Abstract
Cyst nematodes and root-knot nematodes elaborately transform cells within the vascular cylinders of plant roots into enlarged, multinucleate, and metabolically active feeding cells. The giant cells of root-knot nematodes are formed by repeated karyokinesis uncoupled from cytokinesis, whereas the syncytia formed by cyst nematodes arise from coordinated cell wall dissolution and the coalescing of cell cytoplasm of adjacent cells. Both giant cells and syncytia undergo extensive cell wall architectural modifications, including thickening and the formation of numerous ingrowths that increase the plasmalemma surface area for solute uptake. The origin of enzymes involved in these cell wall modifications has been the subject of debate for several decades. Immunolocalization of endo-β-1,4-glucanases (EGases) secreted from cyst nematodes was observed in root cortical tissue during the intracellular migration of the nematodes, but secretion of cyst nematode EGases into developing syncytia was not detected. We have identified five EGase genes from tobacco that are upregulated within plant roots upon infection by both root-knot and cyst nematodes. In situ localization of tobacco EGase transcripts demonstrated that their expression was specifically and developmentally upregulated within giant cells, syncytia, root tips, and lateral root primordia. These data confirm that cell wall modifications within plant-parasitic-nematode feeding cells arise from cell wall–modifying enzymes of plant, rather than nematode, origin.
INTRODUCTION
Feeding cells induced by cyst nematodes (Globodera and Heterodera spp) and root-knot nematodes (RKN; Meloidogyne spp), termed syncytia and giant cells, respectively, are formed from host root cells during parasitism to sustain the growth and reproduction of the nematode (Hussey and Grundler, 1998). Motile second-stage juveniles (J2) of the nematode penetrate plant roots and migrate to the vascular cylinder, where the feeding cells serve as the sole nutritive source for the subsequent sedentary parasitic life stages of these nematodes. Evidence suggests that stylet (hollow feeding spear) secretions originating from three large unicellular esophageal gland cells of RKN and cyst nematodes play essential roles during parasitism of plant roots, including the induction of feeding cells (Hussey, 1989; Davis et al., 2000). Nematode secretions may alter the development of affected host plant cells directly or indirectly (Hussey, 1989). This modification of normal plant cell development causes plant cells to redifferentiate into unique cell types strictly for the benefit of the nematode and is accompanied by multiple changes in plant gene expression (Bird, 1996).
Giant cells and syncytia have several characteristics in common. In both cell types, there is an increase in metabolic activity and cytoplasmic density, the large central vacuole is reduced to several smaller vacuoles, organelles proliferate, individual cells hypertrophy, cell walls thicken, and finger-like protuberances (ingrowths) form along walls adjacent to the xylem vessels to increase membrane surface area for solute uptake (Hussey and Grundler, 1998). The nuclei within these cells enlarge, develop an amoeboid appearance, have a very prominent nucleolus, and are polyploid. In giant cells of RKN, the nuclei are stimulated to divide in the absence of cell division, resulting in enlarged plant root cells containing hundreds of nuclei (Huang and Maggenti, 1969). The syncytium of cyst nematodes also is multinucleate, but it arises by a different mechanism than that of giant cells. Within the initial syncytial cell, the plasmodesmatal openings gradually widen, and wall degradation is initiated at pit fields (Jones, 1981a; Grundler et al., 1998). As the initial syncytial cell enlarges, the cell wall gaps expand and neighboring protoplasms fuse. Progressive cell wall dissolution allows the syncytium to expand longitudinally along the length of the vascular cylinder (extending as far as 2 to 3 mm) and can incorporate up to 200 plant cells (Jones, 1981a).
The extensive cell wall architectural modifications (i.e., thickening, ingrowth, disassembly, and dissolution) that occur in cyst nematode and RKN feeding cells likely are mediated by the activity of both cell wall biosynthetic and cell wall–degrading enzymes. The origin of these enzymes, especially the cell wall–degrading enzymes, has been in question since the earliest microscopic examinations of nematode-induced feeding cells. Early biochemical investigations suggested that plant-parasitic nematodes produced cell wall–degrading enzymes (Dropkin, 1963; Bird et al., 1975), and several investigators hypothesized that the secretion of such enzymes might be responsible for the cell wall dissolution observed in feeding sites (Dropkin, 1969; Bird et al., 1975). Other researchers speculated that the anatomical features of feeding cells were more supportive of a role for cell wall–degrading enzymes of plant origin, based on the observation that cell wall dissolution appears to be a controlled process that does not occur adjacent to the nematode stylet but instead takes place distal to the nematode as the syncytium extends in both directions along the length of the root (Jones, 1981b). Although cell wall–degrading enzymes of plant origin have been implicated in this process (Jones and Dropkin, 1975; Jones, 1981b; Endo, 1986; Grundler et al., 1998), there is no biochemical or molecular evidence to support this hypothesis.
Recently, this issue was revisited by the confirmation of two types of endo-β-1,4-glucanase (EGase) genes, eng-1 and eng-2, that are expressed endogenously within the subventral esophageal gland cells of both cyst nematodes and RKN (Smant et al., 1998; De Boer et al., 1999; Rosso et al., 1999; Goellner et al., 2000a). Developmental expression of the HG-ENG-1 and HG-ENG-2 EGases of the soybean cyst nematode and the GT-ENG-1 and GT-ENG-2 EGases of the tobacco cyst nematode (TCN) supports a role for these enzymes in cell wall degradation during the penetration and intracellular migration of J2 through root tissue (De Boer et al., 1999; Goellner et al., 2000a). The EGases were shown to be regulated developmentally and expressed primarily in J2 within eggs before hatching, through the late J2 parasitic stage, and again only in adult males that regain their vermiform shape and migrate out of the root (De Boer et al., 1999; Goellner et al., 2000a). Immunolocalization of soybean cyst nematode EGases in cross-sections of soybean roots at 24 hr after inoculation with infective J2 showed that the HG-ENG-2 protein is secreted from the nematode stylet into plant tissue and is detectable along the migratory path of the nematode (Wang et al., 1999). In addition to a role in migration, secreted cyst nematode EGases may play a role in the initial stages of syncytia formation, because the sedentary parasitic J2 contain EGase in their subventral esophageal glands (De Boer et al., 1999; Goellner et al., 2000a). A role for cell wall–degrading enzymes of plant origin in developing syncytia is suggested by the fact that nematode EGases are not expressed in later parasitic stages as the syncytium continues to expand along the vasculature. Secretion of RKN EGases has not been evaluated in planta, but they are expressed in J2 and adult female life stages (Rosso et al., 1999). The function of RKN EGases is unclear, but it may be related to the cell wall architecture of giant cells.
In this article, we characterize EGase genes from tobacco and present definitive evidence for the expression and localization of tobacco EGases within plant cells modified for feeding by parasitic nematodes. The expressions of five different tobacco EGase genes were upregulated in plant roots infected by cyst nematodes and RKN, and transcripts of these genes were expressed differentially (both quantitatively and developmentally) within syncytia and giant cells formed by cyst nematodes and RKN, respectively. Secretion of EGases by cyst nematodes into plant root tissues was observed during J2 migration through the root cortex, but no evidence of secretion of cyst nematode EGases into developing syncytia was observed. These data support the hypothesis that a combination of plant cell wall–modifying enzymes is induced by nematode parasitism to form the elaborate plant cell wall modifications observed in nematode feeding cells.
RESULTS
Secretion of TCN EGases in Planta
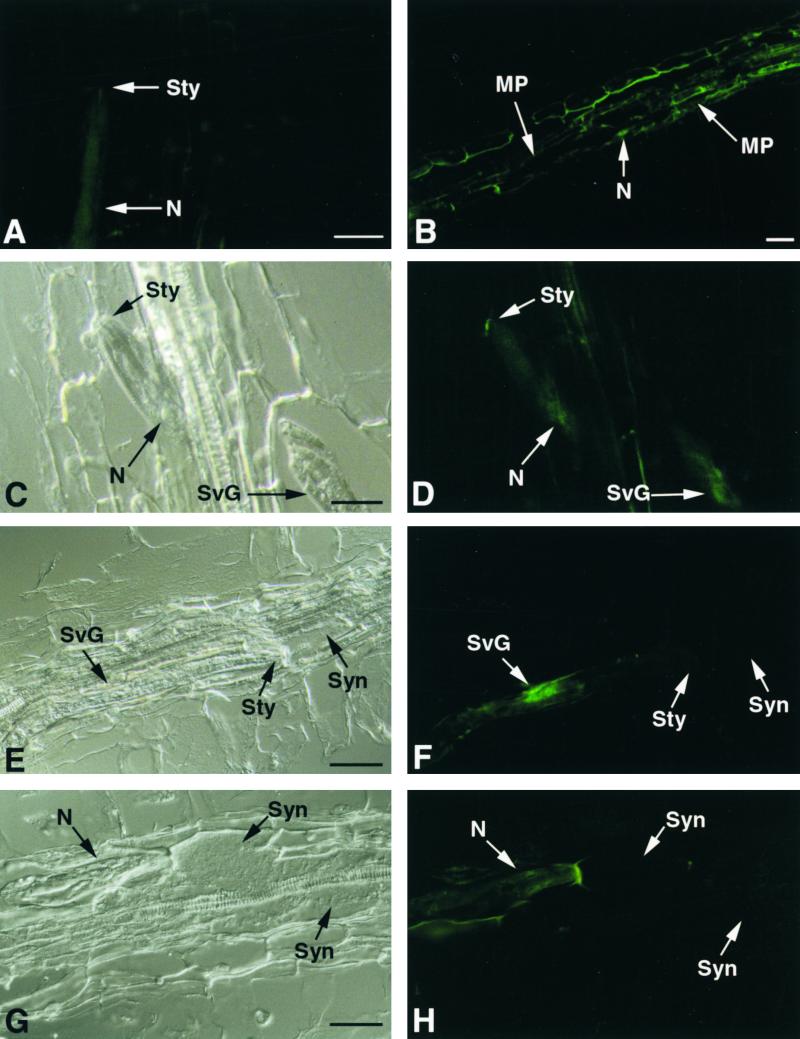
Antiserum raised against recombinant GR-ENG-1 (endoglucanase) of the potato cyst nematode Globodera rostochiensis (Smant et al., 1998) was confirmed to bind to both GT-ENG-1 and GT-ENG-2 of the TCN (Goellner et al., 2000a). GR-ENG-1 antiserum did not bind to total protein preparations from either uninfected or TCN-infected tobacco roots on protein gel blots (data not shown). No staining was observed in plant tissue sections probed with mouse preimmune serum, nor did the preimmune serum bind to the nematode cuticle (Figure 1A). Sections of infected roots at 24 hr after inoculation with J2 of TCN that were probed with GR-ENG-1 antiserum localized TCN EGases within the nematode and secreted into tobacco root cortical tissue (Figure 1B). Within the nematode, EGases were localized throughout the subventral esophageal gland cells, including their extensions, and terminated in the subventral gland ampullae at the base of the metacorpal pump chamber (Figures 1C and 1D). Within plant tissue, GR-ENG-1 antiserum localized TCN EGases that were secreted from the stylet outside the head of the nematode and along the migratory path of the nematode through tobacco root tissue (Figures 1C and 1D). Occasionally, the antiserum bound to the surface of the nematode, which may indicate the binding of secreted EGases to the cuticle as the nematode migrated forward through root tissue (Figure 1H).
Figure 1.
Immunolocalization of TCN EGases during Parasitism of Tobacco Roots.
(A) Longitudinal section through a tobacco root 24 hr after inoculation, with J2 of TCN (N) probed with mouse preimmune serum.
(B) Longitudinal section through a tobacco root 24 hr after inoculation, with TCN J2 probed with GR-ENG-1 antiserum. Binding of the GR-ENG-1 antiserum (green fluorescence) is observed along the migratory path (MP) of multiple migratory juveniles within the root cortical tissue. An arrow points to a nematode tail (N).
(C) Bright-field view of a longitudinal section through a tobacco root 24 hr after inoculation with J2 of TCN (N).
(D) Same section as (C) showing binding of GR-ENG-1 antiserum on the cell wall just outside the head of the nematode and within the nematode's subventral secretory gland cells (SvG).
(E) Longitudinal section through a tobacco root containing a sedentary parasitic J2 of TCN during early syncytium (Syn) development.
(F) Same section as (E) showing specific binding of GR-ENG-1 antiserum to the subventral secretory gland cells but not within the developing syncytium.
(G) Section through a tobacco root containing a sedentary parasitic J2 feeding from a well-developed syncytium.
(H) Same section as (G) showing slight binding of GR-ENG-1 antiserum to the surface of the nematode but not within the syncytium. Autofluorescence of plant cell wall adjacent to mematode head is visible.
Sty, nematode stylet. Bars = 50 μm.
To monitor the secretion of TCN EGases during the initiation of syncytia within host roots, tobacco roots inoculated with infective J2 were fixed for sectioning at 48 to 96 hr after infection. The time of root penetration by J2 was monitored using an inverted microscope, and the stage of nematode development and the extent of syncytium formation were determined in sections. Secreted EGases were not detected by GR-ENG-1 antiserum within initial syncytial cells during the early stages of formation, even when EGases were still detectable within the subventral gland cells of parasitic J2 (Figures 1E and 1F). Secreted TCN EGases also were not detected within well-developed syncytia (Figures 1G and 1H).
Isolation and Characterization of EGase Genes from Tobacco
A 1-kb cDNA product was amplified by reverse transcription–polymerase chain reaction (RT-PCR) from TCN-infected tobacco root tissue (data not shown) using primers designed for two conserved amino acid domains (CWERPEDM and YINAPL) present in plant EGases (Figure 2). No observable products were amplified from identical uninfected root tissue. Five distinct tobacco EGase cDNA sequences representing structurally divergent gene family members were identified after sequencing random clones of the 1-kb product and designated Ntcel2, Ntcel4, NtCel5, Ntcel7, and Ntcel8 based on the nomenclature described by del Campillo (1999). Full-length coding sequences of three of these cDNA clones (Ntcel2, Ntcel7, and Ntcel8) were obtained using 3′ and 5′ rapid amplification of cDNA ends PCR (Figure 2). The 1-kb partial cDNA sequences of Ntcel4 and Ntcel5 showed the highest percentage of nucleotide and amino acid sequence identity with tomato Cel4 and Cel5, respectively (del Campillo and Bennett, 1996; Brummell et al., 1997a), and were not characterized further because of their low expression levels (see below).
Figure 2.
Amino Acid Sequence Comparison of Ntcel2, Ntcel7, and Ntcel8.
Amino acid sequences were aligned using programs of the Wisconsin Package Version 10.0, Seqweb Version 1.1 (Genetics Computer Group, Madison, WI). Dots were introduced by the program to optimize the alignment. Black boxes depict identical amino acids among the three sequences. The vertical arrow designates the putative secretion signal peptide cleavage sites of the proteins as determined by the SignalP Version 1.1 program (Nielson et al., 1997). Horizontal arrows designate two conserved amino acid domains used to amplify tobacco EGases from nematode-infected root tissue. An extra 124–amino acid sequence encoding a putative cellulose binding domain at the C terminus of Ntcel8 is underlined.
A phylogenetic tree constructed using mature amino acid sequences of Ntcel2, Ntcel7, Ntcel8, and 14 other plant EGase sequences depicts the relatedness of the new tobacco EGases with selected members of the plant EGase gene family (Figure 3). The 1500-bp open reading frame (ORF) of the 1674-bp Ntcel2 cDNA clone encodes a 500–amino acid polypeptide, including a 35-mer putative signal peptide (Figure 2). The mature protein has a deduced molecular mass of 51.3 kD and a pI of 8.7. Ntcel2 has 54% nucleotide and amino acid sequence identity with Ntcel7 and 55% nucleotide and 49% amino acid identity with Ntcel8. Ntcel2 shares significant amino acid similarity with pepper Cel3 (89%; Trainotti et al., 1998), tomato Cel2 (86%; Lashbrook et al., 1994), and Arabidopsis Cel1 (73%; Shani et al., 1997) (Figure 3). The 1467-bp ORF of the 1723-bp Ntcel7 cDNA clone encodes a 489–amino acid polypeptide, including a putative signal peptide corresponding to amino acids 1 to 24 (Figure 2). The mature protein has a deduced molecular mass of 51.8 kD and a pI of 8.7. Analysis of the predicted amino acid sequence of Ntcel7 showed 52% nucleotide and 49% amino acid identity with Ntcel8. Ntcel7 shares significant amino acid sequence similarity with tomato Cel7 (86%; Catala et al., 1997), orange Celb1 (71%; J.K. Burns, D.J. Lewandowski, C.J. Nairn, and G.E. Brown, unpublished data), and pea Egl1 (68%; Wu et al., 1996) (Figure 3). The 1872-bp ORF of the 2286-bp Ntcel8 cDNA clone encodes a 624–amino acid polypeptide, including a 28-mer putative signal peptide (Figure 2). The mature protein has a deduced molecular mass of 65.7 kD and a pI of 8.0. This tobacco EGase shares significant amino acid similarity with tomato Cel8 (81%; Catala and Bennett, 1998) and strawberry Eg3 (79%; Trainotti et al., 1999). An amino acid sequence alignment of all three tobacco EGases shows the extra 124 amino acids at the C terminus of Ntcel8 that are absent from Ntcel2 and Ntcel7 (Figure 2).
Figure 3.
Phylogenetic Comparison of Plant EGases.
The evolutionary relationships among mature plant EGase amino acid sequences were calculated using programs of the Wisconsin Package Version 10.0, Seqweb Version 1.1 (Genetics Computer Group) and the Kimura distance correction method (Kimura, 1983). The tree was constructed using the Unweighted Pair Group Method using Arithmetic Averages (UPGMA) method.
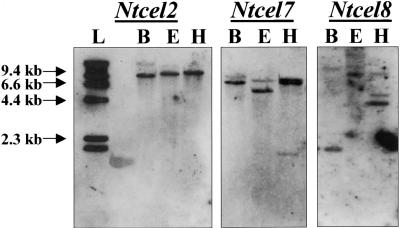
DNA gel blots of BamHI-, EcoRI-, and HindIII-digested tobacco genomic DNA were hybridized separately with 1-kb probes generated to sequences spanning two conserved amino acid domains of Ntcel2, Ntcel7, and Ntcel8 (CWERPEDM and YINAPL; Figure 2). High-stringency genomic analysis suggests that a single gene may encode Ntcel2. In contrast, both Ntcel7 and Ntcel8 appear to belong to small gene families (Figure 4).
Figure 4.
DNA Gel Blot Analysis of Ntcel2, Ntcel7, and Ntcel8 Genes in Tobacco.
Genomic DNA (5 μg) was digested with BamHI (B), EcoRI (E), and HindIII (H), electrophoresed on a 0.7% agarose gel, blotted to nylon membranes, and probed with a 1-kb fragment spanning the conserved amino acid domains CWERPED and YINAPL of Ntcel2, Ntcel7, and Ntcel8. Blots were hybridized in 5 × SSC at 65°C and washed twice in 0.5 × SSC at 68°C and twice in 0.1 × SSC at 68°C. L, 1-kb DNA ladder (Gibco BRL).
Upregulation of Tobacco EGases in Nematode-Infected Roots
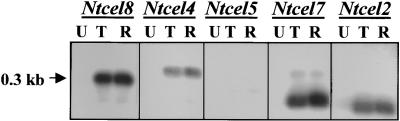
To compare the relative expression of individual tobacco EGase genes in noninfected and nematode-infected (RKN or TCN) tobacco root tissue, we analyzed the presence of EGase mRNA by RT-PCR. Gene-specific primer sets designed for each of the five tobacco EGase cDNA sequences amplified fragments of the following sizes: Ntcel2, 181 bp; Ntcel4, 350 bp; Ntcel5, 245 bp; Ntcel7, 209 bp; and Ntcel8, 300 bp (Figure 5). These same size fragments were not amplified from tobacco genomic DNA or from control reactions that did not contain reverse transcriptase, excluding the possibility of contaminating genomic DNA in the results. Equivalent amounts of either RNA or root tissue were used for the synthesis of first-strand cDNAs, and the relative amounts of amplification products were consistently higher in nematode-infected roots than in uninfected roots for each tobacco EGase gene in three separate experiments (Figure 5). When the entire uninfected root PCR reaction was loaded on a gel, only a faint band could be detected (data not shown). DNA gel blots of amplified fragments hybridized with probes for each tobacco EGase cDNA confirmed the identity of the products (Figure 5). The three genes showing the highest expression levels (Ntcel2, Ntcel7, and Ntcel8) were selected for further analysis.
Figure 5.
Relative RT-PCR Analysis of Tobacco EGase Transcripts in Uninfected and Nematode-Infected Root Tissue.
RT-PCR products were amplified from uninfected (U), TCN-infected (T), or RKN-infected (R) tobacco root tissue 7 to 9 days after infection using tobacco EGase gene-specific primers. A DNA gel blot of the RT-PCR products was probed with tobacco EGase digoxigenin-labeled DNA probes.
In Situ Localization of Tobacco EGase Transcripts in RKN-Infected Roots
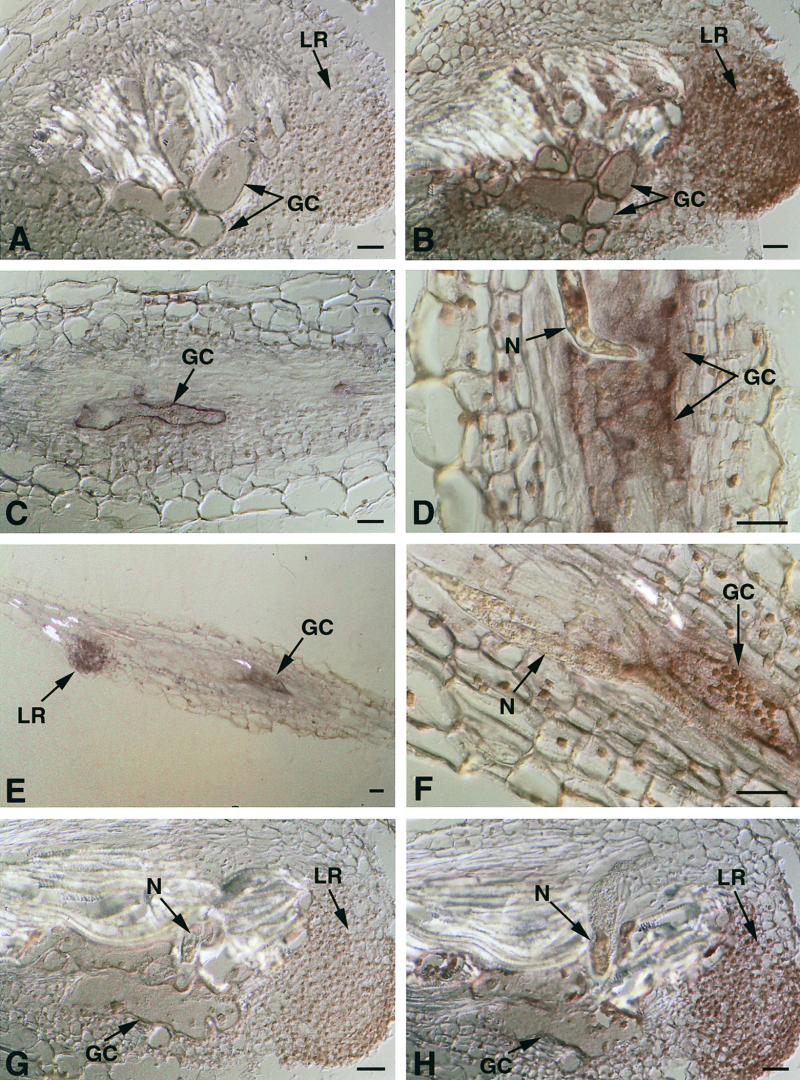
To determine the spatial expression pattern of tobacco Ntcel2, Ntcel7, and Ntcel8 EGases in nematode-infected root tissue, we conducted mRNA in situ hybridizations on sections cut at weekly intervals from developing tobacco galls after inoculation with RKN. RT-PCR experiments indicated that tobacco EGase expression levels were relatively low; therefore, serial sections were cut to a thickness of 20 μm to increase the transcript volume for detecting hybridization signals. Figure 6 depicts longitudinal sections through RKN-infected root galls hybridized with either antisense or sense tobacco EGase riboprobes. Figures 6A and 6B illustrate the giant cell–specific expression of Ntcel7 in serial sections through a RKN-induced tobacco gall 12 to 14 days after infection. Strong labeling was detected within the giant cells and lateral root primordia (Figure 6B) in sections probed with the antisense Ntcel7 riboprobe but not in sections probed with a sense Ntcel7 riboprobe (Figure 6A). Figure 6C also depicts strong expression of Ntcel7 in developing giant cells during the early stages of nematode development (7 to 9 days after infection) but not in surrounding cortical parenchyma cells. Sections probed with tobacco 18S rRNA sense and antisense riboprobes were used as controls. Strong hybridization signal was detected in all gall tissues with the antisense, but not the sense, 18S rRNA probe (data not shown).
Figure 6.
In Situ Hybridization of Ntcel7, Ntcel8, and Ntcel2 mRNA in RKN-Infected Tobacco Roots.
(A) and (B) Longitudinal serial sections through a RKN-induced gall on tobacco roots 12 to 14 days after infection and hybridized with a tobacco Ntcel7 sense (A) or antisense (B) DIG-labeled riboprobe.
(C) Longitudinal section through a RKN-induced gall on tobacco roots 7 to 9 days after infection and hybridized with a tobacco Ntcel7 antisense DIG-labeled riboprobe.
(D) to (F) Longitudinal sections through tobacco roots infected with RKN and hybridized with a tobacco Ntcel8 antisense DIG-labeled riboprobe at 7 to 9 days after infection. (F) is a higher magnification of (E).
(G) and (H) Longitudinal serial sections through a tobacco gall induced by RKN 12 to 14 days after infection and hybridized with a tobacco Ntcel2 sense (G) or antisense (H) DIG-labeled riboprobe.
GC, giant cells; LR, lateral root; N, nematode. Bars = 50 μm.
In situ mRNA hybridizations using tobacco Ntcel8 sense and antisense riboprobes showed a similar expression pattern to that of Ntcel7 in sections of RKN-induced gall tissues (Figure 6). During the early stages of infection (7 to 9 days after infection), Ntcel8 transcripts were localized specifically within developing giant cells and lateral root primordia when probed with an antisense Ntcel8 riboprobe (Figures 6D to 6F) but not a sense riboprobe (data not shown). During the later stages of infection (12 to 14 days after infection), Ntcel8 transcripts still were detected in giant cells (data not shown). Consistently, the signal intensity for Ntcel7 was stronger than that for Ntcel8, but it is unclear if this reflects a true quantitative difference in expression levels.
Tobacco Ntcel2 sense and antisense riboprobes were used to probe sections through RKN-induced gall tissue (Figures 6G and 6H). In sections probed with the Ntcel2 antisense riboprobe, the overall signal intensity was much lower than signal intensities observed in sections probed with either Ntcel7 or Ntcel8 antisense riboprobes. Weak hybridization of Ntcel2 was detected in giant cells and in lateral root primordia at 12 to 14 days after infection with an antisense Ntcel2 riboprobe (Figure 6H) but not a sense Ntcel2 riboprobe (Figure 6G).
In Situ Localization of Tobacco EGase Transcripts in TCN-Infected Roots
To determine the spatial expression pattern of tobacco Ntcel2, Ntcel7, and Ntcel8 EGases in TCN-infected root tissue, we conducted mRNA in situ hybridizations on sections cut from TCN-infected tobacco roots at 7 days after infection (Figure 7). Longitudinal serial sections through TCN-infected tobacco roots were cut to a thickness of 12 μm because roots were much thinner than were RKN-induced galls and we wanted to increase the number of sections containing nematodes. Thinner sectioning may have contributed to a slight loss in hybridization signal intensity (target transcript) compared with in situ hybridizations of RKN-infected root tissue (20 μm thick). In situ hybridizations of tobacco EGase riboprobes to TCN-infected root tissue sections required 6 hr of color development, in contrast to 2 to 4 hr of color development needed for hybridizations of RKN-infected root tissue sections.
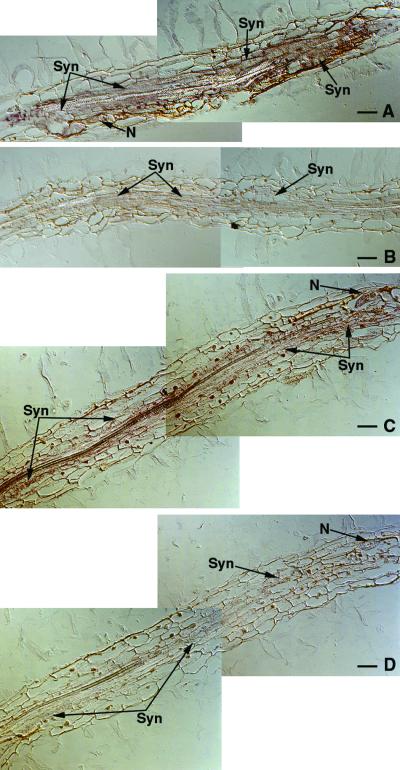
Figure 7.
In Situ Hybridization of Tobacco Ntcel7 and Ntcel8 mRNA in TCN-Infected Tobacco Roots.
(A) and (B) Longitudinal serial sections through tobacco roots infected with TCN and hybridized with a tobacco Ntcel7 antisense (A) or sense (B) DIG-labeled riboprobe 7 days after infection.
(C) and (D) Longitudinal serial sections through tobacco roots infected with TCN and hybridized with a tobacco Ntcel8 antisense (C) or sense (D) DIG-labeled riboprobe 7 days after infection.
N, nematode; Syn, syncytia. Bars = 50 μm.
Longitudinal sections through TCN-infected tobacco roots were treated with antisense or sense tobacco Ntcel2, Ntcel7, Ntcel8, and 18S rRNA riboprobes. Figure 7 shows strong expression of Ntcel7 in TCN-induced syncytia at 7 days after inoculation in sections probed with an antisense (Figure 7A) but not a sense (Figure 7B) Ntcel7 riboprobe. Ntcel7 transcripts also were detected in lateral root tips with the antisense but not the sense Ntcel7 riboprobe, as observed in RKN-infected tobacco root tissue (data not shown).
In situ mRNA hybridizations using tobacco Ntcel8 antisense and sense riboprobes showed a similar expression pattern to that of Ntcel7 in sections of TCN-infected root tissues (Figures 7C and 7D). At 7 days after infection, Ntcel8 transcripts were localized within developing TCN-induced syncytia when probed with an antisense Ntcel8 riboprobe (Figure 7C) but not a sense Ntcel8 riboprobe (Figure 7D). Hybridization signal intensity was much stronger for Ntcel7 than for Ntcel8 and may reflect a quantitative difference in transcript levels, as observed for these probes when hybridized to RKN-infected tobacco root tissue. Ntcel8 transcripts also were detected in lateral root tips with the antisense but not the sense Ntcel8 riboprobes. In situ mRNA hybridizations using tobacco Ntcel2 sense and antisense riboprobes did not show an observable difference in hybridization signal intensity (data not shown). Ntcel2 transcript levels may not be high enough for detection using this method. Sections probed with control tobacco 18S rRNA antisense and sense riboprobes produced strong hybridization signals in all cell types but were more intense within TCN-induced syncytia (data not shown).
DISCUSSION
One of the most distinguishing characteristics of feeding cells in plants induced by RKN and cyst nematode parasitism is the extensive remodeling of their cell walls. Cell expansion, cell elongation, cell wall biosynthesis, and cell wall dissolution are all physiological processes that have been observed indirectly within nematode-induced feeding cells (Jones, 1981a; Endo, 1986). The incorporation of new plant cells via cell wall dissolution is a hallmark of cyst nematode–induced syncytia (Endo, 1986). This report describes the identification of several EGase genes expressed in roots of tobacco, and combined with the temporal and spatial expression of plant EGases and nematode EGase secretion in planta, it provides molecular confirmation of the origin of cell wall–degrading enzymes involved in feeding cell formation. Antiserum specific for TCN EGases confirmed the secretion of nematode EGases into plant tissue at 24 hr after infection during the migration of infective J2 through root tissue. TCN EGases were detected within the secretory gland cells as well as within plant tissue along the migratory path of the J2 through the root cortex. EGase was detected outside the tip of the nematode head on the cell wall in contact with the migrating nematode, similar to what was observed for the soybean cyst nematode (Wang et al., 1999). Once TCN J2 became sedentary adjacent to root vascular tissue (48 hr after infection), no EGase secreted from the nematode was detected outside or inside the initial syncytial cells. By 96 hr after infection, no TCN EGase was detectable within either the nematodes or the expanding syncytia. Although the limits of detection and localization of secreted nematode EGase may warrant further evaluation, the lack of detection in both the nematode and the plant at this stage was consistent with the developmental cessation of EGase expression in TCN observed previously (Goellner et al., 2000a). Because nematode-induced syncytia continue to expand for several weeks after this time (Endo, 1986), it is likely that the cell wall–modifying enzymes involved in this process were of plant origin.
There is considerable evidence of a role for plant EGases during plant developmental processes involving changes in cell wall architecture, ranging from cell wall expansion to dissolution (Brummell et al., 1994; Cosgrove, 1999; Rose and Bennett, 1999). Our report provides direct evidence to support a role for plant EGases during nematode-induced feeding cell development within susceptible host plant roots. Five tobacco cDNA clones (Ntcel2, Ntcel4, Ntcel5, Ntcel7, and Ntcel8), each encoding different EGases, were obtained from nematode-infected roots by RT-PCR using degenerate primers to conserved amino acid domains of plant EGases. Only Ntcel2, Ntcel7, and Ntcel8 were expressed at high enough levels for analyses. Interestingly, all three tobacco EGases appear to be upregulated in response to parasitism by both RKN and TCN. Tobacco Ntcel7 and Ntcel8 were expressed exclusively within RKN-induced giant cells and TCN-induced syncytia and root tips. Initially, this result was somewhat surprising, because extensive cell wall dissolution does not contribute to the formation of RKN-induced giant cells (Huang and Maggenti, 1969), as is the case with syncytia (Endo, 1986). Although giant cells are not formed by the cell wall dissolution of adjacent cells, they must modify their cell wall architecture to expand and elongate.
Our observation that multiple plant EGases are expressed similarly in both giant cells and syncytia, although the formation of the feeding cell phenotypes is very different, may be explained by slight differences in the mechanisms of action and substrate specificities of particular EGase isoforms. It is also possible that the tobacco EGases play a role in similar processes that occur in both developing syncytia and giant cells (e.g., during cell expansion or the formation of elaborate wall ingrowths). The five tobacco EGases isolated in this study may represent a subset of a larger group of tobacco EGases expressed in nematode feeding cells. Additional tobacco EGase gene family members (yet to be identified) may be expressed in syncytia, in which extensive cell wall degradation occurs, but not in developing giant cells. Cellular localization signals for degradative enzymes also likely play a role because the initial cell wall dissolution observed in syncytia is targeted to the plasmodesmata of adjacent cells (Grundler et al., 1998). In addition, EGases may be only one class of cell wall–modifying enzymes recruited to form nematode-induced feeding cells. The coordinated cell wall modifications that occur in developing syncytia and giant cells are complex processes that probably require the synergistic activity of several different types of plant cell wall–modifying proteins (e.g., EGases, expansins, and xyloglucan endotransglycosylases) (Cosgrove, 1999) as well as the coordinated expression of multiple gene family members within a single class of cell wall–modifying proteins (e.g., EGases).
Until now, the tobacco EGase gene family has remained relatively uncharacterized. To date, a single Nicotiana alata sequence encoding an EGase expressed in mature pistils has been reported in GenBank (B.A. McClure and B. Mou, unpublished data) and shares the highest amino acid sequence similarity with Ntcel8 (54% identical). Multiple divergent EGase genes, however, have been reported in a number of different plant species, and simultaneous and overlapping expression of EGases has been observed during different phases of plant development (Lashbrook et al., 1994; del Campillo and Bennett, 1996; Gonzalez-Bosch et al., 1996). In tomato, EGases are encoded by at least eight members of a divergent gene family that exhibit differential patterns of tissue-specific expression and regulation by hormones (Rose et al., 1997). In Arabidopsis, the EGase gene family comprises >12 members (del Campillo, 1999). Primary sequence comparisons and hydrophobic cluster analysis have been used to group EGases according to properties exhibited within the catalytic cores of the proteins (Henrissat et al., 1989). Plant EGases have been assigned to glycosyl hydrolase family 9. Based on primary sequence comparisons, Ntcel2, Ntcel7, and Ntcel8 maintain multiple conserved amino acid domains of plant and microbial EGases and can be assigned to glycosyl hydrolase family 9 (Henrissat et al., 1989). Family 9 glycosyl hydrolases also contain members with and without a putative cellulose binding domain (CBD), suggesting potential differences in substrate specificities. In contrast to Ntcel2 and Ntcel7, Ntcel8 contains a putative CBD. EGases with and without a CBD may have distinct biochemical functions and substrate specificities that contribute to the observed changes in feeding cell wall architecture.
No significant sequence similarity exists between the tobacco EGases and nematode EGases, which are classified into glycosyl hydrolase family 5 (Davis et al., 2000). The cyst nematode EGase catalytic domains are related most closely to bacterial EGases, and the ENG-1 proteins contain a C-terminal CBD similar to type II binding domains of bacteria (Smant et al., 1998; Goellner et al., 2000a).
Nucleotide and amino acid sequence comparisons among the three tobacco EGases revealed between 49 and 55% identity. Amino acid sequence comparisons of Ntcel2, Ntcel7, and Ntcel8 with other plant EGases indicate that tobacco Ntcel2 has the highest percentage of amino acid similarity to pepper Cel3 (89%) and tomato Cel2 (86%), two genes that appear to be regulated by ethylene and that are expressed primarily during fruit ripening (Lashbrook et al., 1994; Trainotti et al., 1998). Ntcel2 also is 73% identical to Arabidopsis Cel1, an elongation-specific endoglucanase (Shani et al., 1997). Interestingly, the Arabidopsis Cel1 promoter recently was shown to be upregulated in RKN-induced giant cells as a Cel1–β-glucuronidase reporter gene fusion in transgenic tobacco (Goellner et al., 2000b). Ntcel7 is most similar to tomato Cel7 and pea Egl1, both of which are induced by auxin and constitute a distinct subfamily of EGases (Wu et al., 1996; Catala et al., 1997). In tomato, Cel7 is induced by auxin and highly expressed in rapidly expanding vegetative tissues and in developing fruit during immature and mature green fruit stages (Catala et al., 1997). Using the tomato mutants Nr (ethylene insensitive) and dgt (auxin insensitive), the observed increase in tomato Cel7 after the application of auxin was shown to be auxin specific rather than an indirect result of auxin-induced ethylene production (Catala et al., 1997). Recently, auxin was shown to mediate the induction and morphogenesis of cyst nematode–induced feeding cells (Goverse et al., 2000). In the tomato dgt mutant, potato cyst nematode–induced feeding cell development was inhibited. On occasion, female development was observed, but the syncytia were half the size of those formed in wild-type plants and cell wall dissolution was severely restricted (Goverse et al., 2000).
Ntcel8 is most similar to the recently identified, yet uncharacterized, tomato Cel8 (Catala and Bennett, 1998) and strawberry Eg3 (Trainotti et al., 1999), both of which contain a putative type-1 CBD (Beguin and Aubert, 1994). It has been proposed that these EGases may function to hydrolyze xyloglucans coating the cellulose microfibrils for subsequent hydrolysis by EGases lacking a CBD (Trainotti et al., 1999). Recently, a divergent class of plant EGases encoding type II integral membrane proteins were localized to the plasma membranes of higher plants (Brummell et al., 1997b; Nicol et al., 1998). These proteins lack the secretory signal peptide normally found in other plant EGases identified to date, do not contain a CBD, and are not hormonally regulated. It has been suggested that this group of plant EGases may have a function in cell wall biosynthesis (Brummell et al., 1997b; Nicol et al., 1998). Interestingly, as additional cells are incorporated into an expanding syncytium via cell wall dissolution, cell wall thickening and ingrowth formation occur in other parts of the syncytium (Dropkin, 1969). In addition to a role in catalyzing cell wall degradation, EGases may play an important role during cell wall biosynthesis in nematode feeding cells.
Nematode feeding cell development is a complex, highly regulated process. The recruitment of putative plant EGases by both cyst nematodes and RKN as components of cell wall modifications in feeding cells is consistent with observations that these nematodes induce numerous changes in plant gene expression during parasitism (Bird, 1996). It is hypothesized that nematodes modify the temporal and spatial expression of plant genes whose wild-type functions have been diverted to serve the parasitic needs of the nematode (Bird, 1996). These gross modifications in plant gene expression highlight the potential value of using plant–nematode interactions as a tool to investigate plant cellular biology.
METHODS
Plant Material
Tobacco (Nicotiana tabacum cv NC95) seed were surface-sterilized with 2.5% sodium hypochlorite for 5 min, followed by several rinses with sterile water, and germinated on Petri plates containing 0.8% Noble agar (Fisher Scientific, Pittsburgh, PA) supplemented with Murashige and Skoog (1962) minimal medium, pH 5.8, and 3% sucrose. Tobacco seedlings were grown in a controlled-temperature growth chamber at 25°C with a 14-hr photoperiod.
Nematode Cultures and Inoculations
The tobacco cyst nematode (TCN; Globodera tabacum ssp solanacearum) (Miller and Gray, 1972) and the root-knot nematode (RKN; Meloidogyne incognita Race 4) (Hartman and Sasser, 1985) were propagated on greenhouse-grown tobacco and tomato (Lycopersicon esculentum cv Rutgers), respectively. To isolate cyst nematode eggs, TCN cysts were crushed gently in a glass homogenizer and the eggs were rinsed onto a 25-μm sieve. Hatching of TCN eggs was stimulated over filter-sterilized tobacco root diffusate (LaMondia, 1995) at 28°C on a Baermann pan. RKN eggs were isolated from egg masses on tobacco roots with 0.5% sodium hypochlorite and then rinsed with water and collected on a 25-μm sieve (Hussey and Barker, 1973). RKN eggs were set up to hatch over water at 28°C on a Baermann pan. After 3 days, hatched second-stage juveniles (J2) of either TCN or RKN were collected on a 25-μm sieve and surface-sterilized with 0.002% mercuric chloride, 0.002% sodium azide, and 0.001% Triton X-100 for 5 min, followed by several washes with sterile water. Surface-sterilized J2 were resuspended in 50 μL of 2 mM penicillin G and 950 μL of 1.5% low-melting-point agarose at 37°C at a concentration of ∼50 J2 / 10 μL for TCN and 5 J2 / 10 μL for RKN. Ten-microliter aliquots of J2 were used to inoculate 2-week-old tobacco root tips.
Isolation and Sequence Characterization of Tobacco Endo-β-1,4-Glucanases
To isolate poly(A+) RNA, 5-cm infected or noninfected tobacco root pieces (excluding root tips) were excised from Petri plates and ground in a small glass homogenizer in 250 μL of lysis binding buffer (100 mM Tris-HCl, pH 7.5, 500 mM LiCl, 10 mM EDTA, pH 8.0, 5 mM DTT, and 1% LiDS [Dynal, Lake Success, NY]). After lysis, the homogenate was centrifuged for 1 min at 13,000g, and the supernatant was transferred to a clean tube. Twenty-five microliters of Dynal magnetic oligo(dT)25 beads equilibrated with lysis binding buffer was added to the supernatant and placed on a rotator for 5 min to allow the mRNA to anneal to the beads. Using a magnetic stand, the beads were washed twice in washing buffer with LiDS (10 mM Tris-HCl, pH 7.5, 0.15 M LiCl, 1 mM EDTA, and 0.1% LiDS) and three times in washing buffer without LiDS. For first-strand cDNA synthesis, the beads were washed several times in 1 × first-strand buffer (25 mM Tris-HCl, pH 8.3, 37.5 mM KCl, and 1.5 mM MgCl2) and then resuspended in 12 μL of RNase-free water. The following components were added to the bead suspension: 4 μL of 5 × first-strand buffer, 2 μL of 0.1 M DTT, 1 μL of 10 mM deoxynucleotide triphosphate (dNTP) mix, and 1 μL of Superscript II reverse transcriptase (200 units/μL; Gibco BRL, Rockville, MD). The reaction mixture was incubated on a rotator at 42°C for 1 hr. After first-strand cDNA synthesis, 2 units of RNase H was added to the reaction and allowed to incubate at 37°C for 20 min. The beads were rinsed twice with TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), resuspended in 25 μL of TE, and stored at −20°C.
Two degenerate primers to conserved amino acid domains of known plant EGase sequences were designed as follows: CWERPED (5′-TGTTGGGARAGRCCHGARGAY-3′) and YINAPL2 (5′-MACHADHGSWGCATTRAYRTAWGT-3′), where R = A + G, Y = C + T, M = A + C, S = G + C, W = A + T, H = A + T + C, and D = G + A + T. A 10-μL aliquot of first-strand cDNA on the beads was washed with 1 × polymerase chain reaction (PCR) buffer (20 mM Tris-HCl, pH 8.4, and 50 mM KCl) before adding the following reaction components: 5 μL of 10 × PCR buffer (200 mM Tris-HCl, pH 8.4, and 500 mM KCl), 1.5 μL of 50 mM MgCl2, 1.0 μL of 10 mM dNTP mix, 2.5 μL of 10 μM 5′-CWERPED, 2.5 μL of 10 μM 3′-YINAPL2, 27 μL of distilled H2O, and 2.5 units of Taq polymerase. The PCR cycles were as follows: 1 cycle at 94°C for 2 min; 5 cycles at 94°C for 1 min, 37°C for 1 min, 72°C for 2 min with a ramp of 14°C / min between the annealing and elongation steps (Compton, 1990); 35 cycles at 94°C for 30 sec, 50°C for 50 sec, 72°C for 2 min; and 1 cycle at 72°C for 10 min. A 1-kb amplified fragment was obtained and cloned into the pCR2.1 TA cloning vector (Invitrogen, Carlsbad, CA). Plasmid DNA was isolated from several individual transformants, and the cDNA inserts were sequenced. Sequencing was performed at the Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Laboratory at the University of Florida (Gainesville). Isolation of the full- length tobacco endo-β-1,4-glucanase (EGase) cDNAs was accomplished using 3′ and 5′ random amplification of cDNA ends systems (Gibco BRL) according to the manufacturer's protocols.
DNA Gel Blot Analysis
Tobacco genomic DNA was isolated from young leaves using the DNeasy Plant Maxi isolation kit (Qiagen, Valencia, CA). Genomic DNA (5 μg) was digested separately with EcoRI, BamHI, and HindIII, electrophoresed on a 0.7% (w/v) agarose gel, and transferred to Hybond-N membrane (Amersham, Arlington Heights, IL). Tobacco genomic DNA was hybridized with digoxigenin (DIG)-labeled tobacco EGase DNA probes corresponding to the 1-kb nucleotide sequence spanning the two conserved amino acid domains, CWERPED and YINAPL, described above. Hybridizations were performed in standard hybridization buffer (5 × SSC [1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate], 0.1% N-lauroylsarcosine, 0.02% SDS, 1% blocking reagent [Roche Molecular, Indianapolis, IN]) at 65°C for 18 hr followed by three 10-min washes in 2 × SSC at room temperature. The membrane was then washed twice at 68°C with 0.5 × SSC for 30 min and twice at 68°C with 0.1 × SSC for 30 min. After incubating the membranes in 1% blocking reagent for 1 hr, the membranes were incubated with a 1:10,000 dilution of sheep anti-DIG alkaline phosphatase (AP) conjugate for 30 min. Unbound antibody was removed by three 15-min washes with maleic acid wash buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5, and 0.3% Tween 20). The membrane was incubated in AP detection buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2) for 10 min followed by a 1:100 dilution of the chemiluminescent substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)pheryl phosphate (CSPD) (Roche Molecular) before wrapping the membrane in Saran Wrap and exposing it to x-ray film for 2.5 hr.
Relative Reverse Transcription–PCR of Tobacco EGases from Infected and Noninfected Roots
Reverse transcription (RT)–PCR was conducted on mRNA extracted from equivalent amounts of total RNA or mRNA isolated from equivalent amounts of nematode-infected tobacco root tissue. The same results were obtained using both methods. Total RNA was isolated using the Puregene RNA isolation kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's instructions and treated with RNase-free DNase I (2 units/μL; Ambion, Austin, TX). The DNase I was removed using DNA-Free (Ambion) according to the manufacturer's protocol. Oligo(dT) magnetic beads (Dynal) were used to extract mRNA from 500 ng of total RNA or directly from tissue ground in lysis binding buffer as described above. The mRNA was eluted from the beads at 65°C for 2 min in 11.5 μL of diethyl pyrocarbonate–treated water. First-strand cDNAs were synthesized using a 3′ degenerate tobacco EGase primer including a BamHI restriction site (3′ Tobcelribo BamHI, 5′-CGCGGATCCGGRTTRTYWCCHARHAWRTARTCHACYTG-3′, where R = A + G, Y = C + T, W = A + T, and H = A + T + C) that recognized all five tobacco EGase cDNA sequences. RT reactions contained the following components: 4 μL of 5 × first-strand buffer (250 mM Tris, pH 8.3, 375 mM KCl, and 15 mM MgCl2), 2 μL of 0.1 M DTT, 1 μL of 10 mM dNTP mix, 0.5 μL of 10 μM 3′ Tobcelribo BamHI primer, 11.5 μL of isolated mRNA from above, and 1 μL of Superscript reverse transcriptase (200 units/μL; Gibco BRL). The reaction was incubated at 42°C for 1 hr, followed by the addition of 2 units of RNase H (Gibco BRL) and an additional 20-min incubation at 37°C. The cDNA was precipitated and resuspended in 10 μL of water. Reactions without the addition of reverse transcriptase were included as a control for contaminating DNA, although amplifications of tobacco genomic DNA using each EGase primer set did not generate fragments of the size predicted for amplification from cDNA.
5′ gene-specific primers containing an EcoRI restriction site and the 3′ Tobcelribo BamHI primer were used in subsequent PCR amplifications. 5′ primer sequences were as follows: 5′ Tobcel2 EcoRI, 5′-CCGGAATTCGTAACATGCAGCATGTGACATCC-3′; 5′ Tobcel4 EcoRI, 5′-CCGGAATTCGCATGTTGGAGCAAGGATCTTC-3′; 5′ Tobcel5 EcoRI, 5′-CCGGAATTCCCAGGCTCACCTAGCTTTCAAGC-3′; 5′ Tobcel7 EcoRI, 5′-CCGGAATTCGGGGGCCTAATTTACAAG-CTAAC-3′; and 5′ Tobcel8 EcoRI, 5′-CCGGAATTCCCATGCACC-TGTGTTTGAGAAGTAC-3′. PCRs contained the following components: 5 μL of 10 × PCR buffer (200 mM Tris, pH 8.4, and 500 mM KCl), 1.5 μL of 50 mM MgCl2, 1.0 μL of 10 mM dNTP mix, 2.5 μL of 5′ primer, 2.5 μL of 3′ primer, 2 μL of cDNA, 35 μL of water, and 0.5 μL of Taq polymerase (5 units / μL). PCR cycles consisted of an initial denaturation step at 94°C for 2 min, followed by 40 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 2 min, and a final 10-min elongation step at 72°C. Ten-microliter aliquots of each RT-PCR reaction were electrophoresed on a 2% agarose gel, transferred to nylon membranes, and hybridized with corresponding tobacco EGase DIG-labeled DNA probes.
Tissue Fixation and Embedding
For immunolocalization, TCN-infected root pieces were excised from Petri plates 24 to 96 hr after inoculation and fixed in 1% paraformaldehyde (PFA) in PBS (137 mM NaCl, 1.4 mM KH2PO4, 2.6 mM KCl, and 1.8 mM Na2HPO4, pH 7.4) for 3 hr at room temperature. After two 15-min washes with PBS, the fixed root pieces were dehydrated in a graded ethanol series (30, 60, 70, 85, 95, and 100%, 15 min each) and then incubated sequentially in ethanol:Histoclear (National Diagnostics, Atlanta, GA) at 75:25, 50:50, and 25:75 for 10 min each. After two 15-min incubations in 100% Histoclear, the root pieces were transferred to molten Paraplast Plus (Fisher Scientific) at 60°C for 2 hr and embedded in blocks. For in situ mRNA localizations, nematode-infected tobacco root pieces were dissected from Petri plates 7 to 9 days or 12 to 14 days after infection and infiltrated with 4% PFA in PBS (130 mM NaCl, 7 mM Na2H2PO4, and 3 mM NaH2PO4) using a short vacuum. The root pieces were then transferred to fresh 4% PFA, incubated an additional 6 hr at room temperature, followed by 17 hr in 4% PFA at 4°C. The root pieces were washed twice in PBS for 20 min each time, dehydrated in a graded ethanol series (30, 60, 70, 85, 95, and 100%), incubated sequentially in Histoclear:ethanol at 25:75, 50:50, and 75:25, and then in 100% Histoclear twice for 30 min each time. The root pieces were incubated in Histoclear:Paraplast (Fisher Scientific) at 75:25 overnight at 60°C and then overnight again in pure Paraplast at 60°C. The Paraplast-embedded root pieces were sectioned to a thickness of 12 μm (TCN-infected tobacco root tissue) or 20 μm (RKN-infected tobacco root tissue) using a rotary microtome (American Optical, Buffalo, NY) and adhered to Superfrost Plus microscope slides (Fisher Scientific) overnight at 40°C on a slide warmer. Three 15-min incubations in Histoclear were used to remove the Paraplast from the sections followed by a graded ethanol series and a final incubation in water to rehydrate the sections.
In Planta Localization of TCN EGases
TCN-infected roots were sectioned to a thickness of 10 μm using a rotary microtome (American Optical) and placed on Superfrost Plus (Fisher Scientific) microscope slides. The sections were adhered to the slides overnight on a 40°C slide warmer. Three 10-min incubations in Histoclear at room temperature were used to remove the paraffin from the sections followed by rehydration in a graded ethanol series and a final incubation in water. Nonspecific binding sites in sections were blocked with 10% normal goat serum containing protease inhibitors (10 μL/mL of stock A = [0.1 mM leupeptin, 100 mM Na2EDTA, and 20 mM iodoacetamide]; 10 μL/mL of stock B = [20 mM phenylmethylsulfonyl fluoride and 0.1 mM pepstatin A]; all chemicals from Sigma, St. Louis, MO) for 3 hr at room temperature. Primary antibody (GR-ENG-1 mouse polyclonal serum; Smant et al., 1998) diluted 1:100 with 10% goat serum in PBS was applied to the sections and incubated overnight at 4°C. After three 5-min rinses in PBS, secondary Alexa 488–goat anti-mouse IgG conjugate (Molecular Probes, Eugene, OR) diluted 1:500 was applied to the sections and allowed to incubate in the dark for 3 hr at room temperature. The sections were rinsed three times for 5 min each in PBS before mounting with antiquenching agent (0.2 M carbonate buffer, pH 8.6, 50% glycerol, and 0.02 mg/mL p-phenylenediamine). Sections were observed and photographed on an epifluorescence microscope (Zeiss, Oberkochen, Germany).
DIG-Labeled RNA Probes
Antisense and sense RNA probes were synthesized to Ntcel2, Ntcel7, and Ntcel8 EGase cDNAs by in vitro transcription. Gene-specific primer sets containing EcoRI and BamHI restriction sites were designed for each of the three tobacco EGase cDNA sequences and used to amplify products ranging in size from 150 to 210 bp. The 5′ Tobcel2 EcoRI and 5′ Tobcel7 EcoRI primers were used together with the 3′ Tobcelribo BamHI primer to amplify products of 181 and 209 bp, respectively of Ntcel2 and Ntcel7. 5′ Tobcel8-2 EcoRI (5′-CCGGAATTCCGTCTTTCGGAACAGCAACCCT-3′) and 3′ Tobcel8-2 BamHI (5′-CGCGGATCCTTCATCTGCGTATCCACTGACAG-3′) were used to amplify a 150-bp fragment from Ntcel8. Two primers designed for conserved regions of plant 18S rRNA genes were used to amplify an 87-bp fragment of the tobacco 18S rRNA gene to be used as a control riboprobe. Primer sequences were as follows: Plant 18S 5′ EcoRI, 5′-CCGGAATTCGAATGATCCGGTGAAGTGTTCGG-3′; and Plant 18S 3′ BamHI, 5′-CGCGGATCCGATAAGGTTTAGTGG-ACTTCTCGC-3′. Each product was digested with both EcoRI and BamHI and cloned into a pBluescript SK+ transcription vector with a truncated multiple cloning site (De Boer et al., 1998) and flanking T3 and T7 promoter sequences. Purified plasmid DNA corresponding to each tobacco riboprobe clone was digested separately with EcoRI and BamHI and column purified, and 1 to 2 μg was added to in vitro transcription reactions containing DIG-UTP (Roche Molecular) and the appropriate polymerase to synthesize RNA probes. Unincorporated nucleotides were removed using mini Quick Spin RNA columns (Roche Molecular), and the incorporation of DIG-UTP was quantified by dot blot analysis.
In Situ RNA Hybridization
Rehydrated tissue sections were pretreated with proteinase K (1 μg/mL [12-μm sections] or 2 μg/mL [20-μm sections] in 100 mM Tris-HCl and 50 mM EDTA, pH 8.0) for 30 min at 37°C, rinsed for 5 min twice with TBS buffer (150 mM NaCl and 10 mM Tris-HCl, pH 7.5), incubated in TBS with glycine (2 mg/mL) for 2 min, and then washed again for 5 min twice with TBS buffer. Sections were postfixed with 4% PFA in PBS for 5 min and washed in TBS for 5 min. After acetylation with acetic anhydride (0.25% in 0.1 M triethanolamine-HCl, pH 8.0) for 10 min, the sections were washed with TBS, dehydrated in a graded ethanol series, and air dried. Hybridization solution (40% formamide, 10% dextran sulfate, 1 mg/mL yeast tRNA, 0.5 mg/mL polyadenylic acid, 0.3 M NaCl, 0.01 M Tris-HCl, pH 6.8, 0.01 M sodium phosphate, pH 6.8, 5 mM EDTA, and 40 units/mL ribonuclease inhibitor) was supplemented with 100 ng of DIG-labeled riboprobe per 100 μL of hybridization solution and dispensed onto slides containing sections. Hybridization was performed for 16 to 18 hr at 45°C in a humid chamber. Posthybridization treatments included a 20-min wash with 2 × SSC at room temperature followed by RNase A treatment (50 μg/mL in 0.5 M NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) for 30 min at 37°C. Slides were then washed in 0.2 × SSC twice for 1 hr at 55°C and once with 0.1 × SSC for 30 min at 55°C. Slides were rinsed in TBST (0.1 M Tris-HCl, pH 8.0, 0.15 M NaCl, and 0.3% Triton X-100) for 10 min on a shaking platform and then blocked overnight at 4°C with 2% BSA fraction V (Fisher Scientific) in TBST. Hybridized DIG-labeled transcripts were immunolocalized by incubating sections with a 1:200 dilution of sheep anti-DIG-AP conjugate (Roche Molecular) in 1% BSA in TBST for 2 hr at room temperature. The slides were washed three times for 15 min each with TBST and once with AP detection buffer (0.1 M Tris-HCl, pH 9.5, 0.1 M NaCl, and 50 mM MgCl2) for 10 min. Substrate buffer (175 μg/mL 5-bromo-4-chloro-3-indolyl phosphate and 350 μg/mL nitroblue tetrazolium chloride in AP detection buffer) was dispensed onto sections on slides, and color development was allowed to proceed for 4 to 8 hr at room temperature in the dark. Slides were washed with water before mounting with 50% glycerol, 7% gelatin, and 1% phenol. Sections were photographed using a Zeiss Axiophot microscope equipped with Nomarski differential interference contrast optics.
GenBank Accession Numbers
GenBank accession numbers for the five tobacco EGase cDNA sequences described here are AF362948 (Ntcel2), AF362950 (Ntcel4), AF362951 (Ntcel5), AF362947 (Ntcel7), and AF362949 (Ntcel8).
Figure 3, sequences used for the analysis (GenBank accession numbers in parentheses) were: Arabidopsis Cel1 (X98544); bean Bac1 (M57400); orange Celb1 (AF000136); pea Egl1 (L41046); pepper Cel3 (X97189); tobacco pistil EGase (AF128404); tobacco Cel1 (AF362949), Cel2 (AF362948), Cel4 (AF362950), Cel5 (AF362951), and Cel7 (AF362947); tomato Cel1 (U13054), Cel2 (U13055), Cel3 (U78526), TPP18/Cel4 (U20590), Cel5 (AF077339), Cel7 (Y11268), and Cel8 (AF098292); and strawberry Eg3 (AJ006349).
Acknowledgments
Appreciation is extended to Dr. Geert Smant of the Laboratory of Nematology, Wageningen University, The Netherlands, for supplying the Globodera rostochiensis ENG-1 antiserum used in this study. This research was supported by the North Carolina Agricultural Research Service and the North Carolina Tobacco Research Commission under Grant 6-64855.
References
- Beguin, P., and Aubert, J.-P. (1994). The biological degradation of cellulose. FEMS Microbiol. Rev. 13, 25–58. [DOI] [PubMed] [Google Scholar]
- Bird, A.F., Dowton, W.J.S., and Hawker, J.S. (1975). Cellulase secretion by second-stage larvae of the root-knot nematode (Meloidogyne javanica). Marcellia 38, 165–169. [Google Scholar]
- Bird, D.M. (1996). Manipulation of host gene expression by root-knot nematodes. J. Parasitol. 82, 881–888. [PubMed] [Google Scholar]
- Brummell, D.A., Lashbrook, C.C., and Bennett, A.B. (1994). Plant endo-1,4-β-glucanases: Structure, properties and physiological function. In Enzymatic Conversion of Biomass for Fuels Production, M.E. Himmel, J.O. Baker, and R.P. Overend, eds (Washington, DC: American Chemical Society), pp. 100–129.
- Brummell, D.A., Bird, C.R., Schuch, W., and Bennett, A.B. (1997. a). An endo-1,4-β-glucanase expressed at high levels in rapidly expanding tissues. Plant Mol. Biol. 33, 87–95. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A., Catala, C., Lashbrook, C.C., and Bennett, A.B. (1997. b). A membrane-anchored E-type endo-1,4-β-glucanase is localized on Golgi and plasma membranes of higher plants. Proc. Natl. Acad. Sci. USA 94, 4794–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala, C., and Bennett, A.B. (1998). Cloning and sequence analysis of TomCel8, a new plant endo-β-1,4-d-glucanase gene, encoding a protein with a putative carbohydrate binding domain. Plant Physiol. 118, 1535. [Google Scholar]
- Catala, C., Rose, J.K.C., and Bennett, A.B. (1997). Auxin regulation and spatial localization of an endo-β-1,4-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J. 12, 417–426. [DOI] [PubMed] [Google Scholar]
- Compton, T. (1990). Degenerate primers for DNA amplification. In PCR Protocols: A Guide to Methods and Applications, M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, eds (San Diego, CA: Academic Press), pp. 39–45.
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Davis, E.L., Hussey, R.S., Baum, T.J., Bakker, J., Schots, A., Rosso, M.N., and Abad, P. (2000). Nematode parasitism genes. Annu. Rev. Phytopathol. 38, 341–372. [DOI] [PubMed] [Google Scholar]
- De Boer, J.M., Yan, Y., Smant, G., Davis, E.L., and Baum, T.J. (1998). In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 30, 309–312. [PMC free article] [PubMed] [Google Scholar]
- De Boer, J.M., Yan, Y., Wang, X., Smant, G., Hussey, R.S., Davis, E.L., and Baum, T.J. (1999). Developmental expression of secretory β-1,4-endoglucanases in the subventral esophageal glands of Heterodera glycines. Mol. Plant-Microbe Interact. 12, 663–669. [DOI] [PubMed] [Google Scholar]
- del Campillo, E. (1999). Multiple endo-1,4-β-d-glucanase (cellulase) genes in Arabidopsis. Curr. Top. Dev. Biol. 46, 39–61. [DOI] [PubMed] [Google Scholar]
- del Campillo, E., and Bennett, A.B. (1996). Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol. 111, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dropkin, V.H. (1963). Cellulase in phytoparasitic nematodes. Nematologica 9, 444–454. [Google Scholar]
- Dropkin, V.H. (1969). Cellular responses of plants to nematode infections. Annu. Rev. Phytopathol. 7, 101–122. [Google Scholar]
- Endo, B.Y. (1986). Histology and ultrastructural modification induced by cyst nematodes. In Cyst Nematodes, F. Lamberti and C.E. Taylor, eds (New York: Plenum Press), pp. 133–146.
- Goellner, M., Smant, G., De Boer, J., Baum, T., and Davis, E. (2000. a). Isolation of β-14-endoglucanase genes from Globodera tabacum and their expression during parasitism. J. Nematol. 32, 154–165. [PMC free article] [PubMed] [Google Scholar]
- Goellner, M., Shani, Z., Shoseyov, O., and Davis, E.L. (2000. b). Plant endoglucanase expression in cyst and root knot nematode-induced feeding cells. Phytopathology 90, S28 (abstr.). [Google Scholar]
- Gonzalez-Bosch, C., Brummell, D.A., and Bennett, A.B. (1996). Differential expression of two endo-1,4-β-glucanase genes in pericarp and locules of wild-type and mutant tomato fruit. Plant Physiol. 111, 1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverse, A., Overmars, H., Engelbertink, J., Schots, A., Bakker, J., and Helder, J. (2000). Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant-Microbe Interact. 13, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Grundler, F.M.W., Sobezak, M., and Golinowski, W. (1998). Formation of wall openings in root cells of Arabidopsis thaliana following infection by the plant-parasitic nematode, Heterodera schachtii. Eur. J. Plant Pathol. 104, 545–551. [Google Scholar]
- Hartman, K.M., and Sasser, J.N. (1985). Identification of Meloidogyne species on the basis of differential host test and perineal-pattern morphology. In Advanced Treatise on Meloidogyne, Vol. 2. Biology and Control, J.N. Sasser and C.C. Carter, eds (Raleigh, NC: North Carolina State University Graphics), pp. 69–77.
- Henrissat, B., Claeyssens, M., Tomme, P., Lemesle, L., and Mornon, J.P. (1989). Cellulase families revealed by hydrophobic cluster analysis. Gene 81, 83–95. [DOI] [PubMed] [Google Scholar]
- Huang, C.S., and Maggenti, A.R. (1969). Mitotic aberrations and nuclear changes of developing giant cells in Vicia faba caused by the root-knot nematode, Meloidogyne javanica. Phytopathology 59, 447–455. [PubMed] [Google Scholar]
- Hussey, R.S. (1989). Disease-inducing secretions of plant-parasitic nematodes. Annu. Rev. Phytopathol. 27, 123–141. [Google Scholar]
- Hussey, R.S., and Barker, K.R. (1973). A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis. Rep. 57, 1025–1028. [Google Scholar]
- Hussey, R.S., and Grundler, F.M.W. (1998). Nematode parasitism of plants. In The Physiology and Biochemistry of Free-Living and Plant-Parasitic Nematodes, R.N. Perry and D.T. Wright, eds (Wallingford, UK: CABI Publishing), pp. 213–243.
- Jones, M.G.K. (1981. a). Host cell responses to endoparasitic nematode attacks: Structure and function of giant cells and syncytia. Ann. Appl. Biol. 97, 353–372. [Google Scholar]
- Jones, M.G.K. (1981b). The development and function of plant cells modified by endoparasitic nematodes. In Plant Parasitic Nematodes, Vol. 3, B.M. Zuckerman and R.A. Rohde, eds (New York: Academic Press), pp. 255–279.
- Jones, M.G.K., and Dropkin, V.H. (1975). Scanning electron microscopy of syncytial transfer cells induced in roots by cyst nematodes. Physiol. Plant Pathol. 7, 259–263. [Google Scholar]
- Kimura, M. (1983). The Neutral Theory of Molecular Evolution. (Cambridge, UK: Cambridge University Press).
- LaMondia, J.A. (1995). Hatch and reproduction of Globodera tabacum tabacum in response to tobacco, tomato, or black nightshade. J. Nematol. 27, 382–386. [PMC free article] [PubMed] [Google Scholar]
- Lashbrook, C.C., Gonzalez-Bosch, C., and Bennett, A.B. (1994). Two divergent endo-β-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell 6, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, L.I., and Gray, B.J. (1972). Heterodera solanacearum n.sp., a parasite of solanaceous plants. Nematologica 18, 404–413. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane–bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson, H., Engelbrecht, J., Brunak, S., and Von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C., and Bennett, A.B. (1999). Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends in Plant Science 4, 176–183. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C., Catala, C., Brummell, D.A., Lashbrook, C.C., Gonzalez-Bosch, C., and Bennett, A.B. (1997). The tomato endo-β-1,4-glucanase gene family: Regulation by both ethylene and auxin. In Biology and Biotechnology of the Plant Hormone Ethylene, A.K. Kanellis, C. Chang, H. Kende, and D. Grierson, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 197–205.
- Rosso, M.N., Favery, B., Piotte, C., Arthaud, L., De Boer, J.M., Hussey, R.S., Bakker, J., Baum, T.J., and Abad, P. (1999). Isolation of a cDNA encoding a β-1,4-endoglucanase in the root-knot nematode Meloidogyne incognita and expression analysis during plant parasitism. Mol. Plant-Microbe Interact. 12, 585–591. [DOI] [PubMed] [Google Scholar]
- Shani, Z., Dekel, M., Tsabary, G., and Shoseyov, O. (1997). Cloning and characterization of elongation specific endo-β-1,4-glucanase (Cel1) from Arabidopsis thaliana. Plant Mol. Biol. 34, 837–842. [DOI] [PubMed] [Google Scholar]
- Smant, G., et al. (1998). Endogenous cellulases in animals: Cloning of expressed β-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl. Acad. Sci. USA 95, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti, L., Ferrarese, L., and Casadoro, G. (1998). Characterization of cCel3, a member of the pepper endo-β-1,4-glucanase multigene family. Hereditas 128, 121–126. [DOI] [PubMed] [Google Scholar]
- Trainotti, L., Spolaore, S., Pavanello, A., Baldan, B., and Casadoro, G. (1999). A novel E-type endo-β-1,4-glucanase with a putative cellulose-binding domain is highly expressed in ripening strawberry fruits. Plant Mol. Biol. 40, 323–332. [DOI] [PubMed] [Google Scholar]
- Wang, X., Meyers, D., Yan, Y., Baum, T.J., Smant, G., Hussey, R.S., and Davis, E.L. (1999). In planta localization of a β-1,4-endoglucanase secreted by Heterodera glycines. Mol. Plant-Microbe Interact. 12, 64–67. [DOI] [PubMed] [Google Scholar]
- Wu, S.C., Blumer, J.M., Darvill, A.G., and Albersheim, P. (1996). Characterization of an endo-β-1,4-glucanase gene induced by auxin in elongating pea epicotyls. Plant Physiol. 111, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]