Abstract
Producing robust and scalable fluid metering in a microfluidic device is a challenging problem. We developed a scheme for metering fluids on the picoliter scale that is scalable to highly integrated parallel architectures and is independent of the properties of the working fluid. We demonstrated the power of this method by fabricating and testing a microfluidic chip for rapid screening of protein crystallization conditions, a major hurdle in structural biology efforts. The chip has 480 active valves and performs 144 parallel reactions, each of which uses only 10 nl of protein sample. The properties of microfluidic mixing allow an efficient kinetic trajectory for crystallization, and the microfluidic device outperforms conventional techniques by detecting more crystallization conditions while using 2 orders of magnitude less protein sample. We demonstrate that diffraction-quality crystals may be grown and harvested from such nanoliter-volume reactions.
In the same way that miniaturization has impacted the electronics industry, microfluidics promises to spark a revolution in fields ranging from analytical chemistry to biology and medicine. In principle, microfluidic devices can increase throughput and decrease cost by densely integrating complex assays and analytical measurements in a chip format. Driven by the early success of separation by capillary electrophoresis (1–6), other applications such as patterned surface deposition (7, 8), DNA analysis (9, 10), and cell sorting (11, 12) have been realized in microfluidic chips. The use of nanoliter reaction volumes and parallel sample processing represent potential advantages of microfluidic devices, making them ideally suited to total chemical analysis, ultra-high-throughput screening applications, and other cases where reagents are precious. However, an obstacle that thus far has hampered development of the field is the lack of a scalable, robust system to manipulate and dispense fluids with subnanoliter precision.
For a fluid metering system to have universal applicability, it must be insensitive to both the specific fluid properties and the surrounding channel architecture. The need to integrate these functions into massively parallel chip architectures further requires that the method be scalable to complex devices. Previous work on microfluidic metering has resulted in the development of valveless electrokinetic and pressure-driven metering systems (13–19). These systems are powerful in that they are able to manipulate nanoliters of fluid and inject these small volumes into downstream components for applications such as electrophoretic analysis. However, these schemes have not been demonstrated to be highly scalable and are not robust for a number of reasons. For example, their performance depends on both the fluid viscosity and the fluidic resistance of the microchannel. Electrokinetic flow additionally depends on the properties of the working fluid, particularly the pH and ionic strength (20, 21). Both pressure-driven and electrokinetic techniques are “leaky”: reagents diffuse through junctions and channels over time, strongly constraining both maximally achievable incubation times and chip complexity.
One strategy to address these issues is to use microelectromechanical system techniques to fabricate mechanical valves on chips. True mechanical valves can be both leak-proof and insensitive to the properties of the working fluid but generally are challenging to fabricate and thus difficult to scale up into complex systems. Another practical issue for all fluidic devices of significant complexity is the problem of priming, or initially filling the device with fluid. For microelectromechanical system devices made from conventional materials such as silicon or glass, this requirement precludes the use of multiply crossed, highly complex fluidic architectures. Devices from such materials must be primed by using a flow-through method and therefore require an outlet through which the ambient atmosphere may be vented. Initial introduction of fluid to a complex structure results in the formation of air bubbles that cannot be easily removed and adversely affect the performance of the device. Furthermore, because the priming fluid must pass through the entire device, it may subsequently contaminate or dilute the sample solutions.
We have shown previously how micromechanical valves and pumps can be fabricated from silicone elastomer by using a relatively simple soft lithography process, and that these plumbing components can be incorporated into devices with a modest degree of integration: cell sorters, PCR machines, and rotary mixers with approximately a dozen valves (19, 22, 23). The technical difficulties associated with priming complex devices can be surmounted because the silicone elastomer is gas-permeable. For example, it has been shown that complex geometries in elastomeric devices can be primed with a single fluid by submersion in a large reservoir of buffer solution followed by exposure to vacuum for several minutes (24).
Here we report a robust and scalable microfluidic metering scheme called barrier interface metering (BIM). BIM has picoliter accuracy, negligible sample waste, and complete insensitivity to the fluid properties. Moreover, BIM is highly scalable in that it allows for massively parallel dispensing strategies to be implemented on a chip with no increase in control complexity. To illustrate the power and flexibility of this metering scheme, we applied it to ultrasmall-volume screening of protein crystallization conditions, a major hurdle in structural biology efforts.
Diffraction data obtained through x-ray crystallography are necessary to understand and model macromolecular structure, protein–ligand interactions, and the physical manifestation of certain diseases and for the rational design of drugs. Despite high demand, determination of protein structure remains an arduous and often unreliable task, principally because of the difficulty in growing diffraction-quality crystals (25). This process is entirely empirical and involves the parallel and combinatorial mixing of hundreds of different solutions containing various salts, buffers, and precipitating agents with target protein samples. Conventional crystallization techniques such as microbatch or hanging-drop vapor diffusion typically use 0.5–1.0 μl of concentrated protein solution per assay, necessitating milligram quantities of sample material for crystallization screening trials. Unfortunately, many interesting biological molecules such as multiprotein complexes and membrane proteins are only available in submilligram quantities. Furthermore, such large-scale screening is impractical for proteins that cannot be expressed in a model organism and instead must be isolated from a bulk sample. As a result, the cost and time associated with producing and purifying a large volume of concentrated protein sample limits the number of experiments that may be performed (26).
Liquid-handling robots have been used to address some of these issues (27–29). Although these machines can dispense volumes as small as 50 nl accurately, their cost and complexity make them inaccessible to the vast majority of researchers. Robotic systems typically require large amounts of dedicated space, can have difficulty dispensing viscous solutions, and are expensive to purchase and maintain. Moreover, because robots have been used solely to automate and reduce the volume of conventional crystallization techniques, they suffer from fundamental limitations intrinsic to these methods.
To overcome metering limitations in existing microfluidic devices and apply this technology and to a fundamental scientific problem, we developed a microfluidic chip that enables the large-scale screening of protein crystallization conditions by using ultrasmall-volume reactions (Fig. 1). The device implements 144 simultaneous metering and mixing reactions while only requiring two hydraulic control lines. The protein crystal screening problem requires robust fluid metering as 48 different solvents of varying viscosity, surface tension, ionic strength, and pH are used in each chip. Significant savings in sample consumption and experimentalist time were achieved through the use of this device without sacrificing crystallization success rates. Moreover, we discovered that the fluid mechanical properties that come into play at small-length scales allow the chip to implement highly efficient protein crystallization kinetics.
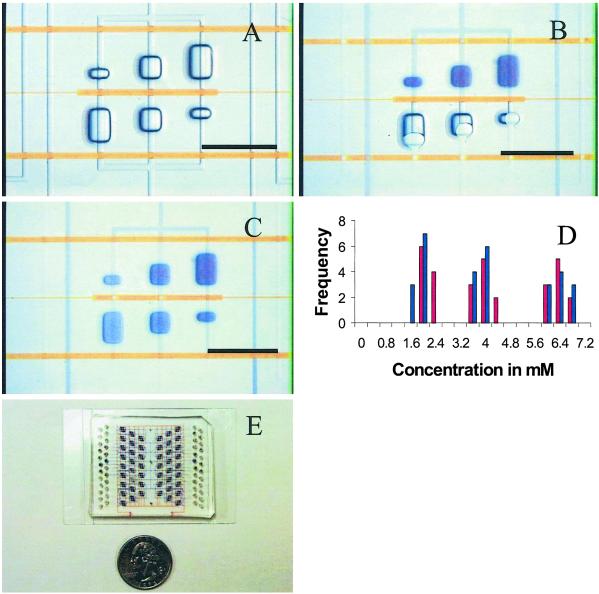
Fig 1.
(A) Section of a device showing three pairs of compound reaction chambers. Control channels are filled with 20 mM Orange G (Aldrich). Buried control channels of the elastomer chip are separated from open-bottom flow recesses by a 15-μm elastomer membrane. Hydraulic actuation of the control channel deflects the membrane and pinches off the flow line, creating a fluidic seal. Containment valves (Upper and Lower) allow isolation of compound wells during incubation. (B) Loading of reagents using pressurized outgas priming method. The interface valve (Center) is actuated, and reagents are loaded into adjacent sides of compound wells. (Lower) Wells are being dead-end-loaded with water. (Upper) Wells have been loaded with 13 mM bromophenol blue sodium salt (Aldrich). (C) A gradient of dye concentration. The containment valves (Upper and Lower) isolate compound wells, and the interface valve is released to allow diffusive mixing. The image shows complete mixing after 2 h. (D) Histogram showing the insensitivity of BIM to fluid viscosity. BIM was used to combine 7 mM bromophenol blue sodium salt with water (η ≅ 1 cP, 1 P = 0.1 Pa⋅sec) or 34% (wt/wt) sucrose (η ≅ 4 cP) 10 times each at mixing ratios (dye:water/sucrose) of 1:4, 1:1, and 4:1. Water measurements are shown in blue, and sucrose is shown in red. The variations in the concentration measurements (≈10%) are comparable to those taken on solutions of known concentrations. (E) Prototype protein crystallization chip with 144 parallel reaction chambers. (Scale bars, 1 mm.)
Materials and Methods
Device Fabrication.
The protein crystallization chip is comprised of a multilayer silicone elastomer (General Electric RTV 615) chip sealed to an etched glass substrate, creating a hybrid glass/elastomer flow structure. The channel and control structures of the chip were fabricated by the technique of multilayer soft lithography (19). Negative master molds were fabricated in photoresist (Shipley SJR 5740) by using conventional photolithography. Photoresist was spun onto silicon wafers at 2,000 rpm for 60 sec to create a 10-μm-thick layer and patterned by using positive high-resolution transparency masks. The mask defining the control structure was printed at 101.5% of the final desired device size to compensate for shrinkage of the elastomer that occurs during the initial curing. After patterning, the channel mold was annealed at 120°C for 20 min to achieve rounded channel geometry.
Liquid silicone elastomer (20 part A:1 part B) was spun onto the channel mold at 2,600 rpm for 60 sec for a thickness of 25 μm. Liquid silicone elastomer (5 part A:1 part B) was poured on the control structure mold to a thickness of ≈1 cm. After both structures were partially cured at 80°C for 1 h, the elastomer was peeled off the control structure mold, and control ports were punched by using a 20-gauge luer stub. The molded control structure was washed with ethanol and aligned to the flow structure. The combined device then was cured for 4 h at 80°C, creating a monolithic elastomer device. Forty-eight wells for the introduction of crystallizing agents were punched on the periphery of the device by using a 15-gauge luer stub.
Microwells were etched into a standard soda-lime glass microscope slide substrate (Corning 2947). The slides were patterned with photoresist (Shipley SJR 5740) by using a negative high-resolution transparency film as a mask. The back of the slides then were masked with an additional layer of photoresist and hard-baked at 125°C for 20 min to protect them during etching. The etch was performed for 90 min at 25°C with propeller agitation in equal parts of deionized water, 1 M hydrochloric acid, and buffered oxide etchant (6 ammonium fluoride:1 hydrofluoric acid, Transene Company). Hydrochloric acid was used to prevent the redeposition of insoluble fluoride salts (30). The etch was performed for 90 min at 25°C for a maximum well depth of 80 μm. The slides were first washed in acetone to remove the photoresist and then cleaned in an acid bath (NanoStrip Cyantek, Fremont, CA). The elastomer chip was aligned to the glass substrate and then baked overnight at 80°C to promote adhesion. The elastomer chips were disposed after a single use, and the glass substrates were cleaned in an acid bath (NanoStrip Cyantek) and reused.
Sample Loading.
Three-microliter aliquots of crystallization agent were dispensed into each of the 48 wells by using a pipette. The chip then was placed in a machined aluminum carrier consisting of a bottom plate and a top plate with observation windows. The top plate additionally has two cavities with a raised lip around their periphery and stainless steel input ports for pressurization. The cavities mate with the 48 reagent wells, creating a seal against the compliant elastomer chip when the plates are pressed together. With the carrier assembled, the cavities were pressurized, causing the 48 crystallizing agents to be pushed simultaneously into the chip. Three microliters of protein sample were loaded onto the device at a single port through a length of tubing connected to the chip via a stainless steel adapter. The injected sample volume was measured by tracking the advance of the protein solution meniscus. All microwells could be filled completely in <4 min by using a loading pressure of 7 psi (1 psi = 6.89 kPa). Using 42 of these devices, a single researcher was able to set up 6,048 crystallization experiments in a period of 30 h, consuming a total of only 150 μl of protein sample.
Absorption Measurements.
Absorption measurements were taken after 12 h of diffusive mixing to determine the concentration of dye in each chamber. The chip was illuminated with white light filtered through a 530-nm long-pass filter and imaged onto a charge-coupled device camera. Intensity measurements averaged over ≈1,000 pixels were taken on the channel connecting the opposing wells and compared with adjacent background. Concentrations were determined by a calibration curve of known concentrations.
Crystallization Protocols.
Control crystallization experiments using standard sparse matrix crystallization screens (Hampton Research, Laguna Niguel, CA; Emerald Biostructures, Bainbridge Island, WA) were performed both in microbatch and hanging-drop formats. In hanging-drop experiments, 1 μl of protein sample was combined with 1 μl of crystallizing agent on a glass coverslip and suspended over a well containing 0.5 ml of crystallizing agent. Vacuum grease was used to create a seal between the well and the coverslip, allowing the sample to equilibrate with the reservoir by vapor diffusion. Microbatch experiments were conducted by combining 1 μl of protein sample with 1 μl of crystallizing agent under paraffin oil.
Microbatch, hanging-drop, and on-chip experiments were incubated at 25°C for up to 3 weeks. Experiments were inspected daily, and hits were recorded. A hit was defined as single crystals, plates, rods, or spherulites. Phase separation and precipitation were not counted as hits. Where crystals were observed, they were confirmed to be protein by staining with IZIT dye (Hampton Research), harvesting and crushing with a glass probe (the “click” test), and/or comparison with negative (no-protein) controls. A list of the conditions that were and were not successful for each method is available as supporting information on the PNAS web site, www.pnas.org.
Crystal Extraction and Diffraction Studies.
Crystals were extracted from the chip under a humidity hood to avoid evaporation. The elastomeric side of the chip was peeled off of the substrate, and 1 μl of cryoprotectant (25% ethylene glycol with mother liquor) was dispensed onto the well. The crystals were extracted by using a CryoLoop (Hampton Research) and flash-frozen in liquid nitrogen. Diffraction data were collected at station 8.3.1 of the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) at an incident wavelength of 1 Å, with a 20-sec exposure and 1° oscillation.
Results and Discussion
The BIM scheme depends crucially on the ability to prime complex devices and in particular on the ability to fill dead-end channels. We developed a simple and convenient technique for priming complex devices with multiple fluids called pressurized outgas priming. By using this technique a fluid is injected into the channel structure, pressurizing the gas ahead of it and forcing that gas to diffuse quickly into the bulk matrix of the elastomer. Despite the low surface energy of the elastomer (22 mN/m), aqueous solutions may be introduced easily at moderate pressures (1–8 psi) into channels having dimensions as small as 1 μm, eliminating the need for surface-modification protocols (31). The pressurized outgas priming technique may be used to fill arbitrarily complex connected fluidic structures in a few minutes. Because no outlet is needed for the venting of gas, dead-end reaction chambers and channels may be used, allowing significant design flexibility. Furthermore, because the priming is selective and integrated valves may be used to direct flow of the fluid, a device can be primed with many different fluids in different channels or chambers.
This latter property is the central idea behind the BIM scheme. The basic principle is to set up a geometry in which two microfluidic chambers can be isolated from each other and the rest of the chip by a set of three microfabricated valves. (Fig. 1). The barrier valve that separates the chambers from each other is closed, and the chambers are primed with two different fluids, respectively, by using pressurized outgas priming. The two containment valves responsible for isolating the chambers from the rest of the chip are then actuated, creating a closed, stable fluidic system in which two volumes of distinct fluids are in close proximity to each other and are separated only by the barrier valve. Once this valve is opened, the two fluids mix by diffusion. The precise volumes that are metered and mixed are determined by the geometry of the chambers. Exquisite precision is possible, because these geometries result from a lithographic process during fabrication.
Fig. 1A shows a set of reaction chambers designed to combine two fluidic samples at three different mixing ratios. The three pairs of coupled microwells form compound reaction chambers that may be isolated from each other by actuating two containment valves. Microwells having approximate volumes of 5, 12.5, and 20 nl are combined to create mixing ratios of 4:1, 1:1, and 1:4 while maintaining a constant reaction volume of 25 nl. A microfluidic channel gated by an interface valve connects each pair, creating a controlled fluidic interface. The interface valve is actuated to create a barrier between the opposing microwells, and the two solutions are dead-end-loaded, completely filling opposite sides of the reaction chamber (Fig. 1B). The containment valves are then actuated while the interface valve is released, creating a fluidic interface between the two solutions and allowing them to mix by diffusion. Fig. 1C shows the complete diffusive mixing of an organic dye with water, creating a set of three distinct concentrations in separate reaction chambers. This method is robust and scalable; Fig. 1D shows a histogram of mixing ratios achieved in 20 sets of three reaction chambers using two fluids of varying viscosity. Similar experiments using solutions of sodium chloride ranging from 0 to 2 M have shown that metering is independent of ionic concentration (data not shown).
In this simple configuration mixing is dominated by diffusion of the reagents, which takes on the order of 1 h for small molecules in an aqueous solution. Several rapid mixing schemes have been developed to overcome the well known microfluidic problem of mixing at low Reynold's number (32–34). In cases for which speed is an issue, it would be straightforward to implement the BIM scheme in another geometry such as a ring and then accelerate mixing by active pumping around the ring by using either a “kneading” (33) or chaotic mixing configuration (32). Purely diffusive mixing removes the sometimes confounding effects of turbulence and convection. One situation where this is particularly useful is the problem of protein crystal growth (35).
The successful crystallization of a protein is determined both by thermodynamic and kinetic considerations. A concentrated solution of the target molecule must be brought to a state of supersaturation in which the crystal phase is energetically favorable and then kept in this state to allow for nucleation and growth to occur. Supersaturation is induced through the addition of a precipitating agent chosen to manipulate thermodynamic variables such as solution pH, dielectric constant, salt concentration, and effective protein concentration. Because currently there is no way to predict a priori which combination of variables will be favorable to crystallization, determination of conditions is done by trial and error. Thorough investigation of phase space is impractical, and initial experimentation is often directed toward a sparse matrix or incomplete factorial sampling of likely crystallization agents (36, 37).
Despite the need for a brute-force attack, it is possible to use the universal phase properties of the precipitant–protein interaction to systematically design experiments that increase the chances of achieving crystal growth (38). A simplified two-dimensional phase space having the concentration of protein and of precipitating agent as variables is shown in Fig. 2. Hypothetical solubility and precipitation curves bound a region of supersaturation in which crystal growth is supported. This region may be divided further into a labile region near the precipitation curve and a metastable region near the solubility curve. The labile region supports rapid nucleation, resulting in the growth of a large number of small, low-quality crystals. The growth of large, high-quality crystals is supported in the metastable region where nucleation is a rare event and thus requires long incubation times (39). Because the three-dimensional nucleation required for critical nucleus aggregation generally has a larger activation energy than that of subsequent one- or two-dimensional nucleation needed for crystal-facet growth, an optimal crystal-growth scheme should provide independent control over the these two phases of crystal growth (40).
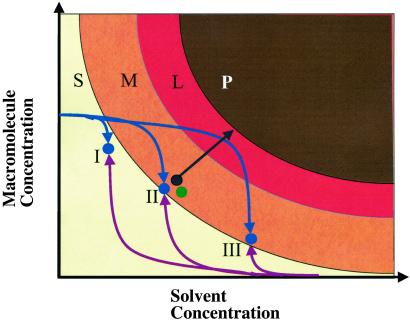
Fig 2.
Schematic diagram of the evolution of hanging-drop, microbatch, and micro free induction-decay experiments through a two-dimensional phase space having macromolecule and precipitating-agent concentrations as variables. The phase space is divided into soluble (S), metastable (M), labile (L), and precipitation (P) regions. The microbatch and hanging drop start at point II, where the target molecule is combined 1:1 with the precipitating agent. Microbatch experiments are incubated under immiscible oil, preventing subsequent concentration of reagents and therefore sampling only a point in phase space (green). In hanging-drop experiments the mixture is allowed to equilibrate through vapor diffusion with a large reservoir of precipitating agent, slowly concentrating the reagents, and driving the sample into the supersaturation region (black). The evolution of a micro free interface diffusion reaction site having three different mixing ratios is shown. Curves represent the average state of both the sample side (blue) and precipitating-agent side (purple) of each compound well. The final states (I–III) are determined by the mixing ratio. The curves are representative of a counterdiffusion between lysosyme and sodium chloride and agree with numerical finite-element simulations. A decrease in protein concentration due to precipitation or crystal growth is not included in the figure. (The figure was adapted from ref. 39.)
The BIM scheme provides exactly this property by implementing “free interface diffusion” between the precipitant and the protein solutions (40). The phase-space trajectory taken by the chip during equilibration depends on the diffusion constants of the species involved. A short time after the chip interface valves are opened, the protein concentration on the protein side changes very little, whereas that of the counter solvent, which typically has a much larger diffusion constant, increases to one of three final values determined by the lithographically defined mixing ratios. Subsequently, over a time of ≈8–24 h the protein concentration equilibrates, increasing on the solvent side and decreasing on the protein side. The final protein concentration is determined once again by the mixing ratios. The chip therefore takes a curved path through phase space, which in principle allows the protein solution to have efficient crystal nucleation in the labile region followed by high-quality growth in the metastable region. This can be contrasted with the kinetics of the microbatch and hanging-drop methods, which are the two most popular methods for crystallization screening but have phase trajectories that are far from ideal (Fig. 2).
The favorable properties of free interface diffusion have been known for a long time (40), yet it is not a popular choice in the protein crystallography community because of the large required sample volume and the confounding effect of gravity (35). It is difficult to set up an interface in a capillary tube, and even if an interface can be created, convective currents due to density differences in the solutions cause complex mixing at the interface. Even worse, after a crystal nucleates it can fall away from the interface and out of the optimal growth conditions. Thus it was thought that free interface diffusion would only realize its practical advantages in microgravity environments; experiments in space are not conclusive but suggest that large, high-quality crystals can be grown in this setting (41, 42). The unusual properties of fluid flow in microfluidic devices make it both possible and practical to implement nearly ideal free interface diffusion conditions in terrestrial devices. The BIM method allows establishment of the fluidic interface without any transient mixing. The interface is parallel to gravity, and thus the nucleated crystals do not fall out of the region of ideal growth. Finally, convection is negligible at the growth interface, because the Grashoff number (43), which measures the ratio of buoyant to viscous forces, is small.
On-chip crystallization experiments were conducted on 11 model macromolecules including 7 commercially available crystallization standards (lysosyme, glucose isomerase, xylanase, thaumatin, protease k, bovine trypsin, and beef liver catalase), 3 proteins with unpublished structures (bacterial primase catalytic core domain, type II topoisomerase ATPase domain, and a mycobacterial RNase), and a bacterial 70S ribosome. Each protein was tested against two or more standard sparse matrices of precipitants. To compare crystallization in chip against standard crystallization methods, crystallization experiments were repeated for nine of the model macromolecules by using the conventional microbatch and hanging-drop techniques; this allowed us to keep precipitant chemistries constant while varying the kinetic scheme for crystal growth.
Crystal growth was observed in the chips for all model macromolecules tested and showed an excellent degree of correlation with successful conditions revealed by more standard screening techniques. Crystals were observed at incubation times as short as 3 h and as long as 7 days. Crystals of six different protein models grown in chips are shown in Fig. 3, and a histogram comparing the number of successful experiments for each method for nine model proteins is shown in Fig. 4.
Fig 3.
Optical micrographs of macromolecular crystals grown in chip. (A) Chicken egg-white lysosyme (Sigma–Aldrich): 50 mg/ml in 0.2 M sodium acetate, pH 4.7; mixing ratio of 4:1 with 0.2 M magnesium chloride hexahydrate/30% (wt/vol) isopropanol/0.1 M Hepes-Na, pH 7.5. (B) Bacterial primase catalytic core domain: 15 mg/ml in 50 mM sodium chloride/20 mM Tris⋅HCl, pH 8.0/1 mM DTT; mixing ratio of 4:1 with 1.4 M potassium/sodium phosphate, pH 6.8. (C) Type II topoisomerase ATPase domain/ADP: 12 mg/ml in 100 mM sodium chloride/20 mM Tris, pH 7.0; mixing ratio of 1:1 with 0.2 M ammonium fluoride/20% (wt/vol) polyethylene glycol 3350, pH 6.2. (D) Thaumatin (Sigma–Aldrich): 50 mg/ml in 0.1 M ADA (Sigma–Aldrich), pH 6.5; mixing ration of 1:1 with 0.8 M potassium sodium tartrate tetrahydrate/0.1 M Hepes, pH 7.5. (E) Xylanase (Hampton Research): 43% (wt/vol) glycerol/180 mM Na/K phosphate, pH 7.0; mixing ratio of 4:1 with 0.2 M calcium chloride dihydrate/28% (vol/vol) polyethylene glycol 400/0.1 M Hepes, pH 7.5. (F) Glucose isomerase (Hampton Research): 31 mg/ml in 10 mM ammonium sulfate; mixing ratio of 1:1 with 0.2 M calcium chloride dihydrate/28% polyethylene glycol 400/0.1 M Hepes, pH 7.5. (Scale bars, 100 μm.)
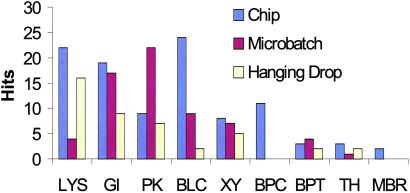
Fig 4.
Histogram of crystallization hits for sparse matrix screens of model proteins. The number of screens tested on each protein are: lysosyme (LYS), 2; glucose isomerase (GI), 2; protease K (PK), 1; bovine liver catalase (BLC), 1; xylanase (XY), 2; bacterial primase catalytic core domain (BPC), 3; bovine pancreas trypsin (BPT), 1; thaumatin (TH), 1; and mycobacterial RNase (MBR), 3.
One surprising outcome of this study was that identical sparse matrix screens led to crystal growth more often in the chip than by conventional techniques in all but two cases. For the bacterial primase catalytic core domain, 11 conditions producing needle crystals of dimensions >100 μm were detected on chip; no hits were initially observed in either macroscopic method. Moreover, an additional on-chip experiment optimizing around the crystallization conditions identified from our initial screens produced crystals whose largest dimension exceeded 400 μm (Fig. 3B). These conditions were subsequently used to reproduce crystallization in microbatch formats, demonstrating that optimized on-chip crystallization conditions can be exported successfully to macroscopic techniques.
In two other cases, the chip also produced crystal forms that were not observed in conventional experiments. A previously unidentified crystal form of the bacterial 70S ribosome was obtained in three conditions of a sparse matrix of precipitants (Hampton Crystal Screen I), demonstrating that large protein–nucleic acid complexes may be crystallized in chip (C.L.H., A. Vila-Sanjurjo, and J. Cate, personal communication). Crystals of a previously uncrystallized mycobacterial RNase also were obtained from a single experimental condition on chip, whereas no crystals had been observed for this sample despite extensive prior trials using traditional methods; subsequent broad-based screening efforts around this condition using hanging-drop vapor diffusion setups proved successful but only after the protein concentration was increased to >40 mg/ml. Together, these data validate the use of BIM-type microfluidic devices for the purposes of macromolecular crystallization screening.
A second area where BIM-based chips show favorable experimental properties is reduced precipitation during initial mixing. In microbatch or hanging-drop experiments, the sudden addition of the precipitating agent to the protein sample induces rapid convective mixing, causing large transient concentration gradients throughout the drop and often resulting in precipitation of the macromolecule. In micro free interface diffusion experiments, large concentration gradients are localized at the fluidic interface, allowing higher levels of supersaturation to be achieved without inducing precipitation of the macromolecule. It was observed that mixing ratios that lead to crystallization on chip often caused the protein to immediately precipitate in hanging-drop and microbatch experiments. In the case of a type II topoisomerase ATPase domain, the final concentration of precipitating agent achievable in chip was four times greater than that possible for microbatch.
Finally, crystal growth in micro free interface diffusion experiments was generally observed to be faster than in microbatch or hanging drop. For the type II topoisomerase ATPase domain, crystal growth in microbatch required ≈1 week, whereas crystals grown on chip with the same conditions appeared after only 4 h of incubation. When crystals grew on chip in less than 12 h, they were always observed on the protein side of the compound well, suggesting that the short crystallization times are due to the high degree of supersaturation achieved in the initial phase of diffusive equilibration.
In certain cases (glucose isomerase, xylanase, thaumatin, and the type II topoisomerase ATPase domain), conditions were optimized sufficiently to grow (>30 μm) crystals large enough for x-ray diffraction studies. To show that crystals grown in the BIM devices could be used in crystallographic structure-determination efforts, we extracted several crystals from different chips, mounted and flash-froze them in cryoloops, and subsequently exposed them to both laboratory and synchrotron radiation x-ray sources. Fig. 5 shows a high-resolution diffraction pattern for a single thaumatin crystal grown from only 5 nl of protein solution (Fig. 5). These results demonstrate that diffraction-quality crystals may be grown and harvested from the ultrasmall-volume reactions performed in these microfluidic devices.
Fig 5.
X-ray diffraction pattern (resolution <1.35 Å) from a single thaumatin crystal grown from 5 nl of sample in chip. (Inset) Clean reflection at 1.35-Å resolution.
In conclusion, we have demonstrated a microfluidic technique that can be used to meter and mix small amounts of reagents in a highly scalable fashion. In applying this technique to protein crystal growth we found that crystallization experiments in chip result in faster crystal growth with a higher hit rate than conventional techniques. Beyond simple screening for crystallization conditions, we have also shown that it is possible to grow and recover diffraction-quality crystals from the chips. Because the soft lithographic techniques used to fabricate the chip are forgiving and inexpensive, we anticipate that microfluidics will provide a robust and affordable approach to crystallization that will have broad accessibility and applicability.
Supplementary Material
Acknowledgments
We thank the members of the J.M.B. lab, as well as Tom Alber, Jamie Cate, and James Holton for advice and materials used in this work. We also thank Kyle Self, Susanna Ng, Philip Lam, Joe Barco, Emerson Quan, and Shelley Godley of Fluidigm Corp. for sharing data and assisting with device fabrication. This work was supported in part by the National Science Foundation (XYZ in a chip program, to S.R.Q. and C.L.H.). C.L.H. was supported partly by Natural Sciences and Engineering Research Council of Canada Julie Payette Fellowship. J.M.B. and E.S. were supported by The David H. and Lucille M. Packard Foundation, The G. Harold and Leila Y. Mathers Charitable Foundation, and National Institutes of Health Grants CA77373 and P50-GM62410.
Abbreviations
BIM, barrier interface metering
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mathies R. A. & Huang, X. C. (1992) Nature 359, 167-169. [Google Scholar]
- 2.Manz A., Harrison, D. J., Verpoorte, M. J., Fettinger, J. C., Paulus, A., Ludi, A. & Widmer, H. M. (1992) J. Chromatogr. 593, 253-258. [Google Scholar]
- 3.Effenhauser C. S., Manz, A. & Widmer, H. M. (1993) Anal. Chem. 65, 2637-2642. [Google Scholar]
- 4.Harrison D. J., Fluri, K., Seiler, K., Fan, Z., Effenhauser, C. S. & Manz, A. (1993) Science 261, 895-897. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson S. C., Hergenroder, R., Koutny, L. B. & Ramsey, J. M. (1994) Anal. Chem. 66, 1114-1118. [Google Scholar]
- 6.Effenhauser C. S., Paulus, A., Manz, A. & Widmer, H. M. (1994) Anal. Chem. 66, 2949-2953. [Google Scholar]
- 7.Chiu D. T., Jeon, N. L., Huang, S., Kane, R. S., Wargo, C. J., Choi, I. S., Ingber, D. & Whitesides, G. M. (2000) Proc. Natl. Acad. Sci. USA 97, 2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delamarche E., Bernard, A., Schmid, H., Michel, B. & Biebuyck, H. (1997) Science 276, 779-781. [DOI] [PubMed] [Google Scholar]
- 9.Volkmuth W. D. & Austin, R. H. (1992) Nature 358, 600-602. [DOI] [PubMed] [Google Scholar]
- 10.Chou H., Spence, C., Scherer, A. & Quake, S. R. (1999) Proc. Natl. Acad. Sci. USA 96, 11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu A. Y., Spence, C., Scherer, A., Arnold, F. H. & Quake, S. R. (1999) Nat. Biotechnol. 17, 1109-1111. [DOI] [PubMed] [Google Scholar]
- 12.Fu A. Y., Chou, H., Spence, C., Arnold, F. & Quake, S. R. (2002) Anal. Chem. 74, 2451-2457. [DOI] [PubMed] [Google Scholar]
- 13.Jensen K. (1998) Nature 393, 735-737. [Google Scholar]
- 14.Jacobson S. C., McKnight, T. E. & Ramsey, J. M. (1999) Anal. Chem. 71, 4455-4459. [Google Scholar]
- 15.Kanholz A. E., Weigl, B. H., Finlayson, B. A. & Yager, P. (1999) Anal. Chem. 71, 5340-5347. [DOI] [PubMed] [Google Scholar]
- 16.Dertinger S. K. W., Chiu, D. T., Jeon, N. L. & Whitesides, G. M. (2001) Anal. Chem. 73, 1240-1246. [Google Scholar]
- 17.Zhang C. & Manz, A. (2001) Anal. Chem. 73, 2656-2662. [DOI] [PubMed] [Google Scholar]
- 18.Qui C. X. & Harrison, D. J. (2001) Electrophoresis 22, 3949-3958. [DOI] [PubMed] [Google Scholar]
- 19.Unger M. A., Chou, H., Thorsen, T., Scherer, A. & Quake, S. R. (1999) Science 288, 113-116. [DOI] [PubMed] [Google Scholar]
- 20.Brechtel R., Hohmann, W., Rudinger, H. & Watzig, H. (1995) J. Chromatogr. A 716, 97-105. [Google Scholar]
- 21.Lucy C. A. & Underhill, R. S. (1996) Anal. Chem. 68, 300-305. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Enzelberger, M. & Quake, S. R. (2002) Electrophoresis 23, 1531-1536. [DOI] [PubMed] [Google Scholar]
- 23.Xia Y. & Whitesides, G. M. (1998) Angew. Chem. 37, 550-575. [DOI] [PubMed] [Google Scholar]
- 24.Monahan J., Gewirth, A. A. & Nuzzo, R. G. (2001) Anal. Chem. 73, 3193-3197. [DOI] [PubMed] [Google Scholar]
- 25.Chayen E. N. (2002) Trends Biotechnol. 20, 98. [DOI] [PubMed] [Google Scholar]
- 26.Stevens R. C. (2000) Structure 8, R177-R185. [DOI] [PubMed] [Google Scholar]
- 27.Abola E., Kuhn, P., Earnest, T. & Stevens, R. C. (2000) Nat. Struct. Biol. 7, 973-977. [DOI] [PubMed] [Google Scholar]
- 28.Nyarsik M. U., Horn, R. M., Rauth, H., Przewieslik, T., Saenger, W., Lehrach, H. & Eickhoff, H. (2001) J. Biotechnol. 85, 7-14. [DOI] [PubMed] [Google Scholar]
- 29.Kuil M. E., Bodenstaff, E. R., Hoedemaeker, F. J. & Abrahams, J. P. (2002) Enzyme Microb. Technol. 30, 262-265. [DOI] [PubMed] [Google Scholar]
- 30.Stjernstrom M. & Roeraade, J. (1998) J. Micromech. Microeng. 8, 33-38. [Google Scholar]
- 31.Chaudhury M. K. & Whitesides, G. M. (1992) Science 255, 1230-1232. [DOI] [PubMed] [Google Scholar]
- 32.Stroock A. D., Dertinger, S. K. W., Ajdari, A., Mezi, I., Stone, H. A. & Whitesides, G. M. (2002) Science 295, 647-651. [DOI] [PubMed] [Google Scholar]
- 33.Hou-Pu C., Unger, M. A. & Quake, S. R. (2001) Biomed. Microdevices 3, 323-330. [Google Scholar]
- 34.He B., Burke, B. J., Zhang, X., Zhang, R. & Regnier, F. E. (2001) Anal. Chem. 73, 1942-1947. [DOI] [PubMed] [Google Scholar]
- 35.McPherson A., (1999) Crystallization of Biological Macromolecules (Cold Spring Harbor Lab. Press, Plainview, NY).
- 36.Jancarik J. & Kim, S. H. (1991) J. Appl. Crystallogr. 24, 409-411. [Google Scholar]
- 37.Carter C. W. & Carter, C. W. (1979) J. Biol. Chem. 254, 12219-12223. [PubMed] [Google Scholar]
- 38.Carter C. W., Baldwin, E. T. & Frick, L. (1988) J. Cryst. Growth 90, 60-73. [Google Scholar]
- 39.Luft J. R. & DeTitta, G. T. (1997) Methods Enzymol. 276, 110-130. [PubMed] [Google Scholar]
- 40.Salemme F. R. (1972) Arch. Biochem. Biophys. 151, 533-539. [DOI] [PubMed] [Google Scholar]
- 41.Koszelak S., Day, J., Leja, C., Cudney, R. & McPherson, A. (1995) Biophys. J. 69, 13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strong R. K. & Stoddard, B. L. (1992) J. Cryst. Growth 119, 200-214. [Google Scholar]
- 43.Bird R. B., Stewart, W. E. & Lightfoot, E. N., (1960) Transport Phenomena (Wiley, New York), pp. 138, 148.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.