Abstract
Telomerase is a ribonucleoprotein (RNP) required for maintenance of telomeres. Although up-regulated telomerase activity is closely linked to the cellular immortality characteristic of late stage carcinogenesis, recently, mutations in the telomerase RNA gene in humans have been associated with dyskeratosis congenita and aplastic anemia, both typified by impaired haemopoietic function. These mutations include base changes in a highly conserved putative telomerase RNA pseudoknot. Here, by using in vitro telomerase assays, NMR, and UV absorbance melting analyses of model oligonucleotides designed to form a “trans-pseudoknot,” we describe functional, structural, and energetic properties of this structure. We demonstrate that the pseudoknot domain exists in two alternative states of nearly equal stability in solution: one is the previously proposed pseudoknot formed by pairing P3 with the loop domain of P2b, and the other is a structured P2b loop alone. We show that the two-base mutation (GC107/8 → AG) present in one gene copy in a family with dyskeratosis congenita abrogates telomerase activity. This mutation hyperstabilizes the P2b intraloop structure, blocking pseudoknot formation. Conversely, when the P3 pseudoknot pairing is hyperstabilized by deleting a conserved bulge in P3, telomerase activity also decreases. We propose that the P2b/P3 pseudoknot domain acts as a molecular switch, and interconversion between its two states is important for telomerase function. Phylogenetic covariation in the P2b and P3 sequences of 35 species provides a compelling set of “natural” compensatory base pairing changes supporting the existence of the crucial molecular switch.
Eukaryotic chromosome stability requires maintenance of telomeric DNA by the ribonucleoprotein (RNP) telomerase (1–3). Telomerase is activated in >85% of malignant human tumors and is closely linked to cellular immortality, a step in carcinogenesis. It is also active in some self-renewing tissues with high regenerative potential, although repressed in other somatic adult tissues (4, 5). Telomerase inactivity contributes to cellular senescence (5). Mutations in the human telomerase RNA gene have recently been associated with dyskeratosis congenita and aplastic anemia (6, 7), both being characterized by haemopoietic function losses. Core components of telomerase are its reverse transcriptase subunit and telomerase RNA (called hTER or hTR in humans), which contains a short template sequence copied into telomeric DNA by telomerase, and other essential domains. One highly conserved domain is a putative pseudoknot required for telomerase activity and assembly of stable RNP (8–10) (Fig. 1a).
Fig 1.
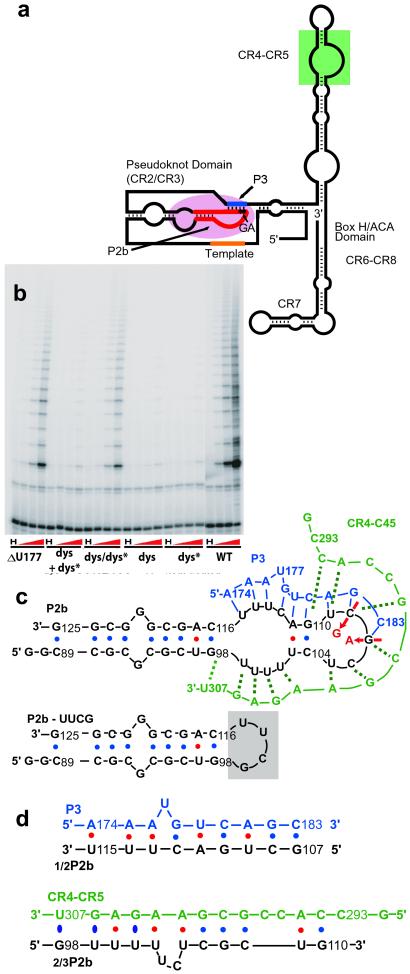
Proposed vertebrate telomerase RNA secondary structure and model oligonucleotide systems studied. (a) Schematic diagram of putative core secondary structure (8). CR2 to CR8 are conserved regions. The pseudoknot domain is formed between CR2, the long stem-loop that includes P2b, and CR3, which includes P3. (b) Representative telomere repeat amplification protocol assay of telomerase reconstituted in vivo in VA13 human cells, from indicated hTR mutants. “H” denotes that telomerase was inactivated by heat (80°C, 10 min), dys + dys* contains both RNA species, and dys/dys* contains RNA with both dyskeratosis and P3 compensatory mutations in the same molecule. (c) P2b sequence (black). Residues 99–115 comprise loop J2b/3 in hTR. Putative pseudoknot formed with P2b and P3 (blue) and a possible pairing with CR5 (green). Residue numbers are from full-length hTR. Two mutations in one family with dyskeratosis, G107A and C108G, are shown by red and arrows. P3-mutant deletes the putative U177 bulge in P3. (d) Small oligonucleotides comprising the P2b moiety proposed to base pair with P3, 1/2P2b, and control 2/3P2b with CR5.
Telomerase function requires the pseudoknot according to phylogenetic studies and in vivo and in vitro mutagenic studies (10, 11); the specific sequence is also important (8–10). Enzymatic and chemical probing of ciliate telomerase RNA in vitro suggested that the pseudoknot is dynamic (11). We present here direct biophysical evidence of the secondary structure, base pairing, and overall topology of the core of the pseudoknot and measurement of its thermodynamics. Effects of the G107A, C108G (GC107/8 → AG) hTR mutation in stem-loop “P2b” that causes dyskeratosis congenita in one family (6) provides the first biophysical explanation of the molecular basis for the disease.
Materials and Methods
RNA Samples.
Oligonucleotides P2b, “P2b-dys,” and “P2b-UUCG” were synthesized in vitro by using T7 RNA polymerase. Each had a dangling 5′ G to improve transcription yields, but otherwise was identical to the natural hTR sequence. Samples were prepared as described (12). Sample conditions were 50 mM NaCl, 10 mM sodium phosphate (pH 6.4) and 0.1 mM EDTA. Oligonucleotides “P3” (wild-type), “P3-mutant,” “CR5,” “1/2P2b,” and “2/3P2b” were chemically synthesized by Dharmacon Research. Complexes were incubated at 55°C with the preannealed P2b stem-loop for 1 h. NMR samples generally had RNA at 1–2 mM, but some were at 275 μM. Samples contained 5 mM MgCl2 for native gels or 1 mM MgCl2 for UV melting. CD revealed no effect of Mg.
In Vivo Reconstituted Telomerase Activity.
Effects of hTR mutations on telomerase activity were analyzed with telomerase assembled in vivo in VA13 immortalized human fibroblast cells, in which neither human telomerase enzyme reverse transcriptase (hTERT) and hTR gene products nor endogenous telomerase activity are detectable (13). Six micrograms each of hTR constructs containing different P3 mutations were transfected, together with 6 μg of hTERT constructs, into 4 × 106 VA13 cells by using Superfect (Qiagen, Valencia, CA). Telomerase activity was assayed 48 h later; telomere repeat amplification protocol (TRAP) assays were carried out by using the TRAPeze kit (Intergen, Purchase, NY) following the manufacturer's directions except for a three-step PCR: 94°C for 10 s, 55°C for 30 s, 72°C for 30 s, 25 cycles. The TRAP reaction product was analyzed by 10% denaturing PAGE followed by phosphoimaging.
UV Melting.
UV melting studies were performed on a Cary 3E UV-Visible spectrometer using heating rates of 0.25–1.6°C per min with 0.25–0.5°C per data point, on 2.4 μM RNA. To address potential linear duplex formation, melting temperatures were determined at 2.75 and 275 μM RNA. Results were independent of RNA concentration indicating monomeric behavior. Thermodynamic parameters were determined by fit to different models using the origin program (OriginLab, Northampton, MA): sequential one-step (A ⇔ B) or two-step (A ⇔ B ⇔ C) models.
NMR Spectroscopy.
All experiments were performed on Varian INOVA 600 MHz and Bruker 500 MHz NMR spectrometers. Data were processed with NMRPipe (14) and analyzed by using sparky (University of California, San Francisco). The one-dimensional (1D) spectra used jump-and-return and presat sequences for H2O and D2O solutions, respectively. Typically, two-dimensional (2D) NMR data were apodized in each dimension with a phase-shifted sine-bell or Gaussian function, followed by zero-filling twice, before Fourier transformation.
H2O-NOESY spectra were recorded at 1, 5, 15, and 20°C, with mixing times of 75, 150, and 250 ms. For water suppression, Watergate or the s136g shape pulse were used on Bruker or Varian spectrometers, respectively. D2O-NOESY and total coherence spectroscopy spectra were acquired at 600 MHz at 5, 20, and 30°C by using mixing times of 80 and 50 ms, respectively. DQF-COSY spectra were acquired at 20°C.
Two types of natural-abundance 13C NMR experiments were performed: heteronuclear multiple quantum coherence NMR (HMQC) at 600 MHz (20 and 25°C) and CT-HSQC at 500 MHz (25°C). Water 15N-HMQC spectra were acquired at 15°C on {15N,13C-U}-labeled P2b samples at 600 MHz.
Results
Effect of Mutations on Telomerase Activity.
The proposed secondary structure of hTR contains eight phylogenetically conserved regions, CR1–CR8 (8). CR2 and CR3 fall within the putative pseudoknot (Fig. 1a). Stem P3 of the pseudoknot contains conserved residues in the P2b loop (region J3/2b, nucleotides 99–115 of CR2) and its paired CR3 residues (8). The GC107/8 → AG dyskeratosis congenita (6) mutation in CR2 thus both disrupts predicted base pairing of CR3 with P2b and alters a conserved primary sequence (Fig. 1b). To understand the mechanism by which correct folding of the pseudoknot affects telomerase activity and hence cellular proliferation, we first examined the activity by using hTR constructed with mutations in the pseudoknot. Mutant “dys” corresponds to mutation GC107/8 → AG (Fig. 1 a and b). Complementary bases in the other strand of the P3 pairing were mutated in the dys* mutant, and dys/dys* combined the two mutated regions in the same RNA molecule, thus restoring the predicted base pairing potential of the pseudoknot. The mutant “ΔU” has U177 in the P3 pairing region deleted (Fig. 1 b and c). In the proposed secondary structure (8), unpaired U177 forms a bulge. The presence and position of this single-base bulge (but not its base identity) is conserved in vertebrates (8). The dys and dys* mutations each reduced telomerase activity >100-fold, whereas dys/dys* partially restored activity, compared with wild-type hTR (Fig. 1b). The ΔU mutation reduced telomerase activity roughly 10-fold (Fig. 1b and data not shown).
Oligonucleotides.
To elucidate the structural basis of the mutation effects on telomerase activity, we studied oligonucleotide structures and interactions: P2b, the wild-type sequence of the stem-loop formed by stem P2b and loop J2b/3 (8) (residues 89–125); P2b-dys, the dyskeratosis sequence with the GC107/8 → AG double mutation (6); and, to aid in assigning NMR resonances, P2b-UUCG, in which a UUCG tetraloop replaces the 17-nucleotide loop of P2b (Fig. 1c). P3 (blue, Fig. 1c) corresponds to CR3 moiety predicted from phylogenetic analysis to form the pseudoknot by base pairing with loop J2b/3. In P3-mutant, bulged U177 is deleted. The control, CR5 (green, Fig. 1c; nucleotides 293–307) encompass the CR5 sequence. CR5 could potentially form a similar number of base pairs with the P2b loop as P3, and thus was chosen rather than an arbitrary oligonucleotide. For assignments of base pairs formed between P2b and P3 and to study the stability of this specific interaction per se, we used the P2b loop sequence proposed to base pair with P3 (1/2P2b; Fig. 1d) and the portion of the P2b loop that could potentially interact with control CR5 (2/3P2b; Fig. 1d).
Gel Electrophoresis.
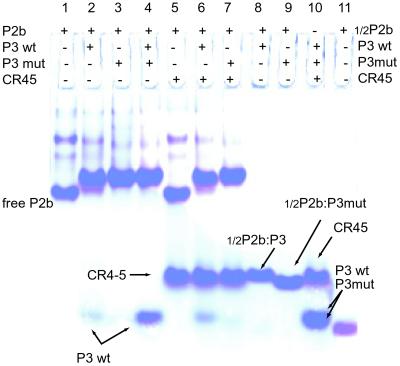
P3 and P2b interact in trans to form a heterodimer (“trans-pseudoknot”). We found direct biophysical evidence that this pseudoknot involves, as previously proposed (8), base pairing between CR3 and its complement in J2b/3. In nondenaturing polyacrylamide gel electrophoresis at a 1:1 molar ratio, P2b and P3 form a stable complex with lower electrophoretic mobility than free P2b (Fig. 2, lanes 1 and 2). P3-mutant also stably associates with P2b (Fig. 2, lane 3). Competition assays with equimolar ratios of P2b, P3, and P3-mutant reveal that P3-mutant binds more tightly to P2b (Fig. 2, lanes 3 and 4) in accord with its higher melting temperature (see below). The P2b–P3-mutant heterodimer gives sharper bands than the P2b–P3 heterodimer, suggesting greater conformational flexibility of the wild-type complex. The gel exhibits no stable association between P2b and CR5 nor does CR5 interfere with the association of P2b and P3 or P3-mutant (Fig. 2, lanes 6 and 7). The short “1/2P2b:P3” and “1/2P2b:P3-mutant” complexes have expected mobilities (Fig. 2, lanes 8, 9, and 11). CR5, P3, and P3-mutant sequences do not associate with each other (Fig. 2, lane 10) nor do any of them adopt a structure (data not shown).
Fig 2.
Complex between P2b and P3 oligonucleotides in nondenaturing polyacrylamide gels. Lane 1, free P2b. Lane 2, P2b and P3 (1:1), with slight P3 excess providing a marker. Lane 3, P2b and P3-mutant (1:1). Lane 4, P3 and P3-mutant competing for binding to P2b (1:1); wild-type P3 is outcompeted. Lane 5, P2b and CR5 (1:1). Lane 6 shows association of P2b and P3, and lane 7 shows association of P2b and P3-mutant in the presence of CR5. Lane 8, 1/2P2b with P3. Lane 9, 1/2P2b with P3-mutant. Lane 10, P3, P3-mutant, and CR5 (1:1:1). P3-mutant runs slightly faster than P3; both bands overlap at high RNA concentrations. Lane 11, free 1/2P2b.
UV and NMR Melting Studies.
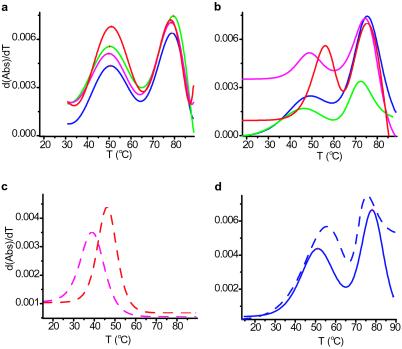
Stem-loop P2b has two ordered domains, and its loop region is internally structured, as shown by UV melting experiments and NMR. Derivatives of UV melting curves reveal two distinct transitions, one at ≈50°C (Tm1) and one at ≈79°C (Tm2). Similarly, the imino proton region in 1D NMR spectra shows two distinct sets of resonances, one that broadens at 35°C and a second set that broadens above 50°C. The first transition corresponds to melting of the loop, which is ordered (see below), and the second transition to melting of the GC-rich stem (Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). The same two melting temperatures were observed over a >100-fold RNA concentration range (2.4–275 μM). This finding is consistent with the above nondenaturing gel electrophoresis results. Note that the loop of the free P2b-dys oligonucleotide melts at higher temperature than wild-type: Tm1 of 54°C vs. 50°C (Fig. 3d). This finding independently indicates that bases in the loop participate in specific interactions.
Fig 3.
Comparison of the relative stabilities of P2b, P2b-dys, and the “trans-pseudoknot.” (a) Derivative of UV melting curves for P2b (blue; Tm = 50.6°C, 78.8°C), P2b:P3 (magenta; Tm = 49.9°C, 78.5°C), P2b:P3-mutant (red; Tm = 51.0°C, 78.5°C), and P2b:CR5 (green; Tm = 50.3°C, 79.5°C) in equimolar ratios, each oligonucleotide at 2.4 μM. (b) Same as in a but RNA at 275 μM. P2b (Tm = 48.7°C, 77.7°C), P2b:P3 (Tm = 46.1°C, 75.5°C), P2b:P3-mutant (Tm = 58.6°C, 79.2°C), and P2b:CR5 (Tm = 46.0°C, 73.4°C). (c) Short double-stranded RNA constructs 1/2P2b:P3 (dashed magenta; Tm = 38.7°C) and 1/2P2b:P3-mutant (dashed red; Tm = 46.0°C). (d) Comparison of melting curve derivatives for P2b (blue, Tm = 50.6°C, 78.8°C) and P2b-dys (dashed blue, Tm = 54.2°C, 77.0°C). After dialysis against NMR buffer, 1 mM MgCl2 was added in each case, resulting in a lower Mg-to-RNA ratio for the high RNA concentration samples. Notice in a and b a lower Tm at a high concentration for each P2b domain.
To form Watson–Crick base pairs with P3, internal base pairs of the P2b loop must melt. Therefore we expected the P3:P2b interaction to have a melting temperature at least comparable to Tm1. These “trans ” interactions with P2b were analyzed with the model oligonucleotides via UV melting curves (Fig. 3). At 2.4 μM RNA, the melting temperature (Tm1) of the internal P2b loop is ≈50°C (Fig. 3a, blue curve). At 275 μM RNA, Tm1 and the melting temperature for the trans complex P2b:P3 are approximately the same (Fig. 3b). The melting temperature of the P2b:P3-mutant trans complex is ≈10°C higher (Fig. 3b). RNA concentration of 275 μM is the same as in our low concentration NMR and nondenaturing gel electrophoresis studies, where the trans-pseudoknot forms. Hence, deleting the bulged U177 stabilizes P3 pairing in the trans-pseudoknot, although it reduces telomerase activity ≈10-fold (Fig. 1b). Thermodynamic parameters obtained via 1/2P2b:P3 and 1/2P2b:P3-mutant melting manifest the extra stability gained by deleting U177 (Fig. 3c). For the mixture of P2b and CR5, the melting curve is broader than with the P3:P2b complexes. In control analyses, 1D NMR and UV melting indicate that P3 and P3-mutant are completely unstructured, and that CR5 makes only one detectable internal base pair at 5°C. Because Tm1 for free P2b and Tm for P2b:P3 are within experimental error, both systems must have comparable stabilities under our experimental conditions.
NMR Spectroscopy.
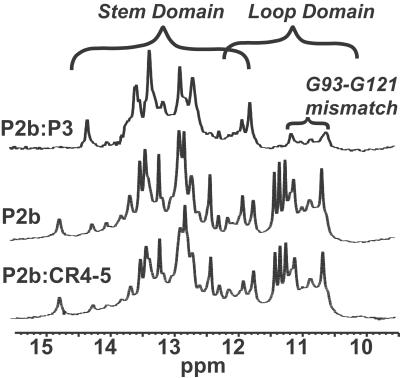
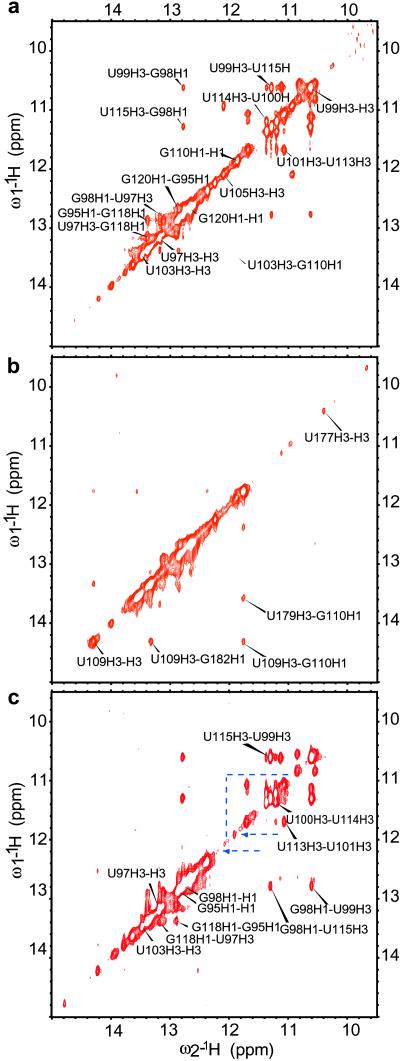
Seven of the U residues in the P2b loop are in a well defined local structure. NMR spectra exhibit several imino proton resonances at 10.5–11.5 ppm, characteristic of U residues structured but not in canonical Watson–Crick base pairs (Fig. 4). Comparison with P2b-UUCG enabled identification of proton imino, amino, and adenine H2 resonances from the stem and confirmed that all resonances between 10.5 and 11.5 ppm correspond to the loop, except for broad signals at 11.12, 10.82, 10.61, and 10.53 ppm from the G93/G121 mismatch in the stem (Figs. 4 and 5a). NOESY imino proton signals from the U-rich loop give several cross-peaks indicative of a well defined structure. The AU base pair, all Cs and the GG mismatch in the P2b stem were identified by comparing P2b with P2b-UUCG (Fig. 9, which is published as supporting information on the PNAS web site). This tetraloop construct is closed by GC base pair G98–C116, but otherwise conforms to well-characterized U1U2C3G4 tetraloops (15–19). The H2 resonance of adenine 117 gives clear inter- and intrastrand connectivities that helped assign the closing base pair of the stem of P2b, G98C116, and base pairs on the 5′ side of the A117U97 pair (Figs. 9 and 10, which are published as supporting information on the PNAS web site). Heteronuclear multiple quantum coherence NMR spectra of a P2b sample double-labeled with 13C and 15N specifically in all uridine residues, confirmed that the first three pairs of Us in the loop form noncanonical base pairs. H2O-NOESY spectra of P2b exhibit clear connectivities between the G98 imino proton, and the noncanonical U99U115 base pair in the loop. A clear pattern of cross-peaks connects all three UU base pairs in the loop: U99U115, U100U114, and U101U113. We also assigned base pairs A111U103 and G110C104 (Fig. 5a; see also Figs. 11–13, which are published as supporting information on the PNAS web site). P2b has a structured internal loop and a 5-residue terminal loop.
Fig 4.
Association of P2b and P3 monitored by NMR. Imino proton region of P2b, P2b:P3, and P2b:CR45.
Fig 5.
Formation of a pseudoknot in trans monitored by NOESY. Imino proton region for P2b (a) and P2b:P3 (b) in 1:1 ratio, showing disappearance of internal loop cross-peaks when P2b associates with P3 and appearance of a new set of cross-peaks. (c) Dyskeratosis mutant of P2b. The three UU base pairs of the loop give cross-peaks identical to P2b. Other cross-peaks disappear, indicated by blue dashed lines.
1D NMR experiments confirm that P2b and P3 interact. Imino proton resonances from the P2b loop disappear on binding P3 (Fig. 4), so the interaction involves bases in the loop. In contrast, imino protons for the ordered loop of theP2b plus CR5 sample are unchanged, although other resonances show some subtle changes relative to free P2b, suggesting nonspecific or transient interactions (NMR spectra in D2O; Fig. 11, see also Figs. 14 and 15, which are published as supporting information on the PNAS web site). Also, peaks of the P2b:P3 complex and the P2b plus CR5 sample are uniformly broadened relative to free P2b from increased size. These observations indicate some interaction between P2b and CR5 that does not involve canonical base pairing with the loop. Without additional stabilizing factors, interaction of J2b/3 with CR4-CR5 seems unlikely to occur in vivo, because in the context of the full-length hTR, the CR5 sequence spans two stable phylogenetically conserved helices that would both have to melt: helix P6 (8) and stem-loop P6.1 (20). Stem-loop P6.1 has a Tm of 55°C under conditions used in this work (data not shown).
Imino proton diagonal- and cross-peaks in NOESY spectra from the P2b loop entirely disappear with P3 added, whereas new imino-to-imino cross-peaks appear (Fig. 5 a and b and Fig. 12). The heterodimer formed between 1/2P2b and P3 approximates the base pairing of P3 and the P2b loop and was used to assign base pairs formed between P2b and P3 (Fig. 12). This heterodimer behaves as a typical double-stranded RNA in native gels (Fig. 2, lanes 8, 9, and 11), UV melting (see below), and proton 2D NMR. The NOESY imino proton region allows the typical sequential connectivity walk in dsRNA, except for base pairs involving the three A's at the 5′ end of P3. Evidence that at least two of these A's form base pairs comes from the existence of downfield imino signals and their characteristic cross-peaks with adenine H2 peaks.
P2b-dys failed to associate with P3 in nondenaturing gel electrophoresis (data not shown). This finding was confirmed by NMR (Figs. 16 and 17, which are published as supporting information on the PNAS web site). The NOESY imino proton cross-peak patterns in free P2b and free P2b-dys are distinctively different (Fig. 5 a vs. c). In P2b-dys, the three UU base pairs in the loop still form, giving rise to exactly the same resonances as in wild-type P2b. However, in P2b-dys GU and GC base pairs in the loop disappear. Different secondary structure underlies the different internal stabilities of the P2b and P2b-dys loops. NMR spectra of these constructs show that the dys mutation prevents pseudoknot formation not only because of loss of two base pairs in the P3 pairing, but also because the P2b loop becomes locked into a novel, hyperstable internal loop structure (Supporting Text, which is published as supporting information on the PNAS web site).
Phylogenetic Sequence Covariation.
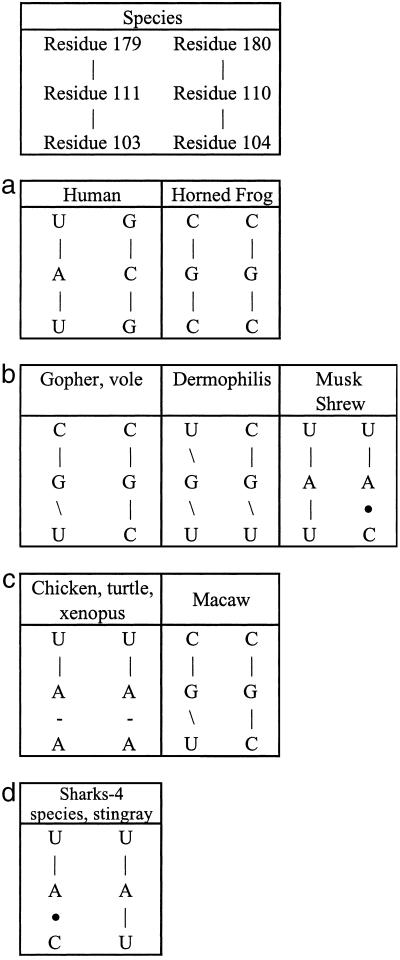
Independent support for the molecular switch is evident in the sequence covariation of 35 published vertebrate telomerase RNA sequences (8) at all three doublets of bases corresponding to the Watson–Crick base pairs in the P2b intraloop and paired P3 of the human sequence (Fig. 6). In each case, sequence variations in P3 that would modify (generally lower) the stability of the P3–P2b interaction are accompanied by base substitutions in P2b with compensatory stability changes of those base pairs in the P2b intraloop. This is a very convincing set of “natural” compensatory base pairing changes and is completely in accord with the notion of the crucial molecular switch.
Fig 6.
Species covariation of residues forming Watson–Crick base pairs in the human sequence of the molecular switch. (a) Watson–Crick base pairs in both P3 (Upper) and P2b intraloop (Lower) interactions. (b) Lower stability in the P3-P2b interaction is more than compensated by lower stability in the P2b intraloop. (c) P3 has a bulged C, rather than a bulged U, and consequently lower stability to balance the loss of Watson–Crick base pairing in the P2b intraloop. (d) P3 has a bulged A, rather than a bulged U, plus an extra bulge with consequently lower stability to balance the loss of Watson–Crick base pairing for the P2b intraloop.
Discussion
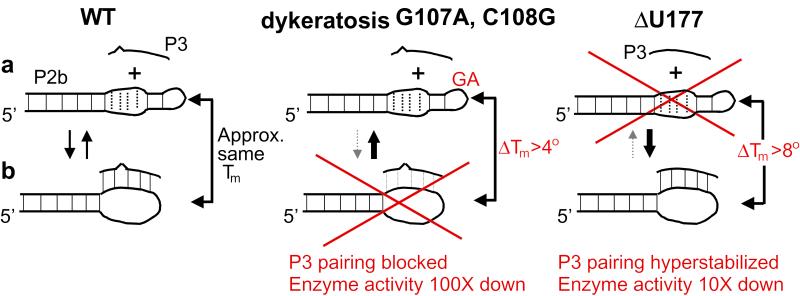
Here we have shown that the naturally occurring human dyskeratosis disease mutant GC107/8 → AG hTR cannot form the pseudoknot with P3 and has lost nearly all in vitro telomerase activity. Enzyme activity is partially restored by compensatory base mutations that restore this pseudoknot pairing. A four-base deletion of residues 110–113, which disrupts the P3 pairing, has recently been associated with human aplastic anemia (7). These in vitro and in vivo mutant phenotypes, together with other data (9, 10), demonstrate the importance of the ability to form a pseudoknot. However, deleting the bulged U177 in P3, which hyperstabilizes the P3 pairing interaction, also caused a significant reduction in telomerase activity. We conclude that an ultrastable pseudoknot also impairs function, and propose that the P2b loop must be able to assume both its P3 paired and internally stabilized conformational states.
The phylogenetic sequence covariation we found provides a compelling set of natural compensatory base pairing changes completely in accord with the molecular switch between P2b stabilized by a “loop” with internal base pairs and, after breaking those internal base pairs, P2b base paired with P3.
Fig. 7 illustrates the two proposed alternative states of the conserved pseudoknot in telomerase function. In the “open” state (Fig. 7a), there are two well-defined, highly ordered domains: the J2b/3 loop and the P2b stem. In the alternative “closed” state (Fig. 7b), J2b/3 bases pair with P3 in forming the pseudoknot, losing their internal noncanonical and Watson–Crick base pairing, because all of the U-rich loop proton imino signals disappear. Under our experimental conditions, the stability of the P3 pairing is comparable to the stability of the loop J2b/3 internal structure. Because the dys mutation increases the stability of the internal loop structure while simultaneously decreasing the number of potential P3 base pairs, the pseudoknot cannot form. Hence, only the open state is achieved by the dys mutant. Conversely, deletion of U177 impairs telomerase function because it increases the stability of the pseudoknot and hence blocks conversion to the open state. If the wild-type pseudoknot was static, present throughout the functional cycle of telomerase, one would expect the U177 deletion to cause little change in telomerase activity. Previous in vitro studies of the counterpart pseudoknot in purified Tetrahymena telomerase RNA showed patterns of protection that were proposed to result from the presence of comparable open and closed states (21). Our results are also consistent with chemical and enzymatic probing of human telomerase RNA in vitro and in vivo (22). In vivo, other factors might stabilize either or both forms.
Fig 7.
A human telomerase RNA molecular switch mediated by the pseudoknot domain. In this model, two states are equally important: (a) in the open state of wild-type human telomerase RNA, the J2b/3 loop is free but internally structured; (b) in the alternative closed state, loop residues base pair with P3. The stability of the P3 interaction is comparable to the stability of the internal structure of loop J2b/3, so both states can interconvert. The dyskeratosis mutation G107A, C108G increases loop stability while simultaneously decreasing the potential number of P3 base pairs, so the pseudoknot does not form. Deletion of U177 hyperstabilizes the pseudoknot. Both mutations impair telomerase activity.
The strong sequence covariation of the P3-P2b pairing is consistent with a need for balance between the open and closed states of the pseudoknot region to be finely controlled for optimal telomerase functioning. Interconversion between the two states is also predicted to be highly sensitive to sequence context and arrangement within the paired region, further explaining why restitution of P3 base pairing in the dys/dys* mutant and in other P3 pairing mutants (9, 10, 23, 24) only partially restores telomerase activity (Fig. 1b). Like RNAs of other complexes such as the spliceosome, telomerase RNA appears to be a dynamic molecule whose function requires it to assume more than one secondary conformation.
Supplementary Material
Acknowledgments
We thank Drs. Nikolai B. Ulyanov and Dudy Tzfati for valuable discussions, and Drs. Jinghua Yu and Zhihua Du for advice on NMR. This work was supported in part by National Institutes of Health grants (to E.H.B. and T.L.J.).
Abbreviations
RNP, ribonucleoprotein
hTR, human telomerase RNA
References
- 1.Blackburn E. H. (2001) Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E. H. (2000) Nat. Struct. Biol. 7, 847-850. [DOI] [PubMed] [Google Scholar]
- 3.Collins K. (2000) Curr. Opin. Cell Biol. 12, 378-383. [DOI] [PubMed] [Google Scholar]
- 4.Bryan T. M. & Cech, T. R. (1999) Curr. Opin. Cell Biol. 11, 318-324. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E. H. (2000) Nature 408, 53-56. [DOI] [PubMed] [Google Scholar]
- 6.Vulliamy T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P. J. & Dokal, I. (2001) Nature 413, 432-435. [DOI] [PubMed] [Google Scholar]
- 7.Vulliamy T., Marrone, A., Dokal, I. & Mason, P. J. (2002) Lancet 359, 2168-2170. [DOI] [PubMed] [Google Scholar]
- 8.Chen J. L., Blasco, M. A. & Greider, C. W. (2000) Cell 100, 503-514. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Rivera L. & Blasco, M. A. (2001) J. Biol. Chem. 276, 5856-5865. [DOI] [PubMed] [Google Scholar]
- 10.Bachand F., Triki, F. & Autexier, C. (2001) Nucleic Acids Res. 29, 3385-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilley D. & Blackburn, E. H. (1999) Proc. Natl. Acad. Sci. USA 96, 6621-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mujeeb A., Clever, J. L., Billeci, T. M., James, T. L. & Parslow, T. G. (1998) Nat. Struct. Biol. 5, 432-436. [DOI] [PubMed] [Google Scholar]
- 13.Reddel R. R., Bryan, T. M. & Murnane, J. P. (1997) Biochemistry (Moscow) 62, 1254-1262. [PubMed] [Google Scholar]
- 14.Delaglio F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 15.Molinaro M. & Tinoco, I. (1995) Nucleic Acids Res. 23, 3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams D. J. & Hall, K. B. (2000) J. Mol. Biol. 297, 1045-1061. [DOI] [PubMed] [Google Scholar]
- 17.Akke M., Fiala, R., Jiang, F., Patel, D. & Palmer, A. G. (1997) RNA 3, 702-709. [PMC free article] [PubMed] [Google Scholar]
- 18.Allain F. H. & Varani, G. (1995) J. Mol. Biol. 250, 333-353. [DOI] [PubMed] [Google Scholar]
- 19.Fourmy D., Yoshizawa, S. & Puglisi, J. D. (1998) J. Mol. Biol. 277, 333-345. [DOI] [PubMed] [Google Scholar]
- 20.Chen J. L., Opperman, K. K. & Greider, C. W. (2002) Nucleic Acids Res. 30, 592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya A. & Blackburn, E. H. (1994) EMBO J. 13, 5721-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antal M., Boros, E., Solymosy, F. & Kiss, T. (2002) Nucleic Acids Res. 30, 912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachand F. & Autexier, C. (2001) Mol. Cell. Biol. 21, 1888-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Autexier C., Pruzan, R., Funk, W. D. & Greider, C. W. (1996) EMBO J. 15, 5928-5935. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.