Abstract
Enterohemorrhagic Escherichia coli O157 is one of the leading worldwide public health concerns, causing large outbreaks of hemorrhagic colitis as well as numerous small outbreaks and sporadic cases. The variability of restriction enzyme-digestion patterns of O157 genomes, which is widely used to distinguish strains in the molecular epidemiology of O157 infections, suggests the presence of some genomic diversity among the strains. Based on the complete genome sequence of O157 Sakai, we analyzed the whole genome structures of eight O157 strains displaying diverse XbaI-digestion patterns by a systematic PCR analysis that we have named whole genome PCR scanning. This analysis identified not only the O157-specific sequences that are highly conserved among the strains, but also revealed an unexpectedly high degree of genomic diversity. In particular, prophages, including Shiga toxin-transducing phages, exhibited extensive structural and positional diversity, implying that variation of bacteriophages is a major factor in generating genomic diversity among the O157 lineage.
Strains of Escherichia coli are genomically and phenotypically highly heterogeneous. Most are commensal inhabitants of the intestinal tract, but various pathogenic strains also exist. Among the pathogenic strains, enterohemorrhagic E. coli (EHEC), O157 is a major worldwide public health concern. It frequently causes large outbreaks of hemorrhagic colitis, as well as numerous small outbreaks and sporadic cases (1). We have recently determined the complete genome sequence of O157 strain RIMD 0509952 (referred to as O157 Sakai), and compared the sequence with a nonpathogenic E. coli strain, K-12 MG1655 (2, 3). The comparison revealed that a total of 4.1-Mb chromosome sequence is conserved in the two strains, though it is interrupted by numerous strain-specific DNA sequences of various sizes (referred to as S-loops for O157 Sakai and K-loops for K-12; details are available at http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/) (2). Because the two strains are distantly related (4), it was assumed that the 4.1-Mb conserved sequence largely represents the chromosome backbone of E. coli. The lengths of strain-specific loops total 1.4 Mb and 0.5 Mb for O157 and K-12, respectively. Numerous virulence-related genes are encoded on S-loops, but a few on K-loops. Importantly, many of the strain-specific loops are of foreign origin, implying that acquisition of a large amount of foreign DNA by means of horizontal gene transfer contributed to generating the genomic diversity of E. coli (2, 5). The presence of a large number of prophages and prophage-like elements in O157 as well as in K-12 suggested that bacteriophages have played key roles in the process (2, 6). In O157 Sakai, 18 prophages (Sp1–18) and 6 prophage-like elements (SpLE1–6) were identified, comprising about two-thirds of the Sakai-specific sequences. Among the 18 prophages, 13 were lambda-like phages resembling each other, and they included Shiga toxin-transducing phages (Stx phages). Various DNA segments with almost identical sequences were shared by these lambda-like phages, suggesting that recombination and duplication of these phages may have occurred in the evolution of O157 (6). If such genetic events frequently occurred, there must exist a certain level of genomic diversity in the O157 lineage. The variability of restriction enzyme-digestion patterns of O157 genomes, which is widely used in molecular epidemiological studies of O157 infections, may partly reflect such a genomic diversity.
In this study, based on the genome sequence of O157 Sakai, we performed a comparative analysis of the whole genome structures of eight O157 strains displaying diverse XbaI-digestion patterns using a systematic PCR analysis that we have named whole genome PCR scanning (WGPScanning). The analysis revealed an unexpectedly high genomic diversity of the O157 strains, particularly in the prophages and prophage-related elements, including Stx phages.
Materials and Methods
Bacterial Strains.
Eight O157 strains examined in this study were selected from a total of 1,798 O157 strains isolated in Japan in 1998. They were collected through the O157 surveillance system organized by National Institute for Infectious Diseases of Japan, and all were stx1- and/or stx2-positive and O157 serotype. To select the strains to be tested, we performed a clustering analysis of the 1,798 strains based on their XbaI-digestion patterns on pulsed-field gel electrophoresis as described (7), and the following 8 strains were selected from distinct clusters: 980938 (referred to as no. 2), 980706 (no. 3), 990281 (no. 4), 980551 (no. 5), 990570 (no. 6), 981456 (no. 7), 982243 (no. 8), and 981795 (no. 9). RIMD 0509952 (Sakai), which has been deposited at the American Type Culture Collection (ATCC BAA-460), was used as a reference (2).
Primer Design.
Based on the Sakai sequence, we prepared 549 pairs of PCR primers that amplified 549 segments covering the whole Sakai chromosome, with overlaps of certain lengths at every segment end. Primers were generally 22 nt long. Among the 549 pairs, both primers of 424 pairs annealed to regions conserved in both O157 Sakai and K-12, and both primers of 53 pairs to S-loops. In the remaining 72 pairs, one primer annealed to the conserved region and the other to the S-loop. For analysis of pO157, a 93-kb virulence plasmid, 11 primer pairs were prepared. All primer sequences are available at http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/pcrscan.html.
PCR Analysis.
PCR reactions were performed using genomic DNA as template and long accurate (LA) PCR kits (Takara Shuzo, Kyoto) with 30 cycles of a two-step amplification program: 20 seconds at 96°C/10 min at 69°C. For amplification of DNA segments longer than 15 kb, the extension time was changed to 16 min. PCR products were routinely analyzed by 0.5% agarose gel electrophoresis (12 cm gel). Field-inversed gel electrophoresis was used for products longer than 15 kb.
Results
Principle of WGPScanning Analysis.
We originally used a systematic PCR analysis, called PCR scanning, to analyze the strain-to-strain difference of the genetic organization of the R/F pyocin locus on the Pseudomonas aeruginosa chromosome (8). In that analysis, we selected primer target regions with spacing of desired lengths and designed a set of PCR primer pairs so that amplified segments overlapped with adjacent segments at both ends and covered the entire region to be examined. By comparing the amplified fragments with that of the reference genome, we could determine whether the target regions were present in each tested strain, whether the target regions were arranged in the same order, and whether the segments between target regions had undergone any structural changes. By extending this approach to the whole genome level (WGPScanning), we have examined the genomic diversity of O157 strains using the Sakai strain as a reference.
PCR Primers and Tested Strains.
For chromosome scanning, we prepared 549 pairs of primers based on the Sakai sequence. The mean size of the amplified segments was 10.4 kb, with the maximum and minimum segment sizes being 20.7 and 3.7 kb, respectively. Overlaps between adjacent segments ranged from 21 to 4,132 bp. For the analysis of pO157, we prepared 11 primer pairs. Because a region of pO157 (position 25,821–28,955; DDBJ accession no. AB011549) prevented long PCR amplification, we divided the region into two short segments.
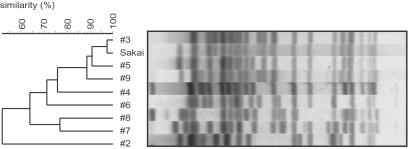
To select strains with potential genomic diversity, we performed a clustering analysis of 1,798 human isolates based on their XbaI-restriction patterns, and selected 8 strains from distinct clusters (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). All were O157:H7-serotype except no. 8 (O157:H−). Strains 4 and 6 contained only stx2, but the others contained both stx1 and stx2. The 8 strains exhibited significant variation in the XbaI-digestion pattern (Fig. 1). Strain 2 was most distantly related to Sakai, but the DNA sequences of their mdh and aroE genes were identical (data not shown).
Fig 1.
XbaI-digestion patterns of the O157 strains tested in this study. Genomic DNAs were digested with XbaI and analyzed by pulsed-field gel electrophoresis followed by the clustering analysis.
WGPScanning and Data Presentation.
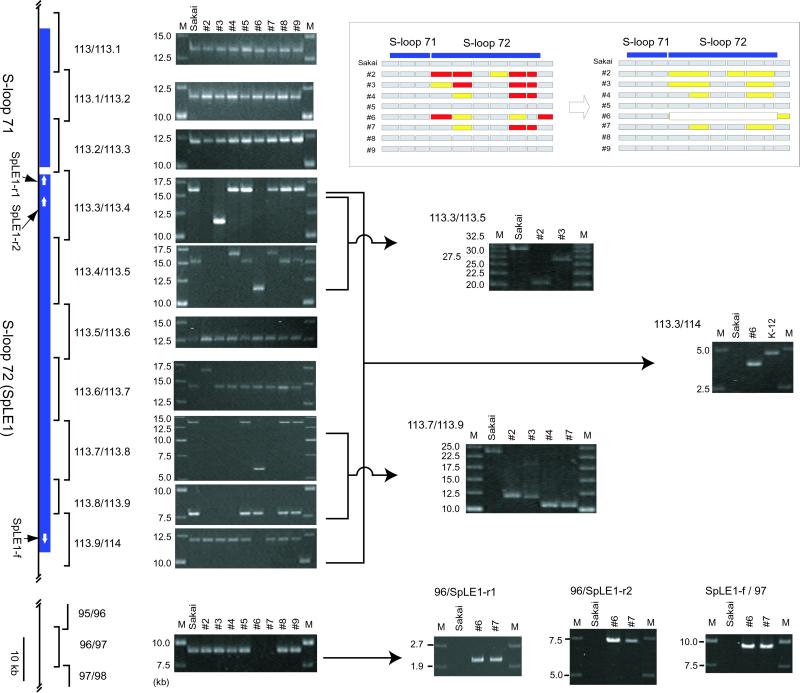
In Sakai, all primer pairs yielded products of the expected sizes. The results of a total of 4,480 PCR reactions from the eight tested strains are summarized in Fig. 2. In this presentation, segments that yielded PCR products of identical and different sizes as those of Sakai are depicted by gray and yellow rectangles, respectively, and those unable to be amplified by red rectangles. When we could not amplify a segment, we performed additional PCR studies using various combinations of primers, considering three types of genomic variation: (i) lack of the target sequence due to deletion or sequence polymorphism, (ii) expansion of the distance between the primers by large insertion, and (iii) improper primer combination as the result of genomic rearrangement, such as translocation or inversion. One example is shown in Fig. 3. By such PCR experiments, most variation of the first type could be discriminated from the latter two types. These results were incorporated in Fig. 2. In some cases, deletions of S-loops were confirmed by PCR using two primers targeted to the flanking DNA. These deletions are indicated by open rectangles in Fig. 2.
Fig 2.
WGPScanning analysis of O157 strains. For each strain, the initial scanning data are presented in the upper line, and the incorporated result with additional PCR analyses is presented in the lower line (see Fig. 3 for examples). Segments yielding products of identical and different sizes from those of Sakai are depicted as gray and yellow rectangles, respectively, and those not amplified are depicted by red. S-loops longer than 500 bp are depicted by blue rectangles, and those deleted in the tested strains are depicted by open rectangles. Positions of stx1 and stx2 in Sakai (220.3/220.4 and 110.2/110.3, respectively), and alternative integration sites for Stx phages (208/209), SpLE1 (96/97), and Mu-like phages (174/175 and 407/408) are indicated. The summary of the initial scanning data for each chromosome is also presented.
Fig 3.
WGPScanning analysis of SpLE1. As an example of the WGPScanning analysis, results of the SpLE1-surrounding region are presented. Segments yielding no PCR products in the initial scanning were subjected to further PCR analysis using different primer combinations. The summary of the initial scanning data and incorporated results of additional analyses are shown in Insets. The presence of SpLE1 in the 96/97 segment of strains 6 and 7 was confirmed by PCR using SpLE1-specific primers (Lower).
Structural Diversity of O157 Chromosomes.
In the initial scanning of the eight chromosomes, we obtained 4,149 PCR products from 4,392 reactions (94%). Among them, 4,005 were indistinguishable in length from those of Sakai, but 144 exhibited size differences (referred to as [Y] segments), indicating that some structural changes have occurred in these segments. We could not amplify the remaining 243 segments ([R] segments). The numbers of [Y] and [R] segments were different from strain to strain, ranging from 15 in no. 5 to 96 in no. 2 (Fig. 2). This difference correlates roughly with the degree of variation in their XbaI-digestion patterns (Fig. 1). These results indicate that significant genomic diversity exists among the O157 strains, and that the variation of XbaI-digestion patterns reflects the genomic diversity.
On the Sakai chromosome, a total of 146 S-loops longer than 500 bp have been identified (2). Among the 549 targeted segments, 210 contained such loops or parts of large loops, whereas the remaining 339 did not. The segments unrelated to the loops were well conserved among the eight strains: 98% of the 2,712 PCR products derived from these segments were indistinguishable in length from those of Sakai (Fig. 2 and Table 2, which is published as supporting information on the PNAS web site), and only 10% of the 339 segments displayed structural polymorphism among the tested strains (Table 1). In contrast, segments associated with loops longer than 500 bp were highly variable. In particular, an extremely high level of variation was observed in those related to prophages: among the 67 Sp-related segments, only 5 were conserved in all tested strains (Table 1). SpLE-related segments also exhibited high variations. These results clearly indicated that the genomic diversity observed in the O157 strains is mainly attributable to the variation of these genetic elements. On the other hand, 114 loop-associated segments exhibited no polymorphism, indicating that at least 95 loops are completely conserved in the eight strains (a list of conserved loops are available in Table 3, which is published as supporting information on the PNAS web site).
Table 1.
Summary of the initial scanning of O157 chromosomes
| Segments
|
S-loop unrelated (n = 339)
|
S-loop related (n = 210) | ||
|---|---|---|---|---|
| Sp-related (n = 67) | SpLE-related (n = 21) | Others (n = 122) | ||
| All conserved | 306 (90%) | 5 (7%) | 9 (43%) | 100 (82%) |
| Including [Y] not [R] | 21 (6%) | 10 (14%) | 4 (19%) | 14 (11%) |
| Including [R] | 12 (4%) | 52 (78%) | 8 (38%) | 8 (7%) |
Conserved, [Y], and [R] segments correspond to gray, yellow, and red rectangles in Fig. 2, respectively.
Because the WGPScanning analysis revealed that variation of prophages and prophage-like elements is deeply involved in the genomic diversity of O157 strains, we analyzed more in details the variations in SpLE1, Stx phages, and Mu-like phage.
SpLE1.
SpLE1 (corresponding to S-loop 72) is integrated into a serine tRNA gene, serX. It encodes 111 Sakai-specific genes, including a urease operon and several potentially virulence-related genes. The scanning data indicated that SpLE1 or SpLE1-like elements are present in all tested strains, but they exhibited various structural and positional variations (Fig. 3). A region containing the 113.8 region was missing in strains 2, 3, 4, and 7. The 113.3/113.4 and 113.4/113.5 segments also displayed various types of structural polymorphism.
PCR products were obtained from the five internal segments of no. 6, but not from the two segments containing the left or right boundary (113.3/113.4 and 113.9/114). Furthermore, the result of PCR examination of no. 6 using the 113.3/114 primer pair indicated that it contained no strain-specific sequence at this locus, and thus, the SpLE1 must exist at some other locus. Considering that two copies of SpLE1 are present in an O157 strain, EDL933, one at the serX locus and the other at serW (9), the most likely candidate for the SpLE1 locus of no. 6 is the serW locus located in the 96/97 segment. In fact, we could not amplify the segment from no. 6. The 96/97 segment was not amplified from no. 7 either, suggesting that no. 7 may also contain an additional copy of SpLE1-like element at the serW locus like EDL933. Thus, we examined the 96/97 segment of the two strains by PCR using SpLE1-specific primers (Fig. 3). The results clearly indicated that each strain contains an SpLE1-like element at this locus.
Stx Phages.
One of the most unexpected findings obtained in the initial scanning was that none or only a few of Sp5 (the Stx2 phage)-related segments (110/110.1 to 110.5/111) were amplified in many strains although all possessed stx2. In strains 5 and 9, all or most of Sp5-related segments were amplified. By PCR using an stx2-specific primer (stx2-r) and the 110.2-f primer, we confirmed that stx2 exists in the 110.2/110.3 segment in the two strains (data not shown). In the remaining 6 strains, PCR analysis using the 110-f/111-r primer pair yielded a product of the same size as that from K-12, indicating that no prophage exists at this locus, and that Stx2 phages are located at some other locus.
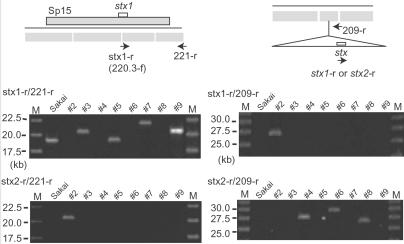
Similarly, none or only one of the Sp15 (the Stx1 phage)-related segments (220.1/220.2 to 220.4/221) were amplified in two stx1-positve strains (nos. 2 and 8), raising again a possibility that their Stx1 prophages exist somewhere else. In fact, PCR analysis using an stx1-specific primer (stx1-r, same as 220.3-f) and the 221-r primer yielded products in Sakai, nos. 3, 5, 7, and 9 (Fig. 4), but not in nos. 2 and 8. The Stx1 phage of no. 7 exists at the same locus as in Sakai, but has significantly diverged from Sp15 in genome structure.
Fig 4.
Identification of alternative integration sites for Stx phages. PCR analyses using the stx1-r or stx2-r primer with the 221-r primer revealed that the Stx2 phage of no. 2 is integrated at the Sp15 (the Stx1 phage of Sakai) locus. Similar analyses using the 209-r primer revealed that the Stx1 phage of no. 2 and the Stx2 phages of nos. 4, 6, and 8 are integrated in the 208/209 region.
To determine alternative integration sites for the Stx phages, we first examined all Sp loci exhibiting different amplification patterns from Sakai by PCR using the stx2-r primer and primers targeted to the outside of each prophage region. By this method, we found that the Stx2 phage of no. 2 exists in the Sp15 locus (Fig. 4). We searched alternative integration sites for the Stx1 phages by the same strategy, but could not find it at any Sp loci. Subsequently, we examined all other [R] segments, which may contain large insertions, by PCR using the stx1-r or stx2-r primer and either primer for each tested segment. With this approach, we found that the Stx1 phage of no. 2 and the Stx2 phages of nos. 4, 6, and 8 are present in the 208/209 region (Fig. 4). Subsequent sequence analyses of the prophage boundary regions revealed that these Stx phages are integrated into the 5′ end of Ecs2813(sbcB) (Fig. 7, which is published as supporting information on the PNAS web site). We could not identify the locations of the Stx1 phage of no. 8 and the Stx2 phages of nos. 3 and 7 by these PCR experiments.
Mu-Like Phages.
Sakai possesses a Mu-like phage (Sp18) that is integrated in the sorbose operon located in the 421/422 region. Sp18-related segments were amplified only from no. 4, indicating its rare distribution in O157 (Fig. 2). In no. 4, we could not amplify either segment that contains the boundary of Sp18. This finding suggested that the phage exists at some other locus in no. 4, most likely on one of the three segments, 98/99, 174/175, and 407/408, which were not amplified specifically in no. 4. Thus, these segments were again examined by PCR using Sp18-specific primers, 421.1-r and 421.3-f. Unexpectedly, we obtained PCR products from two of them, 174/175 and 407/408 (data not shown), indicating that no. 4 contains two copies of Mu-like phage at these loci. Sequencing analyses revealed that they are integrated into Ecs2374 and Ecs4832, respectively (Fig. 7).
pO157.
Possession of a large virulence plasmid, pO157, is one of the characteristics of EHEC. The genome sequence of pO157 from Sakai is almost identical to that from EDL933, the complete sequence of which was also determined (10, 11). As shown in Fig. 2, the genome organization of pO157 is relatively well conserved among the eight strains, but exhibited some diversity. In particular, the plasmid of no. 2 seems to have undergone significant structural changes, consistent with the finding that the chromosome of no. 2 is most diverged from the others.
Discussion
Rapid progress in bacterial genome analysis has explosively expanded our knowledge about the metabolism, virulence, and evolution of each bacterium. Initial bacterial genome projects focused on a single genotype of species, but more attention is now being paid to intraspecific genomic comparisons because they efficiently provide more detailed information on the phenotype–genotype relationships. In this study, by using WGPScanning, a recently developed method for the overall structural comparison of closely related genomes, we analyzed the genomic diversity of O157 strains, and demonstrated that a high level of diversity is present in the O157 lineage. The data clearly indicate that variation of prophages and prophage-related elements is a major determinant in generating such diversity.
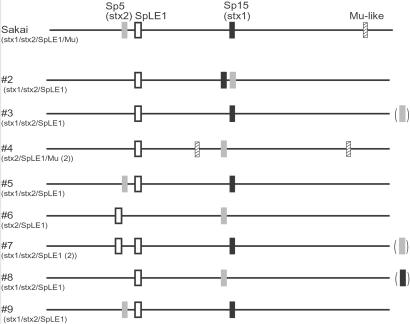
Among the methods for whole genome comparison of closely related strains, whole genome sequencing may be the most powerful in that it provides the most precise information. However, it is economically unrealistic to thoroughly sequence every strain to be compared. DNA microarray comparison is also powerful in that it quickly reveals the gene contents of many strains (12, 13), but does not provide information about gene position or about genes that are absent in the reference strain. In contrast, the presence of such genes can be detected by WGPScanning when a PCR product of a different size from the reference is obtained. Once this PCR fragment is obtained, we can determine the altered genome organization by DNA sequencing. Positional changes of genetic elements can also be detected as demonstrated for SpLE1 and Mu-like phage (Fig. 5).
Fig 5.
Positional diversity of Stx phages, SpLE1, and Mu-like phages among the O157 strains. Locations of these genetic elements on each chromosome are shown. The locations of the Stx1 phage of no. 8 and the Stx2 phages of nos. 3 and 7 were not identified in this study.
When we cannot amplify certain segments because of large insertions or rearrangements, it is difficult to determine the structural changes by WGPScanning alone. This could be resolved by construction of a DNA library with large insert DNA, such as bacterial artificial chromosome (BAC) library. Because the region to be examined is already identified from the scanning data, we can easily isolate the clone to be analyzed. A series of PCR screening, which we used for determining the alternative integration sites of Stx phages, are also useful to identify the location of a translocated gene(s).
Our WGPScanning system is probably applicable to other types of E. coli strains because we designed most primers to anneal to the regions on the E. coli chromosome backbone, where the nucleotide sequences are identical between O157 Sakai and K-12 MG1655. In fact, our primer pairs yielded 408 segments from K-12, and additional 44 segments by altering the primer combinations so that they skipped the S-loops, covering 96% of the K-12 chromosome (data not shown). Considering the phylogenetic distance between O157 and K-12 (4), we expect that these primers would work efficiently in most E. coli strains.
Among the findings obtained in this study, the most striking is the extensive variations of Stx phages. The genome sequences of Stx1 and Stx2 phages from Sakai and EDL933 have been determined (9, 14–16). Each phage from both strains exhibits some variations in limited parts of the phage genomes, but is integrated at the same locus. Among the Stx phages of the 8 strains examined in this study, only the Stx2 phage of strain 5 seems to be identical to that of Sakai. Although minor variations were observed in several phages (the Stx2 phage of no. 9 and the Stx1 phages of nos. 3, 5, and 9), the variations of other phages were extensive (Fig. 2). There are at least two types of Stx1 phages and three types of Stx2 phages using different integration sites (Fig. 5).
Recktenwald and Schmidt (17) recently reported that an Stx2e-encoding phage, ø27, is integrated in yecE located in the 194/195 segment, and that its genome structure markedly differs from either Sp5 or Sp15. Because the phage was isolated from a non-O157 strain, the finding implies that EHEC strains belonging to different lineages have acquired stx by infection with different types of Stx phages. This finding supports the parallel evolution theory of virulence in pathogenic E. coli (18). However, our results clearly indicate that various types of Stx phages exist even in the O157 lineage. Although the molecular mechanism generating such diversity is not known, it is unlikely that different Stx phages infected each O157 strain independently. The presence of so many prophages in O157, especially lambda-like phages resembling each other, may play an important role. These phages can provide many opportunities for recombination among resident prophages or with newly incoming phages (6). In fact, extensive variation in genome structures was observed not only in the Stx phages but also in other lambda-like phages. We are now carrying out sequence determination of the O157 genome regions exhibiting any structural polymorphism detected in this study, including all of the Stx phages. The genome comparison of these phages will provide more insight into the genetic basis underlying the generation of the genomic diversity of O157.
Supplementary Material
Acknowledgments
We thank G. Christie and E. Oswald for their critical reading of the manuscript, H. Wakimoto, A. Yoshida, S. Setsu, M. Takahashi, and S. Satoh for their technical assistance, and Y. Hayashi for her language assistance. This work was supported by Japan Society for the Promotion of Science Research for the Future Programs (00L01411), by Grants-in-Aid for Scientific Research (B) and for Scientific Research on Priority Areas (C) from The Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a grant from the Yakult Foundation.
Abbreviations
EHEC, enterohemorrhagic Escherichia coli
Stx, Shiga toxin
WGPScanning, whole genome PCR scanning
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mead P. S. & Griffin, P. M. (1998) Lancet 352, 1207-1212. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., Han, C. G., Ohtsubo, E., Nakayama, K., Murata, T., et al. (2001) DNA Res. 28, 11-22., and addendum (2001) 28, 47–52. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1462. [DOI] [PubMed] [Google Scholar]
- 4.Pupo G. M., Karaolis, D. K. R., Lan, R. & Reeves, P. R. (1997) Infect. Immun. 65, 2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence J. G. & Ochman, H. (1998) Proc. Natl. Acad. Sci. USA 95, 9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnishi M., Kurokawa, K. & Hayashi, T. (2001) Trends Microbiol. 9, 481-485. [DOI] [PubMed] [Google Scholar]
- 7.Terajima J., Izumiya, H., Iyoda, S., Tamura, K. & Watanabe, H. (2002) Jpn. J. Infect. Dis. 55, 19-22. [PubMed] [Google Scholar]
- 8.Nakayama K., Takashima, K., Ishihara, H., Shinomiya, T., Kageyama, M., Kanaya, S., Ohnishi, M., Murata, T., Mori, H. & Hayashi, T. (2000) Mol. Microbiol. 38, 213-231. [DOI] [PubMed] [Google Scholar]
- 9.Perna N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 10.Makino K., Ishii, K., Yasunaga, T., Hattori, M., Yokoyama, K., Yutsudo, C. H., Kubota, Y., Yamaichi, Y., Iida, T., Yamamoto, K., et al. (1998) DNA Res. 5, 1-9. [DOI] [PubMed] [Google Scholar]
- 11.Burland V., Shao, Y., Perna, N. T., Plunkett, G., Sofia, H. J. & Blattner, F. R. (1998) Nucleic Acids Res. 26, 4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behr M. A., Wilson, M. A., Gill, W. P., Salamon, H., Schoolnik, G. K., Rane, S. & Small, P. M. (1999) Science 284, 1520-1523. [DOI] [PubMed] [Google Scholar]
- 13.Salama N., Guillemin, K., McDaniel, T. K., Sherlock, G., Tompkins, L. & Falkow, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino K., Yokoyama, K., Kubota, Y., Yutsudo, C. H., Kimura, S., Kurokawa, K., Ishii, K., Hattori, M., Tatsuno, I., Abe, H., et al. (1999) Genes Genet. Syst. 74, 227-239. [DOI] [PubMed] [Google Scholar]
- 15.Plunkett G., Rose, D. J., Durfee, T. J. & Blattner, F. R. (1999) J. Bacteriol. 181, 1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama K., Makino, K., Kubota, Y., Watanabe, M., Kimura, S., Yutsudo, C. H., Kurokawa, K., Ishii, K., Hattori, M., Tatsuno, I., et al. (2000) Gene 258, 127-139. [DOI] [PubMed] [Google Scholar]
- 17.Recktenwald J. & Schmidt, H. (2002) Infect. Immun. 70, 1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid S. D., Herbelin, C. J., Bumbaugh, A. C., Selander, R. K. & Whittam, T. S. (2000) Nature 406, 64-67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.