Abstract
Transport of nucleotide sugars across the Golgi apparatus membrane is required for the luminal synthesis of a variety of plant cell surface components. We identified an Arabidopsis gene encoding a nucleotide sugar transporter (designated GONST1) that we have shown by transient gene expression to be localized to the Golgi. GONST1 complemented a GDP-mannose transport–defective yeast mutant (vrg4-2), and Golgi-rich vesicles from the complemented strain displayed increased GDP-mannose transport activity. GONST1 promoter::β-glucuronidase studies suggested that this gene is expressed ubiquitously. The identification of a Golgi-localized nucleotide sugar transporter from plants will allow the study of the importance of this class of proteins in the synthesis of plant cell surface components such as cell wall polysaccharides.
INTRODUCTION
In recent years, the realization of the crucial role of the plant cell surface in growth and development has triggered a renaissance in cell wall research (Roberts, 2001). Much of the renewed interest in this field stems from the potential to alter cell wall composition, which would be of major importance in the food and timber industries. As such, studies of cell wall biosynthesis are crucial. Cellulose synthesis occurs on the plasma membrane, whereas noncellulosic polysaccharides are synthesized in the Golgi apparatus (Dupree and Sherrier, 1998). The protein backbone of cell surface–associated glycoproteins is made in the endoplasmic reticulum (ER) and subsequently transferred to the Golgi apparatus for the addition of glycan moieties. Therefore, the plant Golgi apparatus is an organelle specialized for sugar transfer to polysaccharides and glycoproteins (Fincher et al., 1983) and also possibly for the modification of glycolipids (Fujino et al., 1985; Dupree and Sherrier, 1998).
Noncellulosic polysaccharides and cell surface–associated glycoproteins contain a number of different sugars, such as galactose, arabinose, fucose, xylose, mannose, rhamnose, glucose, galacturonic acid, and glucuronic acid. The glycosyltransferases responsible for the synthesis of these glycans recognize specific donor and acceptor substrates. Recently, several genes encoding glycosyltransferases involved in hemicellulose or glycoprotein synthesis have been cloned (Keegstra and Raikhel, 2001). These genes encode type II single transmembrane domain (TMD) proteins resembling the Golgi glycosyltransferases studied in yeast and animal cells. The catalytic domains of these enzymes in yeast and animals have been shown to face the lumen of the Golgi apparatus (Gibeaut, 2000). A similar topology has been demonstrated for all of the plant glycosyltransferases studied to date (Keegstra and Raikhel, 2001).
Glycosyltransferases use sugars that are activated by the addition of a nucleotide (nucleotide sugars) for donation of the sugar moiety. Most sugars, such as galactose and xylose, are linked to the nucleotide UDP, whereas mannose and fucose are attached to GDP (Seitz et al., 2000). Current evidence suggests that the enzymes for nucleotide sugar synthesis are located in the cytosol (Coates et al., 1980; Bonin et al., 1997). This presents an issue of topology, because both the Golgi glycosylation reactions and their products are luminal. Therefore, it has been proposed that nucleotide sugar transporters (NSTs) provide substrates for the luminally orientated glycosyltransferases, and such activities have been detected in plant Golgi membranes (Muñoz et al., 1996; Neckelmann and Orellana, 1998; Wulff et al., 2000).
Genes encoding NSTs have been cloned recently from a wide variety of organisms, including human (Eckhardt et al., 1996), Kluyveromyces lactis (Abeijon et al., 1996), Leishmania donovani (Ma et al., 1997), Saccharomyces cerevisiae (Dean et al., 1997), and Caenorhabditis elegans (Berninsone et al., 2001). These membrane-spanning transporter proteins appear to function as antiporters, exchanging nucleoside monophosphate for specific nucleotide sugars (Capasso and Hirschberg, 1984). Given their role in providing the substrate for glycosyltransferases, NSTs are a potential control point for glycan synthesis via substrate-level control. For example, mutants in the yeast Golgi GDP-mannose transporter Vrg4p are unable to mannosylate proteins and glycolipids effectively (Dean et al., 1997), and the L. donovani GDP-mannose transporter mutant lpg2 is defective in the mannosylation of cell surface lipophosphoglycan (Ma et al., 1997). The identification and cloning of plant Golgi NSTs will be essential for understanding their role in regulating substrate levels for the luminal glycosyltransferases. In this article, we present the identification and characterization of a gene encoding a plant Golgi nucleotide sugar transporter (GONST1) and demonstrate that the protein encoded by GONST1 is able to transport GDP-mannose.
RESULTS
Molecular Cloning and Preliminary Characterization of GONST1
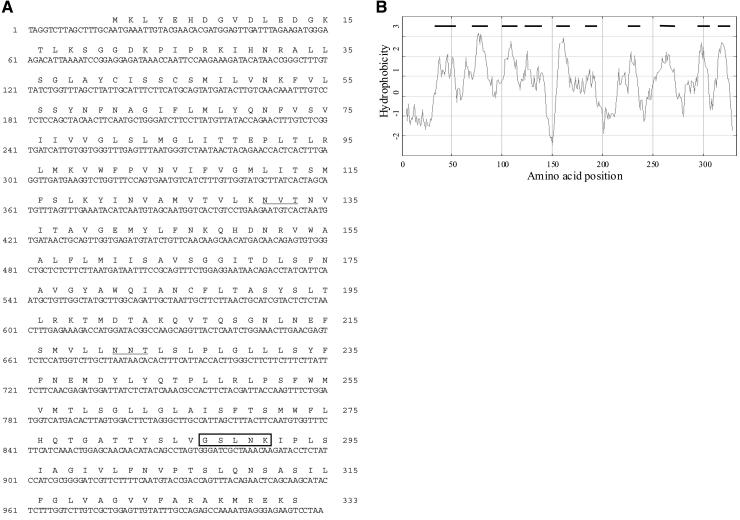
To identify candidate Golgi NST genes, the peptide sequences of the S. cerevisiae Vrg4p and L. donovani LPG2 GDP-mannose transporters were used to search for Arabidopsis expressed sequence tags (ESTs) that might encode homologous proteins. The sequences of two ESTs, 133F12T7 and 139F21T7, both derived from the same gene, showed significant similarity to the C terminus of these proteins. To determine the complete sequence of the gene, the insert from 139F21T7 was used to screen a bacterial artificial chromosome (BAC) genomic library, and the partial sequence of positive BAC clones from chromosome 2 was determined. By using specific primers designed from this genomic sequence, a cDNA was amplified by polymerase chain reaction (PCR) from an Arabidopsis seedling cDNA library. Alignment of the cDNA with the genomic sequence revealed the presence of six introns and seven exons. The predicted peptide sequence is similar to that of the recently deposited predicted protein At2g13650 from the Arabidopsis Genome Sequencing Project, except that there are two additional exons in the cDNA we isolated. The cDNA encodes a predicted protein of ∼37 kD, and we designated the protein Golgi nucleotide sugar transporter 1 (GONST1; Figure 1A). Two putative N-glycosylation sites (amino acid residues 131 to 133 and 221 to 223), based on the consensus glycosylation sequence (NXS/T; Wagh and Bahl, 1981), also are present in the predicted amino acid sequence. Hydrophobicity analysis predicted that the protein contains between eight and 10 TMDs, consistent with a role as a membrane-spanning transporter (Figure 1B). The protein has ∼25% identity with both Vrg4p and LPG2, and the homology extends over the entire sequence (Figure 1C). Importantly, each protein contains a conserved motif containing the amino acids GXLNK. In yeast, the GALNK motif has been shown to be involved in the binding of GDP-mannose by Vrg4p (Gao et al., 2001). Thus, the protein encoded by this cDNA showed all of the characteristics expected of an NST.
Figure 1.
Sequence of GONST1.
(A) Nucleotide sequence of GONST1 cDNA. The two putative N-glycosylation sites are underlined. The GXLNK motif characteristic of GDP-mannose transporters is boxed.
(B) Kyte-Doolittle hydrophobicity plot of GONST1 (Kyte and Doolittle, 1982). Potential TMDs are shown with a line above.
(C) Alignment with the Golgi GDP-mannose transporters of L. donovani LPG2 and S. cerevisiae Vrg4p. The GXLNK motif characteristic of GDP-mannose transporters is boxed.
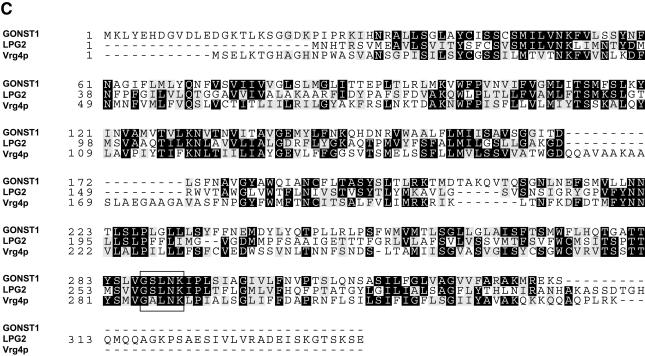
A preliminary investigation of GONST1 expression was performed to ascertain the size and abundance of the GONST1 transcript. Figure 2 shows an RNA gel blot of poly(A)+ RNA from callus and bolt tissue of Arabidopsis hybridized with an antisense RNA probe made from GONST1. A strongly hybridizing transcript of ∼1.2 kb was observed in callus tissue, which is consistent with the length of the cDNA isolated, and a lower level of hybridization to bolt mRNA also was observed. In addition, faint hybridization to a transcript of the same size was observed in leaf mRNA (data not shown). Thus, the RNA gel blots suggested a low level of expression of GONST1 throughout the plant body.
Figure 2.
RNA Gel Blot Probed with GONST1.
(A) Ethidium bromide–stained RNA gel. Lane 1, 2 μg of callus poly(A)+ Arabidopsis RNA; lane 2, 2 μg of bolt poly(A)+ Arabidopsis RNA.
(B) Corresponding RNA gel blot hybridized with digoxigenin-labeled, single-stranded RNA probe of GONST1.
Functional Analysis of GONST1 Expressed in S. cerevisiae
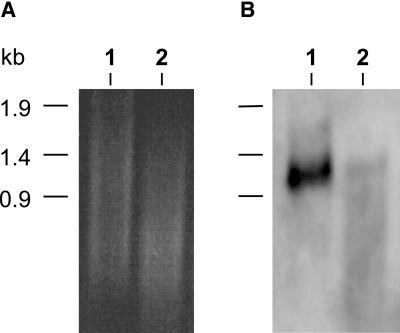
Because GONST1 shows significant homology with Vrg4p and LPG2, we examined whether GONST1 might complement the S. cerevisiae strain JPY26 3d (vrg4-2). This strain is deficient in the ability to transport GDP-mannose because of a mutation in the VRG4 gene (Dean et al., 1997; Gao et al., 2001) and shows both abnormal resistance to vanadate and sensitivity to hygromycin B (Ballou et al., 1991; Dean, 1995), probably as a result of altered cell wall mannan. The GONST1 cDNA cloned in a yeast integrating vector, pRS306 (pSc-GONST1), was used to transform the vrg4-2 mutant. On synthetic complete (SC) medium, vrg4-2 and the transformants grew marginally slower than the wild-type VRG4 strain (Figure 3A). Figure 3B shows that GONST1 complemented the vanadate resistance phenotype of vrg4-2, because the transformant, like the VRG4 wild-type strain (JPY25 6c), was sensitive to 5 mM vanadate. Furthermore, GONST1 could complement the hygromycin B sensitivity of the mutant vrg4-2 strain (Figure 3C), because the transformant, as well as the wild-type strain, grew in the presence of hygromycin B. The vanadate sensitivity and hygromycin B resistance conferred by complementation of the yeast GDP-mannose transporter mutant with GONST1 suggest that the protein encoded by this gene has the ability to transport GDP-mannose.
Figure 3.
Functional Complementation of vrg4-2 yeast by pSc-GONST1.
Yeast strains JPY25 6c (VRG4), JPY26 3d (vrg4-2), and JPY26 3d transformed with pSc-GONST1 (vrg4-2 + pSc-GONST1) were grown on agar plates, with and without supplements, at 30°C.
(A) SC + 0.5 M KCl medium.
(B) SC + 0.5 M KCl medium supplemented with 5 mM sodium orthovanadate.
(C) SC + 0.5 M KCl medium supplemented with 500 μg/mL hygromycin B.
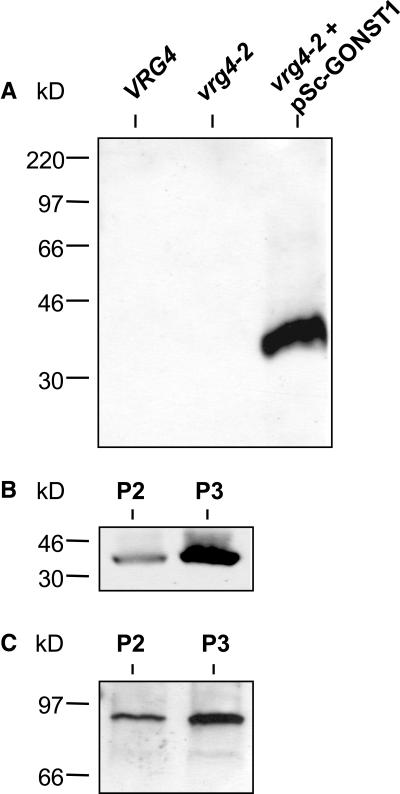
To determine where GONST1 was being expressed in the transformed yeast strain, a rabbit polyclonal antiserum was raised against a synthetic peptide corresponding to the 14 predicted N-terminal amino acids of this protein. Figure 4A shows that the anti-GONST1 antiserum detected a protein that was present in the 125,000g (P3) membrane fraction of the transformed yeast strain but that was absent from the same fraction of wild-type VRG4 and vrg4-2 mutant yeast. The protein detected by the antiserum has a mass of ∼37 kD, consistent with the predicted size of GONST1. To prepare a membrane fraction enriched in GONST1 for transport studies, membranes were separated by differential centrifugation into a larger organelle P2 (7,800g pellet) fraction, which was likely to contain ER membranes, and a smaller vesicle P3 (125,000g pellet) fraction, which was likely to be Golgi rich (Horazdovsky and Emr, 1993; Dean and Poster, 1996). Proteins from the P2 and P3 pellets of the transformant were probed with the anti-GONST1 antiserum (Figure 4B) and an antiserum against Anp1p (Figure 4C), a known Golgi-localized protein (Jungmann and Munro, 1998). Both GONST1 and Anp1p were shown to be enriched in the P3 fraction, consistent with GONST1 being targeted correctly to the Golgi apparatus in the transformed yeast strain.
Figure 4.
The GONST1 Protein Is Expressed in vrg4-2 Yeast Transformed with pSc-GONST1 and Is More Abundant in the Golgi-Rich (P3) Fraction.
(A) Protein (5 μg) of the P3 fractions prepared from yeast strains JPY25 6c (VRG4), JPY26 3d (vrg4-2), and JPY26 3d transformed with pSc-GONST1 (vrg4-2 + pSc-GONST1) were resolved by SDS-PAGE and subjected to protein gel blot analysis using the anti-GONST1 antiserum.
(B) and (C) The P2 and P3 fractions of JPY26 3d transformed with pSc-GONST1 were resolved and subjected to protein gel blot analysis using the anti-GONST1 antiserum ([B]; 7.5 μg of protein) or antiserum against Anp1p, a yeast Golgi marker protein ([C]; 5 μg of protein).
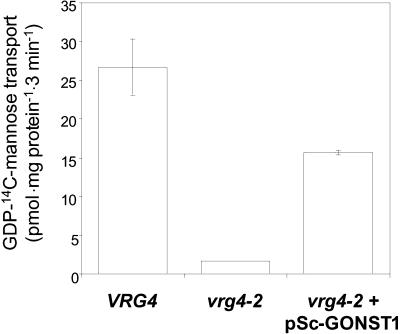
Because GONST1 was present in the yeast Golgi-rich fraction, we used this subcellular fraction to measure GDP-mannose uptake. To avoid the interference produced by the activity of dolichol phosphomannose synthase (Dpm1p) that may mask the measurement of GDP-mannose transporter activity (Abeijon et al., 1989), we used both wild-type VRG4 and vrg4-2 strains that harbor a mutation in the DPM1 gene, resulting in a 90 to 95% decrease in Dpm1p activity (Orlean et al., 1988). Golgi-rich vesicles were prepared from strains VRG4, vrg4-2, and vrg4-2 expressing the GONST1 protein and incubated with GDP-14C-mannose. As shown in Figure 5, the quantity of GDP-mannose incorporated into the Golgi-rich vesicles of vrg4-2 yeast expressing the GONST1 protein was almost 10-fold greater than that incorporated into vesicles of the untransformed vrg4-2 mutant. This result strongly suggests that GONST1 is indeed a GDP-mannose transporter.
Figure 5.
Increased GDP-Mannose Uptake in the Golgi-Rich Vesicle Fraction of vrg4-2 Yeast Expressing GONST1.
Golgi-rich (P3) vesicles were prepared from yeast strains JPY25 6c (VRG4), JPY26 3d (vrg4-2), and JPY26 3d transformed with pSc-GONST1 (vrg4-2 + pSc-GONST1). Membranes were incubated in reaction mix containing 0.18 μM GDP-14C-mannose (224.1 mCi/mmol) for 0 or 3 min at 30°C, and reactions were stopped by filtration. GDP-14C-mannose uptake is calculated as the difference between the two time points. The averages of triplicate assays and standard error bars are shown. The results shown are representative of three independent experiments, and enhanced incorporation of GDP-mannose into the P3 fraction of three other transformants also was observed.
Targeting of the GONST1-YFP Fusion Protein to the Golgi Apparatus
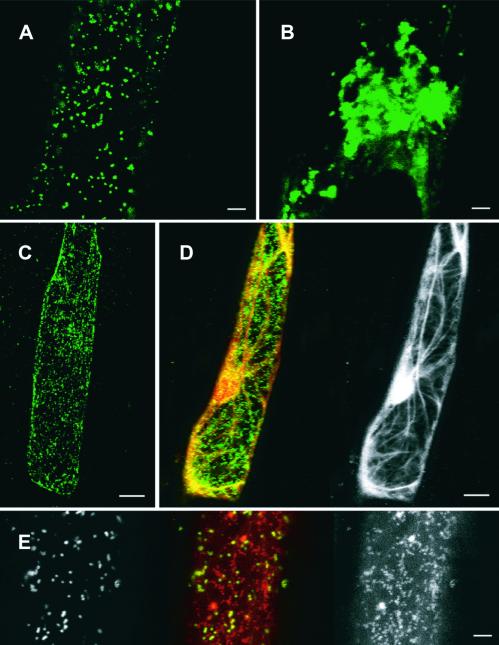
The subcellular localization of GONST1 was investigated in plants by using a fusion protein consisting of GONST1 fused at its C terminus to a modified green fluorescent protein (GFP) named YFP. Fusion of C-terminal tags such as GFP, myc, or hemagglutinin does not affect the localization of NSTs in other organisms (Descoteaux et al., 1995; Ma et al., 1997; Gao and Dean, 2000; Lühn et al., 2001), and GFP does not alter the targeting of the Golgi xylosyltransferase in plants (Essl et al., 1999). The chimeric protein (GONST1-YFP) was expressed transiently in onion epidermal cells after biolistic transformation. Two days after bombardment, many cells displayed a punctate pattern of YFP fluorescence, consistent with a Golgi-localized protein (Figures 6A and 6C). The diameter of the spots was 1 μm, similar to the size of the Golgi apparatus in tobacco cells (Dupree and Sherrier, 1998).
Figure 6.
Distribution of GONST1-YFP in Living Onion Epidermal Cells.
(A) Extended focus confocal image through cortical cytoplasm of a single onion cell showing the punctate distribution of fluorescent GONST1-YFP. Bar = 10 μm.
(B) Extended focus image through cortical cytoplasm of a single cell as shown in (A) showing clustering of GONST1-YFP fluorescence after BFA treatment. Bar = 10 μm.
(C) Extended focus image of a cell as shown in (A). Bar = 25 μm.
(D) Image of a single onion cell as in (C) showing colocalization of the actin cytoskeleton (yellow) and GONST1-YFP (green) (left image) and the actin cytoskeleton alone (right image). Bar = 25 μm.
(E) Localization of mitochondria (red and right image) and GONST1-YFP (green and left image). Bar = 10 μm.
To test further the identification of these fluorescent structures as Golgi stacks, we investigated their sensitivity to brefeldin A (BFA) and colocalization with mitochondria and actin components of the cortical cytoskeleton (Figures 6B, 6D, and 6E). BFA is known to have a variety of effects on the morphology of the Golgi apparatus, causing either clustering or vesiculation, depending on the conditions and tissue studied (Satiat-Jeunemaitre and Hawes, 1992, 1994; Driouich et al., 1993; Satiat-Jeunemaitre et al., 1996; Robinson et al., 1997; Staehelin and Driouich, 1997; Wee et al., 1998; Essl et al., 1999). When transformed epidermal peels were incubated in 100 μg/mL BFA for 2 hr, the pattern of YFP fluorescence within the cells changed dramatically. Rather than the small, numerous punctate bodies noted previously, a few large, aggregated fluorescent structures were observed (Figure 6B). This indicated that the GONST1-YFP fusion protein was localized in a BFA-sensitive compartment, which strongly supports Golgi localization. To exclude the possibility that the punctate fluorescence was mitochondrial in origin, the mitochondria were labeled with MitoTracker Orange (Molecular Probes, Eugene, OR) for 10 min. Mitochondria and GONST1-YFP fluorescence did not colocalize (Figure 6E).
Several recent publications have demonstrated that the plant Golgi apparatus moves on an actin/ER network (Boevink et al., 1998; Nebenfuhr et al., 1999). We used a GFP-mouse talin construct (Kost et al., 1998) to label actin in conjunction with our GONST1-YFP to investigate any colocalization of these structures. The punctate bodies containing the GONST1 protein were associated with and streamed along actin components of the cortical cytoskeleton (Figure 6D; a time lapse movie is available at http://www.bio.cam.ac.uk/proteomics/GONST1plusactin.mov). These data are consistent with previous studies showing the Golgi apparatus interacting with the actin network. Although we cannot formally exclude the possibility that the YFP fusion causes targeting of GONST1 to the Golgi apparatus, we consider this highly unlikely. The observed punctate distribution of the GONST1-YFP fluorescence, the effect of BFA, the lack of colocalization with mitochondria, the mobility on the actin cytoskeleton, and the functional data from yeast indicate that GONST1 is a Golgi-localized GDP-mannose transporter.
β-Glucuronidase (GUS) Histochemical Analyses of GONST1 Expression
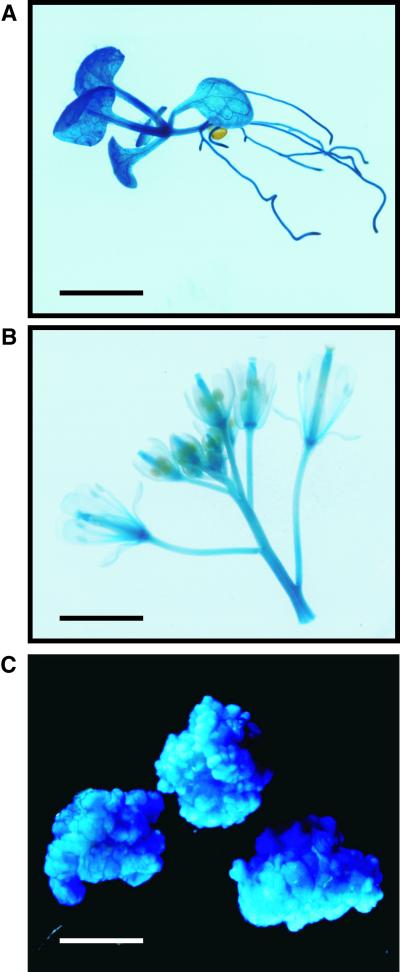
To investigate the expression of GONST1 in plants, we transformed Arabidopsis with a GONST1 promoter::GUS fusion protein (pGONST1::GUS). Histochemical staining for GUS in whole seedlings (Figure 7A) demonstrated that GUS activity was ubiquitous throughout the plant body, with slightly higher levels of expression being observed in the vascular tissues. Staining of Arabidopsis inflorescences demonstrated that all floral organs plus the pedicels were stained evenly (Figure 7B). In RNA gel blot experiments, it was observed that the GONST1 transcript was expressed most abundantly in callus tissue (Figure 2). With this in mind, we made a line of transgenic callus derived from the GONST1-GUS Arabidopsis to confirm that histochemical staining correlated with the results observed in the RNA gel blot experiments. Histochemical staining of the transgenic callus demonstrated intense staining of the callus tissue (Figure 7C) within 30 to 60 min of exposure to the GUS substrate, which was significantly faster than the time required to stain the intact plant tissue. These data show that the expression of the GDP-mannose transporter GONST1 is essentially ubiquitous in these plant tissues.
Figure 7.
Expression of GONST1-GUS in Arabidopsis.
(A) Seedling, stained overnight.
(B) Inflorescence, stained overnight.
(C) Callus, stained for 30 min.
Bars = 0.5 cm.
DISCUSSION
Previous work has shown a requirement for the transport of nucleotide sugars across the Golgi membrane for the synthesis of plant cell surface components (Muñoz et al., 1996; Neckelmann and Orellana, 1998; Wulff et al., 2000). In this article, we present the cloning and preliminary characterization of a Golgi-localized nucleotide sugar transporter (GONST1) from plants and demonstrate that it is able to transport GDP-mannose across Golgi membranes. GONST1 shows significant amino acid identity (∼25%) to the two previously isolated GDP-mannose transporters, Vrg4p and LPG2 (Figure 1; Dean et al., 1997; Ma et al., 1997). Importantly, all three sequences possess a conserved amino acid motif, GXLNK. In yeast, the GALNK motif has been shown to bind to the GDP moiety of GDP-mannose and thereby facilitate transport of this nucleotide sugar into the Golgi lumen (Gao et al., 2001). Hydrophobicity analysis predicts that the GONST1 sequence has between eight and 10 TMDs. By comparing the patterns of hydrophobicity of GONST1 and the functionally related Vrg4p and LPG2, and also of the more distantly related human GDP-fucose transporter (Lühn et al., 2001), we predict that GONST1 contains 10 TMDs. This is more than the predicted six to eight TMDs of Vrg4p, but it would place both the N and C termini on the cytosolic face, as has been shown for this yeast GDP-mannose transporter (Gao and Dean, 2000). In both of these models, the putative GDP binding motif (GXLNK) would be in a cytosolic loop, as required for the binding of cytosolic GDP-mannose.
Complementation of the growth phenotypes of the yeast GDP-mannose transporter mutant vrg4-2 (Dean et al., 1997) indicated that GONST1 can increase GDP-mannose transport into the yeast Golgi apparatus (Figure 3). By using a GDP-14C-mannose uptake assay of a yeast Golgi-enriched vesicle fraction, membranes from the vrg4-2 mutant displayed a severe defect in the ability to transport GDP-mannose, with activity ∼5% of that in the wild-type VRG4 strain, consistent with previous findings (Dean et al., 1997). We showed that vesicles isolated from vrg4-2 expressing the GONST1 protein possessed increased GDP-mannose uptake activity to ∼60% of the wild-type level (Figure 5). Thus, because GONST1 was able to complement a yeast mutant deficient in a GDP-mannose transporter, and also increased the uptake of GDP-mannose in yeast Golgi vesicles that expressed the GONST1 protein, we concluded that GONST1 is an Arabidopsis NST that uses GDP-mannose as its substrate.
Searches of the Arabidopsis genome database reveal that GONST1 is a member of a large family of putative Golgi NSTs in Arabidopsis (P. Dupree, unpublished results). One of these, At1g07290, shows ∼50% amino acid identity to GONST1 and possesses the GXLNK motif; therefore, it is a candidate for a second Arabidopsis GDP-mannose transporter. The other sequences show more distant homology and perhaps are translocators of the many other nucleotide sugars required in the Golgi lumen. Indeed, there is a large family of eukaryotic NSTs with divergent sequences and nucleotide sugar specificities (Berninsone and Hirschberg, 2000). To date, the majority of NSTs have been found to be specific for a single nucleotide sugar (Hirschberg et al., 1998). For example, the UDP-N-acetylgalactosamine transporter isolated from rat liver transports only this nucleotide sugar (Puglielli et al., 1999). Monospecific transporters would permit simple regulation of luminal nucleotide sugar content through the modulation of expression levels or potential post-translational modifications that may lead to regulation of their activities. However, recently, a small number of transporters have been shown to transport more than one nucleotide sugar (Hong et al., 2000; Berninsone et al., 2001). Interestingly, LPG2 is multispecific, transporting GDP-arabinose and GDP-fucose as well as GDP-mannose (Hong et al., 2000). Hence, it is possible that GONST1 also may be involved in the transport of other nucleotide sugars, such as GDP-fucose and GDP-glucose, both of which are required for plant glycan synthesis (Piro et al., 1993; Wulff et al., 2000).
The GONST1-YFP fusion protein was targeted to the Golgi apparatus, displaying a typical punctate pattern of fluorescence when expressed transiently in onion epidermal cells (Figure 6A). This punctate pattern represented Golgi stacks, because BFA treatment caused the fluorescently labeled Golgi apparatus to aggregate (Figure 6B). The Golgi stacks harboring GONST1-YFP also were shown to stream along the actin components of the cortical cytoskeleton (Figure 6D), consistent with previous studies of Golgi streaming (Boevink et al., 1998; Nebenfuhr et al., 1999). It will be interesting to determine by immunoelectron microscopy whether GONST1 is localized to specific cisternae within the Golgi stack, because the glycosyltransferases in the different cisternae require different substrates (Dupree and Sherrier, 1998). Targeting of NSTs to the Golgi apparatus is an interesting but poorly understood phenomenon. Studies on the yeast GDP-mannose transporter indicate that the N-terminal sequence may facilitate export to the Golgi apparatus and that dimerization of Vrg4p also is necessary (Gao and Dean, 2000). Although we have not fully characterized the targeting of GONST1 in yeast, the localization of GONST1 in the Golgi-rich vesicles of transformed yeast cells (Figures 4B and 4C) further suggests some conservation between plants and yeast of targeting of Golgi NSTs.
What are the roles of a GDP-mannose transporter in plants? Mannose is present in a range of cell wall polysaccharides in a variety of plant species. The major mannose-containing polysaccharides are the mannans, with a backbone of β-(1→4)–linked mannose residues, glucomannans, in which glucose partly replaces the mannose, and the closely related galactomannans, with galactose residues α-(1→6) linked to the mannan backbone. Galactomannans function as storage polysaccharides in many legume seed endosperms, and similarly, pure mannans are found in the seed of both monocotyledonous and dicotyledonous plant species (Buckeridge et al., 2000). Glucomannan is a major hemicellulose of gymnosperm secondary cell walls and is a significant component of angiosperm secondary walls (Piro et al., 1993). We know little as yet about the presence of mannan in Arabidopsis cell walls, but mannose constitutes 4 to 9% of the neutral sugars present in leaf walls (Reiter et al., 1997), which may be derived from secondary wall mannan. Our expression studies indicate that GONST1 is expressed throughout the plant (Figure 7), which may suggest that it is required for the synthesis of a variety of mannose-containing macromolecules other than secondary wall mannan. Alternative mannose-containing structures include glucuronomannan, a polymer of very low abundance and unknown function, which has a backbone of α-(1→4)–linked mannose and β-(1→2)–linked glucuronic acid. In addition, arabinogalactan proteins have been reported to contain mannose residues (Fincher et al., 1983), and glycolipids also can be decorated with mannose (Fujino et al., 1985). Based on what we know about the synthesis of other polysaccharides and yeast glycolipids, the addition of mannose to these glycans likely occurs in the Golgi apparatus, but this has yet to be investigated in Arabidopsis. GONST1 is unlikely to be involved in N-linked glycan synthesis, because that occurs on a dolichol-linked precursor in the ER (Schenk et al., 2001). If GONST1 transports other nucleotide sugars such as GDP-fucose, it could be involved in the synthesis of xyloglucan and pectin as well as mannose-containing polysaccharides.
The role of GDP-mannose transport across the Golgi membrane in the substrate control of biosynthesis has been demonstrated in studies of the yeast Vrg4p (Dean et al., 1997). In these studies, Dean and colleagues demonstrated that the glycoproteins and glycolipids synthesized in the vrg4-2 mutant were mannosylated to a much lower level. These studies and those of others (Toma et al., 1996) strongly support the hypothesis that NST activity may be an important control point in glycosylation, via the provision of substrate for the luminal glycosyltransferases (Abeijon et al., 1997). It has been shown that the ratio of glucose and mannose in glucomannan can be altered by varying the GDP-mannose/GDP-glucose concentrations in vitro (Piro et al., 1993). The catalytic domains of all known plant Golgi glycosyltransferases reside within the Golgi lumen (Keegstra and Raikhel, 2001). Therefore, a transporter is required to provide substrates for these enzymes, and we propose that the activity of this transporter might regulate synthesis by limiting substrate to these enzymes. However, the possibility exists that some glycosyltransferases may be multiple membrane-spanning cellulose synthase–like enzymes (Gibeaut, 2000). In this scenario, nucleotide sugars would be delivered directly to the catalytic site of the enzymes on the cytosolic side of the Golgi membrane, and the growing polysaccharide chain would be extruded into the Golgi lumen (Gibeaut, 2000). There is as yet no experimental evidence for this model. Future experiments on Arabidopsis plants with the GONST1 gene disrupted will enable a distinction to be made between these two models for the incorporation of mannose into various glycans in the plant Golgi apparatus.
METHODS
Plant Growth and Transformation
The Arabidopsis thaliana (ecotype Columbia) used in this study was grown according to Wee et al. (1998). Initiation and maintenance of liquid callus cultures were performed according to Prime et al. (2000). Transgenic Arabidopsis lines were generated by Agrobacterium tumefaciens–mediated transformation using the floral dipping technique as described by Clough and Bent (1998). Transformants were selected according to Wee et al. (1998).
Yeast Strains and Growth Conditions
Saccharomyces cerevisiae strains JPY25 6c (MATa ura3–52 his3-Δ200 trp1-Δ901 ade2–101 dpm1) and JPY26 3d (MATα ura3–52 leu2–3 112 ade2–101 vrg4–2 dpm1) (Dean et al., 1997) were grown on synthetic complete (SC) agar (Sherman, 1991) supplemented with 0.5 M KCl (Poster and Dean, 1996) at 30°C. JPY26 3d transformed with the URA3-containing yeast integration plasmid pSc-GONST1 were selected on SC-uracil agar. Hygromycin B and vanadate sensitivity were tested by supplementing SC plates with 500 μg/mL hygromycin B (Dean, 1995) and 5 mM sodium orthovanadate (Ballou et al., 1991), respectively. Plates were photographed 4 or 5 days after streaking. Yeast strain JPY26 3d was transformed by the lithium acetate procedure (Agatep et al., 1998).
Generation of Constructs
DNA manipulations were performed according to standard protocols (Sambrook et al., 1989). Expressed sequence tag (EST) clones, Texas A&M University bacterial artificial chromosome (BAC) filters, and BAC clones were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The BAC filters were screened using the insert from EST clone 139F21T7 labeled with fluorescein, hybridized, and visualized using the Gene Images module from Amersham Pharmacia. The GONST1 gene was predicted from the sequence of the positively hybridizing BAC clones T15K7 and T10F5 using GENSCAN (Burge and Karlin, 1997), GRAIL (Uberbacher and Mural, 1991), and NETGENE2 (Hebsgaard et al., 1996). Polymerase chain reaction (PCR) was used to amplify the GONST1 cDNA from the Arabidopsis ecotype Landsberg cDNA library pFL61 (obtained from the American Type Culture Collection, Rockville, MD) using specific primers designed from the genomic sequence. The upstream primer sequence was 5′-TTAAAGAATTCT-AGGTCTTAGCTTTGCAATG-3′ and the downstream primer was 5′-TATATGTCGACTTAGGACTTCTCCCTCATTT-3′. The 1.0-kb PCR fragment corresponding to the GONST1 cDNA was then ligated into the EcoRI–SalI site of the yeast expression vector p426GPD to create pGONST1.
The construct pGONST1-YFP was created by cloning a PCR-amplified fragment using the upstream primer (5′-TTAGAGGATCCTAGGTCTTAGCTTTGCAATG-3′) and the downstream primer (5′-GAG-AGAATTCGGACTTCTCCCTCATTTGG-3′) into the BamHI and EcoRI sites of the binary vector pBIN 35S Knat YFP (a generous gift of Jim Haseloff, University of Cambridge, UK). For the construction of pGONST1::GUS, an ∼2-kb region of genomic DNA upstream of the GONST1 translation initiation codon was amplified by PCR from the TAMU BAC clone T10F5 using the upstream primer 113 (5′-GAG-ACCCGGGCTCAAGAACAAGATGAGTGTCGTTAAC-3′) located 1868 bp upstream of the translation initiation codon and the downstream primer 114 (5′-TCTCGGATCCGCAAAGCTAAGACCTACCAAAGA-AAC-3′) located immediately adjacent to the initiation codon. The PCR product was digested with SmaI and BamHI and cloned immediately upstream of the β-glucuronidase (GUS) gene in plasmid pBI 101.2 (Jefferson et al., 1987) to create a transcriptional fusion. For the construction of pGEM4Z-GONST1 for RNA probe synthesis, pGONST1 was digested with KpnI–EcoRI to release the GONST1 cDNA, and this DNA fragment then was subcloned into pGEM4Z (Promega). For the construction of pSc-GONST1, the GONST1 cDNA sequence plus the adjoining glyceraldehyde phosphate dehydrogenase promoter were amplified by PCR from pGONST1 using the upstream primer 184 (5′-AATTAACCCTCACTAAAGGG-3′) and the downstream primer 185 (5′-CCTTCCGTCGACTCAATTGAG-GTCTTCCTCGCTGATTAATTTTTGTTCGGACTTCTCCCTCATTTTGGC-TC-3′). The resultant PCR fragment was digested and subcloned as a SacI–SalI fragment into the yeast integration vector pRS306 (Sikorski and Hieter, 1989). EST and BAC clones and all constructs were sequenced on Applied Biosystems sequencers (models 377 and 373; Foster City, CA) using big dye terminator reactions.
RNA Isolation and RNA Gel Blot Analyses
Total RNA was isolated as described by Coen et al. (1990). Poly(A)+ RNA was isolated from total RNA with the PolyATract mRNA Isolation System (Promega). RNA gel blotting was performed as described by Sambrook et al. (1989) with 2 μg of mRNA loaded per lane. The RNA probe synthesized from pGEM4Z-GONST1 was labeled with digoxygenin-11-UTP (Roche) and used according to the kit manufacturer's instructions.
Subcellular Fractionation of Yeast
Yeast strains were grown in MM2 (glucose [2%], yeast nitrogen base without amino acids [0.67%; Difco], uracil [40 mg/L], adenine [59 mg/L], ala, arg, asn, asp, cys, glu, gln, gly, his, myo-inositol, ile, lys, met, p-aminobenzoic acid, phe, pro, ser, thr, trp, tyr, val [all at 76 mg/L], and leu [380 mg/L]) at 30°C to an OD600 of 3 to 5. Yeast cells were fractionated essentially as described previously (Gao et al., 2001) with some modifications. After washing in 10 mM NaN3, cells were resuspended in spheroplast buffer (1.4 M sorbitol, 50 mM potassium phosphate, pH 7.5, 10 mM NaN3, 0.33% β-mercaptoethanol, and 8 units lyticase/OD600) and incubated for 2 hr at 37°C. Spheroplasts were pelleted and resuspended in ice-cold membrane buffer (0.8 M sorbitol, 10 mM triethanolamine, pH 7.2, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride) and lysed by repeated pipetting (20 times) in a wide-bore pipette followed by four strokes in a Dounce homogenizer (Wheaton, Millville, NJ). The lysed cells were centrifuged at 1000g, forming S1 (supernatant) and P1 (pellet) fractions. P1 was lysed another two times by pipetting and homogenization as described above, and the three S1 fractions were pooled. S1 was centrifuged at 5,000g, 7,800g, and 125,000g to obtain fractions P-loose, P2, and P3, respectively. Pellet fractions were resuspended in a small volume of membrane buffer.
Protein Fractionation and Immunoblotting
P2 and P3 fractions were pelleted for 45 min at 100,000g and resuspended in 10 mM Tris-Cl, pH 7.5, and 1 mM EDTA. Protein concentrations were determined by the bicinchoninic acid method according to the manufacturer's instructions (Pierce Chemical Co.). Samples were incubated for 15 min at 37°C in Laemmli buffer containing 2% SDS, and after brief centrifugation, samples were fractionated by 12% SDS-PAGE and transferred to a nitrocellulose membrane by electroblotting (Laemmli, 1970; Towbin et al., 1979). Anti-GONST1 antiserum was raised in rabbits against a synthetic peptide of the 14 N-terminal amino acids (MKLYEHDGVDLEDG) and affinity purified by Abcam (Cambridge, UK). When using the anti-GONST1 antiserum, membranes were blocked overnight at 4°C in PBS, 5% milk, and 0.5% Tween 20 followed by incubation in anti-GONST1 antiserum (1:2000 dilution) overnight at 4°C. The anti-Anp1p antiserum (a kind gift of Sean Munro, MRC-LMB, Cambridge, UK) was used as described previously (Jungmann and Munro, 1998). Antibody binding was detected using anti–rabbit antiserum conjugated to horseradish peroxidase (1:250 dilution; Bio-Rad, Hercules, CA) and enhanced chemiluminescence (Wee et al., 1998).
GDP-Mannose Uptake Assay
The Golgi-rich vesicle fraction (P3; 350 μg of protein) was incubated at 30°C in reaction mixture (1 mL) containing 0.3 M sucrose, 30 mM triethanolamine, pH 7.2, 5 mM MgCl2, 5 mM MnCl2, and 0.18 μM GDP-14C-mannose (224 mCi/mmol; DuPont–New England Nuclear). After 3 min, reactions were stopped with 3 mL of ice-cold stop buffer (0.5 M sucrose and 1 mM EDTA) and the Golgi vesicles were immediately washed onto 0.45-μm nitrocellulose filters (25 mm diameter; Whatman) by vacuum filtration (model FH 225V; Hoefer Amersham Pharmacia Biotech, Little Chalfont, UK). The vesicles were washed again with 5 mL of stop buffer and dried, and the filters were transferred to vials containing 5 mL of scintillation fluid (Ecoscint A; National Diagnostics, Atlanta, GA). The radioactivity was counted by scintillation counting. A time 0 assay was used to determine the amount of radioactivity bound nonspecifically to the outside of the vesicles and to the nitrocellulose filters.
Biolistic Transformation
Plasmid DNA (5 μg) was delivered into onion (Allium cepa) epidermal cells using tungsten particle bombardment. For experiments in which the actin cytoskeleton and the Golgi apparatus were labeled simultaneously, using constructs pGONST1-YFP and the talin–green fluorescent protein (GFP) construct (PSY C14) (Rees et al., 1990; McCann and Craig, 1997; Kost et al., 1998), respectively, 2.5 μg of each plasmid was bound to the tungsten microprojectiles.
Onion bulbs were purchased from a local market, and inner epidermal peels were placed on agar plates containing 1 × Murashige and Skoog (1962) salts, 30 g/L sucrose, and 2% agar (type IV; Sigma, St. Louis, MO), pH 5.7. Peels were bombarded within 1 hr of transfer to agar plates. Tungsten microprojectiles (1.1 μm; Bio-Rad) were coated with DNA according to the manufacturer's instructions. Microprojectiles were bombarded into the onion epidermal cells using a Biolistic PDS-1000/He system (Bio-Rad) with 900-p.s.i. rupture discs under a vacuum of 28 in Hg. After bombardment, the cells were allowed to recover for 48 hr on agar plates at 22°C in continual light before being viewed under the confocal microscope. To monitor the effects of brefeldin A (BFA) on the distribution of pGONST1-YFP, transformed onion epidermal peels were incubated in 100 μg/mL BFA for 2 hr. The effects of BFA on the distribution of the YFP signal then were monitored as described above. For the MitoTracker experiments, sections of transformed onion epidermal peels were stained with 65 μM CM-H2 TMROS (MitoTracker Orange, Molecular Probes, Eugene, OR) by incubation in the dark for 10 min before viewing.
Confocal Microscopy
Imaging of GFP, YFP, and MitoTracker fluorescence was performed on a Leica (Wetzlar, Germany) DMRXA confocal laser scanning microscope. Serial optical sections of 0.5 to 2 μm were obtained using an excitation wavelength of 488 nm for GFP and MitoTracker and 514 nm for YFP, with emission signals being collected at the emission maxima of 510, 527, and 576 nm, respectively. A combined extended focus image then was produced from the confocal series using Leica TCS-NT software.
Detection of GUS Activity
Preliminary screening of 59 independently transformed lines for GUS activity was performed using a fluorometric assay as described by Gallagher (1992). Five independent homozygous lines (giving effectively 100% kanamycin-resistant progeny after selfing for two generations of transformants) identified from the GUS assay screen were selected for GUS histochemical analyses. Transgenic Arabidopsis seedlings, inflorescences, and callus from each of the five lines were stained with 1 mM 5-bromo-4-chloro-3-indoyl-β-d-glucuronide (Melford Laboratories, Kings Lynn, UK) in staining buffer (100 mM NaPO4, pH 7.2, 25 mM EDTA.Na2, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 0.1% Triton X-100) for between 30 min and overnight at 37°C and were cleared in 70% ethanol for 48 hr. Stained plant material was photographed using a Nikon (Tokyo, Japan) F camera body attached to a Nikon Multiphot Macro system with Kodak 64 T tungsten film.
Accession Number
The EMBL accession number for the GONST1 cDNA is AJ314836.
Acknowledgments
We thank Yeu Chung Lee for initial contributions to this project and Neta Dean, Jürgen Stolz, and Lorena Norambuena for help with the yeast studies. We also thank Jim Sullivan for help with confocal microscopy, Jo Cornah for advice on the GUS expression studies, Nam-Hai Chua for providing the PSY C14 plasmid, and Tracy Prime for generous technical assistance. This research was supported by the Biotechnology and Biological Sciences Research Council. M.G.H. was the recipient of a Broodbank Trust Fellowship. A.O. was financially supported by the Fondo Nacional de Desarrollo Cientifico y Tecnológico Grant 1000675 and the Iniciativa Científica Milenio Grant ICM P 99-031-F, and all of the authors thank the Royal Society and Comisión Nacional de Investigación Científica y Technológica Chile for additional financial support.
References
- Abeijon, C., Orlean, P., Robbins, P.W., and Hirschberg, C.B. (1989). Topography of glycosylation in yeast: Characterization of GDP-mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc. Natl. Acad. Sci. USA 86, 6935–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeijon, C., Robbins, P.W., and Hirschberg, C.B. (1996). Molecular cloning of the Golgi apparatus uridine diphosphate-N-acetylglucosamine transporter from Kluyveromyces lactis. Proc. Natl. Acad. Sci. USA 93, 5963–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeijon, C., Mandon, E.C., and Hirschberg, C.B. (1997). Transporters of nucleotide sugars, nucleotide sulfate and ATP in the Golgi apparatus. Trends Biochem. Sci. 22, 203–207. [DOI] [PubMed] [Google Scholar]
- Agatep, R., Kirkpatrick, R.D., Parchaliuk, D.L., Woods, R.A., and Gietz, R.D. (1998). Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol protocol. Technical Tips Online (http://tto.trends.com). September 14, 2001. [Google Scholar]
- Ballou, L., Hitzeman, R.A., Lewis, M.S., and Ballou, C.E. (1991). Vanadate-resistant yeast mutants are defective in protein glycosylation. Proc. Natl. Acad. Sci. USA 88, 3209–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone, P.M., and Hirschberg, C.B. (2000). Nucleotide sugar transporters of the Golgi apparatus. Curr. Opin. Struct. Biol. 10, 542–547. [DOI] [PubMed] [Google Scholar]
- Berninsone, P.M., Hwang, H.-Y., Zemtseva, I., Horvitz, H.R., and Hirschberg, C.B. (2001). SQV-7, a protein involved in Caenorhabditis elegans epithelial invagination and early embryogenesis, transports UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose. Proc. Natl. Acad. Sci. USA 98, 3738–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Bonin, C.P., Potter, I., Vanzin, G.F., and Reiter, W.D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalysing the first step in the de novo synthesis of GDP-l-fucose. Proc. Natl. Acad. Sci. USA 94, 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge, M.S., dos Santos, H.P., and Tiné, M.A.S. (2000). Mobilization of storage cell wall polysaccharides in seeds. Plant Physiol. Biochem. 38, 141–156. [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Capasso, J.M., and Hirschberg, C.B. (1984). Mechanisms of glycosylation and sulfation in the Golgi-apparatus: Evidence for nucleotide sugar/nucleoside monophosphate and nucleotide sulfate/nucleoside monophosphate antiports in the Golgi-apparatus membrane. Proc. Natl. Acad. Sci. USA 81, 7051–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coates, S.W., Gurney, T., Sommers, L.W., Yeh, M., and Hirschberg, C.B. (1980). Subcellular localization of sugar nucleotide synthetases. J. Biol. Chem. 255, 9225–9229. [PubMed] [Google Scholar]
- Coen, E.S., Romero, J.M., Doyle, S., Elliot, R., Murphy, G., and Carpenter, R. (1990). Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63, 1311–1322. [DOI] [PubMed] [Google Scholar]
- Dean, N. (1995). Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. USA 92, 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, N., and Poster, J.B. (1996). Molecular and phenotypic analysis of the S. cerevisiae MNN10 gene identifies a family of related glycosyltransferases. Glycobiology 6, 73–81. [DOI] [PubMed] [Google Scholar]
- Dean, N., Zhang, Y.B., and Poster, J.B. (1997). The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 272, 31908–31914. [DOI] [PubMed] [Google Scholar]
- Descoteaux, A., Luo, Y., Turco, S.J., and Beverley, S.M. (1995). A specialized pathway affecting virulence glycoconjugates of Leishmania. Science 269, 1869–1872. [DOI] [PubMed] [Google Scholar]
- Driouich, A., Zhang, G.F., and Staehelin, L.A. (1993). Effect of brefeldin A on the structure of the Golgi apparatus and on the synthesis and secretion of proteins and polysaccharides in sycamore maple (Acer pseudoplatanus) suspension-cultured cells. Plant Physiol. 101, 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupree, P., and Sherrier, D.J. (1998). The plant Golgi. Biochim. Biophys. Acta 1404, 259–270. [DOI] [PubMed] [Google Scholar]
- Eckhardt, M., Mühlenhoff, M., Bethe, A., and Gerardy-Schahn, R. (1996). Expression cloning of the Golgi CMP-sialic acid transporter. Proc. Natl. Acad. Sci. USA 93, 7572–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essl, D., Dirnberger, D., Gomord, V., Strasser, R., Faye, L., Glossl, J., and Steinkellner, H. (1999). The N-terminal 77 amino acids from the tobacco N-acetylglucosaminyltransferase I are sufficient to retain a reporter protein in the Golgi apparatus of Nicotiana benthamiana cells. FEBS Lett. 453, 169–173. [DOI] [PubMed] [Google Scholar]
- Fincher, G.B., Stone, B.A., and Clarke, A.E. (1983). Arabinogalactan-proteins: Structure, biosynthesis and function. Annu. Rev. Plant Physiol. 34, 47–70. [Google Scholar]
- Fujino, Y., Ohnishi, M., and Ito, S. (1985). Further studies on sphingolipids in wheat grain. Lipids 20, 337–342. [Google Scholar]
- Gallagher, S.R. (1992). GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression (San Diego, CA: Academic Press).
- Gao, X.-D., and Dean, N. (2000). Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J. Biol. Chem. 275, 17718–17727. [DOI] [PubMed] [Google Scholar]
- Gao, X.-D., Nishikawa, A., and Dean, N. (2001). Identification of a conserved motif in the yeast Golgi GDP-mannose transporter required for binding to nucleotide sugar. J. Biol. Chem. 276, 4424–4432. [DOI] [PubMed] [Google Scholar]
- Gibeaut, D.M. (2000). Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol. Biochem. 38, 69–80. [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouze, P., and Brunak, S. (1996). Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg, C.B., Robbins, P.W., and Abeijon, C. (1998). Transporters of nucleotide sugars, ATP and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67, 49–69. [DOI] [PubMed] [Google Scholar]
- Hong, K., Ma, D., Beverley, S.M., and Turco, S.J. (2000). The Leishmania GDP-mannose transporter is an autonomous, multi-specific hexameric complex of LPG2 subunits. Biochemistry 39, 2013–2022. [DOI] [PubMed] [Google Scholar]
- Horazdovsky, B.F., and Emr, S.D. (1993). The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268, 4953–4962. [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann, J., and Munro, S. (1998). Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 17, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Raikhel, N. (2001). Plant glycosyltransferases. Curr. Opin. Plant Biol. 4, 219–224. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lühn, K., Wild, M.K., Eckhardt, M., Gerardy-Schahn, R., and Vestweber, D. (2001). The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 28, 69–72. [DOI] [PubMed] [Google Scholar]
- Ma, D., Russell, D.G., Beverley, S.M., and Turco, S.J. (1997). Golgi GDP-mannose uptake requires Leishmania LPG2. J. Biol. Chem. 272, 3799–3805. [PubMed] [Google Scholar]
- McCann, R.O., and Craig, S.W. (1997). The I/LWEQ module: A conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. USA 94, 5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz, P., Norambuena, L., and Orellana, A. (1996). Evidence for a UDP-glucose transporter in Golgi apparatus–derived vesicles from pea and its possible role in polysaccharide biosynthesis. Plant Physiol. 112, 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for the rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nebenfuhr, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckelmann, G., and Orellana, A. (1998). Metabolism of uridine 5′-diphosphate-glucose in Golgi vesicles from pea stems. Plant Physiol. 117, 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean, P., Albright, C., and Robbins, P.W. (1988). Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263, 17499–17507. [PubMed] [Google Scholar]
- Piro, G., Zuppa, A., Dalessandro, G., and Northcote, D.H. (1993). Glucomannan synthesis in pea epicotyls: The mannose and glucose transferases. Planta 190, 206–220. [DOI] [PubMed] [Google Scholar]
- Poster, J.B., and Dean, N. (1996). The yeast VRG4 gene is required for normal Golgi functions and defines a new family of related genes. J. Biol. Chem. 271, 3837–3845. [DOI] [PubMed] [Google Scholar]
- Prime, T.A., Sherrier, D.J., Mahon, P., Packman, L.C., and Dupree, P. (2000). A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21, 3488–3499. [DOI] [PubMed] [Google Scholar]
- Puglielli, L., Mandon, E.C., Rancour, D.M., Menon, A.K., and Hirschberg, C.B. (1999). Identification and purification of the rat liver Golgi membrane UDP-N-acetylgalactosamine transporter. J. Biol. Chem. 274, 4474–4479. [DOI] [PubMed] [Google Scholar]
- Rees, D.J.G., Ades, S.E., Singer, S.J., and Hynes, R.O. (1990). Sequence and domain-structure of talin. Nature 347, 685–689. [DOI] [PubMed] [Google Scholar]
- Reiter, W.D., Chapple, C., and Somerville, C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12, 335–345. [DOI] [PubMed] [Google Scholar]
- Roberts, K. (2001). How the cell wall acquired a cellular context. Plant Physiol. 125, 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.G., Baumer, M., Hinz, G., and Hohl, I. (1997). Ultrastructure of the pea cotyledon Golgi apparatus: Origin of dense vesicles and the action of brefeldin A. Protoplasma 200, 198–209. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Satiat-Jeunemaitre, B., and Hawes, C. (1992). Redistribution of a Golgi glycoprotein in plant cells treated with brefeldin A. J. Cell Sci. 103, 1153–1166. [Google Scholar]
- Satiat-Jeunemaitre, B., and Hawes, C. (1994). G.A.T.T. (a general agreement on traffic and transport) and brefeldin A in plants cells. Plant Cell 6, 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre, B., Cole, L., Bourett, T., Howard, R., and Hawes, C. (1996). Brefeldin A effects in plant and fungal cells: Something new about vesicle trafficking? J. Microsc. 181, 162–177. [DOI] [PubMed] [Google Scholar]
- Schenk, B., Fernandez, F., and Waechter, C.J. (2001). The ins(ide) and outs(ide) of dolichol phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology 11, 61R–71R. [DOI] [PubMed] [Google Scholar]
- Seitz, B., Klos, C., Wurm, M., and Tenhaken, R. (2000). Matrix polysaccharide precursors in Arabidopsis cell walls are synthesized by alternate pathways with organ-specific expression patterns. Plant J. 21, 537–546. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin, L.A., and Driouich, A. (1997). Brefeldin A effects in plants: Are different Golgi responses caused by different sites of action? Plant Physiol. 114, 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma, L., Pinhal, M.A.S., Dietrich, C.P., Nader, H.B., and Hirschberg, C.B. (1996). Transport of UDP-galactose into the Golgi lumen regulates the biosynthesis of proteoglycans. J. Biol. Chem. 271, 3897–3901. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberbacher, E.C., and Mural, R.J. (1991). Locating protein coding regions in human DNA sequences using a multiple sensor-neural network approach. Proc. Natl. Acad. Sci. USA 88, 11261–11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh, P.V., and Bahl, O.P. (1981). Sugar residues on proteins. Crit. Rev. Biochem. 10, 307–377. [DOI] [PubMed] [Google Scholar]
- Wee, E.G.-T., Sherrier, D.J., Prime, T.A., and Dupree, P. (1998). Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff, C., Norambuena, L., and Orellana, A. (2000). GDP-fucose uptake into the Golgi apparatus during xyloglucan biosynthesis requires the activity of a transporter-like protein other than the UDP-glucose transporter. Plant Physiol. 122, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]