Abstract
Huntington's disease is a progressive neurodegenerative disease caused by a polyglutamine (polyQ) repeat expansion in the huntingtin protein [Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983]. To understand the mechanism by which polyQ repeats cause neurodegeneration and cell death, we modeled polyQ neurotoxicity in Caenorhabditis elegans. In our model, expression of N-terminal fragments of human huntingtin causes polyQ-dependent degeneration of neurons. We conducted a genetic screen to identify proteins that protect neurons from the toxic effects of expanded polyQ tracts. Loss of polyQenhancer-1 (pqe-1) gene function strongly and specifically exacerbates neurodegeneration and cell death, whereas overexpression of a pqe-1 cDNA protects C. elegans neurons from the toxic effects of expanded huntingtin fragments. A glutamine/proline-rich domain, along with a charged domain, is critical for PQE-1 protein function. Analysis of pqe-1 suggests that proteins exist that specifically protect neurons from the toxic effects of expanded polyQ disease proteins.
At least eight hereditary neurodegenerative disorders, including Huntington's disease (HD), have been identified in which the disease locus encodes a protein containing an expanded glutamine tract (1). HD patients carry expanded glutamine repeats in the N terminus of huntingtin, a widely expressed protein of unknown function (2, 3). Although huntingtin is expressed in many cell types, the primary cellular pathology of HD is degeneration of neurons of the striatum and cortex, leading to dramatic personality changes and motor dysfunction in early stages of the disease (4, 5). Generally, the onset of disease symptoms is inversely related to the length of the glutamine expansion. However, additional factors distinct from polyglutamine (polyQ) length influence the age of onset. For example, genotypic variation linked to a region encoding the GluR6 kainate receptor accounts for ≈10% of the variance in the age of onset (6, 7).
Molecular aspects of HD provide clues toward understanding the mechanism by which mutant huntingtin causes neurotoxicity. Expanded glutamine tracts in the huntingtin protein alter its physical properties (8, 9). PolyQ-containing N-terminal fragments of mutant huntingtin form cytosolic and nuclear aggregates in affected tissue (10). Therefore, the expanded glutamine tract in mutant huntingtin may elicit neurotoxicity by altering interactions of huntingtin with critical cellular constituents. For example, in the yeast two-hybrid system, normal huntingtin associates with Hip1, a human homolog of the yeast cytoskeletal protein, Sla2 (11–13). Loss of a normal interaction between Hip1 and mutant huntingtin may disrupt cytoskeletal integrity, leading to degeneration. Furthermore, transcription factors are sequestered by mutant huntingtin, interfering with normal gene transcription (14–16). Thus, the mechanisms of pathogenesis of HD may be triggered by changes or alterations of interactions with normal or abnormal protein partners.
The molecular and functional similarities between Caenorhabditis elegans and Homo sapiens nervous systems suggest that invertebrate genetics can be used to address basic questions regarding polyQ expansion diseases. Several Drosophila and C. elegans model systems have been established in which expression of expanded glutamine tracts in huntingtin and other polyQ disease proteins (e.g., ataxin 1 and ataxin 3), lead to neurodegeneration (17–25). We established a C. elegans model system where expression of the N terminus of human huntingtin was directed to a limited set of neurons, including the well characterized glutamatergic ASH sensory neurons using the osm-10 promoter (17, 25–28). In our model system, expression of the N terminus of human huntingtin carrying a polyQ tract consisting of 150 glutamines (Htn-Q150) leads to progressive degeneration, but not death, of the ASH neurons in aged (8-day-old) animals (17).
To identify cellular pathways involved in polyQ neurotoxicity, we performed a genetic screen for mutations that enhanced degeneration of ASH neurons expressing expanded huntingtin fragments. We demonstrate that loss of polyQ enhancer-1 (pqe-1) gene function dramatically exacerbates polyQ toxicity in C. elegans neurons, whereas its overexpression promotes neuron survival. Overt phenotypes have not been observed in pqe-1 mutant animals other than enhancement of polyQ neurotoxicity. Alternative splicing of pqe-1 yields multiple nuclear localized PQE-1 proteins that contain a glutamine/proline-rich domain and/or an exonuclease domain. Genetic analysis of polyQ neurotoxicity in C. elegans reveals that PQE-1 proteins normally provide dramatic protection against expanded polyQ tracts.
Materials and Methods
Assessment of ASH Degeneration and Survival.
ASH survival was assessed using GFP reporters expressed in ASH. osm-10:GFP (pHA#29) expresses GFP in ASH and three other sensory neurons (28). The presence of pHA#29 in transgenic animals caused dye-filling defects in 0–5% of ASH neurons, independent of age and/or genotype. Aging of animals to 8 days was performed as previously described (17). ASH survival in the L1/L2 stage of pqe-1(rt13) animals was assessed by collecting eggs for 5 h and incubating for 24 h before scoring; Htn-Q150 and GFP were expressed from extrachromosomal arrays by using the osm-10 promoter. All animals were cultured at 25°C unless otherwise stated.
Genetic Screening and Mapping.
dpy-20(e1282); rtEx102 [osm-10:Htn-Q150, dpy-20(+)] were generated by injection of pHA#16 and pDPY20 at 65 and 140 ng/μl, respectively (17). Mutant alleles were isolated as animals with ASH-specific dye-filling defects among the F2 progeny of rtEx102 animals mutagenized with 25 mM ethylmethane sulfonate. Three-day-old F2 rtEx102 animals were incubated with the vital fluorescent dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocynanine perchlorate (DiD) and examined by fluorescence microscopy for ASH-specific dye-filling defects (29). Approximately 29,000 F2 animals were screened.
pqe-1 alleles were backcrossed at least twice to wild type (N2); rt13 was backcrossed four times. Unmutagenized Htn transgenes with or without the osm-10:GFP transgene (pHA#29) were reintroduced into pqe-1 animals by using 75 ng/μl of elt-2:GFP (pJM#67) (30) as a marker for backcrossing (Tables 1 and 2). The six alleles of pqe-1 identified are rt13 (corresponding to Q561→STOP), rt58 (Q632→STOP), rt61 (Q526→STOP), rt62 (Q664→STOP), rt64 (M1I), and rt67 (S709N). The percentages of ASH neurons defective in dye filling were 71%, 70%, 86%, 42%, 76%, and 22%, respectively, in 3-day-old pqe-1 mutant animals expressing Htn-Q150. The percentages of ASH neurons defective in dye filling were 1%, 3%, 0%, 0%, 3%, and 2%, respectively, in 3-day-old pqe-1 mutant animals expressing Htn-Q2. No ASH dye-filling defects were seen in normal animals expressing Htn-Q150 or Htn-Q2 at 3 days of age. More than 150 ASH neurons were scored for each data point. ASH neuron survival and dye filling were unaffected in all pqe-1 strains lacking transgenes.
Table 1.
Progressive enhancement of expanded polyQ toxicity in pqe-1 mutant animals
| Genotype | Lines | % defective, 3 days | % defective, 8 days |
|---|---|---|---|
| Htn-Q150 | 13 | 0 | 13+/−4 |
| pqe-1(rt13) | — | 0 | 0 |
| pqe-1(rt13); Htn-Q2 | 5 | 1+/−1 | 5+/−1 |
| pqe-1(rt13); Htn-Q23 | 5 | 0 | 2+/−1 |
| pqe-1(rt13); Htn-Q95 | 5 | 7+/−2 | 36+/−7 |
| pqe-1(rt13); Htn-Q150 | 4 | 73+/−5 | 88+/−4 |
The genotype and huntingtin transgene are listed in column 1; the number of transgenic lines scored is listed in column 2. The huntingtin fragments were expressed from extrachromosomal arrays by using the osm-10 promoter. The average percentage of ASH neurons affected (dye-filling defective) in adult animals is reported (in columns 3 and 4) at 3 and 8 days. More than 150 ASH neurons were scored for each data point. The SEM is listed.
Table 2.
pqe-1 loss of function specifically enhances expanded polyQ-mediated cell death
| Genotype |
Lines |
% affected at 3 days | % affected at 8 days | ||
|---|---|---|---|---|---|
| Degeneration | Death | Degeneration | Death | ||
| GFP | 5 | 0 | 0 | 1+/−1 | 0 |
| Htn-Q150, GFP | 13 | 0 | 0 | 15+/−4 | 0 |
| pqe-1(rt13); GFP | 3 | 0 | 0 | 3+/−3 | 0 |
| pqe-1(rt13); Htn-Q2, GFP | 5 | 0 | 0 | 8+/−2 | 0 |
| pqe-1(rt13); Htn-Q150, GFP | 5 | 6+/−2 | 76+/−9 | 5+/−3 | 95+/−3 |
The genotype and transgene are listed in column 1; the number of transgenic lines scored is listed in column 2. The huntingtin fragments and the GFP reporter construct were expressed from extrachromosomal arrays by using the osm-10 promoter. The percentage of ASH neurons degenerating (dye-filling defective) but surviving (expressing osm-10:GFP) is reported in subcolumns 3 and 5. The percentage of dead ASH neurons (dye-filling defective and lacking GFP expression) is reported in subcolumns 4 and 6. More than 150 ASH neurons were scored for each data point. The SEM is listed.
sDf121, a large deletion that removes pqe-1 and many adjacent genes (31), was introduced into rt13 mutant animals expressing Htn-Q150. Eighty-four percent of ASH neurons were affected in heterozygous pqe-1(rt13)/sDf121 animals expressing Htn-Q150 (n = 46); a similar number of affected ASH neurons was observed in homozygous pqe-1(rt13) animals expressing Htn-Q150, suggesting that pqe-1(rt13) is a loss-of-function allele. All pqe-1 analyses were done with homozygous pqe-1(rt13) animals unless otherwise stated.
Genetic linkage of pqe-1 to chromosome III was determined using RW7000 (32). dpy-1(e1) pqe-1(rt13) unc-32(e189) and the strain CB4856 were used for recombination mapping by using single-nucleotide polymorphisms identified by Wicks et al. (33) and the C. elegans genome-sequencing project.
Transgenic Strains.
In all transgenic strains described, the Htn fragments were expressed by using the ASH neuron-specific osm-10 promoter. Htn-Q150 was expressed from rtIs11 in cDNA rescue experiments (Table 3). rtIs11 is an integration into chromosome V of the extrachromosomal array rtEx1, which contains pHA#16, pDPY20, and pKP#58 (17). In the overexpression experiment, Htn-Q150 was expressed from rtIs18 (Table 4). rtIs18 is an integration into chromosome I of the extrachromosomal array rtEx362, which contains pHA#16 and elt-2:GFP (30). Cosmids, pqe-1 promoter constructs, or osm-10 promoter constructs (5 ng/μl) were coinjected with elt-2:GFP (30) (Tables 1) or rol-6 (34) (Table 4) at 75 ng/μl as a marker.
Table 3.
PQE-1 proteins containing the Q/P-rich and charged domain suppress polyQ toxicity in pqe-1 animals
| Genotype |
Lines |
% affected at 3 days | %
affected, total |
|
|---|---|---|---|---|
| Degeneration | Death | |||
| pqe-1(rt13); Htn-Q150 | − | 3 | 97 | 100 |
| pqe-1(rt13); Htn-Q150; F52C9 | 7 | 11+/−5 | 30+/−5 | 56+/−14 |
| pqe-1(rt13); Htn-Q150; GFP | 4 | 7+/−3 | 93+/−4 | 99+/−1 |
| pqe-1(rt13); Htn-Q150; PQE-1A:GFP | 6 | 7+/−2 | 19+/−4 | 26+/−4 |
| pqe-1(rt13); Htn-Q150; PQE-1B:GFP | 4 | 4+/−2 | 96+/−9 | 100 |
| pqe-1(rt13); Htn-Q150; PQE-1C:GFP | 5 | 9+/−3 | 23+/−8 | 33+/−8 |
The genotype and transgene are listed in column 1; the number of transgenic lines scored is listed in column 2. Htn-Q150 was expressed from rtIs11, an array expressing Htn-Q150 and OSM-10:GFP by using the osm-10 promoter that has been integrated into chromosome V. The percentage of ASH neurons degenerating (dye-filling defective) but surviving (expressing osm-10:GFP) is reported in subcolumn 3. The percentage of dead ASH neurons (dye-filling defective and lacking GFP expression) is reported in subcolumn 4. The percentage of affected neurons (sum of subcolumns 3 and 4) is indicated. The cosmid F52C9 contains pqe-1(+). More than 150 ASH neurons were scored for each data point. The SEM is listed.
Table 4.
Overexpression of PQE-1C suppresses polyQ toxicity in pqe-1(+) animals
| Genotype |
Lines |
% affected at 8 days | % affected, total |
|
|---|---|---|---|---|
| Degeneration | Death | |||
| Htn-Q150; GFP | 4 | 45+/−6 | 46+/−7 | 91+/−3 |
| Htn-Q150; GFP; PQE-1B | 2 | 37+/−17 | 52+/−5 | 88+/−13 |
| Htn-Q150; GFP; PQE-1C | 5 | 26+/−4 | 23+/−6 | 50+/−3 |
The genotype and transgene are listed in column 1; the number of transgenic lines scored is listed in column 2. Htn-Q150 was expressed from rtIs18, an array expressing Htn-Q150 by using the osm-10 promoter that has been integrated into chromosome I, resulting in a highly penetrant phenotype. PQE-1 proteins and GFP were expressed from extrachromosomal arrays by using the osm-10 promoter. The percentage of ASH neurons degenerating (dye-filling defective) but surviving (expressing osm10:GFP) is reported in subcolumn 3. The percentage of dead ASH neurons (dye-filling defective and lacking GFP expression) is reported in subcolumn 4. The percentage of affected neurons (sum of subcolumns 3 and 4) is indicated. More than 85 ASH neurons were scored for each data point; the SEM is listed.
Immunohistochemistry and Microscopy.
Polyclonal antisera were generated in rabbits (Research Genetics, Huntsville, AL) against an N-terminal peptide VITNNKKKRIDVVTLDEDAPRRVQV and a Q/P-rich domain (amino acids 96–321) GST fusion protein and a C-terminal (amino acids 1546–1670) GST fusion protein. Affinity-purified sera recognized the predicted overexpressed PQE-1B:GFP and PQE-1C:GFP proteins but not PQE-1A:GFP protein in normal animals treated with Bowin's fixative (35). Htn-Q150 was detected by using HP1 (36). PQE-1:GFP fusion proteins and Htn-Q150 were expressed in ASH neurons by using the osm-10 promoter. Images were captured by using openlabs software (Improvisions, Coventry, U.K.) and a Zeiss Axioplan.
Plasmid construction, is presented in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
To identify C. elegans genes that normally act to protect neurons from polyQ toxicity, we screened for mutations that caused early onset degeneration of ASH neurons in animals expressing expanded huntingtin fragments. The sensory endings of ASH neurons are exposed to the environment and can take up DiD, a lipophilic fluorescent dye that allows rapid visualization of the ASH neurons in vivo (28). If the ASH neurons are not intact, ASH neurons will fail to fill with dye. By monitoring the uptake of DiD, we isolated six mutant strains with significant dye-filling defective ASH neurons in young animals, ranging from 86 to 22% at 3 days. No defect was observed in animals lacking the Htn-Q150 transgene, indicating that the dye-filling defect depended on Htn-Q150 expression. The overall levels of Htn-Q150 were unchanged in mutant animals, based on immunohistochemistry. Noncomplementation data, genetic linkage to chromosome III, and the identification of molecular lesions described below revealed that all six of these strains carry a loss-of-function mutation in a gene we designated polyQ enhancer-1 (pqe-1). The analyses presented herein use the representative mutant allele rt13, which generates a premature termination codon in pqe-1. Approximately 75% of the ASH neurons are dye filling defective in young pqe-1(rt13) mutant animals expressing Htn-Q150 from extrachromosomal arrays.
In the analysis presented herein, “affected” neurons refers to the population of ASH neurons that failed to fill with dye (dye-filling defective). Dye-filling defective ASH neurons may be absent (e.g., due to cell death) or have defective sensory endings (e.g., due to degeneration at the sensory endings) (29). To differentiate between these two possibilities, we monitored the expression of a GFP reporter construct expressed in ASH neurons or used the immunohistochemical detection of OSM-10, an endogenous C. elegans protein expressed in ASH neurons (28). The subset of neurons that were dye-filling defective, but expressed GFP and/or OSM-10 immunoreactivity, were classified as “degenerating.” “Dead” neurons were dye-filling defective and did not express GFP and/or OSM-10 immunoreactivity.
To ascertain whether the number of affected ASH neurons depended on the size of the expansion in the glutamine tract, we assessed ASH neuron dye filling in pqe-1 animals expressing huntingtin N-terminal fragments with varying lengths of polyQ tracts (Table 1). The number of dye-filling defective ASH neurons expressing Htn-Q95 or Htn-Q150 but not Htn-Q2 or Htn-Q23 N-terminal fragments was enhanced in young (3-day-old) pqe-1 mutant animals. Furthermore, the number of affected neurons increased in aged (8-day-old) pqe-1 mutant animals expressing Htn-Q95 or Htn-Q150. We conclude that mutations in pqe-1 specifically enhance the toxicity associated with expanded polyQ tracts, and this toxicity increases with age.
To discriminate between cell death and degeneration, Htn-Q150 and Htn-Q2 were coexpressed with a GFP reporter construct in pqe-1 animals (Table 2). The vast majority of ASH neurons were dead in pqe-1 animals expressing Htn-Q150 based on the loss of the GFP signal. A small number of the surviving ASH neurons in these animals degenerated, based on their inability to dye fill. ASH neuron survival and dye filling were unimpaired in pqe-1 animals expressing the GFP reporter construct and Htn-Q2. GFP expression levels were unchanged in pqe-1 mutant animals expressing Htn-Q2, suggesting that mutation of pqe-1 does not alter expression levels of the osm-10 promoter. To determine whether the process of ASH neurodegeneration and cell death in pqe-1 mutant animals expressing Htn-Q150 was progressive, we assessed ASH survival in 1-day-old larvae. We found that 51% of ASH neurons degenerated, whereas 41% were dead, based on dye filling and GFP expression. Thus, ASH neurons degenerate and eventually die, providing evidence for the progressive nature of polyQ degeneration in pqe-1 animals.
Given the highly penetrant ASH cell death in 3-day-old pqe-1 animals expressing Htn-Q150, we speculated that pqe-1 function might be involved in the apoptotic cell death pathway. In C. elegans development, apoptosis depends on the caspase encoded by the ced-3 gene (37). To ascertain whether loss of pqe-1 function enhanced apoptotic death, we assessed programmed cell death during development in two different neuronal cell lineages in pqe-1;ced-3 mutant animals. The aberrant survival of cells that normally die during development in the ventral cord (VC) can be assessed by using a lin-11:GFP reporter construct (38). In ced-3(n2427) animals, extra VC-like neurons expressing lin-11:GFP are observed in L4 animals. The number of extra VC-like neurons observed in pqe-1(rt13);ced-3(n2427) animals was unchanged (data not shown). Similarly, in the ABp lineage, the sister cell of the ASI neuron normally dies during development. In ced-3(n717) animals, 18% of the ASI sister cells survive and differentiate into neurons based on their ability to dye fill. In pqe-1(rt13);ced-3(n717) animals, the number of additional ASI sisters was unchanged (data not shown). These results indicate that pqe-1 does not play a direct role in the apoptotic cell death pathway.
Alternatively, loss of pqe-1 function may have altered the proper development or function of ASH neurons, resulting in the highly penetrant ASH cell death in 3-day-old pqe-1 animals expressing Htn-Q150. To address this possibility, we monitored the expression of two ASH specific cell markers to confirm that ASH neurons were born and differentiated in pqe-1 mutant animals (26, 28). Both sra-6 and osm-10 were expressed normally in the ASH neurons of pqe-1 animals (data not shown). The morphology of the ASH neurons was normal based on dye-filling assays, and pqe-1 mutant animals responded to stimuli detected by ASH neurons (data not shown). We conclude that ASH neurons develop and function normally in pqe-1 animals, but the ASH neurons degenerate and eventually die in the presence of Htn-Q150.
To address the specificity of pqe-1 for polyQ disease proteins, we assessed the effect of loss of pqe-1 function on neurodegeneration caused by unrelated neurotoxic stimuli. Mutations in the C. elegans deg-1 gene, which encodes a degenerin family ion channel expressed in ASH and other neurons, results in a leaky channel causing swelling and necrotic-like cell death (39). We identified a cold-sensitive allele of deg-1 in our screen for polyQ enhancers as a mutation that caused ASH neurodegeneration, but this degeneration did not depend on the presence of the Htn-Q150 transgene. The rt70 allele of deg-1 changed Arg-610 to histidine, causing half of the ASH neurons to die in adult animals raised at 20°C. In pqe-1 animals, the percentage of ASH neurons affected did not increase significantly at 20°C; 53% degenerated in pqe-1(rt13);deg-1(rt70). This result indicates that, whereas mutation of pqe-1 has pronounced effects on the ability of expanded polyQ tracts to induce neurodegeneration and neuronal cell death, loss of pqe-1 function has little effect on unrelated neurotoxic stimuli.
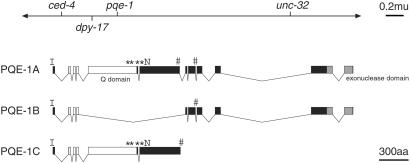
Genetic mapping localized pqe-1 to a 0.3 map unit region of chromosome III (Fig. 1); phenotypic rescue of the pqe-1 mutant phenotype was obtained with two overlapping cosmids, F52C9 and T15B12. The corresponding genomic region was predicted by the C. elegans genome sequencing project to contain multiple genes of a putative operon (40). Amplification and sequencing of genomic DNA from the six pqe-1 mutant strains revealed that all pqe-1 mutant strains contained DNA polymorphisms perturbing the predicted gene F52C9.8 (Fig. 1).
Fig. 1.
Alternative splicing of pqe-1 yields multiple protein isoforms. pqe-1 maps genetically to chromosome III (−1.91 map units) to the right of dpy-17. On the basis of our analysis, pqe-1 corresponds to F52C9.8 and encodes at least three different proteins generated by differential mRNA splicing. The exonic structures of pqe-1 splice forms are depicted. Exons encoding the Q/P-rich domain are illustrated as white boxes. Black boxes represent the exons encoding the charged region. Sequences corresponding to the exonuclease domain are illustrated as gray boxes. The approximate locations of pqe-1 mutations are labeled: * for stop codons, I for rt64 M1I, and N for rt67 S709N. The regions encoding putative nuclear localization signals are marked by #. The PQE-1A protein sequence is shown in Fig. 3, which is published as supporting information on the PNAS web site.
pqe-1 encodes a 1,670-aa protein (PQE-1A) containing three distinct domains (Fig. 1). The N-terminal 568 amino acids of PQE-1A are enriched in glutamine and proline residues, 118 (21%) and 86 (15%), respectively. A region enriched in charged amino acids (38% D, R, E, or K in amino acids 614–1356) that contains three consensus nuclear localization signals follows this Q/P-rich domain (Fig. 1). The PQE-1A C terminus contains a putative exonuclease domain (PFAM PF00929) of the RNase D family (41). In our analysis, we found that alternative splicing resulted in the generation of two shorter PQE-1 proteins: PQE-1B and PQE-1C. In the pqe-1B mRNA, exons 5 and 6 are removed, generating a protein that lacks the Q/P-rich N terminus and most of the charged domain (Fig. 1). In the pqe-1C mRNA, intron 6 is not removed during mRNA splicing. Translation terminates at a stop codon in intron 6, resulting in PQE-1C, a truncation of PQE-1A. PQE-1C contains the Q/P-rich N terminus and most of the charged domain but lacks the putative exonuclease domain (Fig. 1). Three additional splice forms that contain the putative exonuclease domain have recently been identified (42).
The preponderance of pqe-1 mutations in the N-terminal exons of PQE-1C and PQE-1A suggested that these proteins are required for the suppression of polyQ neurotoxicity. This hypothesis was tested in phenotypic rescue experiments by using the individual pqe-1 cDNAs. The pqe-1 promoter was used for expression of PQE-1A, PQE-1B, and PQE-1C in C. elegans. Both PQE-1A and PQE-1C were able to rescue the pqe-1 mutant phenotype, but PQE-1B did not (data not shown). To determine whether pqe-1 functions autonomously in the ASH neurons, PQE-1:GFP fusion proteins were expressed in ASH by using the osm-10 gene promoter (28) (Table 3). Only PQE-1A:GFP and PQE-1C:GFP were able to rescue pqe-1 and thereby restored ASH survival in animals expressing Htn-Q150. These results suggest that PQE-1 proteins containing the Q/P-rich N terminus and the charged domain are critical in protecting ASH neurons from the toxic effects of expanded polyQ tracts. Consistent with this proposed protective function, overexpression of PQE-1C in aged animals with a wild-type pqe-1 gene decreased Htn-Q150 toxicity in ASH neurons, whereas overexpression of PQE-1B had no effect on ASH survival (Table 4).
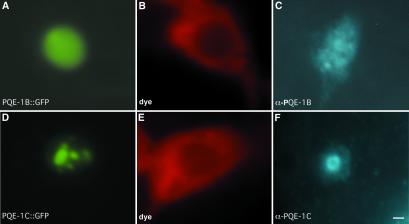
The PQE-1:GFP fusion proteins expressed in ASH using the osm-10 gene promoter were also used to assess the subcellular localization of PQE-1 proteins. Consistent with the presence of canonical nuclear localization signals, PQE-1B:GFP and PQE-1C:GFP were predominantly detected in the ASH nucleus (Fig. 2). PQE-1B:GFP was diffusely distributed in the nucleus with one (or a few clustered) more intense spot/s visible near the perimeter of each nucleus. PQE-1C:GFP was also localized to the nucleus, but patterns of expression ranged from a few spots within the nucleus (predominantly) to diffuse staining throughout the nucleus (less frequently) for all transgenic lines (Fig. 2). No GFP expression was detectable in the ASH neurons of transgenic animals expressing PQE-1A:GFP (despite the ability of this fusion protein to rescue the pqe-1 mutant phenotype). We confirmed the subcellular localization PQE-1B and PQE-1C using antisera that recognized either the PQE-1 N or C terminus. We were unable to detect PQE-1A expressed from either the osm-10 or pqe-1 promoter by using immunohistochemistry. No endogenous PQE-1 protein was detected in normal animals or in those expressing Htn-Q150. Additionally, the expression of GFP using the pqe-1 promoter did not yield detectable GFP protein. Together, these data suggest that the pqe-1 gene is transcribed at low levels and that cellular PQE-1A protein levels may be tightly controlled. Data from microarray analysis suggest that pqe-1 mRNAs are expressed at low or undetectable levels (43). Thus, PQE-1A, PQE-1B, and PQE-1C are likely low-abundance nuclear proteins.
Fig. 2.
PQE-1B and PQE-1C are nuclear proteins. Subcellular localization of PQE-1B:GFP and PQE-1C:GFP was examined by using the osm-10 promoter to drive transgene expression. The PQE-1B:GFP and PQE-1C:GFP fusion proteins were detected in the nucleus of the ASH neurons of live animals (A and D, respectively). The cell bodies of the corresponding ASH neurons were visualized by dye filling with DiD (B and E, respectively). PQE-1B:GFP and PQE-1C:GFP were detected in the ASH nuclei of Bowin's fixed animals (35) by using affinity-purified antisera that recognize the C and N termini of PQE-1 (C and F, respectively). PQE-1B:GFP and PQE-1C:GFP are still localized to the nucleus in animals expressing Htn-Q150 (not shown). (Bar = 0.5 μm.)
It has been previously suggested that inappropriate interactions of expanded glutamine tracts with Q-rich cellular proteins [i.e., cAMP-response element-binding protein (14)] are a critical part of disease pathology (44). Perhaps PQE-1 proteins bind expanded polyQ tracts and shield Q-rich cellular proteins from aberrant interactions with Htn-Q150. Previous work suggests that proteins that bind directly to expanded polyQ tracts can often be detected in polyQ-containing aggregates in vivo (45–48). In normal C. elegans expressing Htn-Q150, cytoplasmic aggregates of Htn-Q150 were detected in 5% of ASH neurons in young animals and in 55% of ASH neurons at 8 days (17). In young pqe-1 animals, the few surviving ASH neurons contain cytoplasmic aggregates (data not shown). Because we do not detect nuclear Htn-Q150 aggregates in our model, we evaluated whether the localization pattern of the PQE-1:GFP proteins would change in the ASH neurons of animals expressing Htn-Q150. By monitoring GFP fluorescence, we found that PQE-1B:GFP and PQE-1C:GFP proteins remained within the nucleus and were not detected in cytoplasmic aggregates (data not shown). We also directly assessed in vitro the ability of a short Q/P-rich N-terminal fragment of PQE-1 to bind to Htn-Q150 or to full-length Htn expressed in lymphoblastoid cell lines in GST pulldown assays (13); no binding was detected. These results, along with in vivo fluorescence studies, suggest that PQE-1 proteins may not normally protect neurons by directly binding to Htn-Q150. Perhaps PQE-1 proteins fulfill their protective function by binding specific low-abundant nuclear Q-rich cellular targets, shielding these proteins from inappropriate interactions with expanded huntingtin fragments.
Discussion
The genetic screen described in this paper used a previously established C. elegans model in which N-terminal fragments of the human huntingtin protein containing short and expanded glutamine tracts were expressed in C. elegans neurons (17). Only expanded huntingtin fragments caused neurodegeneration and formed aggregates. Degeneration was age- and polyQ length-dependent, reminiscent of HD in humans. Given the striking similarities between C. elegans neurons and human neurons at the molecular and functional level, genes that modify polyQ toxicity and cell death in vivo in C. elegans neurons are likely to modify toxicity in humans as well.
Here we describe the identification and analysis of the C. elegans polyQ enhancer-1 (pqe-1) gene. Loss of pqe-1 function strongly and specifically accelerated neurodegeneration and cell death of ASH neurons expressing N-terminal human huntingtin fragments containing expanded polyQ tracts. No obvious phenotypes have been observed in pqe-1 mutant animals other than enhancement of polyQ neurotoxicity. The data presented here support the idea that PQE-1 proteins protect neurons from expanded polyQ tracts, although the relationship between PQE-1 function and polyQ toxicity remains unresolved. PQE-1 proteins may participate in the pathogenic mechanism underlying polyQ-induced neurodegeneration. Alternatively, mutations in pqe-1 may function as a genetic modifier of the age of onset or progression of neurodegeneration.
The molecular structure of PQE-1 proteins may provide insight into how these proteins participate in the neurodegeneration and cell death of ASH neurons expressing the expanded huntingtin fragments. Like many proteins found in eukaryotic organisms (49), the N terminus of both PQE-1A and PQE-1C is rich in glutamines. One popular hypothesis is that expanded polyQ tracts interact with Q-rich cellular targets, sequestering these proteins from their normal function, leading to cellular dysfunction. In our analysis of PQE-1 proteins, we did not detect a direct interaction between a short Q/P-rich N-terminal fragment of PQE-1 and expanded huntingtin fragments. Furthermore, the low level of PQE-1 protein expression makes it unlikely that PQE-1 proteins normally protect neurons by directly binding to expanded polyQ repeats. Instead, we propose that the loss of PQE-1 proteins may expose low-abundant cellular targets to aberrant interactions with expanded huntingtin fragments, accelerating neurodegeneration and eventual cell death. Thus, the overexpression of PQE-1C may provide additional interactions with glutamine-rich proteins, protecting neurons from polyQ toxicity.
Interactions between glutamine- and asparagine-rich proteins have been proposed to play a role in both polyQ disease and prion formation (50). One therapeutic option is to interrupt aberrant protein–protein interactions presumed to cause pathology. Artificial proteins and peptides have been designed that bind to expanded polyQ tracts, thereby preventing aggregation or inappropriate interactions with cellular target proteins (51). An alternative strategy is to introduce a protein that binds to, but does not perturb the function of, a critical target protein. Expression of high levels of PQE-1A and PQE-1C could protect by binding cellular targets of polyQ tracts. Although many Q-rich proteins are found in eukaryotic organisms, a clear human homolog of full length PQE-1A or PQE-1C has not been identified. However, proteins with similar neuroprotective functions are likely to be found in human neurons.
Transcriptional dysregulation has been previously implicated in huntingtin toxicity in several systems (52). It is thought that mutant huntingtin sequesters transcription factors, interfering with normal gene transcription (14–16). Many transcriptional proteins are glutamine-rich, lending credence to this hypothesis. We did not identify any genes in our screen that encode transcription factors. But, given the essential role of CBP and Sp1 transcription factors, mutations in these genes were unlikely to be isolated in our screen of adult animals for polyQ toxicity enhancement.
Because the majority of pqe-1 mutant alleles (including rt13) did not affect the PQE-1B form of the protein, the role of the pqe-1 putative exonuclease domain in polyQ toxicity is unclear. This exonuclease domain is most similar (39% identical) to uncharacterized GOR proteins found in humans and gorillas (GenBank accession nos. XM_172745 and BAA00906) and to Saccharomyces cerevisiae Rex3p (33% identical) (41, 53). Rex3p is one of several related exonucleases involved in 3′ processing of U5 snRNA and Rnase P RNA. Rex3p is also required for the maturation of the RNA of MRP, a ribonucleoprotein enzyme involved in processing precursor rRNA in eukaryotes. Interestingly, mutations in genes involved in RNA binding have been identified as modifiers of polyQ toxicity in a Drosophila model for ataxin-1-induced neurodegeneration, suggesting a relationship between alterations in RNA processing and polyQ toxicity (23).
PolyQ modifiers identified in C. elegans provide candidate genes with therapeutic value for patient populations. The homologous proteins in humans may be directly involved in processes underlying pathogenesis or may modify disease onset or progression. Genetic modifiers of disease can have a major impact on patients (i.e., ApoE and Alzheimer's disease) but are poorly understood in polyQ diseases. For example, despite the rough correlation between polyQ tract length and age of disease onset, variation in the age of onset for HD is large. For a polyQ tract of 41Q, the age of onset ranges from the late 30s to the early 80s (54). PolyQ tract length accounts for only 70% of the variance in the age of onset for patients with less than 52Q (54). The remaining 30% of variance is likely genetic in origin. Two independent genetic linkage analyses suggest that about 10% of the remaining variance in Huntington's age of onset is linked to the GluR6 kainate receptor DNA region (6, 7). The remaining variance suggests that additional unidentified loci may make a significant contribution to the age of onset and progression of HD and other polyQ diseases. Perhaps polymorphisms in human glutamine/proline-rich proteins modify HD onset or progression.
Genetic analysis of polyQ neurotoxicity in C. elegans reveals that the pqe-1 gene normally provides dramatic protection against polyQ neurotoxicity. Perturbing pqe-1 gene function sensitizes the C. elegans ASH neurons to the toxic effects of expanded huntingtin fragments. Perturbation of pqe-1 also enhances the neurotoxicity associated with expression of expanded polyQ tracts in the context of ataxin-1 (unpublished results). Our results suggest that the PQE-1 proteins containing the Q/P-rich domain and the charged region are critical for protecting neurons against polyQ neurotoxicity. We propose that PQE-1 proteins protect neurons by interacting with potential low-abundant nuclear Q-rich cellular target(s) of expanded glutamine tracts. Further studies of PQE-1 function may provide insight into pathogenic mechanisms underlying general polyQ-induced neurodegeneration and cell death and may reveal new molecular targets for the treatment and prevention of the devastating effects of polyQ diseases in humans.
Supplementary Material
Acknowledgments
We thank Marcy MacDonald, Denise Ferkey, Sander van den Heuvel, and members of Sander van den Heuvel's laboratory for advice. M. Vidal (Harvard Medical School) provided the cDNA library, and Y. Kohara (NIG, Mishima) provided additional cDNA clones. The C. elegans Genetics Center provided C. elegans strains; A. Fire (Carnegie Institute) provided pPD95.67; A. Coulson (Sanger Centre) provided C. elegans cosmids; P. Sengupta (Brandeis) provided oyIs14; H. R. Horvitz (MIT) provided mt10113; and M. MacDonald (Massachusetts General Hospital) provided Htn lymphoblastoid cell lines. We thank John Speith (Washington University) at the C. elegans sequencing center for identifying low-abundance pqe-1 transcripts. We gratefully acknowledge the support of Massachusetts General Hospital Fund for Medical Discovery (P.W.F.); the Damon Runyon Cancer Research Fund DRG#1656 (C.V.); the Hereditary Disease Foundation; the Searle Scholar Foundation; and the National Institute of General Medical Sciences (A.C.H.).
Abbreviations
HD, Huntington's disease
polyQ, polyglutamine
DiD, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocynanine perchlorate
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Zoghbi H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23 217-247. [DOI] [PubMed] [Google Scholar]
- 2.Huntington's Disease Collaborative Research Group (1993) Cell 72 971-983. [DOI] [PubMed] [Google Scholar]
- 3.Strong T. V., Tagle, D. A., Valdes, J. M., Elmer, L. W., Boehm, K., Swaroop, M., Kaatz, K. W., Collins, F. S. & Albin, R. L. (1993) Nat. Genet. 5 259-265. [DOI] [PubMed] [Google Scholar]
- 4.Vonsattel J. P., Myers, R. H., Stevens, T. J., Ferrante, R. J., Bird, E. D. & Richardson, E. P., Jr. (1985) J. Neuropathol. Exp. Neurol. 44 559-577. [DOI] [PubMed] [Google Scholar]
- 5.Martin J. B. & Gusella, J. F. (1986) N. Engl. J. Med. 315 1267-1276. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein D. C., Leggo, J., Chiano, M., Dodge, A., Norbury, G., Rosser, E. & Craufurd, D. (1997) Proc. Natl. Acad. Sci. USA 94 3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDonald M. E., Vonsattel, J. P., Shrinidhi, J., Couropmitree, N. N., Cupples, L. A., Bird, E. D., Gusella, J. F. & Myers, R. H. (1999) Neurology 53 1330-1332. [DOI] [PubMed] [Google Scholar]
- 8.Trottier Y., Lutz, Y., Stevanin, G., Imbert, G., Devys, D., Cancel, G., Saudou, F., Weber, C., David, G., Tora, L., et al. (1995) Nature 378 403-406. [DOI] [PubMed] [Google Scholar]
- 9.White J. K., Auerbach, W., Duyao, M. P., Vonsattel, J. P., Gusella, J. F., Joyner, A. L. & MacDonald, M. E. (1997) Nat. Genet. 17 404-410. [DOI] [PubMed] [Google Scholar]
- 10.DiFiglia M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277 1990-1993. [DOI] [PubMed] [Google Scholar]
- 11.Kalchman M. A., Koide, H. B., McCutcheon, K., Graham, R. K., Nichol, K., Nishiyama, K., Kazemi-Esfarjani, P., Lynn, F. C., Wellington, C., Metzler, M., et al. (1997) Nat. Genet. 16 44-53. [DOI] [PubMed] [Google Scholar]
- 12.Wanker E. E., Rovira, C., Scherzinger, E., Hasenbank, R., Walter, S., Tait, D., Colicelli, J. & Lehrach, H. (1997) Hum. Mol. Genet. 6 487-495. [DOI] [PubMed] [Google Scholar]
- 13.Faber P. W., Barnes, G. T., Srinidhi, J., Chen, J., Gusella, J. F. & MacDonald, M. E. (1998) Hum. Mol. Genet. 7 1463-1474. [DOI] [PubMed] [Google Scholar]
- 14.McCampbell A., Taylor, J. P., Taye, A. A., Robitschek, J., Li, M., Walcott, J., Merry, D., Chai, Y., Paulson, H., Sobue, G., et al. (2000) Hum. Mol. Genet. 9 2197-2202. [DOI] [PubMed] [Google Scholar]
- 15.Nucifora F. C. J., Sasaki, M., Peters, M. F., Huang, H., Cooper, J. K., Yamada, M., Takahashi, H., Tsuji, S., Troncoso, J., Dawson, V. L., et al. (2001) Science 291 2423-2428. [DOI] [PubMed] [Google Scholar]
- 16.Dunah A. W., Jeong, H., Griffin, A., Kim, Y., Standaert, D. G., Hersch, S. M., Mouradian, M. M., Young, A. B., Tanese, N. & Krainc, D. (2002) Science 296 2238-2243. [DOI] [PubMed] [Google Scholar]
- 17.Faber P. W., Alter, J. R., MacDonald, M. E. & Hart, A. C. (1999) Proc. Natl. Acad. Sci. USA 96 179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satyal S. H., Schmidt, E., Kitagawa, K., Sondheimer, N., Lindquist, S., Kramer, J. M. & Morimoto, R. I. (2000) Proc. Natl. Acad. Sci. USA 97 5750-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazemi-Esfarjani P. & Benzer, S. (2000) Science 287 1837-1840. [DOI] [PubMed] [Google Scholar]
- 20.Warrick J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N. & Bonini, N. M. (1998) Cell 93 939-949. [DOI] [PubMed] [Google Scholar]
- 21.Jackson G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., MacDonald, M. E. & Zipursky, S. L. (1998) Neuron 21 633-642. [DOI] [PubMed] [Google Scholar]
- 22.Marsh J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J. & Thompson, L. M. (2000) Hum. Mol. Genet. 9 13-25. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Funez P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P. J., et al. (2000) Nature 408 101-106. [DOI] [PubMed] [Google Scholar]
- 24.Parker J. A., Connolly, J. B., Wellington, C., Hayden, M., Daussett, J. & Neri, C. (2001) Proc. Natl. Acad. Sci. USA 98 13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morley J., Brignull, H., Weyers, J. & Morimoto, R. (2002) Proc. Natl. Acad. Sci. USA 99 10417-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Troemel E. R., Chou, J. H., Dwyer, N. D., Colbert, H. A. & Bargmann, C. I. (1995) Cell 83 207-218. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan J. M. & Horvitz, H. R. (1993) Proc. Natl. Acad. Sci. USA 90 2227-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hart A. C., Kass, J., Shapiro, J. E. & Kaplan, J. M. (1999) J. Neurosci. 19 1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins L. A., Hedgecock, E. M., Thomson, J. N. & Culotti, J. G. (1986) Dev. Biol. 117 456-487. [DOI] [PubMed] [Google Scholar]
- 30.Fukushige T., Hawkins, M. G. & McGhee, J. D. (1998) Dev. Biol. 198 286-302. [PubMed] [Google Scholar]
- 31.Stewart H. I., O'Neil, N. J., Janke, D. L., Franz, N. W., Chamberlin, H. M., Howell, A. M., Gilchrist, E. J., Ha, T. T., Kuervers, L. M., Vatcher, G. P., et al. (1998) Mol. Gen. Genet. 260 280-288. [DOI] [PubMed] [Google Scholar]
- 32.Korswagen H. C., Durbin, R. M., Smits, M. T. & Plasterk, R. H. (1996) Proc. Natl. Acad. Sci. USA 93 14680-14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wicks S. R., Yeh, R. T., Gish, W. R., Waterston, R. H. & Plasterk, R. H. A. (2001) Nat. Genet. 28 160-164. [DOI] [PubMed] [Google Scholar]
- 34.Kramer J. M., French, R. P., Park, E. C. & Johnson, J. J. (1990) Mol. Cell. Biol. 10 2081-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nonet M. L., Staunton, J. E., Kilgard, M. P., Fergestad, T., Hartwieg, E., Horvitz, H. R., Jorgensen, E. M. & Meyer, B. J. (1997) J. Neurosci. 17 8061-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persichetti F., Ambrose, C. M., Ge, P., McNeil, S. M., Srinidhi, J., Anderson, M. A., Jenkins, B., Barnes, G. T., Duyao, M. P., Kanaley, L., et al. (1995) Mol. Med. 1 374-383. [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan J., Shaham, S., Ledoux, S., Ellis, H. M. & Horvitz, H. R. (1993) Cell 75 641-652. [DOI] [PubMed] [Google Scholar]
- 38.Reddien P. W., Cameron, S. & Horvitz, H. R. (2001) Nature 412 198-202. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Anoveros J., Ma, C. & Chalfie, M. (1995) Curr. Biol. 5 441-448. [DOI] [PubMed] [Google Scholar]
- 40.The C. elegans Sequencing Consortium (1998) Science 282 2012-2018. [DOI] [PubMed] [Google Scholar]
- 41.Moser M. J., Holley, W. R., Chatterjee, A. & Mian, I. S. (1997) Nucleic Acids Res. 25 5110-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein L., Sternberg, P., Durbin, R., Thierry-Mieg, J. & Spieth, J. (2001) Nucleic Acids Res. 29 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill A. A., Hunter, C. P., Tsung, B. T., Tucker-Kellogg, G. & Brown, E. L. (2000) Science 290 809-812. [DOI] [PubMed] [Google Scholar]
- 44.Perutz M. F. (1996) Curr. Opin. Struct. Biol. 6 848-858. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez I., Xu, C. J., Juo, P., Kakizaka, A., Blenis, J. & Yuan, J. (1999) Neuron 22 623-633. [DOI] [PubMed] [Google Scholar]
- 46.Stenoien D. L., Cummings, C. J., Adams, H. P., Mancini, M. G., Patel, K., DeMartino, G. N., Marcelli, M., Weigel, N. L. & Mancini, M. A. (1999) Hum. Mol. Genet. 8 731-741. [DOI] [PubMed] [Google Scholar]
- 47.Perez M. K., Paulson, H. L., Pendse, S. J., Saionz, S. J., Bonini, N. M. & Pittman, R. N. (1998) J. Cell Biol. 143 1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chai Y., Koppenhafer, S. L., Bonini, N. M. & Paulson, H. L. (1999) J. Neurosci. 19 10338-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michelitsch M. D. & Weissman, J. S. (2002) Proc. Natl. Acad. Sci. USA 97 11910-11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perutz M. F. (1999) Trends Biochem. Sci. 24 58-63. [DOI] [PubMed] [Google Scholar]
- 51.Kazantsev A., Walker, H. A., Slepko, N., Bear, J. E., Preisinger, E., Steffan, J. S., Zhu, Y.-Z., Gertler, F. B., Housman, D. E., Marsh, J. L., et al. (2002) Nat. Genet. 30 367-376. [DOI] [PubMed] [Google Scholar]
- 52.Cha J.-H. (2000) Trends Neurosci. 23 387-392. [DOI] [PubMed] [Google Scholar]
- 53.van Hoof A., Lennertz, P. & Parker, R. (2000) EMBO J. 19 1357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gusella J. F., Persichetti, F. & MacDonald, M. E. (1997) Mol. Med. 3 238-246. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.