Abstract
The in vitro DNA binding activity of the Arabidopsis Tag1 transposase (TAG1) was characterized to determine the mechanism of DNA recognition. In addition to terminal inverted repeats, the Tag1 element contains four different subterminal repeats that flank a transcribed region encoding a 729–amino acid protein. A single site-specific DNA binding domain is located near the N terminus of TAG1, between residues 21 and 133. This domain binds specifically to the AAACCC and TGACCC subterminal repeats, found near the 5′ and 3′ ends of the element, respectively. The ACCC sequence within these repeats is critical for recognition because mutations at positions 3, 5, and 6 abolished binding, yet the first two bases also are important because substitutions at these positions decreased binding by up to 90%. Weak interaction also occurs with the terminal inverted repeats, but no binding was observed to the other two 3′ subterminal repeat regions. Sequence analysis of the TAG1 DNA binding domain revealed a C2HC zinc finger motif. Tests for metal dependence showed that DNA binding activity was inhibited by divalent metal chelators and greatly enhanced by zinc. Furthermore, mutation of each cysteine residue predicted to be a metal ligand in the C2HC motif abolished DNA binding. Together, these data show that the DNA binding domain of TAG1 specifically binds to distinct subterminal repeats and contains a zinc finger.
INTRODUCTION
Transposable elements are ubiquitous residents in the genomes of both prokaryotes and eukaryotes that possess the capacity to move from one chromosomal location to another. The mechanism of conservative (cut-and-paste) transposition involves a series of steps, beginning with recognition of specific sequences, usually near the termini of the transposon, by the element-encoded transposase. Next, synapsis of the transposon ends occurs through protein–protein interactions of DNA-bound transposase. Assembly of this ordered nucleoprotein complex, the transpososome, may involve accessory factors (i.e., host proteins and divalent metal ions) and is required for the subsequent donor strand cleavage and transfer reactions (for review, see Kunze et al., 1997; Haren et al., 1999).
In plants, the most extensively studied transposons are the Mutator (Mu), En/Spm, and Activator (Ac) elements from maize. Each of these elements represents the archetype for a distinct superfamily of DNA-type transposons (reviewed by Kunze et al., 1997). The DNA binding activities of the element-encoded proteins have been identified, and their DNA recognition sites have been described. In the case of Mu, the 823–amino acid MURA transposase has been shown to bind to a conserved 32-bp region in the terminal inverted repeat (TIR) of the element (Benito and Walbot, 1997). For En/Spm, the DNA binding domain of the 621–amino acid TNPA protein is located between residues 122 and 427 and binds to a 12-bp sequence motif repeated near the terminus of the element (Gierl et al., 1988; Trentmann et al., 1993). The 807–amino acid Ac transposase (TPase) contains a basic DNA binding domain between amino acids 159 and 206, which recognizes a repeated hexamer motif, AAACGG, in the subterminal regions (STRs) of the element (Kunze and Starlinger, 1989; Feldmar and Kunze, 1991). Recently, it was shown that the Ac TPase binds weakly to the TIRs and displays high affinity and cooperative binding to A/TCG trinucleotides, which often are contained within the STR hexamer motifs (Becker and Kunze, 1997). Furthermore, the Ac TPase DNA binding domain is bipartite: a C-terminal subdomain, between residues 185 and 206, is sufficient for binding the AAACGG motifs, whereas a larger region between 159 and 206 is necessary for binding the TIRs (Becker and Kunze, 1997).
Although the features described above provide important clues to the mechanism whereby eukaryotic transposases recognize the ends of their elements, the nature of this molecular recognition is not known for plant transposons. However, it has been described for the Tc1/mariner family of elements in animals. The Tc1-like transposases have a bipartite, N-terminal DNA binding domain that resembles the paired domain of some transcription factors (Vos and Plasterk, 1994). This domain is proposed to consist of two helix-turn-helix motifs, the first of which has been crystallized in a complex with Tc3 DNA (van Pouderoyne et al., 1997). Although this motif is predicted to be conserved in the Tc1/mariner family, the amino acid similarity in this region is weak (e.g., 30% identity between the Tc1 and Tc3 transposase DNA binding domains) (Plasterk et al., 1999). Likewise, in plants, the DNA binding domains of TNPA and the Ac TPase show little sequence similarity to any described protein (Trentmann et al., 1993; Kunze et al., 1997). Moreover, the Ac domain shows no homology with the putative transposases of elements (from insects, plants, and fungi) within its own family. Thus, a question remains: how do plant transposases recognize their cognate DNA? To address this question, the DNA binding activity of the transposase from the Arabidopsis transposon Tag1 was examined.
Tag1 was the first of several mobile elements to be discovered in Arabidopsis (Tsay et al., 1993; Miura et al., 2001; Singer et al., 2001). The autonomous Tag1 element is 3.3 kb in length and contains 22-bp TIRs (Tsay et al., 1993; Frank et al., 1997). Sequence analysis indicates that Tag1 is a member of the Ac (or hAT, for hobo, Ac, Tam3) superfamily of transposons (Warren et al., 1994; Frank et al., 1997; Essers et al., 2000). Members of this family create 8-bp target site duplications upon insertion and contain a conserved region of amino acids, termed the signature motif, near the C terminus of the putative transposase (Calvi et al., 1991; Hehl et al., 1991). This motif is crucial for the function of the hobo transposase in vivo (Calvi et al., 1991) and has been shown to be involved in the dimerization of the Ac TPase (Essers et al., 2000).
Previous lines of investigation have shown by RNA gel blot analysis that the Tag1 element produces one major (2.3 kb) and several minor (between 1.0 and 1.2 kb) transcripts (Liu and Crawford, 1998). The major transcript is thought to generate the transposase because it contains a large open reading frame (ORF) encoding a polypeptide of 729 amino acids that shares the Ac signature motif at the C terminus (Liu and Crawford, 1998). Amino acid sequence analysis of the putative TAG1 transposase has revealed that, other than this C-terminal signature motif, there is no significant homology shared with the Ac TPase. Because of this lack of homology, it has not been possible to identify any putative DNA binding domains in the polypeptide.
An interesting feature of Tag1 is that repeated DNA sequences within the subtermini, the potential binding sites for the transposase, are not the same (Liu and Crawford, 1998). This is in contrast to what is observed for other elements, where conserved binding sites are found in both STRs. Contained within the 5′ STR of Tag1 are repeats of the sequence AAACCC, whereas the 3′ end harbors three different sets of repeats: TTATT, TATATA, and TGACCC (Liu and Crawford, 1998) (see Figure 1 for scheme). Recent results have shown that ∼100 bp of distal Tag1 sequence (containing the 5′-AAACCC motif and the 3′-TGACCC motif), with intervening spacer DNA, is sufficient for excision in Arabidopsis (Liu et al., 2001).
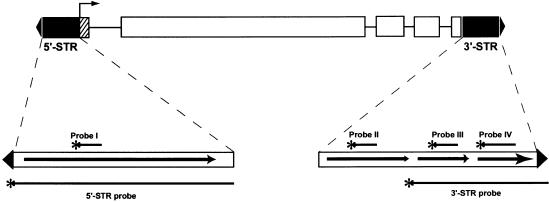
Figure 1.
Structure of the 3.3-kb Tag1 Element.
The ends of the element are defined by the 22-bp TIRs (closed triangles). STRs of the element are shown as closed boxes. The 5′ STR includes nucleotides 23 to 261, which ends at the start of transcription (bent arrow). The 3′ STR (nucleotides 2975 to 3273) begins after the stop codon and extends to the 3′ TIR. Interior to the STRs is the Tag1 coding region, which contains the large ORF (open boxes) interspersed with introns (horizontal lines); the hatched box indicates the 5′ untranslated region. In the expanded views of the STRs, each region containing a distinct repeated sequence is indicated by an arrow. Bars beneath the expanded views indicate the STR probes; the bars above refer to the probes described in Figure 4.
In this study, we set out to determine the in vitro DNA binding activity of the Tag1 transposase. We identified a site-specific DNA binding domain, located near the N terminus of the polypeptide, and show that this domain binds to related, but distinct, hexamer motifs repeated within the STRs. In addition, we report that the observed DNA binding is metal dependent, is enhanced by the addition of zinc, and requires key cysteine residues—properties indicative of a zinc finger DNA binding domain.
RESULTS
Escherichia coli–Expressed TAG1 Protein Binds to the Subterminal Regions of the Tag1 Element
The large ORF of the 2.3-kb Tag1 transcript encodes a protein of 84 kD, the putative TAG1 transposase. To facilitate the purification and biochemical analysis of the transposase, we expressed the Tag1 ORF as a C-terminal fusion with the glutathione S-transferase (GST) 26-kD domain in E. coli, as described in Methods. The resulting GST-TAG1 fusion protein has a predicted molecular mass of 112 kD. Upon induction, a band of ∼115 kD was observed by SDS-PAGE of crude bacterial lysates in both soluble and insoluble fractions (data not shown). Soluble protein, from cells expressing the fusion construct or GST alone, was purified using glutathione–Sepharose and analyzed by immunoblotting. The affinity-purified TAG1 products included the expected 115-kD band as well as a number of smaller polypeptides (Figure 2A). The smaller bands are presumed to be C-terminally truncated versions of the full-length fusion protein because they were purified via the N-terminal GST domain. This idea was supported by immunoblotting, which showed that antibodies directed against the N-terminal 19 amino acids of TAG1 (αNTG1) react with many of these polypeptides (Figure 2A). In an effort to increase the proportion of the full-length product, an additional six strains of E. coli were tested as expression hosts. Only one strain, BL21-CodonPlus (Stratagene), yielded a reproducible improvement, giving a modest increase in the amount of full-length product and a decrease in the accumulation of smaller products (data not shown). Both DH5α and BL21-Codon Plus were used as expression hosts in the experiments described below.
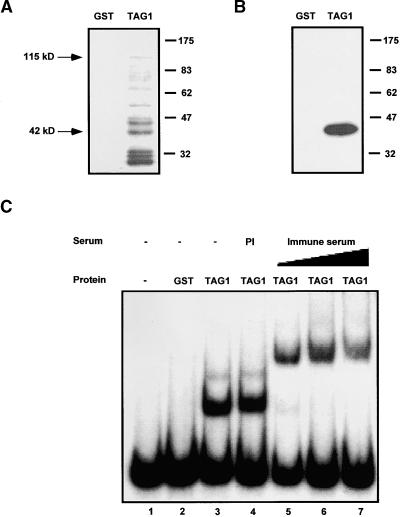
Figure 2.
DNA Binding of GST-TAG1 Fusion Proteins.
(A) Immunoblot of purified fusion products. Recombinant protein (either GST alone or the GST-TAG1 fusion protein) was expressed in E. coli strain DH5α and purified as described in Methods. Eight micrograms of affinity-purified fusion protein was loaded per lane, electrophoresed as described in Methods, and electroblotted onto a nitrocellulose filter. Immunodetection was performed using a 1:1000 dilution of affinity-purified αNTG1 antibodies. Apparent sizes are indicated at right in kilodaltons. The full-length GST-TAG1 fusion product (115 kD) and the 42-kD product are indicated.
(B) Protein gel blot probed with DNA. Extracts were prepared from E. coli expressing GST alone or the GST-TAG1 fusion protein. Approximately 30 μg of affinity-purified protein per lane was resolved by SDS-PAGE and transferred to nitrocellulose. The filter was probed with ∼90 ng of 32P-labeled 5′ STR (nucleotides 1 to 261), washed, and exposed to film at −80°C for 1.5 hr. The DNA binding polypeptide in the TAG1 sample corresponds to a 42-kD fusion product.
(C) DNA binding of recombinant GST-TAG1 in solution. Binding reactions contained 2 μL of affinity-purified protein and ∼20,000 cpm (∼0.5 ng) of 32P-labeled 5′ STR probe. Where indicated, protein was incubated with serum (1.0 μL of preimmune serum [PI]; 0.1, 0.5, or 1.0 μL of αNTG1 immune serum) before addition of the probe. Complexes were separated on a 4% polyacrylamide gel at 4°C, dried, and exposed to film at −80°C.
To test for DNA binding activity of the recombinant transposase, a fragment of the Tag1 element, extending from the 5′ TIR to the start of transcription (nucleotides 1 to 261; 5′ STR probe in Figure 1), was labeled and used to probe a protein gel blot. Figure 2B shows that a single polypeptide (∼42 kD) in the TAG1 sample had a strong affinity for the 5′ STR probe. An abundant band of this size was recognized by the αNTG1 antibodies, as shown by the immunoblot in Figure 2A. No binding was detected for any of the larger TAG1 fusion products.
An electrophoretic mobility shift assay (EMSA) also was used to determine binding of the TAG1 fusion protein to the 5′ STR probe. Figure 2C shows that the migration of this probe was shifted upon the addition of the TAG1 fusion protein, producing two complexes (lane 3); no complex formation was observed when purified GST alone was added (lane 2). This result demonstrates that there is DNA binding activity in the purified TAG1 sample that is not attributable to the GST domain. The presence of TAG1 in the complexes was confirmed by including the αNTG1 antibodies in the binding reaction. Although the addition of preimmune serum caused no change in the electrophoretic mobility (lane 4), the addition of increasing amounts of αNTG1 antiserum resulted in retardation of the complexes (lanes 5 to 7). This experiment also was performed using a 3′ fragment of Tag1 as a probe (nucleotides 3026 to 3295; 3′ STR probe in Figure 1). Results similar to those with the 5′ STR were obtained; that is, binding to the 3′ STR produced two complexes (Figure 3B) that were further shifted upon the addition of αNTG1 antibodies (data not shown). Together, these results indicate that an N-terminal fragment of TAG1 has DNA binding activity.
Figure 3.
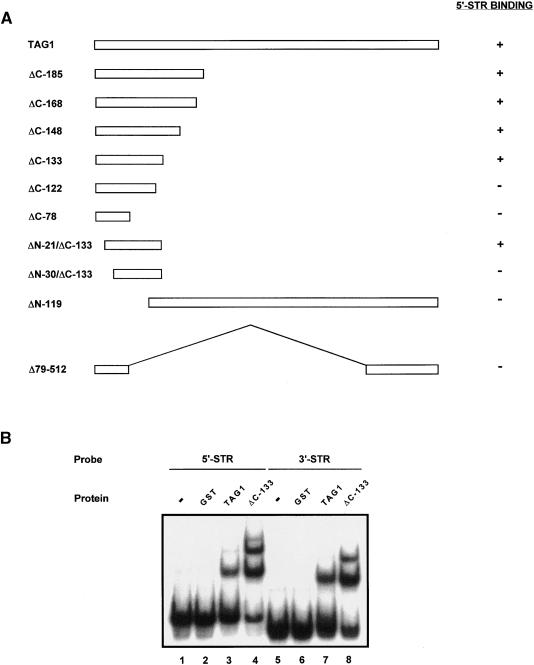
Localization of the TAG1 DNA Binding Domain.
(A) TAG1 deletion derivatives are shown schematically with the amino acid residues of TAG1 indicated. Each derivative was expressed as a GST fusion protein, affinity purified, and assayed by EMSA using the 5′ STR as a probe. Binding of each derivative (present in the protein preparations also containing truncated fusion products) was determined by a shift in the mobility of the probe, as visualized by an autoradiograph of the dried gel.
(B) The TAG1 DNA binding domain interacts with both the 5′ and 3′ STRs. Binding reactions contained 1 μL of the indicated fusion protein (expressed in strain BL21) and ∼22,000 cpm (∼1.4 ng) of 32P-labeled STR DNA. Complexes were separated on a 4% polyacrylamide gel at 4°C, dried, and exposed to film at −80°C.
TAG1 Transposase Has an N-Terminal DNA Binding Domain
Further definition of the DNA binding domain was obtained through a series of deletion derivatives that were expressed in E. coli, affinity purified, and examined by Coomassie blue staining or immunoblotting. Each derivative then was tested for binding to the 5′ STR probe. A summary of these deletion constructs, with their corresponding binding activities, is shown in Figure 3A. By EMSA, we found that DNA binding activity was still retained when amino acids 1 to 20 and 134 to 729 were removed from the TAG1 fusion. Deletion of an additional nine N-terminal or 11 C-terminal amino acids resulted in the complete loss of activity. Thus, a fragment composed of amino acids 21 to 133 of TAG1 is sufficient to bind the 5′ STR. Next, two deletion derivatives were constructed to determine whether any DNA binding activity might be associated with the C terminus of the transposase. The derivatives ΔN-119 and Δ79-512, which contain only portions of the DNA binding region, exhibited no interaction with the 5′ STR probe. These data indicate that TAG1 contains a single DNA binding domain located between residues 21 and 133.
To characterize the activity of the isolated domain, complexes with the 5′ and 3′ STR probes were compared with the complexes formed using protein expressed from the full- length construct. Because the αNTG1 antibodies were generated against the first 19 amino acids of TAG1, we used the epitope-containing ΔC-133 derivative, rather than the ΔN-21/ΔC-133 derivative, for this and subsequent studies. Stronger binding was observed using the ΔC-133–derived protein, albeit the electrophoretic patterns of the faster migrating complexes resembled those obtained with the full- length TAG1 sample (Figure 3B). Even though equivalent amounts of total protein were added to each binding reaction, the stronger signal and additional complex resolved with the ΔC-133 derivative most likely are attributable to higher levels of active protein in this sample.
TAG1 Specifically Recognizes Different Motifs in the STRs
As stated in the Introduction, repeats of the same sequence are not found at each end of Tag1, although the TAG1 DNA binding domain does interact with both STRs in vitro. Therefore, to determine which sequences within the STRs are recognized by TAG1, oligonucleotides containing repeats from each end (Figure 1) were tested as probes by EMSA. The nucleotide sequence of each probe is shown in Figure 4A. The first, probe I, consists of sequence from the 5′ STR, which contains an AAACCC repeat. Probe II contains a portion of the 3′ untranslated region of the Tag1 transcript, in which a repeated TTATT is found. Another repeated sequence, TATATA (probe III), is located 3′ of the transcribed region. Finally, probe IV consists of a sequence from a region closest to the 3′ TIR that contains a TGACCC repeat. As shown in Figure 4B, the ΔC-133 derivative binds to probe I (lane 2) and probe IV (lane 8), which contain the repeats AAACCC and TGACCC, respectively. No complex formation was detected with the A/T-rich probes II (lane 4) and III (lane 6). Because the TIR is identical at each end of Tag1, an oligonucleotide containing this sequence also was tested for binding. Our results indicate that the ΔC-133 derivative forms a very weak complex with the TIR (lane 10). These data suggest that the TAG1 DNA binding domain recognizes two similar, yet distinct, motifs in the STRs of the element as well as a sequence within the TIR.
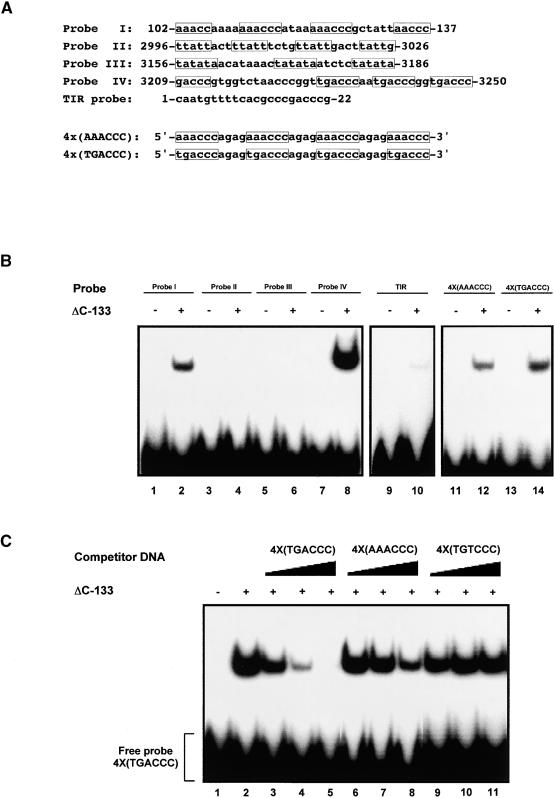
Figure 4.
TAG1 Binds Different Motifs in the STRs.
(A) Sequences of probes. Shown are the top strands of synthetic oligonucleotides that were used as probes in the EMSAs shown in (B). The positions of each sequence relative to the Tag1 element are given; radiolabeling of probes is described in Methods. Repeats within each sequence are boxed. The last two sequences are multimers of the sequence motifs found in probe I and probe IV.
(B) Distinct sequences are bound by TAG1. Each sequence shown in (A) was 32P-labeled for use as a probe in EMSAs with the ΔC-133 derivative (BL21 expressed). Binding reactions contained ∼24,000 cpm (∼0.5 ng) of probe and ∼580 ng of affinity-purified protein. Complexes were separated on 6.5% polyacrylamide gels, dried, and exposed to film at −80°C.
(C) TAG1 specifically binds TGACCC and AAACCC repeats. Binding reactions contained ∼22,000 cpm (∼0.3 ng) of 32P-labeled 4×(TGACCC) probe and excess unlabeled competitor DNA. Specific competitors TGACCC (lanes 3 to 5) and AAACCC (lanes 6 to 8), were added to obtain 10×, 50×, and 250× excess molar ratios. Nonspecific competitor TGTCCC (lanes 9 to 11) was added to obtain 25×, 100×, and 400× molar ratios. Approximately 290 ng of ΔC-133 (BL21 expressed) was incubated with probe and competitor DNA in a buffer containing 10 μg/mL salmon sperm DNA; complexes were resolved on 6.5% polyacrylamide gels. Gels were dried and exposed to film at −80°C.
To test this finding, oligonucleotides containing four direct repeats of the AAACCC or TGACCC motif, separated by four bases, also were used as probes (4× sequences in Figure 4A). The ΔC-133 derivative bound to each of these probes, with a stronger signal resulting from binding to the TGACCC multimer (Figure 4B, lanes 12 and 14). To demonstrate that the observed interaction was specific, EMSAs were performed using unlabeled DNA as a competitor. Binding to the radiolabeled TGACCC multimer was strongly reduced when an unlabeled excess of this same sequence was present (Figure 4C, lanes 3 to 5). A significant reduction in binding to the TGACCC multimer was found with the unlabeled AAACCC multimer (lanes 6 to 8), whereas no competition was observed with an unlabeled multimer containing a single mutation (TGTCCC; Table 1) in the TGACCC motif (lanes 9 to 11). This experiment also was performed using a labeled AAACCC multimer as a probe; again, stronger competition was observed with the unlabeled TGACCC multimer versus the unlabeled AAACCC multimer (data not shown). These results show that the TAG1 DNA binding domain specifically interacts with two different motifs, which are repeated at opposite ends of the element.
Table 1.
Mutational Analysis of DNA Binding Motifs
| Binding Site Sequencea | Percent Bindingb |
|---|---|
| TGACCC | 100 |
| AAACCC | 64 |
| AGACCC | 54 |
| TAACCC | 20 |
| TCACCC | 7 |
| TGAGCC | 14 |
| TGTCCC | <1 |
| TGTGCC | <1 |
| TGACGC | <1 |
| AAACCG | <1 |
| AAACCA | <1 |
a Probes contained four direct repeats of the indicated sequence (top strand is given; substituted bases are underlined) as shown for multimers in Figure 4A. Double-stranded oligonucleotides were labeled with α-32P-dCTP and tested by EMSA for binding by the ΔC-133 derivative.
b The intensity of the signal in the DNA-protein complexes was quantified with Adobe Photoshop (Mountain View, CA), using the signal from the TGACCC binding site probe as 100%.
The requirement of specific bases within these motifs was then investigated by mutational analysis. Because the TGACCC sequence showed stronger binding and competition, nucleotide positions 1 to 5 were substituted within the context of this motif. Position 6 of the AAACCC motif also was substituted, because the 5′ STR contains repeats that are imperfect at this position. Oligonucleotides containing four repeats of each site were tested as probes for binding by the ΔC-133 derivative; these results are compiled in Table 1. Substitution at the first position (T→A), giving rise to an AGACCC binding site, reduced complex formation by ∼50%. A mutation at position 2, to give TCACCC or TAACCC multimers, resulted in a decrease in binding, ranging from 80 to 90%, depending on the substitution. Likewise, a C→G mutation at position 4, resulting in a TGAGCC site, caused a similar decrease in binding. All other mutations (positions 3 and 5 in the TGACCC motif, and position 6 in the AAACCC motif) resulted in no detectable complex formation. In summary, we observed strong binding to the TGACCC motif and moderate binding to the AAACCC and AGACCC motifs. Excluding the first position, alteration of the TGACCC motif reduced binding by at least fivefold, with changes to the third, fifth, and sixth positions completely abolishing binding. Thus, the TAG1 DNA binding domain interacts with the hexamer motifs present at both ends of the Tag1 element, yet it has a preference for the 3′ (TGACCC) motif. In addition, it appears that this interaction is not caused merely by the recognition of a core ACCC sequence but involves the first two nucleotides in the hexamer as well.
TAG1 DNA Binding Activity Is Metal Dependent
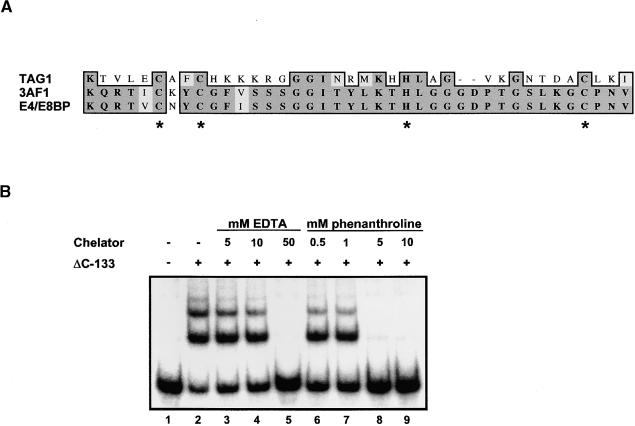
Although searches of available sequences displayed little similarity between TAG1 and other DNA binding proteins, a database search conducted using only the defined DNA binding domain revealed a putative zinc finger motif in the TAG1 transposase. A portion of the TAG1 DNA binding domain aligns with regions from two plant DNA binding proteins, 3AF1 and E4/E8BP, both described as containing putative zinc fingers (Figure 5A). The former is a tobacco protein that binds to a light-responsive promoter (Lam et al., 1990); the latter, from tomato, binds to 5′ flanking sequences of the ethylene-regulated genes E4 and E8 (Coupe and Deikman, 1997). Each protein contains two similar motifs that consist of a series of conserved cysteine and histidine residues; this C2HC-type finger is similar to the canonical zinc fingers found in many transcription factors (Struhl, 1989). Such domains require a zinc ion for activity, because inhibition may be accomplished with the metal chelators EDTA (Kadonaga et al., 1987) and 1,10-phenanthroline (Lam et al., 1990; Coupe and Deikman, 1997; Tsai and Reed, 1998). To determine whether TAG1 DNA binding activity might be metal dependent, EMSAs using the ΔC-133 derivative were performed with increasing amounts of EDTA or 1,10-phenanthroline included in the binding reaction. As shown in Figure 5B, complex formation with the 5′ STR probe was abolished completely with 50 mM EDTA (lane 5) or 5 mM 1,10-phenanthroline (lane 8). Comparable inhibition was observed using the 3′ STR as a probe (data not shown).
Figure 5.
The TAG1 DNA Binding Domain Contains a Putative Zinc Finger.
(A) Amino acid alignment of TAG1 with tobacco 3AF1 and tomato E4/E8BP sequences. The alignment was performed using the ClustalW program. Shown are TAG1 residues 39 to 76, 3AF1 residues 33 to 72, and E4/E8BP residues 213 to 252. Putative metal binding amino acids are indicated with asterisks.
(B) Chelator inhibition of DNA binding. Approximately 300 ng (0.1 μL) of affinity-purified ΔC-133 (BL21 expressed) was incubated with increasing quantities of EDTA or 1,10-phenanthroline in 2% ethanol. Although 1 mM EDTA was present in all EMSA binding reactions (see Methods), only the additional EDTA quantities are indicated (lanes 3 to 5); the reaction in lane 2 contained 1 mM EDTA and is labeled as containing no chelator merely as a reference for the inhibition by excess chelator. Approximately 24,000 cpm of 32P-labeled 5′ STR probe (∼1.4 ng) was added, and complexes were resolved on a 4% polyacrylamide gel; the gel was dried and exposed to film at −80°C.
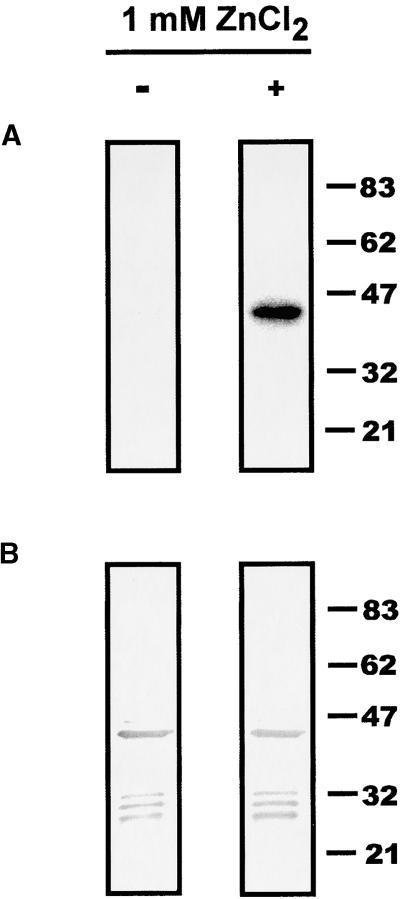
Because the results described above indicated that the binding of TAG1 to DNA required metal ions, we sought to examine directly the effect of zinc on TAG1 binding activity. A protein gel blot was probed with the 5′-STR in the absence or presence of added zinc. As illustrated in Figure 6A, DNA binding by the ΔC-133 derivative was improved dramatically with 1 mM ZnCl2. Immunodetection revealed comparable amounts of the ΔC-133 polypeptide on each filter (Figure 6B), indicating that the variation in signal was not the result of a significant difference in the amounts of protein. This result demonstrates that zinc promotes DNA binding and corroborates the metal dependence of TAG1 binding activity.
Figure 6.
Zinc Promotes TAG1 Binding to DNA.
(A) Effect of added zinc on DNA binding. Approximately 60 μg of BL21-expressed ΔC-133 derivative was loaded onto two lanes of an 8% SDS-PAGE gel, electrophoresed, and transferred to a nitrocellulose filter as described in Methods. After Ponceau S staining, the filter was cut into two pieces, and half was processed with 1 mM ZnCl2 added to all buffers (preincubation, hybridization, and wash; see Methods), whereas the other half was processed without added zinc. Filters were probed with ∼35 ng of 32P-labeled 5′ STR DNA, washed, and exposed to film at −80°C for 20 min.
(B) Immunoblot of the ΔC-133 derivative. Immunodetection of polypeptides present on the filters in (A) was performed after removal of the probe, using a 1:5000 dilution of affinity-purified αNTG1 antibodies.
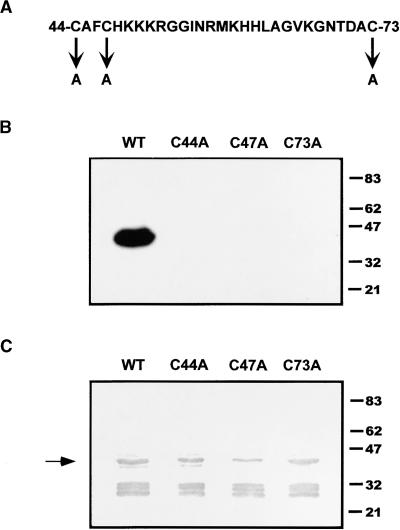
To further examine the role of the zinc finger, we mutated each cysteine residue in the C2HC motif, which is predicted to participate in the coordination of the metal ion and thus is required for activity. Site-directed mutagenesis of the ΔC-133 derivative was performed to replace each cysteine (i.e., Cys-44, Cys-47, and Cys-73) with an alanine residue, as depicted in Figure 7A. Protein from two E. coli transformants for each mutation was affinity purified and tested for DNA binding activity by EMSA. Unlike the wild-type ΔC-133 protein, no detectable complexes were formed using the mutant polypeptides (data not shown). Then, DNA binding by protein gel blot analysis was performed using a single transformant for each Cys→Ala mutation and the 5′ STR probe. A strong signal was observed from the lane containing wild-type ΔC-133 protein (Figure 7B); no signal was detected in lanes containing mutant proteins (Figure 7B), even upon a long exposure of this filter (data not shown). The immunoblot in Figure 7C confirmed the presence of ample amounts of each mutant polypeptide on the filter after removal of the probe. Thus, we conclude that the indicated cysteines are essential for binding activity. Given the observed metal dependence and enhancement with zinc, we suggest that these cysteine residues may act as zinc ligands, forming a zinc finger in the functional DNA binding domain of TAG1.
Figure 7.
Site-Directed Mutational Analysis of Cysteine Residues.
(A) The TAG1 putative zinc finger motif. Amino acids 44 to 73 of TAG1 are shown with cysteine-to-alanine (C→A) substitutions indicated.
(B) DNA binding analysis of C→A mutants. The ΔC-133 derivative and individual mutants were expressed in DH5α and affinity purified as described in Methods. Approximately 90 μg of each was loaded per lane onto an 8% SDS-PAGE gel, electrophoresed, and transferred to nitrocellulose as described in Methods. The protein blot was probed with ∼280 ng of 32P-labeled 5′ STR DNA, washed, and exposed at −80°C for 15 min. The filter then was exposed for longer periods to detect any weak signal (data not shown). WT, wild type.
(C) Immunoblot of C→A mutants. The filter in (B) was stripped of probe and subjected to immunodetection using a 1:5000 dilution of affinity-purified αNTG1 antibodies. The polypeptide (∼43 kD) responsible for the signal in (B) is indicated by the arrow. WT, wild type.
DISCUSSION
Description of the Transposase Binding Sites
By current models, an initial step in conservative transposition involves the synapsis of the transposon ends mediated by element-encoded transposase bound to specific sites near these ends (reviewed by Kunze et al., 1997; Haren et al., 1999). Similar to other plant DNA-type transposons such as Ac and En/Spm, the Tag1 element is composed of a coding region flanked by multiple copies of transposase binding sites. However, in contrast to these other elements, the specific subterminal sequences recognized by the transposase are different in the case of Tag1. Although the hexamer motifs of Tag1 are seemingly related, with a consensus of T/A-G/ A-A-C-C-C, the affinity of the transposase for each clearly is not the same. As demonstrated by competition experiments, our results indicate a stronger affinity for the 3′-TGACCC site versus the 5′-AAACCC site. Although this dissimilarity in binding site sequence apparently is uncommon, such a structure is compatible with the proposed mechanism of end synapsis by the transposase, because the TAG1 transposase binding domain does bind to both termini of the element.
It is noteworthy that the 5′ STR of Tag1 contains more potential binding sites than the four direct TGACCC repeats at the 3′ end. Six perfect copies, as well as an additional six imperfect copies of the AAACCC motif, are distributed in both orientations at the 5′ end. Based on our mutational analysis, we predict that three of the imperfect motifs (substituted in the last position) would not be recognized by the transposase and that two are unlikely to serve as binding sites, being substituted in the second and third position, respectively. The last motif, TAACCC, is a possible site, because we have shown that the recombinant TAG1 DNA binding domain does bind to this motif in vitro. Although the relevant in vivo occupancy of these sites is not known, it is tempting to speculate that the near doubling of sites at the 5′ end of the intact Tag1 element may compensate for the lower affinity of the transposase for this sequence motif. Yet, deletion analysis has shown that the minimal cis requirements of excision in vivo are ∼100 bp of distal sequence containing four motifs at each end (Liu et al., 2001). Within the first 0.1 kb of Tag1, two motifs are substituted in the last position, and based on the results of this present study, these would not be bound by the transposase. By this reasoning, no more than two 5′ sites and four 3′ sites are required for excision at rates comparable to those for the intact element. Thus, the number of binding sites necessary for efficient binding, and subsequent cleavage, is minimal for Tag1.
Elucidation of the Transposase DNA Binding Activity
Previous sequence analyses identified two putative nuclear localization signal sequences near the N terminus of the TAG1 transposase (Liu and Crawford, 1998). The results presented here also position the DNA binding domain in this same region of the polypeptide. One of the unifying themes among transposases is the arrangement of functional domains, in which site-specific DNA binding activities generally are associated with the N terminus of the protein (Kunze et al., 1997; Haren et al., 1999); TAG1 fits with this generality. On the other hand, although divalent metal ions have an established role in the catalytic functions of transposases (Polard and Chandler, 1995), we are unaware of any documented metal-dependent DNA binding activity. Therefore, the observed metal dependence of TAG1 DNA binding, as indicated by the enhancement with zinc and the inhibition by metal chelators, is a novel finding for transposases.
Consistent with this metal requirement is the finding of a C2HC zinc binding motif in the TAG1 DNA binding domain. By site-directed mutagenesis, we found that the three cysteine residues in the TAG1 zinc finger motif are essential individually for DNA binding activity. Whereas critical cysteine residues have been identified in other transposases, none has been shown to participate in metal-dependent DNA binding. The TNPA protein of the En/Spm element contains cysteine residues that are important for DNA binding; however, in this case, it was proposed that the formation of disulfide bonds between these residues was required for the overall structure of the domain (Trentmann et al., 1993). Although it is possible that our cysteine-to-alanine mutations might have disrupted disulfide bond formation, our chelation experiments clearly demonstrate a divalent metal requirement. Moreover, we found that the DNA binding activity of the ΔC-133 derivative is strongly affected by the presence (or absence) of zinc. In fact, all evidence thus far supports a zinc coordination role for these cysteines.
Potential zinc fingers have been proposed for two other eukaryotic transposases. A sequence with four cysteines has been found in the TnpD ORF of En/Spm (Vodkin and Vodkin, 1989). The Drosophila P-element transposase and KP repressor proteins both contain a zinc finger motif of the C2HC type in their DNA binding domains (Lee et al., 1998). The cysteine and histidine residues are required in the C2HC motif, because mutation of the first two residues or the last two residues to alanines abolishes site-specific binding; nonetheless, a metal dependence for DNA binding has not been detected (Lee et al., 1998). A comparison of the TAG1 and P transposase C2HC motifs reveals a difference in spacing between the putative zinc coordinating residues. In TAG1, the arrangement is CX2CX14HX10C (where X indicates any amino acid), whereas a motif of CX3CX9HX3C has been described for the P transposase (Lee et al., 1998). A survey of other C2HC motifs revealed that the number of residues between each ligand is variable (Berkovits and Berg, 1999; Fox et al., 1999); thus, it seems plausible that each of these sequences could adopt a zinc finger conformation. Curiously, the second cysteine in the P motif is preceded by another cysteine residue (i.e., CXXCC). If one considers the cysteine in position 4, rather than position 5, to be the second ligand, an interesting homology with the TAG1 motif arises, with the following consensus: CXFCH/CKX3GX(4-8)HX(3-10)C. Further inquiry is needed to determine whether this homology has any significance.
Zinc finger domains are involved in both nucleic acid recognition and protein–protein interactions (reviewed by Mackay and Crossley, 1998; Takatsuji, 1998); thus, there are at least two possible functions that the TAG1 zinc finger domain could perform. Clearly, it is important for DNA binding; however, we have not determined if it interacts directly with DNA. Another possibility is that it is involved in protein–protein interactions. If this is the case, homodimerization could be required for DNA binding, which would suggest an indirect involvement in DNA binding. This appears to be the situation with HIV-1 integrase, which contains a H2C2 zinc binding motif near the N terminus. Binding of a zinc atom to the integrase appears to stabilize the structure of the N-terminal domain and promote multimerization, rather than being involved directly in DNA binding (Zheng et al., 1996; Heuer and Brown, 1997).
One puzzling observation from our work is the lack of DNA binding by the full-length TAG1 transposase. Although a truncated GST-TAG1 fusion protein (42 kD) showed strong activity, binding by the full-length protein was not detected by any of our protein gel blot analyses (Figure 2B and data not shown). Although it is possible that irreversible denaturation is responsible for the lack of DNA binding by the full-length protein, the results of our EMSAs indicate otherwise. The electrophoretic patterns of the DNA–protein complexes obtained with the full-length TAG1 sample and the isolated DNA binding domain (ΔC-133 derivative) are similar (Figure 3B), suggesting that the binding activities present in these samples are of comparable molecular weights. Thus, the full-length recombinant protein may be unable to bind DNA when in solution as well as when immobilized, perhaps because of the absence of eukaryote-specific modifications and/or improper folding. Alternatively, amino acid sequences adjacent to the DNA binding domain may be inhibitory. This phenomenon of higher DNA binding activity of a truncated version has been reported for the Tc1 transposase: recombinant full-length Tc1A showed very weak affinity for the ends of Tc1, whereas an N-terminal derivative bound strongly (Vos et al., 1993). Furthermore, the C terminus of the prokaryotic Tn5 transposase is reported to interfere with DNA binding that is mediated by an N-terminal domain (Wiegand and Reznikoff, 1994; Davies et al., 1999). Indeed, the description of cryptic DNA binding domains of both plant (VP1; Suzuki et al., 1997) and mammalian (Ets-1; Jonsen et al., 1996) transcription factors has led to the possibility of intramolecular regulatory mechanisms in vivo. Further work is needed to determine if such mechanisms apply to TAG1.
METHODS
Recombinant DNA
The TAG1 open reading frame (ORF) (Liu and Crawford, 1998) was subcloned into the NcoI and XbaI sites of pSPUTK (Stratagene), giving rise to the plasmid pSPTG. The ORF was then subcloned into the SmaI site of pGEX-2TK (Amersham Pharmacia) by NcoI-XbaI digestion of pSPTG, filling in with Klenow and blunt-end ligation. Expression from this construct, pGXTG, results in an additional five amino acids being placed between the 26-kD glutathione S-transferase (GST) domain and the first methionine of TAG1. All subsequent expression constructs were based on either pSPTG or pGXTG. Deletion constructs ΔN-119, Δ79-512, and ΔC-185 were generated by digestion with EcoRV, PstI, and EcoRI, respectively, followed by religation of the plasmid. Other deletions were generated by polymerase chain reaction amplification of the desired region, with a stop codon placed after the indicated TAG1 residue, followed by cloning into the NcoI-EcoRI sites of pGXTG. DNA sequences were verified using the dideoxy chain termination method.
Site-directed mutagenesis was performed using the QuickChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. DNA sequences were confirmed by dideoxy sequencing.
Protein Expression and Purification
Escherichia coli cells (strain DH5α or BL21-CodonPlus-RIL; Stratagene) harboring GST expression constructs were grown overnight with shaking at 35 to 37°C in Luria-Bertani medium containing 100 μg/mL carbenicillin. Luria-Bertani plus carbenicillin medium was seeded with a 1:50 dilution of the overnight culture, and cells were grown to an OD600 between 0.6 and 0.8. Cultures were shifted to 30°C, and expression of fusion proteins was induced for 1.5 hr with 0.4 mM isopropylthio-β-galactoside. Cells were pelleted by centrifugation at 4°C, frozen in liquid nitrogen, and stored at −80°C.
All subsequent steps were performed on ice or at 4°C except where noted. Cells were resuspended in HPK buffer (25 mM Hepes, 0.7 mM Na2HPO4, 135 mM NaCl, and 5 mM KCl, pH 7.34) and incubated with 1 mg/mL lysozyme and 1 mM phenylmethylsulfonyl fluoride for 30 min. Cells were sonicated until the suspension was slightly translucent (usually two or three pulses, ∼30 sec each). A protease inhibitor cocktail (2 μg/mL each antipain, chymostatin, leupeptin, pepstatin, and bestatin and 0.01 trypsin inhibitor unit/mL aprotinin) was added between pulses. Triton X-100 was added to 1%, and the mixture was rotated for 45 min and then centrifuged at ∼12,000g to remove cellular debris. The soluble fraction was diluted 1:1 in HPKE buffer (HPK buffer, 5 mM EDTA, 20 mM Na2HPO4, 1 mM β-mercaptoethanol, and 1% Triton X-100) and incubated with HPKE-equilibrated glutathione–Sepharose 4B (Amersham Pharmacia) for 1 hr. The resin with bound proteins was washed extensively with HPKE and HPK. Fusion proteins were eluted from the resin at 22°C in GSH buffer (10 mM reduced glutathione, 10 mM Hepes, pH 8.0, and 60 mM KCl), concentrated in a Centricon C-30 (Millipore, Bedford, MA), and stored in 10% glycerol at −20°C (short term) or −80°C (long term). Protein concentrations were determined using the Micro BCA protein assay (Pierce Chemical Co.).
Antibody Production, Purification, and Immunoblotting
Polyclonal antibodies were generated against TAG1 residues 1 to 19. The sequence of this peptide, METEHDENYEDIAAANRSI, had a cysteine residue added to the C terminus for eventual conjugation. Peptide synthesis and antibody production were performed by Genemed Synthesis (San Francisco, CA).
Affinity purification of TAG1-specific antibodies was performed using the SulfoLink Kit (Pierce Chemical Co.) with minor modifications. Briefly, 3 mg of the peptide described above was dissolved in sample preparation buffer (0.1 M sodium phosphate and 5 mM EDTA, pH 6.0) plus 1% SDS and reduced with 143 mM β-mercaptoethanol by heating to 70°C for 5 min. The reduced peptide was lyophilized, and reductant was removed by washing with 50% ethanol followed by evaporation with N2. After three washes, the reduced peptide was lyophilized and coupled to SulfoLink Coupling Gel according to the manufacturer's recommendations. Antibodies were eluted from the column in 100 mM glycine, pH 2.75; eluates were neutralized with 0.1 volume of 1 M Tris, pH 9.0.
Immunoblotting was performed as follows. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose filter (Immobilon-NC; Millipore) in a Mini Trans-Blot apparatus (Bio-Rad) using 25 mM Tris, 192 mM glycine, pH 8.0 to 8.3, and 20% (v/v) methanol. Blots were stained with Ponceau S (5 mg/mL in 5% trichloroacetic acid) and then blocked in Tris-buffered saline (20 mM Tris and 150 mM NaCl, pH 7.2) plus 5% (w/v) nonfat milk. Filters were incubated in Tris-buffered saline with a 1:1000 dilution (except where noted) of affinity-purified αNTG1 antibodies. Immunodecorated proteins were detected with a 1:1000 dilution of a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (DAKO Corp, Carpinteria, CA). Development was performed with 0.5 mg/mL diaminobenzidine in 100 mM imidazole, pH 7.0, plus 3.75 mM NiCl2 and 0.001% H2O2.
DNA Probes
Subterminal region (STR) DNA was amplified by polymerase chain reaction and cloned into pBluescript KS+ as XbaI-BamHI (5′ STR) and BamHI-SmaI inserts (3′ STR). Nucleotide sequences were confirmed by dideoxy sequencing. After digestion with XbaI-XmaI or BamHI-XmaI, fragments were separated on agarose gels and purified through a glass wool column.
Each oligonucleotide pair had an additional 4 bp added to the 5′ end for labeling purposes: 5′-GAGC-3′ on the top strand and 5′-GATC-3′ on the complementary strand. Oligonucleotides were purified on 12% polyacrylamide/7 M urea gels and eluted overnight in 0.3 M sodium acetate, pH 7, at 37°C. Double-stranded oligonucleotide DNA was prepared by annealing complementary oligonucleotides as follows: 125 ng of each oligonucleotide was heated to 70°C for 2 min in AR buffer (10 mM Tris, pH 7.5, 20 mM NaCl, 6 mM MgCl2, 1 mM DTT, and 1.5 ng of BSA) and allowed to cool to 37°C.
Double-stranded DNA probes were labeled by filling in with α-32P-dCTP for 1 hr at 30°C using Klenow polymerase (2 units/100 ng DNA) in AR buffer with 6.7 μM each dATP, dTTP, and dGTP. The ends were extended for an additional 2 min with unlabeled deoxynucleotide triphosphates. DNA was extracted with phenol, purified through a ProbeQuant G-50 Micro column (Amersham Pharmacia), and precipitated.
Electrophoretic Mobility Shift Assays
Recombinant proteins were preincubated for 5 min at 22°C in HKG buffer (10 mM Hepes, pH 7.6, 60 mM KCl, 1 mM EDTA, 1 mM DTT, 2 mg/mL BSA, 0.1 mg/mL double-stranded salmon sperm DNA, and 5% glycerol) with 3 mM MgCl2 (except where noted). DNA probes were added, and the binding reaction was allowed to proceed for 25 to 30 min at ∼22°C. Gel loading buffer (25 mM Tris, pH 7.6, 0.02% bromphenol blue, 0.02% xylene cyanol, and 4% glycerol) was added before electrophoresis on 4 or 6.5% polyacrylamide (29:1) gels. Gels were run at 4°C in 0.5 × Tris borate EDTA at 8 V/cm for 1.5 to 2 hr, dried, and exposed at −80°C.
For supershift assays, either preimmune serum or antiserum was added during the preincubation of protein before addition of the probe. In competition experiments, molar ratios of labeled and unlabeled DNA were mixed, and protein preincubated in HKG buffer (plus 3 mM MgCl2) with 10 μg/mL salmon sperm DNA was challenged with this mixture.
For the metal-dependent assays, protein was preincubated with the designated chelator (final concentration as noted) in HKG buffer (without added MgCl2). Because 1,10-phenanthroline was dissolved in ethanol, binding reactions contained a final concentration of 2% ethanol.
DNA Binding on Protein Gel Blots
Protein gel blotting and DNA analyses were performed according to Ciceri et al. (1997) with modifications. Recombinant proteins were heated to 95°C in SDS sample buffer (60 mM Tris, pH 6.8, 1.5% SDS, 4% β-mercaptoethanol, 0.02% bromphenol blue, and 10% glycerol) and separated by 8% SDS-PAGE. Proteins then were transferred to nitrocellulose (Immobilon-NC; Millipore) and stained with Ponceau S to visualize bands. Before hybridization, protein blots were incubated by rocking in 30 mL of SW buffer (20 mM Hepes, 60 mM KCl, 1 mM EDTA, 1 mM DTT, and 3 mM MgCl2, pH 7.6) with 5% (w/v) nonfat milk for 3 to 4 hr at 4°C. Hybridization with probes (107 to 108 cpm/μg DNA) was performed in 10 mL of SW buffer with 5% nonfat milk and 1 μg/mL double-stranded salmon sperm DNA and rocking overnight at 4°C. Filters were washed three times in 50 mL of SW buffer with 0.25% nonfat milk and exposed to film at −80°C. Probes were stripped from nitrocellulose filters by washing in SW buffer (without MgCl2) plus 200 mM KCl and 0.25% nonfat milk.
For the zinc addition experiment, 1 mM ZnCl2 was added to (or omitted from) all incubation buffers (except the stripping buffer) after electroblotting.
Acknowledgments
We thank Dr. Pietro Ciceri and Dr. Robert Schmidt for advice on DNA binding studies, Dr. Victor Vacquier for technical advice, and Mary Galli for technical assistance in the antibody purification. This work was supported by Grant MCB-9808215 from the National Science Foundation.
References
- Becker, H.-A., and Kunze, R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254, 219–230. [DOI] [PubMed] [Google Scholar]
- Benito, M.-I., and Walbot, V. (1997). Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17, 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits, H.J., and Berg, J.M. (1999). Metal and DNA binding properties of a two-domain fragment of neural zinc finger factor 1, a CCHC-type zinc binding protein. Biochemistry 38, 16826–16830. [DOI] [PubMed] [Google Scholar]
- Calvi, B.R., Hong, T.J., Findley, S.D., and Gelbart, W.M. (1991). Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator and Tam3. Cell 66, 465–471. [DOI] [PubMed] [Google Scholar]
- Ciceri, P., Gianazza, E., Lazzari, B., Lippoli, G., Genga, A., Hoscheck, G., Schmidt, R.J., and Viotti, A. (1997). Phosphorylation of Opaque2 changes diurnally and impacts its DNA binding activity. Plant Cell 9, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe, S.A., and Deikman, J. (1997). Characterization of a DNA-binding protein that interacts with 5′ flanking regions of two fruit-ripening genes. Plant J. 11, 1207–1218. [DOI] [PubMed] [Google Scholar]
- Davies, D.R., Braam, L.M., Reznikoff, W.S., and Rayment, I. (1999). The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9-A resolution. J. Biol. Chem. 274, 11904–11913. [DOI] [PubMed] [Google Scholar]
- Essers, L., Adolphs, R.H., and Kunze, R. (2000). A highly conserved domain of the maize activator transposase is involved in dimerization. Plant Cell 12, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmar, S., and Kunze, R. (1991). The ORFA protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 13, 4003–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, A.H., Liew, C., Holmes, M., Kowalski, K., Mackay, J., and Crossley, M. (1999). Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, M.J., Liu, D., Tsay, Y.-F., Ustach, C., and Crawford, N.M. (1997). Tag1 is an autonomous transposable element that shows somatic excision in both Arabidopsis and tobacco. Plant Cell 9, 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl, A., Lutticke, S., and Saedler, H. (1988). TnpA product encoded by the transposable element En-1 of Zea mays is a DNA-binding protein. EMBO J. 7, 4045–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Ton-Hoang, B., and Chandler, M. (1999). Integrating DNA: Transposases and retroviral integrases. Annu. Rev. Microbiol. 53, 245–281. [DOI] [PubMed] [Google Scholar]
- Hehl, R., Nacken, W.K.F., Drause, A., Saedler, H., and Sommer, H. (1991). Structural analysis of Tam1, a transposable element from Antirrhinum majus, reveals homologies to the Ac element from maize. Plant Mol. Biol. 16, 369–371. [DOI] [PubMed] [Google Scholar]
- Heuer, T.S., and Brown, P.O. (1997). Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry 36, 10655–10665. [DOI] [PubMed] [Google Scholar]
- Jonsen, M.D., Petersen, J.M., Xu, Q.-P., and Graves, B.J. (1996). Characterization of the cooperative function of inhibitor sequences in Ets-1. Mol. Cell. Biol. 16, 2065–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga, J.T., Carner, K.R., Masiarz, F.R., and Tjian, R. (1987). Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51, 1079–1090. [DOI] [PubMed] [Google Scholar]
- Kunze, R., and Starlinger, P. (1989). The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8, 3177–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., Saedler, H., and Lonnig, W.-E. (1997). Plant transposable elements. Adv. Bot. Res. 27, 332–470. [Google Scholar]
- Lam, E., Kano-Murakami, Y., Gilmartin, P., Niner, B., and Chua, N.H. (1990). A metal-dependent DNA-binding protein interacts with a constitutive element of a light-responsive promoter. Plant Cell 2, 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.C., Beall, E.L., and Rio, D.C. (1998). DNA binding by the KP repressor protein inhibits P-element transposase activity in vitro. EMBO J. 17, 4166–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., and Crawford, N.M. (1998). Characterization of the putative transposase mRNA of Tag1, which is ubiquitously expressed in Arabidopsis and can be induced by Agrobacterium-mediated transformation with dTag1 DNA. Genetics 149, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Mack, A., Wang, R., Belk, J., Ketpur, N.I., and Crawford, N.M. (2001). Functional dissection of the cis-acting sequences of the Arabidopsis transposable element Tag1 reveals dissimilar subterminal sequence and minimal spacing requirements for transposition. Genetics 157, 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, J.P., and Crossley, M. (1998). Zinc fingers are sticking together. Trends Biochem. Sci. 1, 1–4. [DOI] [PubMed] [Google Scholar]
- Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214. [DOI] [PubMed] [Google Scholar]
- Plasterk, R.H.A., Izsvak, Z., and Ivics, Z. (1999). Resident aliens: The Tc1/mariner superfamily of transposable elements. Trends Genet. 15, 326–332. [DOI] [PubMed] [Google Scholar]
- Polard, P., and Chandler, M. (1995). Bacterial transposases and retroviral integrases. Mol. Microbiol. 15, 13–23. [DOI] [PubMed] [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. (2001). Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, K. (1989). Helix-turn-helix, zinc-finger and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem. Sci. 14, 137–140. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., and McCarty, D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji, H. (1998). Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54, 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentmann, S.M., Saedler, H., and Gierl, A. (1993). The transposable element En/Spm-encoded TNPA protein contains a DNA binding and a dimerization domain. Mol. Gen. Genet. 238, 201–208. [DOI] [PubMed] [Google Scholar]
- Tsai, R.Y.L., and Reed, R.R. (1998). Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 18, 6447–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay, Y.-F., Frank, M.J., Page, T., Dean, C., and Crawford, N.M. (1993). Identification of a mobile endogenous transposon in Arabidopsis thaliana. Science 260, 342–344. [DOI] [PubMed] [Google Scholar]
- van Pouderoyne, G., Ketting, R.F., Perrakis, A., Plasterk, R.H.A., and Sixma, T.K. (1997). Crystal structure of the specific DNA-binding domain of Tc3 transposase of C. elegans in complex with transposon DNA. EMBO J. 16, 6044–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin, M.H., and Vodkin, L.O. (1989). A conserved zinc finger domain in higher plants. Plant Mol. Biol. 12, 593–594. [DOI] [PubMed] [Google Scholar]
- Vos, J.C., and Plasterk, R.H. (1994). Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J. 79, 6125–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, J.C., van Luenen, H.G.A.M., and Plasterk, R.H.A. (1993). Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 7, 1244–1253. [DOI] [PubMed] [Google Scholar]
- Warren, W.D., Atkinson, P.W., and O'Brochta, D.A. (1994). The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac and Tam3 (hAT) element family. Genet. Res. 64, 87–97. [DOI] [PubMed] [Google Scholar]
- Wiegand, T.W., and Reznikoff, W.S. (1994). Interaction of Tn5 transposase with the transposon termini. J. Mol. Biol. 235, 486–495. [DOI] [PubMed] [Google Scholar]
- Zheng, R., Jenkins, T.M., and Craigie, R. (1996). Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 93, 13659–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]