Abstract
The National Institutes of Health Mammalian Gene Collection (MGC) Program is a multiinstitutional effort to identify and sequence a cDNA clone containing a complete ORF for each human and mouse gene. ESTs were generated from libraries enriched for full-length cDNAs and analyzed to identify candidate full-ORF clones, which then were sequenced to high accuracy. The MGC has currently sequenced and verified the full ORF for a nonredundant set of >9,000 human and >6,000 mouse genes. Candidate full-ORF clones for an additional 7,800 human and 3,500 mouse genes also have been identified. All MGC sequences and clones are available without restriction through public databases and clone distribution networks (see http://mgc.nci.nih.gov).
The gene content of the mammalian genome is a topic of great interest. While draft sequences are now available for the human (1, 2), mouse (www.ensembl.org/Mus_musculus), and rat (http://hgsc.bcm.tmc.edu/projects/rat) genomes, the challenge remains to correctly identify all of the encoded genes. Difficulty in deciphering the anatomy of mammalian genes is due to several factors, including large amounts of intervening (noncoding) sequence, the imperfection of gene-prediction algorithms (3), and the incompleteness of cDNA-sequence resources, many of which consist of gene tags of variable length and quality. Full-length cDNA sequences are extremely useful for determining the genomic structure of genes, especially when analyzed within the context of genomic sequence. To facilitate gene-identification efforts and to catalyze experimental investigation, the National Institutes of Health (NIH) launched the Mammalian Gene Collection (MGC) program (4) with the aim of providing freely accessible, high-quality sequences for validated, complete ORF cDNA clones. In this article, we describe our progress toward the goal of identifying and accurately sequencing at least one full ORF-containing cDNA clone for each human and mouse gene, as well as making these fully sequenced clones available without restriction.
Materials and Methods
cDNA Library Production.
MGC cDNA libraries were prepared from a diverse set of tissues and cell lines, in several different vector systems, by using a variety of methods. Vector maps and details of library construction are available at http://mgc.nci.nih.gov/Info/VectorMaps. The complete sequences for each of the MGC vectors can be found at http://image.llnl.gov/image/html/vectors.shtml. The catalog of MGC cDNA libraries can be accessed at http://mgc.nci.nih.gov.
Library Characterization.
Each new cDNA library initially was characterized by generating 5′ and 3′ ESTs (5) from ≈700 clones. The 3′ ESTs give information about the fraction of clones with polyadenylation sites and/or poly(A) tails, thereby providing an indication of the extent of inappropriate, internal priming that occurred during library construction. The 5′ ESTs give an indication of the likely frequency of full-ORF clones in each library, which we estimated by aligning the 5′ ESTs with the existing RefSeq collection (6) and assessing the fraction of alignments that overlap known translational start sites. At this stage, and subsequently during the generation of additional ESTs, the approximate gene diversity in each library was assessed by monitoring the number of distinct UniGene clusters (7) containing at least one EST from that library relative to the total number of generated ESTs.
Library Screening.
Each library deemed to be of high quality then was examined on a larger scale to identify candidate full-ORF cDNA clones for complete sequencing. First, 5′ ESTs were generated from ≈10,000 clones. After removal of recognizable contaminating sequences, these ESTs were deposited into dbEST; the associated cDNA clones for all of these characterized sequences are available through the I.M.A.G.E. consortium (http://image.llnl.gov). After analysis of these sequences, libraries found to be particularly useful for identifying unique, full-ORF clones were sequenced more deeply, generally in increments of 10,000 clones.
Identification of Putative Full-ORF cDNAs.
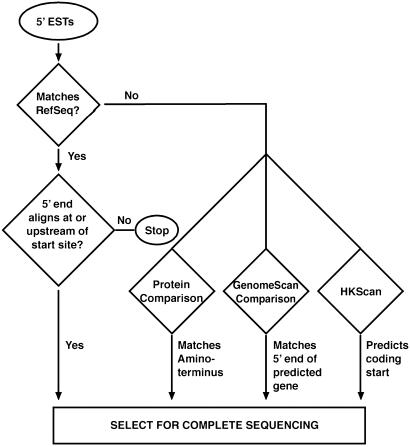
As described in Fig. 1, four tests were used to select candidate full-ORF clones starting from 5′ end sequences. One of these tests, hkscan, was developed specifically for the MGC program (S. Altschul and L. Wagner, unpublished results, using data kindly supplied by C. Burge). hkscan identifies all possible ORFs in a query sequence, allowing the possibility that the sequence is noncoding or that it is truncated at either the 5′ or 3′ end. For each candidate ORF, the hexamer frequencies of the putative coding and noncoding sequences are separately recorded and compared with known hexamer frequencies for coding and noncoding sequence. In addition, the putative coding sequence (CDS) start is compared with the Kozak consensus sequence. Applying Bayesian analysis to these data, a probability is estimated for each of the possible ORFs. These probabilities then are used to assess whether the query contains a transition from noncoding to coding sequence.
Fig. 1.
Tests for identifying putative full-ORF cDNA clones. In the first test, 5′ ESTs first were compared with all available ORF-complete mRNA sequences from the same organism (human or mouse) in the RefSeq collection. When a 5′ EST aligned (>95% homology for 100 or more base pairs) at or upstream of an annotated translation start site, that clone was considered to contain a candidate full-ORF cDNA. However, if the 5′ EST aligned downstream from an annotated translational start site, that clone was eliminated from consideration, although some of these may be full-ORF clones with an alternate 5′ translational start site. Any 5′ ESTs that did not match a RefSeq sequence were subjected to additional tests. In the second test, six possible frame translations were compared with the subset of GenBank protein records originating from Protein Information Resource (15), Protein Data Base (16), or SwissProt (17) that begin with methionine. This test identifies ESTs from genes with an N terminus similar but not identical to a known protein. Thus, in cases where a protein match (<90% identity but with an E value of less than or equal to 10−6) was detected and incorporated the known initiating methionine, the associated cDNA clone was considered a candidate to have a complete ORF. In the third test, we compared each 5′ EST to a collection of predicted genes derived from the human genome sequence by genomescan (18). When a 5′ EST aligned (95% identity for 100 or more bp) to a gene prediction that begins with ATG, the associated clone was considered a candidate. In the fourth test, we used the new program hkscan, which looks for evidence of a transition from noncoding to coding sequence (described in Materials and Methods).
Full-Length Sequencing.
Several different strategies were used for full-insert cDNA sequencing, including primer walking (http://www-shgc.stanford.edu/Seq/cdnapages/maincdna.html), transposon insertions (8, 9), and concatenated shotgun sequencing (10, 11). Importantly, the DNA sequence quality of all full-insert sequences produced by the MGC Program is extremely high. Each single contiguous sequence has no uncertain base calls (“N's”) and has an estimated average error rate of <1 in 50,000 bp.
Results and Discussion
cDNA Libraries.
More than 100 cDNA libraries, derived from a wide variety of tissues and cell lines and prepared by using several different vector systems and library-construction techniques (complete list at http://mgc.nci.nih.gov), were used to select the putative full-ORF clones. Some libraries were produced by standard methods, with the resulting cDNA clone frequencies approximately proportional to the transcript population in the cells used to make the library. In contrast, other libraries were constructed by using normalization (12) and/or size-selection methods that enhance the identification of large transcripts and transcripts expressed at lower levels. EST (5) sequences were generated to evaluate all cDNA libraries for gene diversity and proportion of full-ORF clones. Classification as full-ORF signifies that, as far as is possible to ascertain, the cDNA sequence contains a complete and authentic protein-coding sequence.
Identification and Characterization of Candidate Full-ORF Clones.
For this study, we identified and categorized genes based on the National Center for Biotechnology Information UniGene database (7). In UniGene, GenBank sequences are partitioned into a nonredundant set of clusters, where each cluster contains related sequences and aims to represent a unique gene. Within a cluster are transcripts of various lengths and alternatively processed transcript variants. The common feature linking the cDNAs and ESTs within a cluster is the 3′ sequence adjacent to the poly(A) tail. Because we characterized cDNAs by initially producing 5′ ESTs, in some cases these ESTs did not cluster within UniGene, as intervening sequence data connecting the 3′ and 5′ sequences was not available. For those cases, it was not possible to determine whether two nonoverlapping 5′ ESTs were derived from the same or from a different mRNA. For this and other reasons, we developed criteria to allow us to identify the subset of 5′ ESTs that likely originate at or upstream of the translational start site. Each 5′ EST was subjected to four tests, and clones deemed to be good candidates for having a full-ORF by at least one of the four tests were assigned a reliability score (Fig. 1). The score is based on the false-positive rates that were established for each test by comparing a known set of ESTs and the genes from which they were derived. The 5′ EST with the highest reliability score was selected from each cluster, and the corresponding cDNA clone then was completely sequenced.
When a fully sequenced clone was found to not contain a complete ORF, was found to be chimeric, was associated with a frameshift, or was incompletely processed, then another clone from that cluster was selected for complete sequencing. To date, the MGC Program has sequenced to “finished” standards 12,419 full-ORF human cDNA clones that correspond to 9,530 distinct genes, and 7,456 full-ORF mouse cDNA clones that correspond to 6,368 distinct genes. The MGC includes ≈1,300 human and 1,100 mouse full-ORF cDNA sequences that either did not previously exist or were represented only by partial cDNA sequences in GenBank. The complete inventory of clones and genes sequenced to completion by the MGC Program is available at http://mgc.nci.nih.gov/.
We analyzed the fully sequenced cDNA clones for the presence of complete ORFs by two approaches. First, we computationally translated all of the potential ORFs in each cDNA sequence and compared the resulting amino acid sequences to all proteins in GenBank. If the start codon of an ORF aligned with an initiating methionine of a GenBank protein, then that ORF was deemed to be the most likely one. If the sequence did not match that of a known protein, or did not align with a known initiating methionine, the most likely ORF was selected based on hexamer frequencies and the presence of a Kozak consensus sequence.
The Efficacy of MGC Clone Selection Algorithms.
Our initial generation of large sets of highly accurate cDNA sequences allowed us to study the efficacy of the algorithms we used for selecting candidate full-ORF clones. For each test, we identified the fraction of completely sequenced full-ORF MGC clones identified by that test, regardless of whether the clone is identified by any of the other tests. Also, we assessed each test's false positive rate by examining results for a set of 6,510 ESTs whose CDS-completeness was known. This test set is composed of one CDS-complete and one CDS-incomplete EST for each of 3,255 RefSeq (6) genes. RefSeq is the National Center for Biotechnology Information database that provides curated sequences for nucleic acids, including cDNAs, and proteins. genomescan identified 35% of the genes actually sequenced, with a false positive rate of 6%, from this test set, whereas hkscan identified 50% of genes, with a false positive rate of 23%. Protein comparisons alone identified 25% of genes actually sequenced, with a false positive rate of 5%. For each of these methods, adjusting the reporting threshold allows some control over the tradeoff between a higher rate of true positives and a lower rate of false positives.
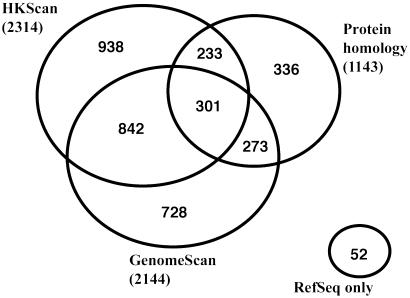
We also determined the performance of these three tests (genomescan, hkscan, and protein homology) in identifying genes matching existing human RefSeq sequences (Fig. 2). Although each test identified only a minority of the RefSeq-matching clones, they successfully identified >99% of the 5,653 RefSeq sequences among the initial set of MGC full-ORF clones when used in combination. Because genes not represented in RefSeq might have substantially different characteristics, such as a weaker similarity to known proteins, than those that are present, the comprehensiveness of these tests for identifying mammalian genes still needs to be established.
Fig. 2.
Efficacy of cDNA clone selection algorithms used by the MGC Program. Three of the tests (protein homology, genomescan, and hkscan), were retroactively assessed for their ability to identify full-ORF clones within a set of 5,653 established full-ORF RefSeq sequences. Only 301 of the RefSeq sequences were identified by all three of the tests, whereas 2,002 were identified by only one of the three tests. When used in combination, the three tests were effective in identifying 5,601 (>99%) of the RefSeq sequences.
Characteristics of the MGC Clones.
The availability of cDNA sequences, particularly from full-length cDNAs, greatly improves the quality of genome annotation, which otherwise is based on gene predictions and EST alignments. To gain insights to the value of MGC sequences for genome annotation, we chose a set of human MGC full-ORF clones that were unique full-ORF cDNAs at the time they were deposited within the National Center for Biotechnology Information RefSeq database. We compared these sequences with gene predictions from the International Protein Index (IPI) model protein set (1), which were derived before the MGC sequences were generated. Because gene models are identified in part by alignment of mRNA and genomic sequences, we did not want to compare the MGC clones to a set of IPI proteins that included MGC sequences. Therefore, we used only those novel cDNAs in the MGC set that we sequenced after the initial publication of the IPI set for this comparison. The genes represented by the sequenced MGC clones are, on average, 29% longer than those encoding the corresponding IPI predicted proteins. Moreover, for 34% of the MGC-unique full-ORF cDNAs, no corresponding IPI prediction was identified. Among the MGC full-ORF sequences are five [MGC:16635 (MGC unique identifier), BC009980 (GenBank accession no.); MGC:17507, BC011204; MGC:26816, BC022546; MGC:17330, BC011049; and MGC:10963, BC004346] that represent genes not annotated on the finished human chromosome 22 sequence (13), which has been carefully curated. Indeed, there are two MGC clones that are novel even with respect to the most current, unpublished annotation. [These annotation data (Release 3.1b, March 5, 2002) were produced by the Chromosome 22 Gene Annotation Group at the Sanger Institute (Hinxton, U.K.) and were obtained from http://www.sanger.ac.uk/HGP/Chr22.] These clones are BC001801, encoding a spliced expressed gene, and BC011679, encoding an unspliced mRNA with a putative 303-aa protein product. A version of this latter clone also has been sequenced independently by another full-insert cDNA sequencing project (http://cdna.ims.u-tokyo.ac.jp/). Moreover, five MGC clones (BC011362, BC014896, BC016737, BC025927, and BC029822) extend annotated genes on chromosome 22 by at least 80 nt. These findings demonstrate that full-length cDNA sequences result in the identification of novel transcripts and may help in improving existing gene models even in regions of the genome that have been extensively characterized, although the numbers of such new gene models will likely be modest.
The carefully annotated chromosome 22 sequence allows another means of estimating the completeness of the current MGC clone set. Of the 546 CDSs annotated on chromosome 22, 287 are present in MGC clones that are fully sequenced with an apparently full-ORF, and another 15% of these genes are in the current MGC pipeline. This evidence suggests that the current MGC collection consists of ≈52% of all genes, and that it will grow to 67% in the near future.
We also looked at the presence of conserved protein domains in novel cDNAs from the MGC program by recording the frequency of occurrence of strong (E value at most 1e−6) reverse psi-blast (14) matches to conserved protein domains in the SMART and Pfam datasets of conceptual translations. We found that 29% of these translation products have matches to known conserved domains, compared with 52% of the proteins that are not unique to the MGC collection. The smaller fraction of conserved domains in the MGC collection is not surprising, as the protein domains in SMART and Pfam are derived in part from known human genes. Therefore, conserved domains found in genes only recently sequenced may be underrepresented in SMART and Pfam. For example, BC004556 encodes a protein with strong matches to Drosophila, rat, and mouse genes for which the conserved domain (pfam03676) postdates and cites the MGC sequence submission. Therefore, in addition to novel mRNA and predicted protein sequences, the MGC sequences can be used to identify novel domains.
The MGC-unique full-ORF sequences include novel human members of important gene families. For example, reverse psi-blast comparison of these sequences with SMART or Pfam serine/threonine or tyrosine kinase domains, at an E value of 0.01, reveals three new candidate kinases, including MGC:22688, BC021666 (similar to serine threonine kinase 32); MGC:26673, BC022530 (member of the activin receptor-like family); and MGC:23665, BC015792. In addition, among the MGC clones are novel splice forms of previously known kinases, such as MGC:9320, BC016285 (similar to protein kinase, cAMP-dependent, catalytic, beta) and MGC:13661, BC012622. Both of these clones have previously unidentified 3′ terminal exons.
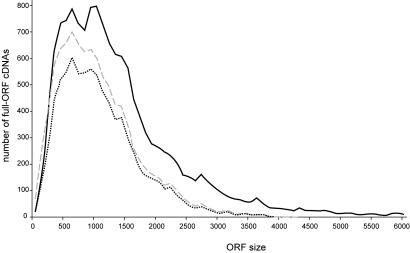
To assess the effectiveness of the current MGC strategy for generating full-ORF clones corresponding to a range of sizes, we compared the ORF distribution of human MGC full-sequenced clones with RefSeq (Fig. 3). Overall, MGC full-ORF clones have been generated for 57% of all human RefSeq sequences. However, as shown in Fig. 3, the MGC strategy has been most effective for ORFs that are <3 kb. Of the 14,161 RefSeq genes, 5,669 (40%) have ORFs of 1 kb or less. Of these RefSeq genes with ORFs of 1 kb or less, 4,188 (74%) have an MGC full-ORF clone. In the 1–3 kb ORF size range, there are 7,236 RefSeq genes, including 3,895 (54%) with an MGC full-ORF clone. However, for RefSeq genes with ORFs of >4 kb, only 120 of 1,256 (9%) have an MGC full-ORF clone. In addition, 65% of the MGC full-ORF clones not currently in RefSeq have ORFs of 1 kb or less.
Fig. 3.
ORF sizes of MGC full-ORF genes compared with RefSeq genes. The ORFs of MGC full-ORF genes and RefSeq genes were binned in 100-nt increments. The absolute numbers of MGC and RefSeq genes are compared for each size increment. RefSeq genes are represented by a solid line, the total of MGC genes is shown with the dashed lines, and MGC genes within the RefSeq set are depicted with dotted lines.
Future Directions of the MGC Program.
The goal of the MGC Program is to obtain a full-ORF cDNA sequence and clone for each human and mouse gene. Our production pipeline currently has putative full-ORF clones corresponding to several thousand additional human and mouse genes. Many of these clones were obtained from high-quality cDNA libraries prepared by standard protocols. The use of specialized approaches for constructing cDNA libraries, including size-selection, subtraction, and normalization, will likely help approach the goal of a full repertoire of human and mouse genes. However, alternative strategies, such as directed cloning based on known or predicted gene sequences, may be needed for constructing full-length cDNAs for genes in which application of the EST strategy has not been successful. The free availability of all these clones, both as in silico sequence and as easily procured clones, should be a boon to the public and private research communities. Furthermore, partnerships are now developing to transfer these cDNA collections to expression vectors for various applications in large-scale proteomics and systems biology, which will even further enhance the utility of this resource.
Supplementary Material
Acknowledgments
We express our gratitude to the following individuals for assistance: Ryan G. Martin, Carla R. Kowis, Vivienne Yoon, Hermela Loulseged, Sarah L. Norris, Shannon M. Lawrence, Natalie E. Walsham, Graham B. Scott, Keelan A. Hamilton, Peter R. Blyth, and Michelle C. Rives (Baylor Human Genome Sequencing Center); Rachel Dickhoff and Julia Greene (Institute for Systems Biology); Keith Wetherby, Russell Pearson, Nicole Dietrich, Peggy Kwong, and Stephen Granite (NIH Intramural Sequencing Center); Eidelyn Gonzalez and Chenier Caiole (Stanford Human Genome Center); J. Asano, S. Chan, N. Girn, R. Guin, R. Kustsche, S. Lee, K. MacDonald, C. Mathewson, T. Olson, P. Pandoh, A.-L. Prabhu, L. Spence, J. Stott, S. Taylor, K. Teague, M. Tsai, G. Yang, and S. Zuyderduyn (University of British Columbia Genome Sciences Centre); and C.B. Burge (Massachusetts Institute of Technology). We also thank Peter Good of the National Human Genome Research Institute and Daniela Gerhard of the National Cancer Institute for helpful discussions during the preparation of the manuscript. The Mammalian Gene Collection Program is an NIH interinstitute effort receiving financial and scientific support from the National Cancer Institute; National Center for Research Resources; National Eye Institute; National Human Genome Research Institute; National Heart, Lung, and Blood Institute; National Institute on Alcohol Abuse and Alcoholism; National Institute on Aging; National Institute of Allergy and Infectious Diseases; National Institute of Arthritis and Musculoskeletal and Skin Diseases; National Institute of Child Health and Human Development; National Institute on Deafness and Other Communication Disorders; National Institute on Drug Abuse; National Institute on Dental and Craniofacial Research; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of Environmental Health Sciences; National Institute of General Medical Sciences; National Institute of Mental Health; National Institute of Neurological Disorders and Stroke; and National Library of Medicine. The Program has received excellent guidance from members of the External Scientific Committee, including Barbara Wold, Philip Sharp, Geoffrey Duyk, Connie Cepko, Stewart Scherer, Lincoln Stein, Ronald Davis, and Edward Harlow.
Abbreviations
NIH, National Institutes of Health
MGC, Mammalian Gene Collection
CDS, coding sequence
IPI, International Protein Index
Data deposition: All MGC sequences have been deposited in the GenBank database (accession nos. can be found in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org) and can be accessed through the MGC web site (http://mgc.nci.nih.gov).
References
- 1.The International Human Genome Sequencing Consortium (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 3.Rogic S., Mackworth, A. K. & Ouellette, F. B. F. (2001) Genome Res. 11, 817-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strausberg R. L., Feingold, E. A., Klausner, R. D. & Collins, F. S. (1999) Science 286, 455-457. [DOI] [PubMed] [Google Scholar]
- 5.Adams M. D, Dubnick, M., Kerlavage, A. R., Moreno, R., Kelley, J. M., Utterback, T. R., Nagle, J. W., Fields, C. & Venter, J. C. (1992) Nature 355, 632-634. [DOI] [PubMed] [Google Scholar]
- 6.Pruitt K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler D. L., Church, D. M., Lash, A. E., Leipe, D. D., Madden, T. L., Pontius, J. U., Schuler, G. D., Schriml, L. M., Tatusova, T. A., Wagner, L. & Rapp, B. A. (2001) Nucleic Acids Res. 29, 11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevchenko Y., Bouffard, G. G., Butterfield, Y. S., Blakesley, R. W., Hartley, J. L., Young, A. C., Marra, M. A., Jones, S. J., Touchman, J. W. & Green, E. D. (2002) Nucleic Acids Res. 30, 2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butterfield Y. S., Marra, M. A., Asano, J. K., Chan, S. Y., Guin, R., Krzywinski, M. I., Lee, S. S., MacDonald, K. W., Mathewson, C. A., Olson, T. E., et al. (2002) Nucleic Acids Res. 30, 2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson B., Lu, J., Shen, Y., Wentland, M. A. & Gibbs, R. A. (1997) DNA Seq. 7, 63-70. [DOI] [PubMed] [Google Scholar]
- 11.Yu W., Andersson, B., Worley, K. C., Muzny, D. M., Ding, Y., Liu, W., Ricafrente, J. Y., Wentland, M. A., Lennon, G. & Gibbs, R. A. (1997) Genome Res. 7, 353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonaldo M. F., Lennon, G. & Soares, M. B. (1996) Genome Res. 6, 791-806. [DOI] [PubMed] [Google Scholar]
- 13.Dunham I., Shimizu, N., Roe, B. A., Chissoe, S., Hunt, A. R., Collins, J. E., Bruskiewich, R., Beare, D. M., Clamp, M., Smink, L. J, et al. (1999) Nature 402, 489-495. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C. H., Huang, H., Arminski, L., Castro-Alvear, J., Chen, Y., Hu, Z.-Z., Ledley, R. S., Lewis, K. C., Mewes, H. W., Orcutt, B. C., et al. (2002) Nucleic Acids Res. 30, 35-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., et al. (2002) Acta Crystallogr. D 58, 899-907. [DOI] [PubMed] [Google Scholar]
- 17.Gasteiger E., Jung, E. & Bairoch, A. (2001) Curr. Issues Mol. Biol. 3, 47-55. [PubMed] [Google Scholar]
- 18.Yeh R. F., Lim, L. P. & Burge, C. B. (2001) Genome Res. 11, 803-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.