Abstract
An enzyme that uses inorganic polyphosphate (poly P) as a donor to convert GDP to GTP has been purified 1,300-fold to homogeneity from lysates of Pseudomonas aeruginosa PAOM5. Poly P chains of 30–50 residues are optimal; those of 15–700 residues can also serve. GDP is preferred over ADP among nucleoside diphosphate acceptors. This nucleoside diphosphate kinase (NDK) activity resides in the same protein isolated for its synthesis of poly P from GTP and designated PPK2 in an accompanying report. The reaction that synthesizes poly P and the reaction that utilizes poly P differ in their kinetic features. Especially notable is the catalytic potency of the NDK activity, which is 75-fold greater than that of poly P synthesis. PPK2 appears in the stationary phase of growth and reaches NDK levels of 5–10% that of the classic NDK; both kinase activities may figure in the generation of the guanosine precursors in the synthesis of alginate, an exopolysaccharide essential for the virulence of P. aeruginosa.
Among its several functions, inorganic polyphosphate (poly P) serves as a kinase donor to generate ATP and GTP (1–3). Universally distributed in all cells, these chains of phosphate residues, usually hundreds long, linked by phosphoanhydride (metaphosphate) bonds, as in ATP, can be used to phosphorylate glucose, nucleoside diphosphates, and protein (3). In this capacity, poly P was found in extracts of Pseudomonas aeruginosa to be a donor to GDP and a potent source of GTP (4, 5). Of particular physiologic and clinical interest are the sources of GTP needed by P. aeruginosa to provide the intermediates for the synthesis of alginate, its exopolysaccharide associated with mucoidy and virulence (6–8).
The present study was undertaken to purify the poly P donor activity designated earlier as PNDK (poly P-driven nucleoside diphosphate kinase; ref. 5) to characterize the enzyme, identify its gene, and, with that, explore the physiologic consequences of overexpression and removal of the gene. At the same time, we were also in pursuit of a putative poly P kinase (PPK2) activity in extracts of P. aeruginosa in addition to the known PPK1 (9, 10) that converts ATP to poly P. The PNDK and PPK2 activities, ostensibly different because of different kinetic features, have proved on isolation of the homogeneous proteins in each case to be the same and to be encoded by the same gene, now designated ppk2 (11). In this report, we describe the isolation and structural properties of PPK2 and its kinetic features based on its activity as a nucleoside diphosphate kinase (NDK). The accompanying report (11) describes the genetic sequence of PPK2 and its conservation in many other bacteria, as well as features of the enzyme in the synthesis of poly P from GTP or ATP.
Materials and Methods
Strains and Materials.
P. aeruginosa PAOM5 (12), the Δppk:tet derivative of non-mucoid strain PAO1, was used in this study. All of the reagents including poly P15 were obtained from Sigma. Purified PPK1 was a generous gift from S. Lee (ICOS Corporation, Bothell, WA).
Assay for PPK2.
To measure PNDK activity, the assay mixture (10 μl) contained the following: 50 mM Hepes-KOH (pH 8.0), 80 mM (NH4)2SO4, 10 mM MgCl2, 0.1% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), 1 mM [8-3H]GDP (20 cpm/pmol), and 3 mM poly P15 (Sigma, expressed as phosphate residues). After incubation at 37°C for 10 min, 1 μl from each sample was applied in a line along the bottom of a polyethyleneimine-cellulose TLC plate and developed with 1 M HCOOH and 1 M LiCl. Each GTP and GDP position was cut out and quantified in a liquid scintillation counter.
Assay for the Other Enzymatic Activities.
To measure the classic ATP-driven NDK activity, the assay mixture contained the following: 50 mM Tris⋅HCl (pH 7.6), 10 mM MgCl2, 1 mM DTT, 1 mM [8-3H]GDP (20 cpm/pmol), and 3 mM ATP. After incubation at 37°C for 10 min, the product was quantified as in the PPK2 assay. The assays for poly P-using (reverse) activity of PPK1 from P. aeruginosa were performed as follows. The assay mixture contained the following: 50 mM Hepes-KOH (pH 7.2), 40 mM (NH4)2SO4, 4 mM MgCl2, 1 mM [8-3H]ADP or -GDP (20 cpm/pmol), 3 mM poly P, and purified PPK1 from P. aeruginosa. Poly P15 (Sigma) and poly P750 (prepared as described in ref. 13), were used in this assay. After incubation at 37°C for 10 min, the product was quantified by the same method as in the PPK2 assay.
Results
Assay of Poly P-Driven GTP Synthesis.
In an earlier study (5), GTP synthetic activity, which is PPK1 independent and driven by short-chain poly P (designated PNDK), was observed in lysates of P. aeruginosa PAO1. The assay uses [8-3H]GDP as acceptor, polyP15 (Sigma) as donor, and a TLC separation to measure the GTP product. This NDK activity in the lysate or the purified enzyme (see below) has kinetic features that distinguish it from those of the activity that converts GTP to poly P. As described in this and the accompanying report (11), both activities reside in the same protein, PPK2.
Growth Phase-Dependent Expression of the Activity.
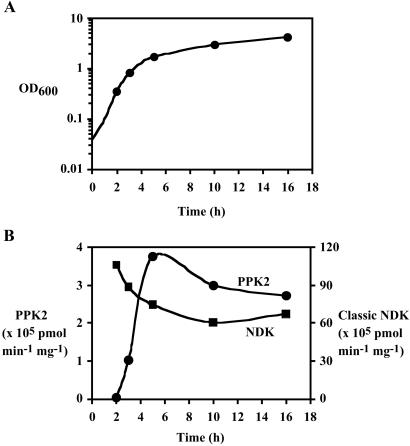
Expression of the enzymatic activity is growth phase dependent (Fig. 1) in P. aeruginosa PAOM5, the strain lacking PPK1 (12); the activity reached maximum at the end of exponential to early stationary phase. In contrast, the level of the classic ATP-driven NDK (14) was not induced and remained unchanged through all of the growth stages.
Fig 1.
Growth-phase dependency of PPK2 and the classic NDK activities. After a 1% inoculation with an overnight preculture, P. aeruginosa PAOM5 was grown in 500 ml of LB media at 37°C with vigorous shaking and its OD600 was monitored. At time points indicated as symbols (corresponding to middle log, late log, early stationary, middle stationary, and late stationary phases, respectively), cells were harvested from 10-ml culture samples. After suspending cells to 1 ml of TED buffer (50 mM Tris⋅HCl, pH 8.0/0.5 mM EDTA/1 mM DTT), cells were sonically disrupted and centrifuged (10,000 × g, 10 min) to give crude lysates, which were assayed for both PPK2 and the classic NDK activities as described in Materials and Methods. (A) Growth curve of P. aeruginosa PAOM5. (B) Both NDK activities in crude lysates of the cells from various growth phases. Circles represent PNDK (PPK2) activity and squares represent the classic ATP-driven NDK (NDK) activity.
Stabilization of the Activity.
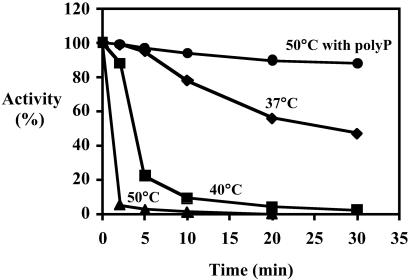
To overcome enzyme instability, which had limited earlier purification efforts (5), factors were sought that would increase its thermostability. For example, exposure to 37°C for 30 min resulted in loss of half of the activity; 96% was lost in 2 min at 50°C. However, with poly P present, the activity was fully retained even after 30 min at 50°C (Fig. 2). The state of oligomerization of the enzyme as a possible explanation for the poly P effect will be considered below.
Fig 2.
Thermostabilization of PPK2. Crude lysate (Table 1, Fraction 1) was incubated at stated temperatures and assayed for PPK2 activity: diamonds at 37°C, squares at 40°C, triangles at 50°C, and circles at 50°C with poly P15 (Sigma; expressed as phosphate residues).
Purification of PPK2.
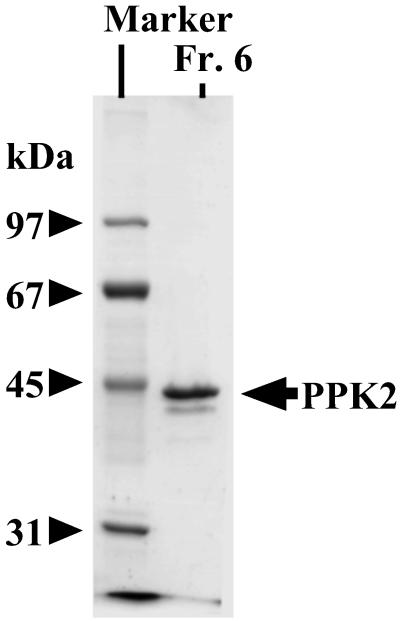
Poly P-driven GTP synthesis was used to purify the enzyme 1,300-fold from lysates of P. aeruginosa PAOM5 cells harvested at early stationary phase by fractionation on columns of phosphocellulose, heparin, and Superdex (Table 1). All steps were carried out in the presence of poly P15 (Sigma) at a concentration of 2 mM expressed as phosphate residues. The molecular mass of the protein band compared with markers was ≈44 kDa (Fig. 3). Analysis of the NH2-terminal sequence and matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry identified the isolated protein with a sequence starting 177 bp upstream and in phase with ORF (GenBank accession no. NP_248831) annotated as a “conserved hypothetical protein” encoded by PA0141 gene (15). Thus, this protein of 357 aa with a mass of 40,786 Da and a predicted isoelectric point of 9.85 is encoded by the same sequence and origin as that described as PPK2 (GenBank accession no. AY168003) (11).
Table 1.
Purification of PPK2
| Fraction | Protein, mg | Total activity, ×108 pmol⋅min−1 | Specific activity, ×105 pmol⋅min−1⋅mg−1 | Purification, fold | Recovery, % |
|---|---|---|---|---|---|
| 1. Crude lysate | 749 | 2.79 | 3.72 | 1 | 100 |
| 2. Soluble fraction | 602 | 2.64 | 4.34 | 1.17 | 94 |
| 3. Phosphocellulose | 8.50 | 2.40 | 281 | 75.5 | 86 |
| 4. Heparin | 2.61 | 2.40 | 920 | 247 | 86 |
| 5. Superdex | 0.617 | 2.06 | 3340 | 898 | 74 |
| 6. Heparin (second) | 0.422 | 2.00 | 4740 | 1270 | 72 |
All operations were at 0–4°C. P. aeruginosa PAOM5 was grown in 2 liters of LB at 37°C aerobically, and harvested when OD600 reached 1.8 (early stationary phase). Cells were suspended into 100 ml of buffer P [50 mM Tris·HCl, pH 8.0/0.5 mM EDTA/1 mM DTT/2 mM poly P15 (Sigma, in phosphate residues)] containing 0.5 mM PMSF. After a lysozyme treatment (0.25 mg/ml, on ice for 30 min), the cells were sonically oscillated and centrifuged (10,000 × g, 10 min) to give the crude lysate (Fraction 1). The crude lysate was further centrifuged (80,000 × g, 1 h) to give the supernatant (Fraction 2). To the lysate supernatant, 300 mM KCl was added, and the mixture was applied to a 20-ml phosphocellulose P-11 column (2.5 × 4 cm, Whatman) equilibrated with buffer P containing 300 mM KCl. The column was washed with 3 column-volumes of equilibration buffer and 8 column-volumes of buffer P containing 150 mM potassium phosphate buffer (pH 8.0). Then the activity was eluted with a 20 column-volume gradient [buffer P, 150–750 mM potassium phosphate buffer (pH 8.0)] at a flow rate of 0.5 ml/min. Peak fractions were pooled and dialyzed against buffer P (Fraction 3). To Fraction 3, 0.1% CHAPS was added, and the mixture was applied to a HiTrap Heparin column (1 ml, Amersham) equilibrated with buffer PC (buffer P containing 0.1% CHAPS). After washing of the column with 5 column-volumes of equilibration buffer, the activity was eluted with a 20 column-volume gradient (buffer PC, 0–500 mM KCl) at a flow rate of 0.25 ml/min, and peak fractions were pooled (Fraction 4). To Fraction 4, 800 mM KCl was added. Each one-third of the mixture was independently applied to a HiLoad 16/60 Superdex 200 pg column (Amersham) equilibrated with buffer PC containing 800 mM KCl, and the activity was eluted with equilibration buffer at a flow rate of 0.28 ml/min. After the three independent Superdex column chromatographies, peak fractions were pooled (Fraction 5). Fraction 5 was dialyzed against buffer PC and then applied to HiTrap Heparin equilibrated with buffer PC. The activity was eluted with buffer PC containing 500 mM KCl. Peak fractions were pooled (Fraction 6).
Fraction numbers for use in legend and in Fig. 3.
Fig 3.
SDS/PAGE analysis of PPK2. SDS/PAGE (10%) was performed as described (33), and proteins were visualized by Coomassie blue staining: 1 μg of Fraction 6 (Fr. 6) in Table 1; marker proteins were phosphorylase b (97 kDa), BSA (67 kDa), ovalbumin (45 kDa), and carbonic anhydrase (31 kDa).
Features of PNDK.
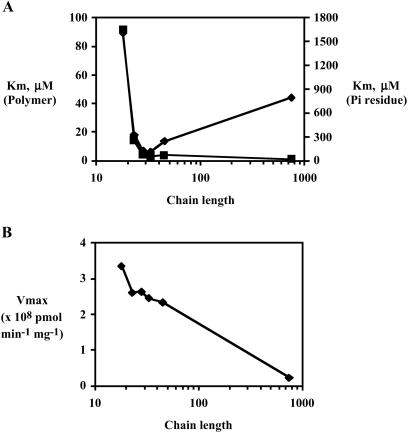
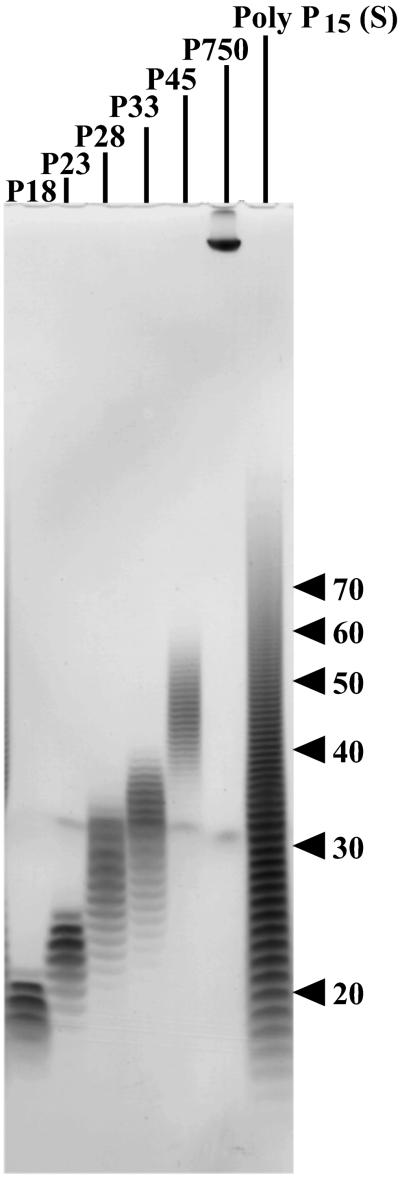
The NDK activity of PPK2 has kinetic features that differ from those of PPK1 and also differ from those of the PPK2 enzyme in its activity as a poly P synthetase. Most striking is the rate of poly P utilization, nearly 100-fold greater than that of poly P synthesis (Table 2). In addition, Mg2+ is favored over Mn2+ by 5-fold at optimal concentrations (10 mM), and GDP is preferred over ATP. The respective Km values are 300 μM for GDP and 750 μM for ADP; the respective Vmax values (pmol/min/mg) are 5.0 × 108 for GTP and 4.6 × 108 for ADP. Poly P chains of 28–45 residues are used more effectively than poly P18 and poly P750 (Fig. 4). With regard to the commonly used commercial poly P15, its wide heterogeneity between 10 and 100 is notable (Fig. 5).
Table 2.
Comparison of polyP-utilizing activity of PPK2 and several PPK1s
| Vmax, ×107 pmol⋅min−1⋅mg−1 | Utilization/synthesis of poly P | GDP/ADP | Poly P750/poly P15 | |
|---|---|---|---|---|
| P. aeruginosa PPK2 | 50 | 75 | 1.1 | 0.1 |
| P. aeruginosa PPK1 | 0.051 | 0.25 | 0.03 | 12 |
| E. coli PPK1 | 0.37 | 0.073 | 0.015 | 11 |
| V. cholerae PPK1 | 9.5 | 59 | 0.00016 | ND |
| H. pylori PPK1 | 12 | 41 | 0.00046 | ND |
For PPK2, poly P15 (Sigma) and GDP were used as a donor and an acceptor, respectively. For PPK1, poly P750 and ADP were used as a donor and an acceptor, respectively.
The Vmax values for poly P-utilizing reactions divided by that for poly P-synthesizing reactions. For determining the Vmax value for poly P-synthesizing reaction of PPK2, GTP and Mn2+ instead of Mg2+ were used as a donor and a divalent cation, respectively. For determining the Vmax value for poly P-synthesizing reaction of PPK1, ATP was used as a donor. For more details, see accompanying report (11).
The Vmax value with GDP divided by that with ADP. Poly P15 (Sigma) or poly P750 was used for determining Vmax values for PPK2 or PPK1, respectively.
The Vmax value with poly P750 divided by that with poly P15 (Sigma). GDP was used as an acceptor for PPK2; ADP was used for PPK1.
Data from Ahn and Kornberg (32).
Data from Ogawa et al. (31).
Not determined.
Fig 4.
Km and Vmax values with poly P of various chain lengths. The assay mixture contained 50 mM Hepes-KOH (pH 8.0), 80 mM (NH4)2SO4, 10 mM MgCl2, 1 mM [8-3H]GDP (20 cpm/pmol), various concentrations of poly P, and various amounts of PPK2 (Fraction 6 in Table 1). Poly P chain lengths were poly P18 (18 ± 2), poly P23 (23 ± 3), poly P28 (28 ± 5), poly P33 (33 ± 6), poly P45 (45 ± 8), and poly P750 (as described in ref. 13). For more details, see Fig. 5 legend. In each case, the Km and Vmax were determined by a Lineweaver–Burk plot (data not shown). (A) Km values for poly P: squares for molar concentration, diamonds for Pi residue concentration. (B) Vmax values with various chain lengths.
Fig 5.
PAGE analysis of poly P samples. Commercial poly P15 (Sigma, 250 mg) was solubilized in 100 ml of 25 mM Hepes-KOH (pH 7.6) and applied to HiTrap Q (5 ml, Amersham Pharmacia) equilibrated with 25 mM Hepes-KOH (pH 7.6). Bound poly P was eluted with a 20 column-volume gradient (0–1 M NaCl in the same buffer) and fractionated. PAGE analysis using 20% acrylamide gel containing 7 M urea was carried out as described (34). Poly P was visualized by toluidine blue staining. Applied samples and chain lengths were: P18 (7.5 μg, 18 ± 2); P23 (5 μg, 23 ± 3); P28 (5 μg, 28 ± 5); P33 (5 μg, 33 ± 6); P45 (3.5 μg, 45 ± 8); P750 (1.5 μg; as described in ref. 13); and commercial poly P15 (S) (80 μg, obtained from Sigma). These poly P samples were also used in Fig. 4.
Processivity of PPK2.
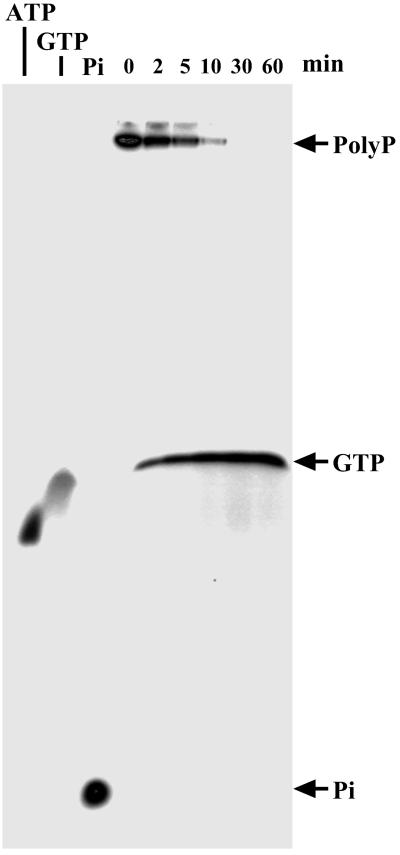
During utilization of poly P750 to convert GDP to GTP, no poly P of intermediate chain length was detected at any time during the course of the reaction (Fig. 6); processivity is also true of the poly P-synthesizing reaction from GTP (11) and with the several PPK1 enzymes (3).
Fig 6.
PPK2 as a processive enzyme. The assay mixture (10 μl) contained 50 mM Hepes-KOH (pH 8.0), 80 mM (NH4)2SO4, 4 mM MgCl2, 1 mM GDP, 100 μM [32P]poly P (88 cpm/pmol), and 11 ng of PPK2 (Fraction 6). After incubation of the mixture at 37°C, the products were separated by PAGE using 20% acrylamide gel containing 7 M urea (34) and visualized by PhosphorImager (Molecular Dynamics). [32P]ATP, [32P]GTP, and [32P]inorganic phosphate were used as standards.
Oligomers of PPK2.
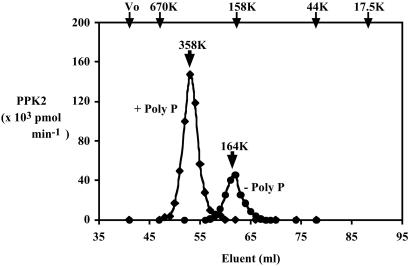
Measured by size-exclusion chromatography, the migration of PPK2 is about that of a 164-kDa unit, presumably a tetramer of the 41-kDa subunit (Fig. 7). A small amount of this protein was detected in the crude lysate (data not shown). In the presence of poly P15 (Sigma, 2 mM in Pi residues), PPK2 migrates as a unit of 358 kDa, ostensibly an octamer. This octamer, on dilution in the absence of poly P, is unstable and breaks up into tetramers (data not shown).
Fig 7.
Mass of PPK2 by size-exclusion chromatography. To measure the molecular mass of PPK2 complexed with poly P, the assay mixture (500 μl) contained 50 mM Tris⋅HCl (pH 8.0), 50 mM (NH4)2SO4, 5 mM MgCl2, 6 μg of PPK2 (Fraction 6 in table 1), and 2 mM poly P15 (Sigma). After incubation for 10 min at 37°C, 0.8 M KCl was added. The mixture was applied to a HiLoad 16/60 Superdex 200-pg column (Amersham Pharmacia) equilibrated with buffer PC [50 mM Tris⋅HCl, pH 8.0/0.5 mM EDTA/1 mM DTT/2 mM poly P15 (Sigma)/0.1% CHAPS] containing 0.8 M KCl. PPK2 was eluted with equilibration buffer and monitored by PNDK activity (diamonds). For measuring molecular mass of PPK2 without poly P, poly P was omitted from the incubation mixture and the equilibration (elution) buffer (circles). Positions of molecular mass standards are shown at the top of the panel: blue dextran (Vo), thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), and equine myoglobin (17.5 kDa).
Discussion
A potent activity in P. aeruginosa converts GDP to GTP by using poly P as donor (5). The present study was designed to isolate this NDK activity, to distinguish it from the well-characterized ATP-dependent NDK (8, 14, 16–19), and to evaluate its metabolic role. Coincident with this study was the discovery in our laboratory of an activity that converts GTP to poly P. This activity might account for the persistence of poly P in cells that lack the reported PPK (now designated PPK1; ref. 11). [The absence of PPK1 results in cells profoundly deficient in motility, quorum sensing, biofilm formation, and virulence in mice (12, 20, 21).] Although these two activities, that is, poly P utilization to make GTP and poly P synthesis from GTP, differed in some kinetic features, they have proved, on purification of each of the activities to homogeneity, to reside in the same protein (designated PPK2) encoded by the gene ppk2.
PPK2 of P. aeruginosa differs from PPK1 in two major features (Table 2). One is that PPK2 utilizes poly P to make GTP at a rate 75-fold greater than the synthesis of poly P from GTP. With PPK1 the reverse is true: poly P synthesis is favored 4-fold over poly P utilization. The other feature is selectivity for guanosine and adenosine nucleotides as donors and acceptors: PPK2 uses GTP and ATP equally well in poly P synthesis, but PPK1 is strictly specific for ATP. In the utilization of poly P as a donor to nucleoside diphosphates, PPK2 prefers GDP over ADP, but PPK1 has a >30-fold preference for ADP over GDP. Thus, PPK2, at least in isolated form, seems to be designed for synthesis of GTP from poly P in contrast to PPK1, which strongly favors synthesis of poly P and exclusively from ATP.
PPK2 in P. aeruginosa is induced >100-fold from barely detectable levels when the culture approaches stationary phase. This is the very stage when GTP is needed for the synthesis of alginate (22, 23), the exopolysaccharide that envelops the bacterium, and possibly for other functions of GTP. Thus, the predominant activity of isolated PPK2 to use poly P for the generation of GTP is reflected in its appearance just when needed in cells in culture. Whether this induction of PPK2 also occurs in clinical isolates of P. aeruginosa and in the biofilms of infected tissues may be revealed when the phenotypes of PPK2 mutants are examined.
By contrast with PPK2, the cellular levels of the classic ATP-driven NDK remain near constant in log and stationary phases of P. aeruginosa. However, the classic NDK undergoes a remarkable transformation in stationary phase when the 16-kDa form is cleaved by elastase to a 12-kDa form that is membrane localized and strongly selective for GDP to form GTP (8, 14, 16–19). However, these events occur at a stage after alginate synthesis is underway. Disruption of algR2, the master regulatory gene for alginate synthesis, depresses not only alginate but also poly P levels (7, 14). These phenotypes are suppressed by an oversupply of gene encoding the classic NDK. It would seem that both NDK and PPK2 share in the supply of massive amount of GTP, and further work is needed to sort out their separate roles and interdependence.
One characteristic of PPK2 in the utilization of poly P as an NDK donor is the efficiency of short-chain-length poly P (Fig. 4). The Km values for chains with 30–750 residues were similar, but, when measured by Vmax values and expressed as Pi residues, the shorter poly P chains were more efficient than the long ones. Yet the abundance and origin of short poly P chains (<100 residues) in P. aeruginosa remain in doubt. Poly P chains synthesized by PPK1 and PPK2 (11) are well in excess of 100 residues. Generation of short chains from long ones does not occur with PPK1 and PPK2, which are highly processive in using poly P and produce no short-chain intermediates. The same is true of the exopolyphosphatases in P. aeruginosa (24), Escherichia coli (25), and yeast (13). Conversion of long-chain poly P to a range of chains can be achieved by a nonprocessive endopolyphosphatase (PPN) abundant in yeast and animal cells; such an activity has yet to be identified in prokaryotes (26–29).
The structure-function relationships of PPK2 are clearly evident from the effects of oligomerization on its stability and activity. From a thermolability manifested by a loss of half the activity after 30 min at 37°C, the enzyme remains fully active even after 30 min at 50°C in the presence of poly P (Fig. 2). This preservation function by poly P is associated with a conversion from a tetrameric state of 164 kDa to an octamer of 358 kDa (Fig. 7). The structures of the tetramer and the octamer of the isolated enzyme complexed with poly P need to be examined as well as determining the oligomeric state of PPK2 in cells at various stages of cultural growth.
The importance of PPK1 in bacterial metabolism and virulence of pathogens (21, 30, 31) has been exploited as a target for the discovery of antibiotics that inhibit the enzyme (S. Lee, personal communication). Now, with the recognition that PPK2 may contribute to the synthesis of alginate and the mucoidy associated with virulence, PPK2 may prove to be another target for the discovery of novel antibiotics.
Acknowledgments
We thank Leroy Bertsch for advice and assistance in preparation of the paper. We thank the National Institute of General Medical Science of the National Institutes of Health for the support of our research and the Yamasa Corporation for fellowship support of K.I.
Abbreviations
CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
NDK, nucleoside diphosphate kinase
poly P, inorganic polyphosphate
PNDK, poly P-driven NDK
PPK, polyphosphate kinase
References
- 1.Kulaev I. S., (1979) The Biochemistry of Inorganic Polyphosphate (Wiley, New York).
- 2.Kornberg A. (1995) J. Bacteriol. 177, 491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornberg A., Rao, N. N. & Ault-Riché, D. (1999) Annu. Rev. Biochem. 68, 89-125. [DOI] [PubMed] [Google Scholar]
- 4.Kuroda A. & Kornberg, A. (1997) Proc. Natl. Acad. Sci. USA 94, 439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishige K. & Noguchi, T. (2001) Biochem. Biophys. Res. Commun. 281, 821-826. [DOI] [PubMed] [Google Scholar]
- 6.May T. B. & Chakrabarty, A. M. (1994) Trends Microbiol. 2, 151-157. [DOI] [PubMed] [Google Scholar]
- 7.Kim H.-Y., Schlictman, D., Shanker, S., Xie, Z., Chakrabarty, A. M. & Kornberg, A. (1998) Mol. Microbiol. 27, 717-725. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarty A. M. (1998) Mol. Microbiol. 28, 875-882. [DOI] [PubMed] [Google Scholar]
- 9.Ishige K., Kameda, A., Noguchi, T. & Shiba, T. (1998) DNA Res. 5, 157-162. [DOI] [PubMed] [Google Scholar]
- 10.Zago A., Chugani, S. & Chakrabarty, A. M. (1999) Appl. Environ. Microbiol. 65, 2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Ishige, K. & Kornberg, A. (2002) Proc. Natl. Acad. Sci. USA 99, 16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rashid M. H., Rao, N. N. & Kornberg, A. (2000) J. Bacteriol. 182, 225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurst H. & Kornberg, A. (1994) J. Biol. Chem. 269, 10996-11001. [PubMed] [Google Scholar]
- 14.Sundin G. W., Shanker, S., Chugani, S. A., Chopede, B. A., Kavanaugh-Black, A. & Chakrabarty, A. M. (1996) Mol. Microbiol. 20, 965-979. [DOI] [PubMed] [Google Scholar]
- 15.Stover C. K., Pham, X.-Q. T., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S. L., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 16.Shanker S., Kamath, S. & Chakrabarty, A. M. (1996) J. Bacteriol. 178, 1777-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chopede B. A., Shanker, S., Sundin, G. W., Mukhopadhyay, S. & Chakrabarty, A. M. (1997) J. Bacteriol. 179, 2181-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay S., Shanker, S., Walden, W. & Chakrabarty, A. M. (1997) J. Biol. Chem. 272, 17815-17820. [DOI] [PubMed] [Google Scholar]
- 19.Kamath S., Kapatral, V. & Chakrabarty, A. M. (1998) Mol. Microbiol. 30, 933-941. [DOI] [PubMed] [Google Scholar]
- 20.Rashid H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatnell P. J., Russell, N. J. & Gacesa, P. (1993) J. Gen. Microbiol. 139, 119-127. [DOI] [PubMed] [Google Scholar]
- 23.Hassett D. J. (1996) J. Bacteriol. 178, 7322-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyake T., Shiba, T., Kameda, A., Ihara, Y., Munekata, M., Ishige, K. & Noguchi, T. (1999) DNA Res. 6, 103-108. [DOI] [PubMed] [Google Scholar]
- 25.Akiyama M., Crooke, E. & Kornberg, A. (1993) J. Biol. Chem. 268, 633-639. [PubMed] [Google Scholar]
- 26.Krishnan P. S. (1952) Arch. Biochem. Biophys. 37, 224-234. [DOI] [PubMed] [Google Scholar]
- 27.Kritskii M. S., Chernysheva, E. K. & Kulaev, I. D. (1972) Biochemistry 37, 822-827. [Google Scholar]
- 28.Kumble K. & Kornberg, A. (1996) J. Biol. Chem. 271, 27146-27151. [DOI] [PubMed] [Google Scholar]
- 29.Sethuraman A., Rao, N. N. & Kornberg, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinsley C. R. & Gotschlich, E. C. (1995) Infect. Immun. 63, 1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa N., Tzeng, C.-M., Fraley, C. D. & Kornberg, A. (2000) J. Bacteriol. 182, 6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn K. & Kornberg, A. (1990) J. Biol. Chem. 265, 11734-11739. [PubMed] [Google Scholar]
- 33.Leammli U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa N., DeRisi, J. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]