Abstract
Auxin rapidly induces auxin/indoleacetic acid (Aux/IAA) transcription. The proteins encoded are short-lived nucleus-localized transcriptional regulators that share four conserved domains. In a transient assay measuring protein accumulation, an Aux/IAA 13–amino acid domain II consensus sequence was sufficient to target firefly luciferase (LUC) for low protein accumulation equivalent to that observed previously for full-length PSIAA6. Single amino acid substitutions in these 13 amino acids, corresponding to known auxin response mutants, resulted in a sixfold to 20-fold increase in protein accumulation. Naturally occurring variant amino acids had no effect. Residues identified as essential by single alanine substitutions were not sufficient when all flanking amino acids were alanine, indicating the importance of flanking regions. Using direct protein degradation measurements in transgenic Arabidopsis seedlings, full-length IAA1, PSIAA6, and the N-terminal 73 PSIAA6 amino acids targeted LUC for rapid degradation with 8-min half-lives. The C-terminal 109 amino acids did not affect LUC half-life. Smaller regions containing domain II also targeted LUC for rapid degradation, but the rates were not equivalent to those of the full-length protein. A single domain II substitution in the context of full-length PSIAA6 increased half-life 30-fold. Proteasome inhibitors affected Aux/IAA::LUC fusion protein accumulation, demonstrating the involvement of the proteasome.
INTRODUCTION
The class of plant hormones called auxins plays an important role in a variety of developmental processes. Auxin acts as a signal for cell division, elongation, or differentiation, depending on the cell type, and it plays an important role in root formation, apical dominance, and tropic responses (Went and Thimann, 1937; Theologis, 1986; Estelle, 1992). The transcriptional activation of a select set of genes, called primary auxin-responsive genes, early in response to auxin might contribute to subsequent auxin-induced growth and developmental responses. These genes can be divided into several groups based on amino acid sequence identity and auxin induction kinetics, the best characterized of which are the auxin/indoleacetic acid (Aux/IAA) gene family, the GH3 gene family, and the SAUR (small auxin-up RNA) gene family (Abel and Theologis, 1996).
The Aux/IAA family of genes encode nuclear proteins (Abel et al., 1994) sharing four highly conserved domains (I, II, III, and IV). Aux/IAA proteins have been postulated to act as transcription factors that mediate the auxin response cascade. Initially, this hypothesis was based on the presence of predicted secondary structural elements similar to those found in a class of prokaryotic transcriptional regulators (Abel and Theologis, 1995). In support of this model, Aux/IAA proteins have been shown to function as negative regulators of auxin response when coexpressed with an auxin response element–β-glucuronidase reporter in transient assays (Ulmasov et al., 1997b). This effect most likely results from interaction with the endogenous DNA binding proteins called auxin response factors, because no specific in vitro binding of Aux/IAA proteins to DNA has been observed (Ulmasov et al., 1997a). The exact mode of action of Aux/IAA proteins remains unclear. Direct evidence for the importance of Aux/IAA proteins in auxin signaling was demonstrated when axr3, a semidominant mutant in Arabidopsis with an altered auxin response, was shown to encode the Aux/IAA protein IAA17 (Rouse et al., 1998). Additional semidominant mutations in Arabidopsis Aux/IAA proteins have been identified subsequently in a variety of genetic screens (Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001).
The in vivo half-lives for two pea Aux/IAA proteins, PSIAA4 and PSIAA6, have been determined by pulse-chase analysis to be 8 and 6 min, respectively (Abel et al., 1994). These half-lives are among the shortest known for eukaryotic proteins and are characteristic of regulatory proteins. PSIAA6 in translational fusion with firefly luciferase (LUC) targeted the fusion protein for rapid proteolysis in transgenic Arabidopsis seedlings, and the N-terminal region was sufficient for low protein accumulation, suggesting that it was sufficient for this effect (Worley et al., 2000). This is consistent with the behavior of degradation signals from other short-lived proteins. Degradation signals act in a transferable fashion such that when fused to a long-lived protein, the fusion protein has a half-life of similar magnitude as does the short-lived protein from which the degradation signal was derived (Sadis et al., 1995; Lenk and Sommer, 2000; Szewczyk et al., 2000). However, no universal consensus sequence for a degradation signal has been identified, requiring empirical identification, although functionally related proteins, such as mitotic cyclins, have degradation signals with significant amino acid identity.
Controlled proteolysis in eukaryotes occurs most often by a mechanism in which the highly conserved protein ubiquitin is conjugated to a substrate through a series of enzymatic activities: E1/ubiqutin-activating enzyme, E2/ubiquitin-conjugating enzyme, and E3/ubiquitin ligase (Hochstrasser, 1996; Ciechanover et al., 2000). Multiple ubiquitin moieties then are attached via an E2/E3 complex through a lysine residue within ubiquitin itself, forming a ubiquitin polymeric chain (Chau et al., 1989). This polyubiquitinated substrate then is targeted for degradation by the 26S proteasome (Hochstrasser, 1996; Ciechanover et al., 2000; Thrower et al., 2000). The ubiquitin–proteasome pathway has been implicated in the degradation of several short-lived proteins in plants, such as B-type cyclins (Genschik et al., 1998), phytochrome A (Clough and Vierstra, 1997), and the transcription factor HY5 (Osterlund et al., 2000).
Here, we describe our studies identifying the Aux/IAA degradation signal sufficient for targeting LUC for low protein accumulation in cultured cells and for rapid proteolysis in transgenic Arabidopsis seedlings. In both assays, the presence of conserved domain II is required, and although a central core of five amino acids is necessary, it is not sufficient for wild-type accumulation. Finally, we show the importance of the proteasome in Aux/IAA degradation.
RESULTS
Transient Assays Suggest That a Fusion Protein Containing IAA17 Accumulates to Much Lower Levels Than Does LUC Alone
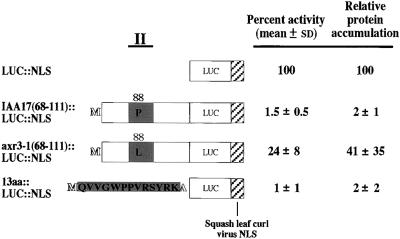
Previously, we developed a transient assay system in tobacco cultured cells to rapidly assess the effect of changes in Aux/IAA amino acids on the accumulation of Aux/IAA::LUC fusion proteins (Worley et al., 2000). We demonstrated that LUC expressed as a fusion protein with either of two different Aux/IAA proteins, PSIAA6 or IAA1, resulted in low protein accumulation in these assays (Worley et al., 2000). The smallest peptide region of PSIAA6 shown in that study to result in low protein accumulation contained amino acids 18 to 73 (Worley et al., 2000). This region contains conserved domain II, a conserved basic KR dipeptide in the intervening region between domains I and II as well as additional nonconserved sequences. To determine whether low protein accumulation was a property of this region only from PSIAA6 or was characteristic of comparable regions from other Aux/IAA proteins, a plasmid for the expression of a similar region from an additional Aux/IAA protein was constructed. IAA17, an Aux/IAA protein from Arabidopsis, was chosen for these studies because IAA17 is important in normal auxin responses, as revealed by identification of an auxin response mutant in this protein (Rouse et al., 1998). A transferable nuclear localization signal (NLS) was added to the C terminus of the IAA17::LUC fusion protein (Worley et al., 2000) because IAA17 has been shown to be localized to the nucleus (Ouellet et al., 2001), but its functional NLS has not been determined. The coding region for IAA17 containing amino acids 68 to 111, similar to PSIAA6 amino acids 18 to 73, was placed in frame upstream of LUC with the added NLS (LUC::NLS) and downstream of the Arabidopsis polyubiquitin promoter, creating UBQ10::IAA17(68-111)::LUC:: NLS. Relative protein accumulation was determined by transient assays as described previously (Worley et al., 2000). IAA17(68-111)::LUC::NLS accumulated to 2% of the level of LUC::NLS, not significantly different from that observed for PSIAA6(18-73)::LUC::NLS (Figure 1) (Worley et al., 2000).
Figure 1.
Relative LUC Activity and Protein Accumulation of Constructs Expressing Deletions of IAA17::LUC::NLS Fusions after Transient Transfection into Tobacco Protoplasts.
Diagram of amino acids 68 to 111 of Arabidopsis IAA17, axr3-1, and the 13–amino acid conserved consensus sequence as fusions with LUC with an added squash leaf curl virus NLS (Worley et al., 2000). The thick line at top (II) and the corresponding shaded regions represent the approximate location and length of conserved domain II found in Aux/IAA proteins. Activities for the fusion proteins after transfection of the respective DNA constructs were calculated as normalized percent activity ±sd relative to the mean normalized activity for LUC::NLS. Relative specific activities were determined by quantitative protein gel blot analyses after expression of the same proteins in yeast, and activity levels were divided by the specific activities to yield the relative protein accumulation. LUC::NLS is not drawn to scale.
The importance of conserved residues in domain II in protein accumulation observed previously (Worley et al., 2000) was confirmed by determining the relative protein accumulation of IAA17(68-111)P88L::LUC::NLS. This amino acid change corresponds to that found in axr3-1 (Rouse et al., 1998). The substitution of the second absolutely conserved proline to leucine in domain II (Figure 2) increased protein accumulation ∼20-fold (Figure 1).
Figure 2.
Amino Acid Sequence Alignment of Aux/IAA Proteins across Conserved Domain II.
Multiple sequence alignment of amino acids spanning conserved domain II, equivalent to amino acids 68 to 111 from IAA17, of 22 Aux/IAA proteins from Arabidopsis and two Aux/IAA proteins from pea. The shaded regions encompass amino acids outside of the conserved domain II, with the 13–amino acid consensus sequence noted at the bottom of the alignment. Invariant residues among all Aux/IAA proteins shown are underlined in the consensus sequence. Numbers below the 13–amino acid consensus sequence are reference points for mutants and variants tested in Figure 3.
Alignment of Aux/IAA Sequences around Conserved Domain II Reveals a Consensus Sequence That Is Sufficient for Low Protein Accumulation in Transient Assays
An alignment of the sequences equivalent to IAA17(68-111) from 22 Arabidopsis Aux/IAA proteins, along with PSIAA6 and PSIAA4/5, two Aux/IAA proteins from pea, was performed to identify the conserved sequence in this region (Figure 2). A 13–amino acid consensus sequence was revealed in this analysis, and the ability of this sequence to direct low protein accumulation was tested in the transient assay (Figure 1). The coding region for this consensus 13–amino acid sequence, which is equivalent to IAA17 domain II with the addition of a translation initiator methionine codon and an alanine codon at its C terminus as a junction amino acid, was placed in translational fusion with the LUC::NLS coding region, creating 13aa::LUC::NLS. After transient introduction, 13aa::LUC::NLS accumulated to 2% of the level of LUC::NLS alone, not significantly different from the value reported for IAA17(68-111)::LUC::NLS (Figure 1). This finding indicates that the 13–amino acid consensus sequence is sufficient for low protein accumulation equivalent to that seen with full-length Aux/IAA proteins.
Mutants and Natural Variants of Aux/IAA Proteins Identify Important Residues within the 13–Amino Acid Degradation Signal
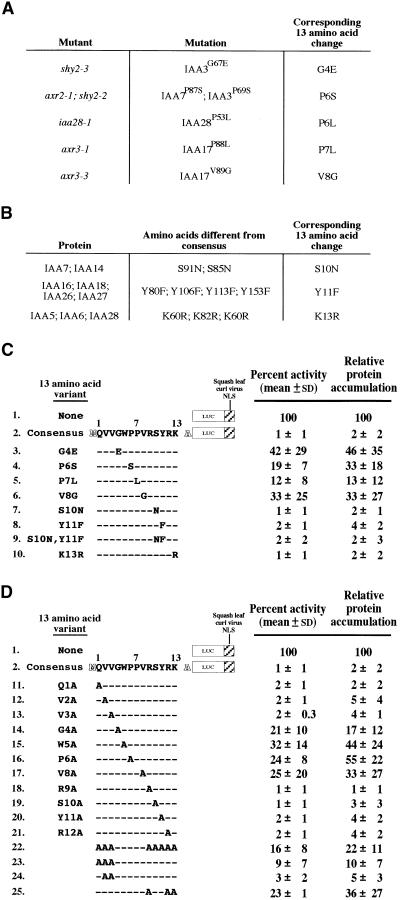
As evident from the alignment of domain II from multiple Aux/IAA proteins, not all 13 amino acids of the consensus sequence are conserved completely (Figure 2). In addition, there have been multiple semidominant auxin response mutants identified to date that encode Aux/IAA proteins with point mutations within domain II (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). To determine whether changes in amino acid sequence from the consensus and whether changes found in the auxin response mutants affect protein accumulation in the context of LUC fusions, consensus codons in the IAA17 13aa::LUC::NLS coding region were replaced by codons for amino acids in the auxin response mutants and some of the natural variants (Figures 3A and 3B, respectively). Relative protein accumulation was determined by transient assay as described above.
Figure 3.
Relative LUC Activity and Protein Accumulation of Constructs Expressing 13–Amino Acid Consensus, Mutant, and Variant LUC::NLS Fusions after Transient Transfection into Tobacco Protoplasts.
(A) Semidominant mutations found in genes that encode Aux/IAA proteins and their corresponding change in the 13–amino acid consensus sequence. References are as follows: axr2-1, Nagpal et al., 2000; axr3-1 and axr3-3, Rouse et al., 1998; iaa28-1, Rogg et al., 2001; shy2-2 and shy2-3, Tian and Reed, 1999.
(B) Natural variants among the Aux/IAA proteins and their corresponding change in the 13–amino acid consensus sequence.
(C) Diagrams of 13–amino acid mutants and natural variants as fusions with LUC::NLS and their respective relative LUC activities and protein accumulation as described for Figure 1.
(D) Diagrams of 13–amino acid processive alanine substitutions as fusions with LUC::NLS and their respective relative LUC activities and protein accumulation as described for Figure 1.
The results for the auxin response mutations revealed the importance of these amino acids in conferring low protein accumulation for the LUC::NLS fusion. Mutations at four positions have been identified through genetic screens that correspond to amino acids 4, 6, 7, and 8 in the 13–amino acid consensus sequence (Figure 3A). When each position was substituted singly to mimic mutations found among the Aux/IAA proteins, these amino acid substitutions caused sixfold to 20-fold increases in protein accumulation (Figure 3C, lines 3 to 6).
Three amino acid substitutions corresponding to those found in at least one wild-type Arabidopsis Aux/IAA protein (Figure 3B) were tested for their effect on LUC::NLS protein accumulation. Single substitutions in the IAA17 13–amino acid sequence similar to those found in other Aux/IAA proteins at positions 10, 11, or 13 did not have an effect on protein accumulation (Figure 3C, lines 7, 8, and 10). In addition, a multiple substitution using natural variant amino acids (positions 10 and 11 simultaneously) in the context of the 13–amino acid LUC fusion, removing both possible phosphorylation sites, did not affect low protein accumulation (Figure 3C, line 9).
To identify additional residues that may be important for low protein accumulation of LUC fusions, we performed processive alanine substitutions whereby a single codon in the 13–amino acid consensus sequence was substituted with the codon for alanine (Figure 3D). The accumulation of each alanine-substituted 13–amino acid fusion protein was determined in the transient assay. From this study, positions 4, 6, and 8 (Figure 3D, lines 14, 16, and 17, respectively) were shown to be important for low protein accumulation, consistent with the auxin response substitutions (Figure 3C, lines 3, 4, and 6, respectively). In addition, position 5 was discovered to be essential for low protein accumulation (Figure 3D, line 15). There is no corresponding Aux/IAA protein with a mutation at this position.
In contrast, single alanine substitutions at the other positions, 1 to 3 and 9 to 12, had no effect on protein accumulation (Figure 3D, lines 11 to 13 and 18 to 21, respectively). Interestingly, two of these positions are conserved absolutely among Aux/IAA proteins (Figure 2), the valine at position 3 and the arginine at position 12, yet substitution of each individually to alanine did not significantly increase protein accumulation (Figure 3D, lines 13 and 21, respectively). Combined with data in Figure 3C, the absolutely conserved residues GWPP along with the valine immediately following, GWPPV (positions 4 to 8 in the 13–amino acid IAA17 consensus), were required for low protein accumulation of IAA17 13–amino acid LUC fusions.
Domain II Core Amino Acids GWPPV Are Required but Not Sufficient for Low Protein Accumulation
To determine whether the amino acids GWPPV essential for low protein accumulation are sufficient for this effect, all other amino acid residues were substituted with alanine, as described above. This LUC fusion protein accumulated ∼10-fold higher than did the wild-type IAA17 13–amino acid LUC fusion (Figure 3D, line 22). These substitutions show that the GWPPV sequence is not sufficient to target LUC:: NLS for the low protein accumulation seen with the wild-type 13–amino acid LUC fusion. Substituting the first three residues with alanine (positions 1, 2, and 3) caused a smaller but significant increase in protein accumulation (Figure 3D, line 23). In contrast, substituting two of them simultaneously with alanine at positions 2 and 3 had no effect (Figure 3D, line 24). At least one of the consensus amino acids at position 2, 3, or 4 appears to be required.
The effects of substitutions C terminal to GWPPV were tested. Positions 10 and 11 can be substituted with nonconserved amino acids (Figure 3, lines 7, 19, and 20), suggesting that these specific amino acids are not required. Position 13, a lysine, can be substituted conservatively with arginine with no effect (see above), indicating that the lysine is not essential. However, all 13–amino acid sequences contain at least one basic residue and often more than one (Figure 2). In IAA17, there are three basic positions C terminal to the necessary core at positions 9, 12, and 13. When positions 9 and 12 were singly substituted with alanine, there was no effect on protein accumulation (Figure 3D, lines 18 and 21). To test the effect of removal of all basic residues, positions 9, 12, and 13 were substituted with alanines simultaneously. This change increased protein accumulation ∼20-fold (Figure 3D, line 25). Combined, these data show that in addition to the required GWPPV sequence, additional components flanking this core are required for low protein accumulation. However, in contrast to the core, conservation of specific residues is not required.
Aux/IAA Proteins Target a Heterologous Protein for Rapid Degradation in Vivo in Transgenic Plants
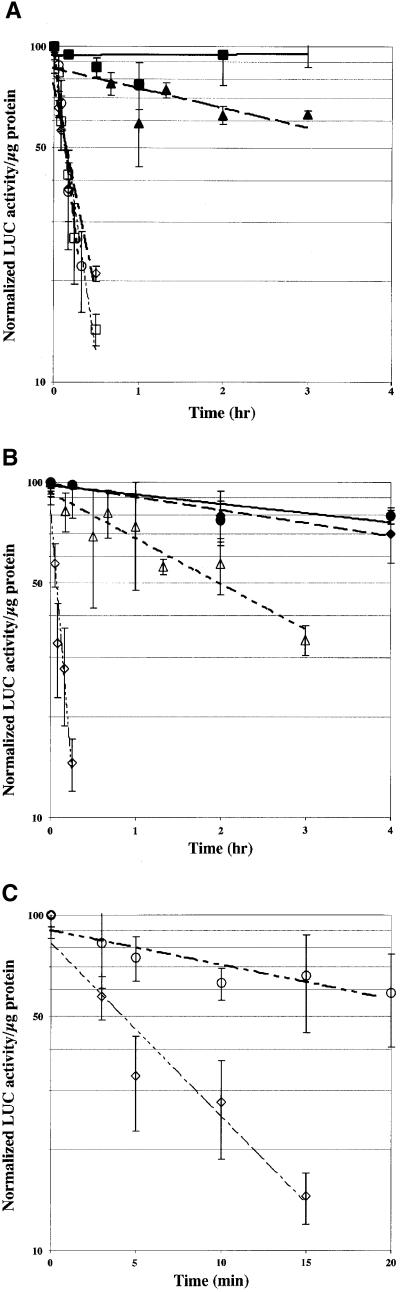
The degradation rate of the PSIAA6::LUC fusion protein in Arabidopsis seedlings was determined previously by radiolabeling and pulse-chase analysis (Worley et al., 2000). To more conveniently determine protein half-life for multiple Aux/IAA fusion proteins, they were stably expressed in transgenic plants and a cycloheximide chase assay for determining the degradation rates of these proteins was used. Transgenic plants were obtained expressing LUC alone (no fusion) or LUC in fusion with full-length PSIAA6, full-length PSIAA6 containing a point mutation corresponding to the axr3-1 mutation in IAA17 (PSIAA6P61L), or full-length IAA1. Transgenic seedlings were incubated in cycloheximide to halt protein synthesis. LUC assays in extracts were performed to determine fusion protein activity at various times, and the values were normalized to a time 0 measurement. In transgenic LUC-expressing seedlings, there was no significant difference in LUC protein level after 3 hr (Figure 4A, closed squares), indicating that the half-life of LUC was significantly longer than 3 hr and could not be measured accurately in these experiments.
Figure 4.
Cycloheximide Chase Analysis of LUC and Aux/IAA::LUC Proteins in Transgenic Arabidopsis Seedlings.
Cycloheximide chase assays were performed on transgenic Arabidopsis lines expressing their respective proteins. After cycloheximide treatment at the times indicated, extracts were prepared and assayed as described in Methods. Data points represent the mean of at least two separate experiments each with multiple independent transgenic lines, and error bars represent standard deviations.
(A) LUC activity per microgram of protein, normalized to time 0 measurement, of LUC (closed squares), PSIAA6P61L::LUC (closed triangles), PSIAA6::LUC (open diamonds), PSIAA6(1-73)::LUC (open circles), and IAA1::LUC (open squares) after the addition of cycloheximide, a protein synthesis inhibitor.
(B) LUC activity per microgram of protein, normalized to time 0 measurement, of LUC::NLS (closed circles), PSIAA6(71-179)::LUC::NLS (closed diamonds), PSIAA6::LUC::NLS (open diamonds), and 13aa:: LUC::NLS (open triangles) after the addition of cycloheximide.
(C) LUC activity per microgram of protein, normalized to time 0 measurement, of PSIAA6::LUC::NLS (open diamonds) and PSIAA6(18-73)::LUC::NLS (open circles) after the addition of cycloheximide. Note the difference in time scale from that of (A) and (B).
In contrast, LUC activity was lost very quickly in plants expressing the fusion proteins PSIAA6::LUC and IAA1::LUC (Figure 4A, open diamonds and open squares, respectively) and was lost slowly in PSIAA6P61L::LUC–expressing plants (Figure 4A, closed triangles). Plotting the data and finding the best-fit curve allowed us to calculate equivalent half-lives of ∼8 min for PSIAA6::LUC and IAA1::LUC and 4 hr for PSIAA6P61L::LUC. Thus, wild-type Aux/IAA fusion proteins are degraded rapidly. The point mutation within conserved domain II in PSIAA6P61L::LUC results in a protein with a half-life that is 30-fold longer than that of PSIAA6::LUC but still significantly shorter than that of LUC alone.
Using transient assays, we had determined previously that the Aux/IAA region C terminal to domain II had a small but statistically significant effect on LUC protein accumulation (Worley et al., 2000). To determine whether this difference resulted from a difference in protein half-life, LUC fusions with either PSIAA6 or a truncated form of PSIAA6 containing all amino acids C terminal to domain II (PSIAA6[71-179]) were expressed in Arabidopsis and their in vivo half-lives determined. In these experiments, LUC contained a C-terminal NLS (see above) to ensure that the Aux/IAA::LUC fusions could localize to the nucleus. In transgenic LUC::NLS–expressing seedlings, there was only a slight decrease in LUC::NLS protein level after 4 hr (Figure 4B, closed circles), with a calculated half-life of 11 hr. In the context of this fusion, the half-life of PSIAA6::LUC::NLS was 4 min (Figure 4B, open diamonds). In contrast, the calculated half-life of PSIAA6(71-179)::LUC::NLS was ∼10 hr (Figure 4B, closed diamonds). The half-life for PSIAA6(71-179)::LUC::NLS was not significantly different from that of LUC::NLS as determined by linear regression analysis (data not shown), indicating that there was no degradation signal contained within the C-terminal portion of PSIAA6 that functions in these fusions.
The consensus 13–amino acid sequence from conserved domain II of IAA17 was shown to be sufficient for low protein accumulation in a transient assay (Figure 1). 13aa:: LUC::NLS was expressed in transgenic Arabidopsis seedlings to determine its in vivo half-life. The calculated half-life for 13aa::LUC::NLS was 2 hr (Figure 4B, open triangles). Although this half-life was much shorter than that of LUC:: NLS, which was 11 hr (Figure 4B, closed circles), it was not as short as that of full-length PSIAA6::LUC::NLS (Figure 4B, open diamonds), suggesting that a component in the cis-acting degradation signal may be missing for proper recognition by the proteolytic machinery in Arabidopsis.
To identify Aux/IAA sequences in addition to the 13–amino acid sequence required in plants for rapid proteolysis equivalent to that seen for full-length Aux/IAA proteins, other LUC fusions with additional Aux/IAA amino acids were expressed in transgenic plants and their half-lives determined in seedlings. The half-life of a LUC::NLS fusion with PSIAA6 amino acids 18 to 73 was 25 min (Figure 4C, open circles). Although this was a relatively short half-life, indicating that these sequences can act as a transferable degradation signal, it was not as short as that for full-length PSIAA6:: LUC::NLS (Figure 4C, open diamonds). In contrast, the half-life of a LUC fusion with PSIAA6 amino acids 1 to 73 was 8 min, not different from that of PSIAA6::LUC and IAA1::LUC (Figure 4A, open circles versus open diamonds, respectively). Thus, amino acids 1 to 73 from PSIAA6 were sufficient to target LUC for rapid degradation in Arabidopsis seedlings equivalent to the full-length Aux/IAA LUC fusions.
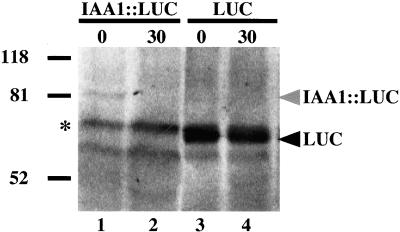
A protein gel blot with equal total protein was prepared to verify that in our cycloheximide chase experiments, LUC activities were reflective of the fusion protein levels (Figure 5). After 30 min, the level of IAA1::LUC protein decreased (Figure 5, lanes 1 and 2), whereas LUC levels were not significantly different from time 0 values (Figure 5, lanes 3 and 4), correlating exactly with the cycloheximide chase LUC activity data (Figure 4A, open and closed squares, respectively). These results validate LUC activity assays as a means of determining degradation rates of proteins.
Figure 5.
Anti-LUC Protein Gel Blot Analysis of Cycloheximide Chase Samples.
Treatment with cycloheximide causes the level of IAA1::LUC to decrease after 30 min, whereas the LUC level is not affected. Extracts were prepared from IAA1::LUC–expressing seedlings after treatment with cycloheximide at time 0 or 30 min (lanes 1 and 2, respectively) or LUC-expressing seedlings after treatment with cycloheximide at time 0 or 30 min (lanes 3 and 4, respectively). The asterisk denotes a band cross-reactive to the anti-LUC antibodies present in all plant extracts. This cross-reactive band was used to normalize loading for quantification of IAA1::LUC and LUC bands, using the STORM system.
Aux/IAA Degradation Is Mediated by the Proteasome Pathway
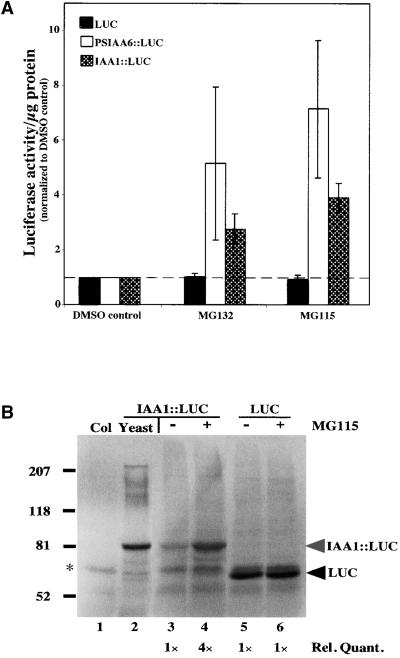
To reveal the nature of the proteolytic machinery responsible for Aux/IAA degradation, the ability of different protease inhibitors to stabilize Aux/IAA proteins in vivo was tested. Although a cell-permeable general inhibitor of serine proteases (4-[2-aminoethyl]-benzenesulfonyl-flouride hydrochloride) had no effect on Aux/IAA degradation (data not shown), the steady state levels of IAA1::LUC and PSIAA6:: LUC were affected by the addition of two cell-permeable proteasome-specific inhibitors, cbz-leu-leu-norvalinal (ZL2NVaH or MG115) and cbz-leu-leu-leucinal (ZL3H or MG132) (Figure 6A). These peptide aldehydes have been shown to strongly inhibit the chymotrypsin activity of the eukaryotic proteasome (Rock et al., 1994) and to inhibit in vivo proteolysis of a B-type cyclin (Nicta;CycB1;1) in tobacco BY2 cells (Genschik et al., 1998). Two hours was determined experimentally to be the time when maximal increase in PSIAA6:: LUC or IAA1::LUC occurred (data not shown). A threefold to fourfold increase in IAA1::LUC levels and a fivefold to sevenfold increase in PSIAA6::LUC levels were seen in the presence of the proteasome inhibitors (Figure 6A). Seedlings expressing LUC alone did not respond to treatment with proteasome inhibitors (Figure 6A) even after a 4-hr incubation (data not shown).
Figure 6.
The Proteasome Pathway Mediates Aux/IAA Degradation.
(A) LUC assays on transgenic plants expressing LUC, PSIAA6::LUC, or IAA1::LUC in the absence (DMSO control) or presence of proteasome inhibitors (MG115 or MG132). Values are normalized to DMSO treatment alone (dashed line). Error bars represent the standard deviation of the mean.
(B) Presence of the proteasome inhibitors in the in vivo LUC assay results in the accumulation of IAA1::LUC but not LUC alone. Extracts were prepared from transgenic IAA1::LUC seedlings treated for 2 hr with DMSO or MG115 (lanes 3 and 4, respectively), transgenic LUC seedlings treated for 2 hr with DMSO or MG115 (lanes 5 and 6, respectively), wild-type (Columbia [Col]) seedlings treated for 2 hr with MG115 (lane 1), or yeast expressing IAA1::LUC (lane 2; size marker for the migration of IAA1::LUC). The asterisk denotes a band cross-reactive to the anti-LUC antibodies present in all plant extracts. This cross-reactive band was used to ensure equal loading. Relative quantification data are shown at the bottom of the blot, quantified using the STORM system.
A protein gel blot was prepared (Figure 6B) to verify that the increase in LUC activity seen in the presence of MG115 was attributable to increased IAA1::LUC protein (Figure 6B, lanes 3 and 4). Equal total protein was loaded in each lane. IAA1::LUC expressed in yeast (Figure 6B, lane 2), in which the Aux/IAA proteins are not degraded (data not shown), was used as a size marker for the migration of IAA1::LUC expressed in Arabidopsis seedlings on SDS-PAGE. The bands equivalent in size to IAA1::LUC (Figure 6B, lanes 3 and 4) were quantified, and there was a fourfold increase from the DMSO to the MG115 treatment, as seen in the LUC assay for IAA1::LUC (Figure 6A). In contrast, there was no increase in LUC protein between MG115-treated and DMSO control seedlings, confirming the LUC assay data for these seedlings (Figure 6B, lanes 5 and 6). Cycloheximide chase experiments in the presence of MG115 demonstrated that the increase seen for IAA1::LUC results from a change in half-life (data not shown). Together, these data indicate that proteasome activity is required for wild-type rates of Aux/IAA protein degradation.
DISCUSSION
Two different methods were used to study the degradation rates of Aux/IAA amino acid sequences in fusion with LUC. Relative protein accumulation in cultured cells after transient transfection of DNA as an indirect measure of protein degradation rates had been used previously (Worley et al., 2000). Although indirect, transient assays allow for rapid analysis of multiple fusion proteins, and steady state levels are reflective of degradation rates under the assumption of equivalent synthesis. The other method, the cycloheximide chase assays, represents a direct measure of protein degradation rates by determining LUC activity in transgenic seedlings after inhibition of protein synthesis. To further strengthen the validity of the latter approach, we demonstrated that LUC activity levels were reflective of protein content. When compared directly, these two methods in general gave similar results (see below for exception) and are in agreement with previously published results. Two Aux/IAA proteins, IAA1 from Arabidopsis and PSIAA6 from pea, were analyzed by both methods (Worley et al., 2000; this study). The 8-min half-lives of IAA1::LUC and PSIAA6::LUC determined by cycloheximide chase assays are almost identical to the 6-min half-life of PSIAA6 in pea seedlings (Abel et al., 1994), consistent with the transient assay results for each fusion protein (Worley et al., 2000), and they closely match the ∼13-min half-life for PSIAA6::LUC in transgenic Arabidopsis seedlings determined by pulse-chase analysis (Worley et al., 2000).
Transient assay results show that the 13–amino acid conserved sequence from domain II of Aux/IAA proteins targets LUC for low protein accumulation. This low protein accumulation is of the same order as that seen for other Aux/IAA proteins in this assay, PSIAA6 and IAA1 (Worley et al., 2000). In Arabidopsis, several semidominant mutations within the conserved domain II of the Aux/IAA proteins have been shown to result in an auxin-related phenotype (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). An analysis of the corresponding mutants within this 13–amino acid sequence revealed the importance of these residues in conferring low protein accumulation.
This result was confirmed using the cycloheximide chase assay for PSIAA6 with a point mutation corresponding to iaa17/axr3-1 (PSIAA6P61L) fused to LUC. This protein has a half-life 30-fold longer than that of PSIAA6::LUC. The magnitude of difference between this mutant and its corresponding wild type is comparable to the steady state level differences observed in the transient assays (Worley et al., 2000). Also in agreement is the 20-fold protein accumulation difference observed in transient assays between wild type and the same mutation in the context of IAA17 with amino acids 68 to 111. In contrast, only a sevenfold increase in half-life was seen in iaa17/axr3-1 compared with that of wild-type IAA17/AXR3 (Ouellet et al., 2001). The differences seen in fold increase over wild type most likely are the result of differences in experimental approach, but the results are qualitatively similar, revealing the importance of this absolutely conserved proline residue in the degradation of Aux/IAA proteins. shy2-2 encodes another Aux/IAA protein with a domain II point mutation. shy2/iaa3 has a higher steady state level, although the fold difference from wild type could not be determined because the wild-type protein was not detectable (Colón-Carmona et al., 2000). Our results indicate that all of the auxin response mutations tested affect proteolysis and suggest that the auxin-response phenotypes result from this slowed degradation.
To determine if other residues within domain II not identified through genetic screens were important for low protein accumulation, we performed processive alanine substitutions of residues in the 13–amino acid consensus sequence. The results revealed the importance of the absolutely conserved tryptophan residue at position 5 in targeting LUC for low protein accumulation. No other single alanine substitution had an effect, indicating that GWPPV at positions 4 to 8 is the required core. Although amino acids GWPP are conserved absolutely, the valine at position 8 is not found in all Aux/IAA proteins. The substitution of isoleucine or leucine at position 8, found in natural Aux/IAA variants, is conservative. Although the effect of isoleucine or leucine was not tested directly, we predict that amino acids with large hydrophobic R groups would substitute for valine, because elimination of a hydrophobic side group (Figure 3C, line 6; Figure 3D, line 17) alters the ability of the fusion to be targeted for low protein accumulation. Curiously, alanine substitution at two completely conserved positions, for valine at position 3 or for arginine at position 12, had no effect. Although the former change can be considered somewhat conservative, the latter is not. Substitutions with nonconservative amino acids at position 3 could have an effect and need to be tested to determine the importance of this position.
Although the GWPPV residues are necessary for low protein accumulation, they are not sufficient when surrounded by all alanine residues. Removing all basic residues C terminal to GWPPV in the 13–amino acid sequence affected protein accumulation, suggesting the importance of an adjacent basic amino acid(s). N terminal to GWPPV in the 13–amino acid sequence, at least one amino acid with a large hydrophobic side chain appears to be required. Additional studies using the cycloheximide chase assays to determine the effect of amino acid substitutions directly and to better quantify differences are required to further elucidate the role of flanking sequences.
Substituting the 13–amino acid consensus sequence with any of the natural variants in fusion with LUC::NLS caused no change in protein accumulation in the transient assay compared with that in the 13–amino acid consensus sequence. This result, along with the transient assay data seen previously for IAA1 (Worley et al., 2000), suggests that targeting a long-lived protein for low protein accumulation is a trait shared among domain II–containing Aux/IAA proteins. However, given the importance of flanking sequences, direct determination of protein half-lives of multiple Aux/IAA proteins should be performed before this can be stated as a generalization for the entire Aux/IAA family. In addition, subtle differences in Aux/IAA protein degradation may be important in auxin signaling.
Previous transient assay data indicated that PSIAA6 might contain an additional degradation signal within its C terminus; although the effect on protein accumulation was small, it was significant (Worley et al., 2000). Cycloheximide chase experiments in transgenic plants showed that the region C terminal to domain II had no effect on the degradation rate of LUC::NLS, revealing that this portion of the protein does not contain a transferable degradation signal. This is further evidence that the degradation signal must reside wholly within the N-terminal portion of the protein.
When assayed in the cycloheximide chase assay, PSIAA6(18-73)::LUC::NLS and 13aa::LUC::NLS half-lives, although shorter than that of LUC alone, were not as short as that of full-length PSIAA6::LUC::NLS. On the basis of the transient assay results for these constructs, their half-lives should be equivalent (Worley et al., 2000; this study). This suggests that other components within the cis-acting degradation signal are necessary to enhance the interaction with the proteolytic machinery in Arabidopsis, whereas tobacco protoplasts either do not require these sequences or can use LUC sequences in the context of the 13–amino acid or PSIAA6(18-73) sequences for recognition by the proteolytic machinery. Alternatively, the transient assay may not have reflected the true relative degradation rates for these fusions. Another explanation may be a difference in the post-translational modifications of Aux/IAA proteins between the two systems (see below). In the cycloheximide chase assay, the additional sequences required are contained within the N-terminal 73 amino acids. Identification of the additional required amino acids within this region is in progress.
Ubiquitination of proteins provides a selective means to degrade regulatory proteins important in a variety of cellu-lar processes, such as signal transduction, cell cycle progression, and transcriptional regulation (Hershko and Ciechanover, 1998). Once multiple ubiquitin moieties are conjugated to a substrate, it is recognized and degraded by the 26S proteasome (Hochstrasser, 1996; Ciechanover et al., 2000), which is made up of the 20S catalytic core with a 19S regulatory complex (Hershko and Ciechanover, 1998; Ciechanover et al., 2000). The increase in Aux/IAA::LUC fusion abundance in the presence of proteasome inhibitors, although indirect evidence, suggests that Aux/IAA proteins are degraded by the proteasome. Because most known substrates of the 26S proteasome are ubiquitinated, it is likely that Aux/IAA proteins are substrates of the ubiquitin pathway. Aux/IAA domain II could serve as a recognition element for ubiquitination or for modifications required for ubiquitination. Because of the low abundance of these proteins, direct visualization of ubiquitinated Aux/IAA proteins has not been achieved.
The TIR1 gene is required for normal auxin response in Arabidopsis (Ruegger et al., 1998). tir1 mutants are defective in several growth processes regulated by auxin, including lateral root formation, auxin-dependent hypocotyl elongation, and auxin-induced cell division (Ruegger et al., 1998). TIR1 encodes an F-box protein and likely functions within an SCF complex (Gray et al., 1999). The COP9 signalosome also was shown to interact with components of the SCFTIR1 complex in vivo (Schwechheimer et al., 2001). It has been suggested that the SCFTIR1 complex, an E3 or ubiquitin ligase, degrades Aux/IAA proteins (Gray and Estelle, 2000). A mutant with reduced COP9 signalosome components was impaired in Aux/IAA degradation (Schwechheimer et al., 2001), linking Aux/IAA degradation to the SCFTIR1 complex. All SCF targets identified to date are phosphorylated before recognition by the complex (Patton et al., 1998). It is interesting to note that iaa3/shy2-2 is phosphorylated in vivo and phyA phosphorylates the N-terminal half of PSIAA4 in vitro (Colón-Carmona et al., 2000). This region encompasses the 13–amino acid stretch identified as necessary and sufficient to target LUC for low protein accumulation (Figure 1) and the portion of PSIAA6, containing amino acids 1 to 73, shown to be degraded rapidly (Figure 4). Our 13–amino acid transient assay data, in which the serine and tyrosine were substituted with asparagine and phenylalanine, respectively (Figure 3C), indicate that phosphorylation within domain II is not necessary for the proper degradation of Aux/IAA proteins. Tobacco protoplasts, although containing an auxin response pathway (Worley et al., 2000), may be less stringent in terms of their ability to recognize and phosphorylate Aux/IAA proteins and target them for low protein accumulation. These cells may be able to use residues of LUC as potential phosphorylation and/or ubiquitination sites, whereas Arabidopsis seedlings expressing 13aa::LUC::NLS are incapable of this altered phosphorylation and/or ubiquitination, leading to a longer lived protein. The direct interaction with TIR1 and its involvement in Aux/IAA degradation are currently under investigation.
METHODS
Molecular Techniques
DNA manipulations used standard molecular protocols (Sambrook et al., 1989). IAA1 and PSIAA6 coding regions were as described by Worley et al. (2000). IAA17 coding regions were obtained by polymerase chain reaction using cDNA as a template using the same methodology. Complementary oligonucleotides encoding the 13–amino acid sequence were ligated into KpnI–NcoI overhangs upstream of the coding region for LUC::NLS, as described previously for other auxin/indoleacetic acid (Aux/IAA) constructs (Worley et al., 2000). The DNA sequence for all Aux/IAA coding regions was verified by automated sequencing. Plant expression cassettes containing the coding regions for both the experimental luciferase (LUC) fusion proteins and an unmodified β-glucuronidase, used for normalization, were cloned into a single plasmid, as described previously (Worley et al., 2000). Plasmids for yeast expression and in planta transformation were constructed by ligating the coding region of interest into pYES2 (Invitrogen, Carlsbad, CA) or into a pBIN19-derived plasmid, respectively (Bevan, 1984).
Transient Expression, Protein Extracts, and Enzyme Activity Assays
Plasmids used for transfecting tobacco protoplasts were isolated by alkaline lysis of large-scale preparations and purified by polyethylene glycol precipitation (Sambrook et al., 1989). A 100-μg aliquot of a single plasmid encoding both the experimental and the normalization proteins was introduced into protoplasts derived from BY2 tobacco cells using polyethylene glycol–mediated DNA transfer (Altman et al., 1992). Protoplasts were incubated for 40 hr, a period in which LUC levels were determined to be at steady state (Worley et al., 1998). Cells were lysed and assayed as described previously (Worley et al., 2000).
Yeast Expression and Immunoblot Analysis
Plasmids encoding the desired fusion proteins were transformed into Saccharomyces cerevisiae strain WCG4α (Richter-Ruoff et al., 1992) using the lithium acetate method (Ito et al., 1983) with URA3 selection. After galactose induction, cells were lysed and assayed as described (Worley et al., 2000).
Yeast extracts containing equal LUC activity and equal total protein were prepared for SDS-PAGE and protein gel blotting as described (Beers et al., 1992). Anti-LUC polyclonal antibodies (Cortex Biochem, San Leandro, CA) were used to visualize their respective immunogens by chemiluminescence (Gallagher et al., 1994). Chemiluminescent visualization was performed using STORM 860 (Molecular Dynamics, Sunnyvale, CA). The signal was quantified using ImageQuant (Molecular Dynamics), and a dilution series on the same gel verified the linearity of the signal. Each construct was expressed in two independently transformed yeast strains for specific activity determination, with at least two separate blots made for each construct, and their quantified signals averaged.
Cycloheximide Chase Experiments
Seedlings were grown under continuous white light in liquid culture (1 × Murashige and Skoog [1962] salts, pH 6.5) for 6 days after a 2-day imbibition at 4°C. Liquid culture media were replaced 4 days after imbibition. On day 6, liquid culture media were removed and fresh liquid media without (time 0 control) or with cycloheximide (200 μg/mL) were added. This concentration of cycloheximide was sufficient to inhibit >92% of the 35S-methionine incorporation (data not shown). The seedlings were placed back under continuous white light for the times indicated in the figures and immediately frozen in liquid nitrogen. Protein extracts were made by grinding seedlings in liquid nitrogen and resuspending in buffer (100 mM KPO4, pH 7.8, 1 mM EDTA, 7 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor tablet [Roche Molecular Biochemicals, Indianapolis, IN] at one tablet per 10 mL). Cell debris was pelleted by centrifugation, and extracts were assayed for LUC activity (Worley et al., 2000). Protein concentration was determined by Bradford assay (Bio-Rad). Equal amounts of protein from each extract were analyzed by protein gel blotting. Data shown represent the average from at least two independent homozygous transgenic lines for each construct with at least three replicates at each time point measured. Degradation rates were identical between independent transgenic plant lines that differed eightfold in LUC activity per microgram of protein (data not shown).
Inhibitor Studies
For proteasome inhibitor studies, seedlings were grown for 7 days on germination medium in continuous white light. These seedlings were then transferred to water for 2 hr in the presence of DMSO control, protease inhibitors (4-[2-aminoethyl]-benzenesulfonyl-flouride hydrochloride [Pefabloc SC; Roche Molecular Biochemicals]), or proteasome-specific inhibitors (MG115 and MG132 [Peptides International, Louisville, KY]). After this treatment, the seedlings were ground in liquid nitrogen and assayed as described above. Data shown represent the average from at least three separate experiments with two replicates for each experiment.
Immunoblot Analysis
Seedling extracts containing equal total protein from each extract were prepared for SDS-PAGE and protein gel blotting (Beers et al., 1992). Equal protein between extracts was achieved by adding negative control Columbia extracts to samples with lower protein content. Anti-LUC polyclonal antibodies (Cortex Biochem) were used to visualize their respective antigens by chemiluminescence (Gallagher et al., 1994). The signal was visualized and quantified on the STORM system (Molecular Dynamics). Two separate blots were made for each construct.
Acknowledgments
We thank Steffen Abel, Charles Gasser, and members of the Callis laboratory for helpful discussions and critical reading of the manuscript. We thank Kirstin Summerfelt for assistance with the BY-2 cells. We thank Colin Leasure, Adria Honda, Dean Rouse, and Jessica Brown for technical assistance. The assistance of the University of California–Davis Controlled Environment Facility in the propagation of transgenic plants is acknowledged. This material is based on work supported by the National Science Foundation under Grant No. 9808791 to J.C. and Jastro Shields Fellowships to J.A.R. and N.Z.
References
- Abel, S., and Theologis, A. (1995). A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J. 8, 87–96. [DOI] [PubMed] [Google Scholar]
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, T., Damm, B., Helfter, U., Willmitzer, L., and Morris, P.C. (1992). Protoplast transformation and methods to create mutants in Arabidopsis thaliana. In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific), pp. 310–330.
- Beers, E.P., Moreno, T.N., and Callis, J. (1992). Subcellular localization of ubiquitin and ubiquitinated proteins in Arabidopsis thaliana. J. Biol. Chem. 267, 15432–15439. [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, V., Tobias, J.W., Bachmair, A., Marriott, D., Ecker, D.J., Gonda, D.K., and Varshavsky, A. (1989). A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A., Orian, A., and Schwartz, A.L. (2000). Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays 22, 442–451. [DOI] [PubMed] [Google Scholar]
- Clough, R.C., and Vierstra, R.D. (1997). Phytochrome degradation. Plant Cell Environ. 20, 713–721. [Google Scholar]
- Colón-Carmona, A., Chen, D.L., Yeh, K.-C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle, M. (1992). The plant hormone auxin: Insight in sight. Bioessays 14, 439–444. [DOI] [PubMed] [Google Scholar]
- Gallagher, S., Winston, S.E., Fuller, S.A., and Hurrell, J.G.R. (1994). Immunoblotting and immunodetection. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: Greene Publishing Associates/Wiley-Interscience), pp. 10.8.1–10.8.17.
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle–dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell 10, 2063–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (1996). Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk, U., and Sommer, T. (2000). Ubiquitin-mediated proteolysis of a short-lived regulatory protein depends on its cellular localization. J. Biol. Chem. 275, 39403–39410. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Richter-Ruoff, B., Heinemeyr, W., and Wolf, D.H. (1992). The proteasome/multicatalytic-multifunctional proteinase: In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett. 302, 192–196. [DOI] [PubMed] [Google Scholar]
- Rock, K.L., Gramm, C., Rothstein, L., Clark, K., Stein, R., Dick, L., Hwang, D., and Goldberg, A.L. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and generation of peptides presented on MHC class I molecules. Cell 78, 761–777. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an Aux/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadis, S., Atienza, C.J., and Finley, D. (1995). Synthetic signals for ubiquitin-dependent proteolysis. Mol. Cell. Biol. 15, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Estelle, M., Lyapina, S., Deshaies, R.J., and Deng, X.-W. (2001). The COP9 signalosome interacts with the E3 ubiquitin ligase SCFTIR1 and mediates auxin-response in Arabidopsis. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Szewczyk, N.J., Hartman, J.J., Barmada, S.J., and Jacobson, L.A. (2000). Genetic defects in acetylcholine signalling promote protein degradation in muscle cells of Caenorhabditis elegans. J. Cell Sci. 113, 2003–2010. [DOI] [PubMed] [Google Scholar]
- Theologis, A. (1986). Rapid gene regulation by auxin. Annu. Rev. Plant Physiol. 37, 407–438. [Google Scholar]
- Thrower, J.S., Hoffman, L., Rechsteiner, M., and Pickart, C.M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J. (1999). Control of auxin-regulated root development by Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went, F.W., and Thimann, K.V. (1937). Phytohormones. (New York: Macmillan).
- Worley, C.K., Ling, R., and Callis, J. (1998). Engineering in vivo instability of firefly luciferase and Escherichia coli β-glucuronidase in higher plants using recognition elements from the ubiquitin pathway. Plant Mol. Biol. 37, 337–347. [DOI] [PubMed] [Google Scholar]
- Worley, C., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]