 2001). In plant disease resistance responses, Ca2+ acts as a signaling molecule, communicating primary recognition events to multiple downstream responses (Grant et al., 2000) such as phytoalexin production, induction of defense genes (Zimmermann et al., 1997), and the hypersensitive response (HR) (Xu and Heath, 1998). In parsley cells, an elicitor-responsive calcium channel has been identified and characterized (Zimmermann et al., 1997), and a transient influx of Ca2+ and H+ and an efflux of K+ and Cl− occur within minutes of fungal elicitor addition (Hahlbrock et al., 1995; Yang et al., 1997). Pharmacological studies with tobacco By2 cells revealed that the HR requires serine proteases, calcium, and protein kinases (Sasabe et al., 2000).
The HR is a programmed cell death response: it is an active process involving transcription and translation, is under genetic control, and has some similarities to mammalian apoptosis (Mittler et al., 1997; del Pozo and Lam, 1998; Maleck and Lawton, 1998). Because of the strong association of the HR with plant disease resistance, the HR is postulated to play an important role in resistance to at least some kinds of pathogens and may serve to localize a pathogen (Goodman and Novacky, 1994). A number of mutants have been identified in Arabidopsis that exhibit precocious cell death, such as the lesion mimic mutants lsd1-lsd7, acd2, acd6, and cpr6 (Greenberg and Ausubel, 1993; Dietrich et al., 1994, 1997; Greenberg et al., 1994; Weymann et al., 1995; Rate et al., 1999). Many lesion mimic mutants are dependent on certain environmental conditions such as light, daylength, and RH for lesion development (Dietrich et al., 1994; Chamnongpol et al., 1996; Morel and Dangl, 1999; Yoshioka et al., 2001). Interestingly, the DND1 gene, which eliminates the HR and can cause a microscopic lesion phenotype when mutated, encodes a putative Ca2+ channel, providing another potential link between Ca2+ and cell death (Clough et al., 2000).
Many lesion mimic mutants exhibit a state of increased disease resistance called systemic acquired resistance (SAR) and show high, constitutive levels of pathogenesis-related (PR) gene expression (Dietrich et al., 1994). It is possible that lesion mimic mutants represent genes that regulate the HR. However, disruptions of cellular physiology apparently unrelated to disease defense also can trigger cell death and SAR (Mock et al., 1999; Molina et al., 1999). This fact can make it difficult to determine whether or not genes defined by lesion mimic mutants play a direct role in cell death signaling and control. In addition, many Arabidopsis mutants (cpr1, cpr5, edr1, mpk4) have been identified that show constitutive expression of defense-related genes in the absence of cell death (Bowling et al., 1994, 1997; Clarke et al., 1998; Frye et al., 2001; Petersen et al., 2000). This indicates that although the HR can trigger systemic PR gene expression and SAR, cell death is not always required for PR gene expression and SAR to occur.
Here, we describe a humidity-sensitive Arabidopsis mutant that exhibited precocious cell death, increased resistance to virulent bacterial and oomyceteous pathogens, and morphological abnormalities. On the basis of molecular analyses and complementation studies, we demonstrate that the phenotype of the mutant is caused by a mutation in a copine gene, CPN1.
RESULTS
cpn1-1 Is a Humidity-Sensitive Lesion Mimic Mutant
The cpn1-1 mutant was identified from a population of Arabidopsis mutagenized by T-DNA insertion (Weigel et al., 2000). The cpn1-1 mutant exhibited clear humidity-dependent developmental and growth abnormalities, as shown in Figure 1. cpn1-1 plants grown in low-humidity (LH; 35 to 45% RH), short-day (SD; 8-hr-light/16-hr-dark) growth conditions were smaller than their wild-type Columbia (Col-0) counterparts and had a more compact growth habit (Figures 1A and 1B). The leaves of LH-grown cpn1-1 mutants were curled, thicker than wild-type leaves, somewhat irregular in shape with short petioles, and occasionally had indentations at the margins. When cpn1-1 mutants were grown in high- humidity (HH; 75 to 85% RH), SD conditions, these developmental defects were absent and the cpn1-1 plants were morphologically indistinguishable from wild-type plants (Figures 1C and 1D). The phenotype was dependent on the age of the plants: cpn1-1 seedlings were indistinguishable from wild-type seedlings for the first 1 to 2 weeks of growth in LH, SD conditions; the developmental defects manifested after that time. When grown under LH, long-day (LD; 16-hr- light/8-hr-dark) conditions, cpn1-1 mutants were much smaller than Col-0 wild-type plants and had dramatically reduced floral apical dominance, shorter bolts, fewer flowers, and reduced seed yield (data not shown). Growth in HH, LD conditions rescued the cpn1-1 mutant, although the mutant was still slightly smaller than the wild type under these conditions (data not shown).
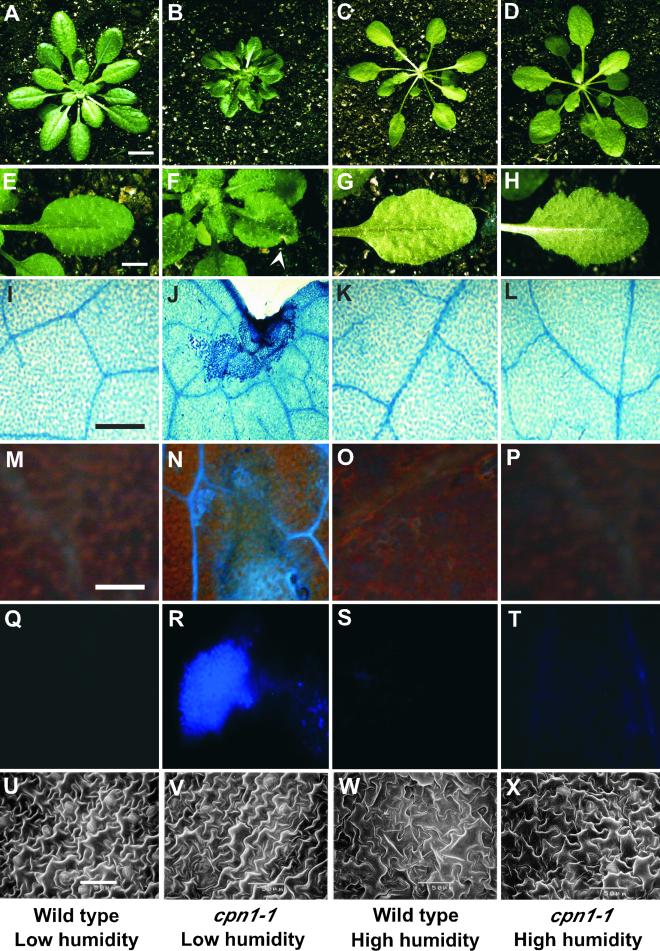
Figure 1.
Humidity-Dependent Developmental and Lesion Mimic Phenotypes of cpn1-1.
The four columns of images, from left to right, represent Col-0 wild type grown in LH, cpn1-1 grown in LH, Col-0 wild type grown in HH, and cpn1-1 grown in HH, respectively. All plants were grown in SD light conditions.
(A) to (D) Four-week-old whole plants. Bar in (A) = 1 cm for (A) to (D).
(E) to (H) Close-up images of leaves. Arrowhead in (F) denotes a lesion. Bar in (E) = 0.5 cm for (E) to (H).
(I) to (L) Sectors of leaves stained with the vital stain trypan blue. Bar in (I) = 200 μm for (I) to (L).
(M) to (P) UV autofluorescence images of leaf sectors. Bar in (M) = 100 μm for (M) to (P).
(Q) to (T) Leaf sectors stained with aniline blue to detect callose accumulation. The scale is the same as in (M) to (P).
(U) to (X) Scanning electron micrographs of the abaxial surfaces of leaves. Bar in (U) = 50 μm for (U) to (X).
The leaves of LH-grown cpn1-1 plants displayed small necrotic lesions (Figure 1F). The largest of these lesions usually occurred at or near the margins of the leaf. No lesions were observed in HH-grown cpn1-1 plants (Figure 1H). Lesions began to appear on the third set of true leaves after 2 weeks of growth in LH, SD conditions. These lesions occurred in cpn1-1 plants that had not been inoculated with a pathogen, and they occurred consistently in all cpn1-1 plants grown in LH. However, it was not clear whether or not these lesions were truly spontaneous, because it was not feasible to grow cpn1-1 mutants under axenic, LH conditions and monitor lesion development. The cpn1-1 lesions stained with trypan blue, indicating the presence of dead cells (Figure 1J). In addition, trypan blue staining revealed numerous microscopic lesions consisting of clusters of a few dead cells throughout LH-grown cpn1-1 leaves (Figure 1J and data not shown). No lesions or dead cells were observed in LH- or HH-grown Col-0 wild-type leaves or in HH-grown cpn1-1 leaves (Figures 1I, 1K, and 1L).
The leaves of LH-grown cpn1-1 plants showed accumulation of autofluorescent phenolic compounds in the areas near the lesions and in scattered patches throughout the leaf (Figure 1N). Autofluorescent phenolic compounds were not observed in LH- or HH-grown Col-0 plant leaves or in HH-grown cpn1-1 plant leaves (Figures 1M, 1O, and 1P). LH-grown cpn1-1 leaves also showed substantial accumulation of callose, as determined by aniline blue staining (Figure 1R), whereas LH- and HH-grown Col-0 wild-type plant leaves and HH-grown cpn1-1 plant leaves did not (Figures 1Q, 1S, and 1T). The accumulation of autofluorescent phenolic compounds and callose has been observed in a number of lesion mimic mutants and is a hallmark of plant stress, including pathogen stress (Bowling et al., 1994; Dietrich et al., 1994).
Because the cpn1-1 mutant was smaller than the Col-0 wild type when grown in LH, we speculated that the cell size might be reduced in cpn1-1. Light microscopic analysis of LH-grown cpn1-1 mutant leaves indicated that the cell size was reduced (Figure 1J and data not shown). In contrast, however, scanning electron microscopy of LH-grown cpn1-1 and Col-0 wild-type abaxial leaf epidermal cells showed no dramatic difference in cell size (Figures 1U and 1V). These observations imply that the mesophyll cells may be smaller in LH-grown cpn1-1 than in LH-grown wild type, whereas the abaxial epidermal cells in LH-grown cpn1-1 may be normal in size but fewer in number. The borders separating cells appeared somewhat thicker in LH-grown cpn1-1 leaves compared with LH-grown Col-0 wild-type leaves, possibly indicating the presence of thicker cell walls in LH-grown cpn1-1 plants. Abaxial epidermal cell size in HH-grown Col-0 wild-type and cpn1-1 mutant plant leaves was similar and larger than that observed in LH-grown wild-type and cpn1-1 plants (Figures 1W and 1X).
cpn1-1 Has an Accelerated Hypersensitive Cell Death Response
The development of necrotic lesions in cpn1-1 plants implies that the cpn1-1 mutation causes aberrant regulation of cell death. To investigate this possibility further, the time of HR onset after inoculation with avirulent Pseudomonas syringae pv maculicola (avrRpt2) (P.s.m. [avrRpt2]) was compared in cpn1-1 and Col-0 wild-type plants. The macroscopic HR can be induced by infiltrating a high concentration (108 colony-forming units [cfu]/mL) of avirulent bacteria into the intercellular spaces of the mesophyll of resistant plant leaves (Klement et al., 1963). The HR is observed as a massive collapse of infiltrated tissue within ∼12 to 36 hr after infiltration and involves the loss of membrane integrity and electrolyte leakage (Goodman, 1968, 1972).
Col-0 wild-type plants possess the RPS2 disease resistance gene and are able to recognize and resist P. syringae bacteria bearing the corresponding avrRpt2 avirulence gene through gene-for-gene–mediated disease resistance (Kunkel et al., 1993). cpn1-1 was isolated from the Col-0 genetic background and appeared to have a functional RPS2 gene (see below). cpn1-1 and Col-0 wild-type leaves were infiltrated with high concentrations of P.s.m. (avrRpt2) bacteria, and the appearance of the HR was monitored over time. Interestingly, both LH- and HH-grown cpn1-1 plants exhibited a faster HR than Col-0 wild-type plants grown under the same conditions, as measured by both the onset of macroscopic leaf collapse and electrolyte leakage, as shown in Figure 2.
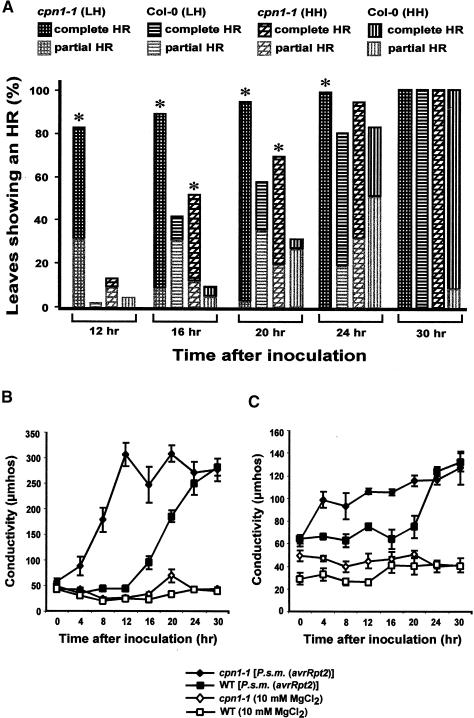
Figure 2.
Accelerated HR in cpn1-1.
The macroscopic HR was first observed at ∼12 hr after inoculation. Fifty to 100 leaves of each type were inoculated for each experiment. For details, see Methods.
(A) Proportions of inoculated Col-0 wild-type and cpn1-1 leaves exhibiting partial and complete collapse over time after inoculation. Asterisks indicate percentages of leaves with partial or complete HR collapse in cpn1-1 plants that are significantly higher than Col-0 wild-type plants grown under the same humidity conditions, at the same time during the experiment, using Student's t test. A leaf was scored as showing complete HR when the leaf had collapsed totally; a leaf was scored as showing partial HR when from 10 to 90% of the infiltrated leaf had collapsed. Leaves showing <10% collapse were not scored as having an HR.
(B) Electrolyte leakage from inoculated LH-grown Col-0 wild-type and cpn1-1 leaves over time after infiltration with P.s.m. (avrRpt2) or 10 mM MgCl2 (control).
(C) Electrolyte leakage from inoculated HH-grown Col-0 wild-type and cpn1-1 leaves over time after infiltration with P.s.m. (avrRpt2) or 10 mM MgCl2 (control).
WT, wild type.
Bars in (B) and (C) indicate standard errors.
LH-grown cpn1-1 plants had significantly more HR responses than LH-grown Col-0 wild-type plants at 12, 16, 20, and 24 hr after inoculation, whereas HH-grown cpn1-1 plants had significantly more HR responses than HH-grown Col-0 wild-type plants at 16 and 20 hr after infiltration (Figure 2A). By 30 hr after infiltration, all cpn1-1 and Col-0 wild-type leaves had collapsed. Significant electrolyte leakage could be detected from LH-grown cpn1-1 tissues infiltrated with P.s.m. (avrRpt2) within 4 to 8 hr after inoculation, whereas similar leakage from LH-grown Col-0 wild-type tissues did not occur until 16 to 20 hr after inoculation (Figure 2B). The peak of electrolyte leakage occurred at 12 hr after inoculation in LH-grown cpn1-1, whereas in LH-grown Col-0, the peak was observed at 24 hr after inoculation. In HH-grown cpn1-1, an increase in ion leakage was observed at 4 hr after inoculation, whereas a similar increase in electrolyte leakage in Col-0 wild type was not observed until 24 hr after inoculation. Some of the electrolyte leakage from HH-grown Col-0 wild-type and cpn1-1 leaves may have been attributable to tissue damage from the infiltration, because the HH-grown leaf tissues were very tender and easily damaged. Together, these results indicated that not all aspects of the cpn1-1 mutant phenotype were humidity sensitive.
Some lesion mimic mutants have a “hair trigger” HR, meaning that they will develop a macroscopic HR at lower bacterial inoculum concentrations than do wild-type plants (Dietrich et al., 1994). cpn1-1 was tested for a hair trigger HR phenotype by determining the threshold concentration of P.s.m. (avrRpt2) that could trigger a macroscopic HR in cpn1-1 compared with Col-0 wild-type plants. The minimum concentration of P.s.m. (avrRpt2) that was sufficient to evoke a macroscopic HR was 5 × 107 cfu/mL in LH- and HH-grown Col-0 wild-type and cpn1-1 plants (data not shown). Therefore, although cpn1-1 plants had a faster HR than Col-0 wild-type plants, they did not appear to have increased sensitivity to low titers of P.s.m. (avrRpt2).
cpn1-1 Has Normal Stomatal Conductance
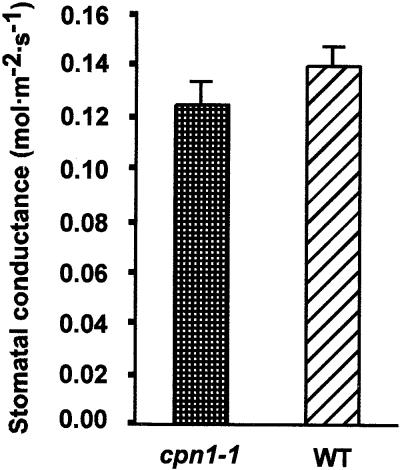
The sensitivity of cpn1-1 to low RH led us to speculate that cpn1-1 might be defective in responding to low humidity. For example, if cpn1-1 were defective in controlling water loss in low humidity, this might trigger stress, cell death, and other physiological problems. However, the stomatal conductance of LH-grown cpn1-1 and Col-0 wild-type plants was similar, as shown in Figure 3. This finding suggests that cpn1-1 is not defective in the regulation of stomatal conductance. Also, cpn1-1 did not appear to be particularly sensitive to drought stress, because it did not have a wilty phenotype or a lower tolerance than Col-0 wild type for low soil water potential (data not shown).
Figure 3.
Stomatal Conductance in Col-0 Wild-Type and cpn1-1 Leaves.
Data represent the average stomatal conductance for LH-grown cpn1-1 and Col-0 wild-type (WT) plants. Stomatal conductance represents the rate of passage of water vapor and carbon dioxide through the stomata of the plant. Bars indicate standard errors.
cpn1-1 Plants Exhibit Strong, Humidity-Dependent Resistance to Virulent P. syringae
Many lesion mimic mutants have increased resistance to virulent pathogens. LH-grown cpn1-1 had dramatically increased resistance to virulent P. syringae pv tomato (P.s.t.) bacteria compared with Col-0 wild-type plants, as shown in Figure 4. Four days after dip inoculation with 5 × 108 cfu/mL P.s.t., wild-type plants showed severe disease symptoms typical of bacterial speck disease, including massive tissue collapse, formation of water-soaked lesions, and chlorosis (Figure 4A). In contrast, the cpn1-1 mutant showed virtually no symptoms of the disease after inoculation (Figure 4B). The leaf tissues of inoculated cpn1-1 mutant plants remained green and healthy, although symptoms occasionally occurred on older leaves, especially when the bacterial inoculum was >108 cfu/mL (Figure 4B). HH-grown cpn1-1 plants did not have increased resistance to P.s.t. compared with Col-0 wild-type plants, as measured by visible symptoms (Figures 4C and 4D).
Figure 4.

Humidity-Dependent Resistance of cpn1-1 to Virulent P.s.t.
(A) LH-grown Col-0 wild-type plants 4 days after inoculation with virulent P.s.t.
(B) LH-grown cpn1-1 plants 4 days after inoculation with virulent P.s.t.
(C) HH-grown Col-0 wild-type plants 4 days after inoculation with virulent P.s.t.
(D) HH-grown cpn1-1 plants 4 days after inoculation with virulent P.s.t.
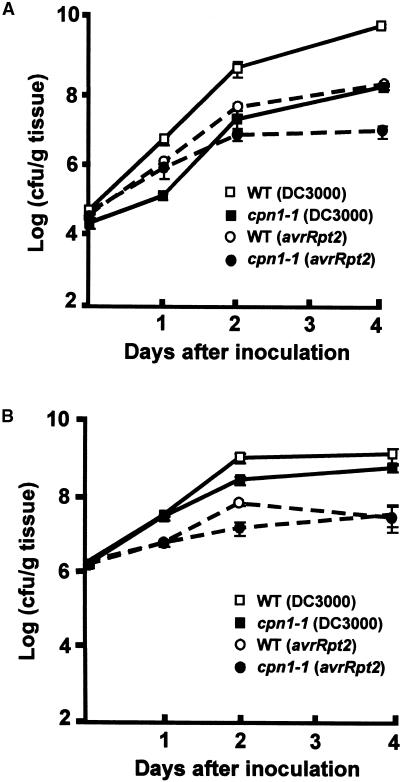
The ability of virulent P. syringae bacteria to grow within LH-grown cpn1-1 mutant leaf tissue was sharply restricted compared with that observed in LH-grown Col-0 wild-type tissue, as shown in Figure 5A. When Col-0 wild-type leaves were infiltrated with 105 cfu/mL virulent P.s.t., the bacterial population increased by several orders of magnitude over the course of 4 days. In contrast, when the leaves of LH-grown cpn1-1 mutant plants were infiltrated with virulent P.s.t., the bacteria were able to multiply only to a much lower level, approximately two or three orders of magnitude less than that observed in wild-type plants. The restriction of virulent P.s.t. multiplication in cpn1-1 mutant leaf tissues was consistent with the dramatic reduction of bacterial speck disease symptoms observed in inoculated LH-grown cpn1-1 mutant plants. Virulent P.s.t. grew to similar levels in HH-grown Col-0 wild-type and cpn1-1 mutant plant leaves after the plants were dip inoculated with a P.s.t. bacterial suspension of 2 × 107 cfu/mL (Figure 5B). These results were consistent with the strong bacterial speck disease symptoms observed in inoculated HH-grown Col-0 wild-type and cpn1-1 mutant plants and indicated that the increased resistance of cpn1-1 to virulent P.s.t. was a humidity-sensitive phenotype. Growth curves determined by infiltration inoculation or dip inoculation produced similar results (data not shown).
Figure 5.
Humidity-Dependent Inhibition of Bacterial Growth in cpn1-1.
(A) Growth of virulent P.s.t. (DC3000) and avirulent P.s.t. (avrRpt2) bacteria in LH-grown Col-0 wild-type (WT) and cpn1-1 plants. Plants were inoculated with the bacterial strains (105 cfu/mL) by syringe infiltration of the leaf mesophyll. For details, see Methods.
(B) Growth of virulent P.s.t. (DC3000) and avirulent P.s.t. (avrRpt2) bacteria in HH-grown Col-0 wild-type (WT) and cpn1-1 plants. Plants were inoculated with the bacterial strains (2 × 107 cfu/mL) by dipping, as described in Methods. This accounts for the higher initial bacterial count in (B) compared with that in (A).
Bars in (A) and (B) indicate standard errors.
To gauge the relative level of resistance of LH-grown cpn1-1 plants to virulent P.s.t., the degree of restriction of virulent P.s.t. growth in cpn1-1 was compared with the restriction of avirulent P.s.t. growth in Col-0 wild-type plants (Figure 5A). Growth of avirulent P.s.t. (avrRpt2) was sharply restricted in both Col-0 and cpn1-1 plants grown under LH conditions. The degree of restriction of virulent P.s.t. growth in LH-grown cpn1-1 plants was similar to the restriction of avirulent P.s.t. (avrRpt2) bacterial growth in LH-grown Col-0 wild-type plants. This indicates that the level of resistance of LH-grown cpn1-1 plants to virulent P.s.t. was similar to the level of resistance of LH-grown Col-0 wild-type plants to avirulent P.s.t. (avrRpt2) bacteria. Interestingly, the growth of avirulent P.s.t. (avrRpt2) bacteria in LH-grown cpn1-1 was moderately but significantly lower than the growth of virulent P.s.t. in LH-grown cpn1-1 plants, suggesting that there may be a slight additive effect of the cpn1-1 mutation and RPS2-mediated gene-for-gene resistance responses. The growth of avirulent P.s.t. (avrRpt2) bacteria was restricted in both Col-0 and cpn1-1 plants grown under HH conditions (Figure 5B). This indicates that RPS2-mediated gene-for-gene recognition and resistance was intact in cpn1-1 and that disease resistance mechanisms functioned normally in plants grown under high RH.
cpn1-1 Plants Constitutively Express Defense-Related Genes
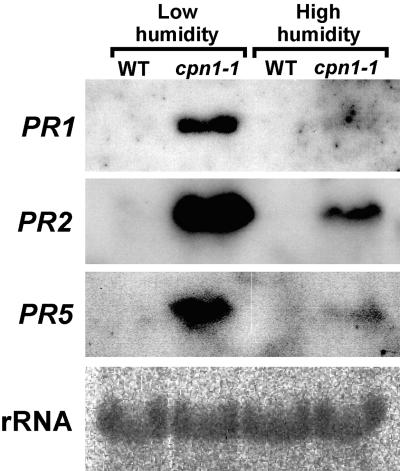
Because cpn1-1 mutant plants had increased resistance to P.s.t. and also had a lesion mimic phenotype, we reasoned that LH-grown cpn1-1 plants might be exhibiting SAR. Therefore, the expression of defense-related genes was investigated in cpn1-1 plants. LH-grown cpn1-1 plants showed high-level accumulation of PR1, PR2, and PR5 transcripts, whereas these transcripts were not detectable in LH-grown Col-0 wild-type plants, as shown in Figure 6 (Uknes et al., 1992). This result suggests that the cpn1-1 mutant was undergoing constitutive SAR in LH. Interestingly, there was moderate accumulation of PR2 transcript and very low but detectable accumulation of PR5 transcript in HH-grown cpn1-1 plants. In contrast, transcripts were not detected for any of these genes in HH-grown Col-0 wild-type. This finding indicates that growth in HH did not completely abolish the effects of the cpn1-1 mutation on PR gene expression.
Figure 6.
PR Gene Transcript Accumulation in cpn1-1 and Col-0 Wild Type.
RNA gel blot analyses of the transcript levels of PR1, PR2, and PR5 in LH- and HH-grown Col-0 wild-type (WT) and cpn1-1 leaf tissues. Twenty micrograms of leaf total RNA were loaded in each lane. rRNA, 28S rRNA stained with methylene blue to show equal RNA loading in each lane.
cpn1-1 Has Enhanced Resistance to an Oomyceteous Pathogen
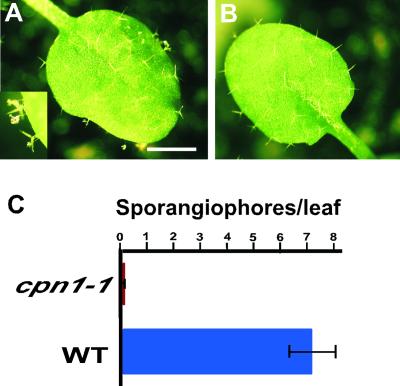
The increased resistance to virulent P.s.t. and the constitutive PR gene expression observed in cpn1-1 implied that cpn1-1 was undergoing salicylic acid–dependent SAR in LH. This suggested that the mutant also might be resistant to other pathogens that are controlled by salicylic acid–dependent SAR, such as the oomyceteous pathogen Peronospora parasitica (Bowling et al., 1994; Nawrath and Métraux, 1999). Two-week-old LH-grown cpn1-1 plants had dramatically increased resistance to virulent P. parasitica (Ahco 2) compared with LH-grown Col-0 wild-type plants, as shown in Figure 7. Virulent P. parasitica was able to sporulate on LH-grown Col-0 wild-type plants, but virtually no sporulation was observed on LH-grown cpn1-1 plants. This finding indicated that LH-grown cpn1-1 had improved resistance to both virulent P. syringae and virulent P. parasitica. This supports the interpretation that LH-grown cpn1-1 was undergoing SAR.
Figure 7.
Resistance of cpn1-1 to P. parasitica.
(A) LH-grown Col-0 wild-type leaf 7 days after inoculation with P. parasitica. Bar = 2 mm. Inset, close-up of sporangiophore at double magnification.
(B) LH-grown cpn1-1 leaf 7 days after inoculation with P. parasitica. The scale is the same as in (A).
(C) Sporangiophore counts on LH-grown Col-0 wild-type (WT) and cpn1-1 leaves 7 days after inoculation. Bars represent standard errors.
Genetic Analysis of cpn1-1
The cpn1-1 mutation was isolated in the homozygous state. The original homozygous cpn1-1/cpn1-1 mutant was outcrossed to Col-0 wild type (CPN1/CPN1). The CPN1/cpn1-1 F1 plants were allowed to self-pollinate, and the segregation of the cpn1-1 phenotype was monitored in the F2 generation. The cpn1-1 phenotype segregated in a mendelian fashion as a single, recessive locus, as indicated in Table 1. The cpn1-1 mutant phenotype was assessed in the F2 generation based on plant morphology, lesion development, and resistance to virulent P.s.t. The mutant phenotypes associated with cpn1-1 were observed to cosegregate in all 103 F2 cpn1-1/cpn1-1 individuals examined. This result suggested that the cpn1-1 mutant phenotype was most likely to be caused by a mutation in one gene or possibly by mutations in two or more very tightly linked genes.
Table 1.
Genetic Segregation Analysis of the cpn1-1 Mutation
| Female × Male | Generation | No. of Plants | Strong Mutant Phenotype | None | χ2 (P) |
|---|---|---|---|---|---|
| Col-0 × cpn1-1/cpn1-1 | F1 | 4 | 4 | ||
| Col-0 × cpn1-1/cpn1-1 | F2 | 394 | 103a | 291 | 0.27 (0.7) |
The lesion mimic, disease resistance, and leaf curling phenotypes segregated together as a recessive trait.
Identification of the CPN1 Gene
Genetic analysis indicated that the cpn1-1 mutation likely was T-DNA tagged. The T-DNA contained the bar gene conferring resistance to the herbicide ammonium glufosinate as a dominant trait (D'Halluin et al., 1992; Weigel et al., 2000). Glufosinate resistance in the F2 generation of the CPN1/CPN1 × cpn1-1/cpn1-1 cross-segregated in a 3:1 ratio of herbicide-resistant to herbicide-sensitive plants in a group of more than 500 F2 individuals (data not shown). This finding indicated the presence of a single T-DNA integration locus in the cpn1-1 mutant line. To determine if the T-DNA was linked to the cpn1-1 mutation, 88 of the 103 cpn1-1/cpn1-1 F2 plants from the CPN1/CPN1 × cpn1-1/cpn1-1 cross were selected and tested for the presence of the T-DNA using a polymerase chain reaction (PCR) assay to amplify the bar gene. All 88 plants tested had the T-DNA, indicating that the T-DNA cosegregated with the cpn1-1 mutant phenotype and that the cpn1-1 mutation was potentially T-DNA tagged (data not shown).
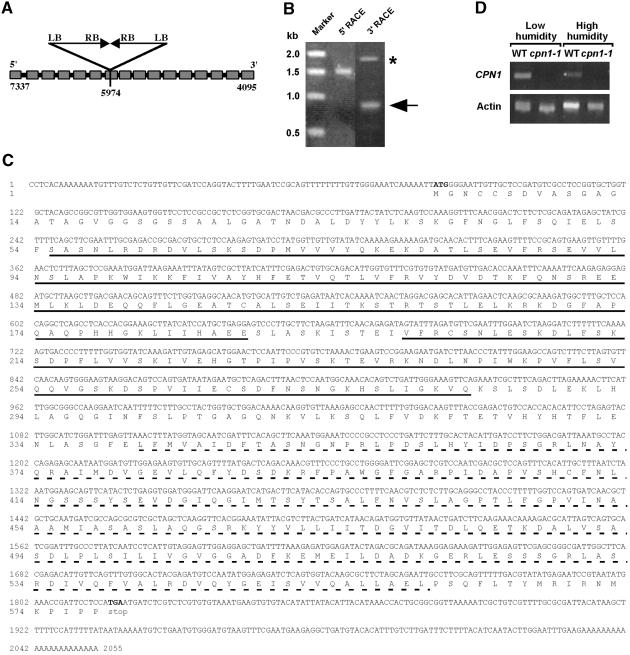
DNA gel blot analysis was used to confirm that the cpn1-1 mutant line had a single T-DNA insertion locus. The locus consisted of two copies of T-DNA in a “head-to-head” configuration, with the right T-DNA borders together and the left T-DNA borders flanking the plant DNA, as shown in Figure 8A. Plasmid rescue of the two left T-DNA borders was performed (Dilkes, 1998). The sequence of the plant DNA flanking the two left T-DNA borders was determined and used in a BLAST search of the Arabidopsis genome database. This revealed that the T-DNA was inserted into the seventh exon of a copine-like gene, CPN1 (Figure 8A). CPN1 is located at the bottom of chromosome 5 distal to EIN2 and proximal to HY5. The Transformation-competent artificial chromosome (TAC) clone containing CPN1 has GenBank accession number AB022212 (Liu et al., 1999).
Figure 8.
Molecular Identification of the CPN1 Gene.
(A) Scheme of the CPN1 gene with the 16 exons depicted as boxes. In cpn1-1, two copies of the T-DNA are inserted in a head-to-head configuration into exon 7 of CPN1. The numbers indicate the base pair positions in Transformation-competent artificial chromosome (TAC) clone K22G18 for the start and stop codons of CPN1 and the exact site of the T-DNA insertion in cpn1-1. RB, right T-DNA border; LB, left T-DNA border.
(B) 5′ and 3′ RACE PCR products resolved on a 1% agarose gel and visualized with ethidium bromide. The 5′ RACE reaction produced a single 1.5-kb band corresponding to the 5′ end of the CPN1 gene transcript. The 3′ RACE reaction produced two bands. The 750-bp band, denoted by the arrow, represents the 3′ end of the CPN1 gene transcript. The 1.8-kb band, denoted by the asterisk, was determined by DNA sequencing to represent a portion of one of the other copine homologs in the Arabidopsis genome (accession number AL163912).
(C) Complete CPN1 cDNA sequence and predicted amino acid translation. The C2 domains are solid underlined. The A domain is dashed underlined.
(D) Reverse transcriptase–mediated PCR analysis of CPN1 transcript accumulation in LH- and HH-grown Col-0 wild-type (WT) and cpn1-1 leaf tissue. CPN1, a 467-bp portion of the CPN1 transcript; Actin, a 900-bp portion of an actin gene.
The complete cDNA sequence of the CPN1 gene was determined by 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR. The 5′ and 3′ RACE PCR amplified single bands corresponding to the CPN1 gene transcript (Figure 8B). This indicated that the CPN1 gene produced a single transcript species. The 5′ and 3′ RACE PCR products were cloned and sequenced to obtain the full-length CPN1 cDNA sequence (Figure 8C). The splicing of the CPN1 cDNA was exactly as predicted by the annotation of the Arabidopsis genome database. The transcription start site was 82 bp upstream of the ATG, and the cDNA had a 192-bp 3′ untranslated region after the TAG stop codon. The CPN1 gene was expressed in leaf tissue of both LH- and HH-grown Col-0 wild-type plants (Figure 8D). As expected, no complete CPN1 transcript was detected in the cpn1-1 mutant leaves (Figure 8D).
The predicted protein product of the CPN1 gene is composed of 578 amino acids, has a molecular mass of 63.1 kD, and has homology with copine proteins (Creutz et al., 1998). The putative CPN1 protein, like other copines, has two C2 domains in the N-terminal portion of the protein and an A domain in the C-terminal region of the protein. In CPN1, the two C2 domains are composed of amino acids 55 to 188 and 199 to 283; the A domain is composed of amino acids 341 to 560 (Figure 8C). The CPN1 protein has a high degree of homology with other copines in Arabidopsis and other species, as indicated in Table 2.
Table 2.
Homology of CPN1 with Other Copines
To prove that the disruption of the CPN1 gene was responsible for the mutant phenotype in cpn1-1 mutant plants, the cpn1-1 mutant was complemented with a wild-type copy of the CPN1 gene. An 8-kb genomic DNA fragment containing the CPN1 gene coding regions plus 3.4 kb of upstream and 1.3 kb of downstream DNA was introduced into the genome of cpn1-1/cpn1-1 mutant plants by Agrobacterium-mediated dip transformation (Clough and Bent, 1998). The 8-kb CPN1 genomic fragment extended partly into the neighboring open reading frames on either side of CPN1. cpn1-1/cpn1-1 plants transformed with the 8-kb CPN1 genomic fragment had a wild-type phenotype, indicating that the T-DNA insertion into the CPN1 gene is respon-sible for the mutant phenotype of cpn1-1 (data not shown).
DISCUSSION
The biological functions of genes defined by lesion mimic mutations are difficult to determine with certainty. It is likely that at least some lesion mimic mutants represent defects in genes that directly regulate the HR defense response. However, it is also likely that many lesion mimic mutants represent perturbations of plant cell physiology unrelated to disease defense responses. For example, physiological alterations caused by overexpression of genes such as pyruvate decarboxylase in potato and a bacterial proton pump in tobacco can trigger lesion formation and increased disease resistance, although the overexpressed genes do not appear to have any direct role in disease resistance responses (Mittler et al., 1995; Tadege et al., 1998). In addition, salicylic acid appears to potentiate cell death triggered by free radicals, implying that plants with increased salicylic acid might be more sensitive to the generation of free radicals by stresses (Mazel and Levine, 2001). The complexity of cell death signaling in plants makes it difficult to determine the specific function of the CPN1 protein. Nevertheless, speculation on the function of CPN1 can be made based on the results presented here and biochemical analyses of related copines, assuming that CPN1 has similar biochemical characteristics. One possibility is that CPN1 plays a direct role in defense cell death signaling and has indirect effects on LH adaptation. Alternatively, CPN1 may have a direct function in LH adaptation and may have indirect effects on plant defense signaling.
CPN1 as a Regulator of Defense and Cell Death
The improved disease resistance and accelerated HR phenotypes of cpn1-1 imply that CPN1 may play a direct role in regulating defense responses, including the HR. Because the cpn1-1 mutation is recessive and probably eliminates CPN1 gene function, CPN1 could function as a repressor of cell death. An interesting possibility is that a pathogen-triggered Ca2+ flux may activate the repressive function of CPN1, possibly by triggering the localization of CPN1 to a membrane where it could exert a repressive influence on cell death signal transduction. This might seem counterintuitive at first, but genetic evidence suggests that the HR is under negative control as well as positive control to prevent uncontrolled cell death and lesion expansion after HR initiation (Dietrich et al., 1994). The accelerated HR in cpn1-1 plants supports a role for CPN1 as a negative regulator of the avirulent pathogen-induced HR defense response.
However, our data indicate that CPN1 also is required for the repression of cell death and PR gene expression in the absence of a pathogen infection. In particular, CPN1 is essential for the repression of cell death in LH growth conditions. It is possible that some aspect of LH signaling or LH adaptation, such as a Ca2+ flux, has the potential to trigger the HR. CPN1 may have a genetically nonredundant role in preventing LH-potentiated cell death from occurring. In plants, increases in intracellular calcium have been observed in response to a wide range of biotic and abiotic stimuli, including osmotic stress, cold, touch, and drought (for reviews, see Knight, 2000; Reddy, 2001). Although the calcium signatures differ for each stimulus, the calcium molecule appears to be a key shared component of signal transduction pathways controlling responses to various environmental stimuli (for reviews, see McAinsh and Hetherington, 1998; Bowler and Fluhr, 2000). CPN1 may play a role in defining Ca2+ signaling specificity by preventing cell death and other defense responses from being triggered by Ca2+ fluxes that are not induced by pathogens. According to this model, cell death occurs in the cpn1-1 mutant in LH because there is no CPN1 protein present to prevent LH signaling events from triggering cell death.
CPN1 also may play a direct role in suppressing PR gene expression in the absence of pathogen attack. Low levels of PR gene expression were observed in HH-grown cpn1-1, indicating a partial derepression or activation of PR gene expression in cpn1-1 mutants even in the absence of cell death and SAR. Thus, PR gene expression in cpn1-1 may occur by different mechanisms in LH and HH: in LH-grown cpn1-1, cell death occurs, triggering SAR and high-level PR gene expression; in HH-grown cpn1-1, low-level PR gene expression occurs because the repressive influence of CPN1 on PR gene expression is absent.
CPN1 as a Regulator of LH Adaptation
A low air moisture level coupled with adequate soil water potential results in high rates of water uptake in the roots and transpiration in the leaves (Mohr and Schopfer, 1995). Under these conditions, the plant must regulate cellular ion balance by reducing transpiration, controlling ion uptake, extruding ions, and sequestering ions in subcellular compartments such as the vacuole. In cpn1-1, the humidity-sensitive phenotype is unlikely to be attributable simply to increased or uncontrolled water loss, because stomatal conductance in cpn1-1 was normal compared with that in Col-0 wild-type plants. Instead, CPN1 may be a key player in LH adaptation.
It is possible that cpn1-1 is defective in controlling ion balance in LH or is defective in responding to altered ion balance in LH. As a Ca2+-dependent phospholipid binding protein, CPN1 could regulate ion transport at a membrane in response to Ca2+ fluxes, for instance. Defective ionic homeostasis in cpn1-1 could trigger cell death by a number of mechanisms in LH, including hyperaccumulation of essential metals or oxidative stress (Wang et al., 1996; Briat and Lebrun, 1999; Hagemeyer, 1999). For example, manganese toxicity causes leaf crinkle and lesion formation in cotton, possibly as a result of increased production of reactive oxygen species (Foy et al., 1978). Ionic imbalances in LH-grown cpn1-1 could trigger cell death and lesion formation, which in turn could trigger SAR. Thus, the increased disease resistance phenotype of cpn1-1 might be an indirect effect of the mutation. It is interesting, however, that HH-grown cpn1-1 plants also exhibited an accelerated HR phenotype and very low-level expression of PR genes. This suggests that CPN1 might be necessary to control cellular homeostasis even in HH conditions, although the defect of the cpn1-1 mutant is much more pronounced in LH.
Conclusion
Plants must integrate signals from a wide range of biotic and abiotic sources to optimize their developmental pattern and physiology for their environment. The growing interconnectedness of plant signaling pathways reflects the integrated nature of plant responses to the environment. The presence of common regulatory components during signal transduction responses to abiotic and biotic stresses suggests that cellular signaling is a web, with proteins receiving inputs from multiple partners and pathways (Bent, 2001). Slight perturbations in a particular signaling pathway could easily deregulate other signaling events in the plant as a result of interconnections between them. The phenomenon of cross-tolerance may reflect this kind of signaling interconnection (for review, see Bowler and Fluhr, 2000). Our results suggest that the CPN1 gene may represent an important component of the plant's ability to monitor and respond to its environment. The CPN1 protein may act as a Ca2+-sensitive modulator of plant responses to environmental signals, including humidity and pathogens.
METHODS
Plant Growth Conditions and Mutant Screening
Arabidopsis thaliana plants were grown in soil (Scotts Redi-Earth Plug and Seedling Mix [E.C. Geiger, Inc., Harleysville, PA]) and watered with distilled water. For low-humidity (LH) growth, plants were maintained at 24°C under either short-day (SD) or long-day (LD) conditions with 75 to 100 μmol·m−2·sec−1 light intensity and 35 to 45% relative humidity. For high-humidity (HH) growth, plants were maintained at 24°C under either SD or LD conditions with 55 to 65 μmol·m−2·sec−1 light intensity and 75 to 85% RH.
Pools of activation-tagged Arabidopsis seed (Columbia [Col-0] ecotype) (Weigel et al., 2000) were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). We screened this population for mutants with improved resistance to the virulent Pseudomonas syringae pv tomato (P.s.t.) strain DC3000 (Cuppels, 1986). The cpn1-1 mutant was identified from among ∼10,000 independent T-DNA lines screened for mutants with increased resistance to P.s.t. For mutant screening, 4-week-old plants grown under LH, SD conditions were screened for increased resistance to virulent P.s.t. by dipping the plants into a bacterial suspension of 5 × 108 colony-forming units [cfu]/mL in 10 mM MgCl2 with 0.00025% (v/v) Silwet L-77 surfactant (Lehle Seeds, Round Rock, TX). After inoculation, plants were maintained at 24°C under SD conditions with 55 to 65 μmol·m−2·sec−1 light intensity and 75 to 95% RH for the development of bacterial speck disease symptoms. Bacterial speck disease symptoms were assessed 4 days after inoculation.
Bacterial Strains and Growth Conditions
For mutant screening, virulent P.s.t. strain DC3000 bacteria were grown on Pseudomonas Agar F (PA) (Sigma) supplemented with 100 μg/mL rifampicin (Sigma) at 28°C. For growth curves, P.s.t. strain DC3000 bacteria were used carrying either the plasmid pVSP61 (empty vector) or pV288 (bearing avrRpt2) and were grown on PA supplemented with 100 μg/mL rifampicin and 25 μg/mL kanamycin (Sigma) (Whalen et al., 1991). For hypersensitive response (HR) assays, P. syringae pv maculicola (P.s.m.) (strain 4326) bacteria were used bearing either plasmid pVSP61 or pV288 and were grown on PA supplemented with 100 μg/mL rifampicin and 25 μg/mL kanamycin. The P.s.m. (4326) strain was used for the HR assays because it is less virulent on Arabidopsis than P.s.t. and allows for a clearer distinction between the HR and disease symptoms.
Bacterial Growth Curves and Peronospora parasitica Inoculations
In planta bacterial growth assays were performed by inoculating 5-week-old plants by either infiltration or dipping. For infiltration inoculations, leaves were inoculated with a suspension of 105 cfu/mL bacteria in 10 mM MgCl2; for dip inoculations, plants were inoculated with a suspension of 2 × 107 cfu/mL bacteria in 10 mM MgCl2 plus 0.00025% (v/v) Silwet L-77. Plants were maintained in SD, 55 to 65 μmol·m−2·sec−1 light intensity, 75 to 90% RH conditions for the duration of the experiment after inoculation. Bacterial populations were assessed at 0, 1, 2, and 4 days after inoculation. Three samples were taken for each data point. For each sample, three whole leaves were harvested, weighed, and ground in 1 mL of 10 mM MgCl2. Bacteria per unit tissue weight was determined by serial dilution plating essentially as described by Whalen et al. (1991), except that bacteria were plated on PA medium supplemented with 100 μg/mL rifampicin and 25 μg/mL kanamycin. Each growth curve experiment was replicated three times. Statistical analyses were performed using Student's t test of the differences between means of log-transformed data.
Resistance to P. parasitica (Ahco 2) was tested in 18-day-old plants. The infected plants were scored after 7 days. More than 100 leaves from 20 plants were chosen at random, and the number of conidiophores per leaf was counted. Statistical significance was measured by Student's t test.
Histochemistry and Microscopy
Leaf samples were taken from 3-week-old plants and stained for dead cells using trypan blue (125 μg/mL) (Vogel and Somerville, 2000). Samples for autofluorescence and callose examination were prepared as described by Dietrich et al. (1994). For scanning electron microscopy, leaves were fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, rinsed in 0.1 M cacodylate buffer, pH 7.4, and then postfixed with OsO4. Samples were dehydrated by a series of increasing concentrations of ethanol, and critical point drying was performed using CO2. Samples were then sputter coated with a gold-palladium mixture and viewed in a scanning electron microscope at a 15° angle.
HR Tests
Plants grown under LH and HH conditions were inoculated with the P.s.m. strains described above at a concentration of 108 cfu/mL in 10 mM MgCl2. Fifty to 100 leaves were infiltrated for each plant–bacterial combination per experiment. Infiltrated leaves were scored for HR development as 0 for no HR, 1 for partial HR, and 2 for complete HR at 4, 8, 12, 16, 20, 24, and 30 hr after inoculation. Statistical analysis was performed using Student's t test. This experiment was repeated three times with similar results.
Electrolyte Leakage Assay
Electrolyte leakage was measured essentially as described by Goodman (1968). LH- and HH-grown cpn1-1 and Col-0 wild-type plants were inoculated with P.s.m. strains as described for the HR tests, and control plants of both genotypes were inoculated with 10 mM MgCl2. Measurements were performed at 0, 4, 8, 12, 16, 20, 24, and 30 hr after inoculation. For each sample at each time point, five 0.07-cm2 leaf discs were excised from infiltrated leaves using a number 1 cork borer and immersed in 3 mL of distilled water and agitated gently in a shaker for 8 min at 28°C. The conductivity of the solution was measured using a Traceable Conductivity Resistivity TDS Salinity Concentration meter (Control Company, Friendswood, TX). Three independent samples were taken at each time point for each plant–inoculum combination.
Stomatal Conductance and Drought Tolerance Measurements
Leaves of 30-day-old LH-grown cpn1-1 and wild-type plants were sealed in the gas exchange cuvette of a Li-Cor 6400 photosynthesis meter. The following parameters were maintained during the measurements: leaf temperature, 24 ± 0.5°C; CO2 concentration, 350 μL· L−1; air flow, 0.1 L·min−1; white light illumination, 400 μmol·m−2· sec−1; RH, 34 to 35%. The plants had been exposed to normal growth light for 3 to 4 hr before the beginning of the experiment. One leaf was measured at a time. Measurements were taken 5 min after leaves were sealed in the cuvette under the conditions described above, at which time the leaves had attained a steady state. The cpn1-1 leaves were too small to fill the leaf chamber completely. Therefore, the area of each cpn1-1 leaf used in the experiment was determined to obtain the conductance per unit area. Leaf area was determined by excising the leaf and measuring its area using NIH Image 1.62f software. The data represent the average stomatal conductance for 15 randomly selected leaves of each genotype. To test plant responses to low soil water potential, the soil was simply allowed to dry out, and the time at which 4-week-old LH- and HH-grown cpn1-1 and Col-0 wild-type plants began to show a visible decline in turgor pressure was observed.
RNA Analyses
Leaf tissue samples were collected from 4-week-old plants grown under LH, SD or HH, SD conditions. Samples were flash frozen in liquid nitrogen, and RNA was extracted using the RNeasy Maxi kit (Qiagen, Valencia, CA). Twenty micrograms of total RNA was separated in a formaldehyde-agarose gel (Ambion Northern Max kit) and transferred to a Bright Star Plus membrane (Ambion, Austin, TX). After RNA transfer, the membranes were stained with methylene blue to verify RNA transfer and compare the relative levels of rRNA in each lane. Plasmids for the PR1, PR2, and PR5 probes were obtained from Dr. Ramesh Raina (Pennsylvania State University). Probe labeling with 32P, hybridization, blot washing, and exposure were performed as described previously (McNellis et al., 1998).
Genetic Analyses
Backcrosses were performed with Col-0 wild type using cpn1-1 mutant as both pollen donor and receiver. F1 and F2 generation plants were tested for the cpn1-1 mutant phenotype by growing them in LH, SD conditions and inoculating with virulent P. syringae. F2 plants with lesions and increased resistance to P. syringae were tested for the presence of the T-DNA insert by polymerase chain reaction (PCR) analysis. The primers used were designed to amplify the bar gene (BARF, 5′-CTCTAGGATCGATCCCCCGGG-3′; BARR, 5′-GAA-TTCCTCGAGTATAAGAGC-3′; annealing at 60°C) and produce a 650-bp product.
DNA Gel Blotting and Plasmid Rescue
Genomic DNA was isolated using a modification of the method of Shure et al. (1983) as described previously (McNellis et al., 1998). DNA gel blot analysis was performed as described previously (McNellis et al., 1998). DNA gel blot analyses of genomic DNA of the cpn1-1 mutant were performed using enzymes that cut the pSKI015 T-DNA once, including BamHI, SpeI, EcoRI, and HindIII (New England Biolabs, Beverly, MA). The pBluescript KS+ (Stratagene) plasmid was used as a probe for the pSKI015 T-DNA. Probe labeling and detection were performed using the Phototope-Star Chemiluminescent Detection Kit (New England Biolabs) according to the manufacturer's instructions. DNA gel blotting of cpn1-1 DNA cut with BamHI revealed 8- and 10-kb fragments hybridizing to pBluescript. Plasmid rescue was done by digestion of 5 μg of cpn1-1 genomic DNA with 40 units of BamHI, self-ligation using T4 DNA polymerase (New En-gland Biolabs), and transformation into Epicurian Coli XL-2 Blue Escherichia coli cells (Stratagene) according to the manufacturer's instructions, except that Luria-Bertani medium supplemented with 100 μg/mL ampicillin (Sigma) was used to select transformants. Both the 8-and 10-kb fragments were rescued as plasmids. A partial sequence of the plant DNA flanking the T-DNA left borders in the rescued clones was obtained by cycle sequencing at the Nucleic Acid Facility at Pennsylvania State University using the T7 primer. The DNA sequence flanked by the T-DNA insert was identified by BLAST search. The exact location of the T-DNA insert was determined by sequencing of the rescued plasmids using the primers 5′-GGA-ACTCCAATTCCCGTGTC-3′ (forward) and 5′-ACCTAAAATGACCATAAATGG-3′ (reverse).
Reverse Transcriptase–Mediated PCR
First-strand cDNA synthesis was performed using 5 μg of total RNA and the RETROscript kit (Ambion). As a positive control, PCR was performed to amplify the cDNA of a constitutively active actin gene using primers 5′-GTTGGGATGAACCAGAAGGA-3′ (forward) and 5′-GAACCACCGATCCAGACACT-3′ (reverse) (annealing at 58°C). cDNA synthesized from the total RNA of cpn1 and Col-0 wild-type plants was tested for copine expression by PCR using the primers 5′-TCT-AGATCATGGAGGAATCGGTTTCAT-3′ (forward) and 5′-ATCATGACT-TCATACACCAGTGC-3′ (reverse) (annealing at 58°C), which produce a 467-bp product.
Genetic Complementation of cpn1-1
An 8-kb BamHI fragment from Transformation-competent artificial chromosome (TAC) clone K22G18 (Liu et al., 1999) consisting of the copine gene coding regions plus upstream and downstream sequences was cloned into the BamHI site of the pCLD04541 binary vector to create pCLD04541-CPN1 (Bent et al., 1994). pCLD04541-CPN1 was introduced into Agrobacterium tumefaciens strain GV3101 by heat shock transformation, and transformants were selected on Luria-Bertani medium supplemented with 50 μg/mL gentamycin, 3 μg/mL tetracycline, and 25 μg/mL kanamycin (Sigma). cpn1-1 mutant plants were transformed with pCLD04541-CPN1 using the floral dip method (Clough and Bent, 1998). Transformants were selected on GM medium (1 × Murashige and Skoog [1962] salts, pH 5.7 [Gibco BRL], 1 × B5 vitamins, solidified with 0.8% phytagar [Gibco BRL]) supplemented with 50 μg/mL kanamycin. Transformants were grown in LH, SD conditions and observed to have a wild-type phenotype.
CPN1 Sequence Analysis
The complete sequence of the CPN1 cDNA, including the 5′ and 3′ untranslated regions, was determined by performing 5′ and 3′ rapid amplification of cDNA ends (RACE) reactions using the SMART RACE cDNA kit (Clontech, Palo Alto, CA) and cloning the resulting PCR products using the Original TA Cloning Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The 5′ RACE gene-specific primer was 5′-GCGACGCGCTGGCGATCATTGCAGCAGCG-3′. The 3′ RACE gene-specific primer was 5′-TCCCTG-CCTGGGGATTCGGAGCTCGTCC-3′. The domains of the CPN1 protein were identified using Pfam and Prosite Profile Scan databases (http://hits.isb-sib.ch/cgi-bin/hits_motifscan). Pairwise alignment of copine sequences was performed using the SIM Alignment Tool (www.expasy.ch/tools/sim-prot.html).
Acknowledgments
We thank Daniel Cosgrove, Sendil Devadas, Ramanand Dixit, and David Geiser for assistance with histochemistry and microscopy; Seogchan Kang for assistance with RNA blot hybridizations; S. Devadas and Ramesh Raina for providing PR gene probes; Philip Jensen and Gro Torsethaugen for help with stomatal conductance experiments; Nina Fedoroff and Ramamurthy Mahalingam for help with the P. parasitica infection assay; Rosemary Walsh for help with scanning electron microscopy; and Sally Assmann and Sona Pandey for providing the actin primers for reverse transcriptase–mediated PCR assays. We thank C. Peter Romaine, S. Kang, and two anonymous reviewers for critical comments on the manuscript. We thank R. Raina and S. Kang for many helpful discussions.
References
- Azzi, A., Boscoboinik, D., and Hensey, C. (1992). The protein kinase C family. Eur. J. Biochem. 208, 547–557. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (2001). Plant mitogen-activated protein kinase cascades: Negative regulatory roles turn out positive. Proc. Natl. Acad. Sci. USA 98, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bowler, C., and Fluhr, R. (2000). The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci. 5, 241–246. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat, J.-F., and Lebrun, M. (1999). Plant responses to metal toxicity. C. R. Acad. Sci. Paris Life Sci. 322, 43–54. [DOI] [PubMed] [Google Scholar]
- Brose, N., Hofmann, K., Hata, Y., and Sudhof, T.C. (1995). Mammalian homologues of Caenorhabditis elegans unc-13 gene define a novel family of C2-domain proteins. J. Biol. Chem. 270, 25273–25280. [DOI] [PubMed] [Google Scholar]
- Caudell, E.G., Caudell, J.J., Tang, C.H., Yu, T.K., Frederick, M.J., and Grimm, E.A. (2000). Characterization of human copine III as a phosphoprotein with associated kinase activity. Biochemistry 39, 13034–13043. [DOI] [PubMed] [Google Scholar]
- Chamnongpol, S., Willekens, H., Langebartels, C., Van Montagu, M., Inze, D., and Van Camp, W. (1996). Transgenic tobacco with a reduced catalase activity develops necrotic lesions and induces pathogenesis-related expression under high light. Plant J. 10, 491–503. [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I.C., Lippok, B., Smith, R.K., Jr., and Bent, A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97, 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz, C.E., Tomsig, J.L., Snyder, S.L., Gautier, M.-C., Skouri, F., Beisson, J., and Cohen, J. (1998). The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding protein conserved from Paramecium to humans. J. Biol. Chem. 273, 1393–1402. [DOI] [PubMed] [Google Scholar]
- Cuppels, D.A. (1986). Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 52, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, O., and Lam, E. (1998). Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol. 8, 1129–1132. [DOI] [PubMed] [Google Scholar]
- D'Halluin, K., Block, M.D., Denecke, J., Janssens, J., Leemans, J., Reynaerts, A., and Botterman, J. (1992). The bar gene as selectable and screenable marker in plant engineering. Methods Enzymol. 216, 415–416. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dietrich, R., Richberg, M.H., Schmidt, R., Dean, D., and Dangl, J.L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of cell death. Cell 88, 685–694. [DOI] [PubMed] [Google Scholar]
- Dilkes, B.P. (1998). Cloning genes from T-DNA tagged mutants. In Methods in Molecular Biology: Arabidopsis Protocols, J.S. Martinez-Zapater, ed (Totowa, NJ: Humana Press), pp. 339–351. [DOI] [PubMed]
- Duncan, R.R., Shipston, M.J., and Chow, R.H. (2000). Double C2 protein: A review. Biochimie 82, 421–426. [DOI] [PubMed] [Google Scholar]
- Essen, L.O., Perisic, O., Cheung, R., Katan, M., and Williams, R.L. (1996). Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature 380, 595–602. [DOI] [PubMed] [Google Scholar]
- Foy, C.D., Chaney, R.L., and White, M.C. (1978). The physiology of metal toxicity in plants. Annu. Rev. Plant Physiol. 29, 511–567. [Google Scholar]
- Frye, C.A., Tang, D., and Innes, R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, R.N. (1968). The hypersensitive reaction in tobacco: A reflection of changes in host cell permeability. Phytopathology 58, 872–873. [Google Scholar]
- Goodman, R.N. (1972). Electrolyte leakage and membrane damage in relation to bacterial population, pH, and ammonia production in tobacco leaf tissue inoculated with Pseudomonas pisi. Phytopathology 62, 1327–1331. [Google Scholar]
- Goodman, R.N., and Novacky, A.J. (1994). The Hypersensitive Reaction in Plants to Pathogen: A Resistance Phenomenon. (St. Paul, MN: American Phytopathological Society Press).
- Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Ausubel, F.M. (1993). Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4, 327–341. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Hagemeyer, J. (1999). Ecophysiology of plant growth under heavy metal stress. In Heavy Metal Stress in Plants, M.V.N. Prasad and J. Hagemeyer, eds (Berlin: Springer-Verlag), pp. 157–181.
- Hahlbrock, K., Scheel, D., Logemann, E., Nurnberger, T., Parniske, M., Reinold, S., Sacks, W.R., and Schmelzer, E. (1995). Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc. Natl. Acad. Sci. USA 92, 4150–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement, Z., Farkas, G.L., and Lovrekovich, L. (1963). Hypersensitive reaction induced by phytopathogenic bacteria in the tobacco leaf. Phytopathology 54, 474–477. [Google Scholar]
- Knight, H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195, 269–324. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., Bent, A.F., Dahlbeck, D., Innes, R.W., and Staskawicz, B.J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., and Shibata, D. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck, K., and Lawton, K. (1998). Plant strategies for resistance to pathogens. Curr. Opin. Biotechnol. 9, 208–213. [Google Scholar]
- Mazel, A., and Levine, A. (2001). Induction of cell death in Arabidopsis by superoxide in combination with salicylic acid or with protein synthesis inhibitors. Free Radical Biol. Med. 30, 98–106. [DOI] [PubMed] [Google Scholar]
- McAinsh, M.R., and Hetherington, A.M. (1998). Encoding specificity in Ca2+ signalling systems. Trends Plant Sci. 3, 32–36. [Google Scholar]
- McNellis, T.W., Mudgett, M.B., Li, K., Aoyama, T., Horvath, D., Chua, N.H., and Staskawicz, B.J. (1998). Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 14, 247–257. [DOI] [PubMed] [Google Scholar]
- Mittler, R., Shulaev, V., and Lam, E. (1995). Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Del Pozo, O., Meisel, L., and Lam, E. (1997). Pathogen-induced programmed cell death in plants: A possible defense mechanism. Dev. Genet. 21, 279–289. [DOI] [PubMed] [Google Scholar]
- Mock, H.P., Heller, W., Molina, A., Neubohn, B., Sandermann, H., Jr., and Grimm, B. (1999). Expression of uroporphyrinogen decarboxylase or coproporphyrinogen oxidase antisense RNA in tobacco induces pathogen defense responses conferring increased resistance to tobacco mosaic virus. J. Biol. Chem. 274, 4231–4238. [DOI] [PubMed] [Google Scholar]
- Mohr, H., and Schopfer, P. (1995). Plant Physiology. (Berlin: Springer-Verlag).
- Molina, A., Volrath, S., Guyer, D., Maleck, K., Ryals, J., and Ward, E. (1999). Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 17, 667–678. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., and Dangl, J.L. (1999). Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 151, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nawrath, C., and Métraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001). Calcium: Silver bullet in signaling. Plant Sci. 160, 381–404. [DOI] [PubMed] [Google Scholar]
- Sasabe, M., Takeuchi, K., Kamoun, S., Ichinose, Y., Govers, F., Toyoda, K., Shiraishi, T., and Yamada, T. (2000). Independent pathways leading to apoptotic cell death, oxidative burst and defense gene expression in response to elicitin in tobacco cell suspension culture. Eur. J. Biochem. 267, 5005–5013. [DOI] [PubMed] [Google Scholar]
- Shure, M., Wessler, S., and Fedoroff, N.V. (1983). Molecular identification and isolation of the Waxy locus of maize. Cell 35, 225–233. [DOI] [PubMed] [Google Scholar]
- Tadege, M., Bucher, M., Stahli, W., Suter, M., Dupuis, I., and Kuhlemeier, C. (1998). Activation of plant defense responses and sugar efflux by expression of pyruvate decarboxylase in potato leaves. Plant J. 16, 661–671. [Google Scholar]
- Tomsig, J.L., and Creutz, C.E. (2000). Biochemical characterization of copine: A ubiquitous Ca2+-dependent, phospholipid-binding protein. Biochemistry 39, 16163–16175. [DOI] [PubMed] [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Oppedijk, B.J., Lu, X., Van Duijn, B., and Schilperoort, R.A. (1996). Apoptosis in barley aleurone during germination and its inhibition by abscisic acid. Plant Mol. Biol. 32, 1125–1134. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Sugita, S., and Sudhof, T.C. (2000). The RIM/NIM family of neuronal C2 domain proteins: Interactions with Rab3 and a new class of Src homology 3 domain proteins. J. Biol. Chem. 275, 20033–20044. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann, K., Hunt, M., Uknes, S., Neuenschwander, U., Lawton, K., Steiner, H.Y., and Ryals, J. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S.C., Hinshelwood, J., Perkins, S.J., and Sim, R.B. (1999). Production and functional activity of a recombinant von Willebrand factor-A domain from human complement factor B. Biochem. J. 342, 625–632. [PMC free article] [PubMed] [Google Scholar]
- Xu, H., and Heath, M.C. (1998). Role of calcium in signal transduction during the hypersensitive response caused by basidiospore-derived infection of the cowpea rust fungus. Plant Cell 10, 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]
- Zimmermann, S., Nurnberger, T., Frachisse, J.M., Wirtz, W., Guern, J., Hedrich, R., and Scheel, D. (1997). Receptor-mediated activation of a plant Ca2+-permeable ion channel involved in pathogen defense. Proc. Natl. Acad. Sci. USA 94, 2751–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]