Abstract
Flavescence dorée (FD) is a grapevine disease that afflicts several wine production areas in Europe, from Portugal to Serbia. FD is caused by a bacterium, “Candidatus Phytoplasma vitis,” which is spread throughout the vineyards by a leafhopper, Scaphoideus titanus (Cicadellidae). After collection of S. titanus specimens from FD-contaminated vineyards in three different areas in the Piedmont region of Italy, we performed a survey to characterize the bacterial microflora associated with this insect. Using length heterogeneity PCR with universal primers for bacteria we identified a major peak associated with almost all of the individuals examined (both males and females). Characterization by denaturing gradient gel electrophoresis confirmed the presence of a major band that, after sequencing, showed a 97 to 99% identity with Bacteroidetes symbionts of the “Candidatus Cardinium hertigii” group. In addition, electron microscopy of tissues of S. titanus fed for 3 months on phytoplasma-infected grapevine plants showed bacterial cells with the typical morphology of “Ca. Cardinium hertigii.” This endosymbiont, tentatively designated ST1-C, was found in the cytoplasm of previtellogenic and vitellogenic ovarian cells, in the follicle cells, and in the fat body and salivary glands. In addition, cell morphologies resembling those of “Ca. Phytoplasma vitis” were detected in the midgut, and specific PCR assays indicated the presence of the phytoplasma in the gut, fat body and salivary glands. These results indicate that ST1-C and “Ca. Phytoplasma vitis” have a complex life cycle in the body of S. titanus and are colocalized in different organs and tissues.
Europe represents the main wine-producing area in the world. In this area, Flavescence Dorée (FD), an insect-borne plant yellow disease of grapevine, causes major concerns (2, 4, 8, 9, 26). Direct control strategies against the etiological agent of FD are indeed not available. In 1967, Japanese scientists reported that plant pathogen phytoplasmas (previously termed mycoplasma-like organisms) were the probable causes of plant yellow diseases (13). FD is caused by a bacterium lacking a cell wall, which has recently been proposed as “Candidatus Phytoplasma vitis” (15, 16, 21, 29, 32). The insect vector of FD is Scaphoideus titanus, a leafhopper (Hemiptera: Cicadellidae) that is widespread in vineyards. S. titanus transmits the bacterium by inoculating it into the phloem of the plant while feeding on the lymph. The control of FD is a high priority in European wine producing areas and is currently achieved through the destruction of infected vineyards and by extensive insecticide treatments against S. titanus. These control strategies have obvious economic and ecological impacts, and the treatment of vineyards with insecticides is incompatible with organic production.
In recent years there has been an increasing interest in the potential use of biological control agents (5, 6, 33, 37). Through the study of microorganisms associated with insects, different biocontrol strategies might be developed, first through the exploitation of microorganisms which are pathogenic to the insect (37). A further possible strategy is the exploitation of symbiotic microorganisms, with the aim of reducing vector competence (5). This reduction of the capacity of an insect to vector a disease agent could be based on microorganisms that naturally interfere with the presence of the pathogen (e.g., the unharmful Rickettsia peacocki, whose presence in the tick Dermacentor andersoni appears to reduce the prevalence of Rickettsia rickettsii [3]) or could be achieved through the genetic manipulation of insect symbiotic microorganisms. Microorganisms which possess the capacity to spread into insect host populations (e.g., by inducing cytoplasmic incompatibility [CI]) are particularly promising, since they could also be exploited to “hitch” a desired genetic trait (43). More in general, microorganisms that manipulate host reproduction, such as those belonging to the genera Wolbachia and Cardinium, offer intriguing clues for the development of future control strategies of insect populations and insect-vector competence (43, 44). In the field of insect-borne grapevine diseases, promising results toward control strategies based on symbiotic bacteria (symbiotic control) have been obtained in the sharpshooter Homalodisca coagulata, the vector of Pierce's disease caused by Xylella fastidiosa (10). It has been recently shown that cell-to-cell signaling controls the insect vector foregut colonization by Xylella fastidiosa, and hence it could be a good target for the disease control (30).
A prerequisite for the development of future strategies for the symbiotic control of insect populations and insect-vector competence is the identification of insect-associated microorganisms whose characteristics appear promising. These characteristics include the presence of the symbiont in organs that also host the pathogen and its potential to spread rapidly into the host populations. Despite the economic impact of FD on European wine production, no studies have thus far been published on the bacterial community associated with its vector S. titanus. In addition, the life cycle of the FD phytoplasma and its distribution throughout the body of the insect vector are not known.
We describe here a symbiont of S. titanus, found during a survey of bacteria stably associated with this insect. Our results show that this endosymbiont and “Ca. Phytoplasma vitis” have a partially overlapping colonization pathway in the insect body, colocalizing in the fat body and salivary glands.
MATERIALS AND METHODS
Insect collection.
A total of 118 S. titanus individuals (69 females, 34 males, and 15 nymphs) were collected between 2002 and 2004 in vineyards with heavy symptoms of FD from seven different areas in the Piedmont region (Table 1). Fifteen insects recovered in Tortona and Valleandona were taken to the Dipartimento di Valorizzazione e Protezione delle Risorse Agroforestali laboratory of the University of Turin and caged on FD-symptomatic grapevine plants (cultivar Barbera) in the greenhouse for a maximum period of 3 months. Insects were preserved frozen or in ethanol or acetone.
TABLE 1.
Insects sampled, their origin, and prevalence of “Ca. Phytoplasma vitis” and ST1-C endosymbiont as determined by specific PCR assays
| Vineyard (province)a | Insect IDb | Grapevine cultivar(s)c | Insect no. and typed | “Ca. Phytoplasma vitis”e | ST1-C endosymbionte |
|---|---|---|---|---|---|
| Boglietto (At) | 1, 7, 8*, 13, 18, 70, 73*, 74, 75, 92, 93, 116, 156, 244*, 251*, 393 | Barbera, Dolcetto, Cortese | 16 (F, 4; M, 0; N, 12) | 0/16 (F, 0/4; M, 0/0; N, 0/12) | 15/16 (M, 0/0; F, 4/4; N, 11/12) |
| Vignale (Al) | 125, 131, 136, 145, 146, 149, 152, 153, 154, 223, 225, 227*, 228*, 229, 230, 231, 233, 234, 236, 237, 238, 263, 270, 278, 307, 328, 338, 340, 344, 346, 348, 350, 367*, 369, 374*, 377, 378, 379, 381, 382, 390, 428, 431, 437 | Nebbiolo | 44 (F, 25; M, 16; N, 3) | 13/44 (F, 9/25; M, 4/16; N, 0/3) | 42/44 (F, 25/25; M, 14/16; N, 3/3) |
| S. Giorgio (At) | 176, 187, 196*, 352, 357, 386*, 396 | Nebbiolo, Rouchet | 7 (F, 5; M, 2) | 0/7 (F, 0/5; M, 0/2) | 6/7 (F, 4/5; M, 2/2) |
| Valleandona (At) | 449, 452, 453, 455, 456, 457, 458, 460, 461, 462, 463, 464, 466, 467, 468, 469, 471, 472**, 473, 474, 475, 476, 477 | Barbera | 23 (F, 14; M, 9) | 6/23 (F, 4/14; M, 2/9) | 22/23 (F, 13/14; M, 9/9) |
| S. Damiano (At) | 439, 440, 441, 447, 451 | Dolcetto | 5 (F, 5) | 1/5 (F, 1/5) | 4/5 (F 4/5) |
| Viarigi (At) | 478 | Grignolino | 1 (F, 1) | 0/1 (F, 0/1) | 1/1 (F, 1/1) |
| Tortona (Al) | 445 | Cortese | 1 (F, 1) | 0/1 (F, 0/1) | 1/1 (F, 1/1) |
| Laboratory plants | 480, 481, 482, 483, 484, 485 | Barbera | 6 (F, 3; M, 3) | 2/6 (F, 2/3; M, 0/3) | 6/6 (F, 3/3; M, 3/3) |
At, Asti; Al, Alessandria.
Individuals marked with single and double asterisks were used in the first LH-PCR and PCR-DGGE screening and for cloning and sequencing, respectively. ID, identification number.
Plant cultivars in the sampled vineyard.
Numbers indicate the individuals sampled for each category: M, males; F, females; N, nymphs-neanides.
Number of individuals positive for the specific PCR assay with respect to the number of total individuals tested. M, males; F, females; N, nymphs.
Insect dissection, TEM, and DNA extraction.
Male and female individuals of S. titanus were killed with ethyl acetate. The 20 insects to be studied by transmission electron microscopy (TEM) were dissected with sterile scalpels and small pliers in a sterile saline solution. The following insect parts were isolated: salivary glands, gut, fat bodies, and ovaries. These samples were fixed in 0.1 M cacodylate buffer (pH 7.2) containing 2.5% glutaraldehyde for 3 h at 4°C. The samples were then washed and postfixed in 1% OsO4 in 0.1 M cacodylate buffer (pH 7.2) for 1.5 h at 4°C. Successively all samples were dehydrated in ethanol and embedded in Epon 812. Thin sections (80 nm) were stained with uranil acetate and lead citrate and examined under a Zeiss EM900 transmission electron microscope (7, 35).
Fifteen S. titanus individuals were also dissected, and total DNA was extracted (from the salivary glands, the intestine, the fat bodies, and the ovaries). DNA extraction was performed according to a method previously described by Doyle and Doyle (14).
Molecular techniques to study the microflora in S. titanus.
Microflora associated with S. titanus was characterized by using two methods independent from bacteria cultivation.
A preliminary screening of the bacterial population associated with adult insects was performed by length heterogeneity PCR (LH-PCR) (34, 39). For LH-PCR, purified DNA was amplified by using the forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and the reverse primer 338R (5′-GCTGCCTCCCGTAGGAGT-3′) (34). Both primers are considered specific for bacteria. The 27F primer was labeled at its 5′ end with the phosphoramidite dye 6-FAM. The reaction mixture contained 1× PCR buffer (Pharmacia, Milan, Italy), 2.5 mM MgCl2, 0.10 mM deoxynucleoside triphosphates, 0.5 μM concentrations of each primer, 5% dimethyl sulfoxide, 1 U of Taq polymerase (Pharmacia), and 50 ng of DNA extracted from the whole insect in a final volume of 50 μl. Initial denaturation at 95°C for 5 min was followed by 35 cycles of 95°C for 1 min, annealing at 55°C for 45 s, and extension at 72°C for 2 min. A final extension at 72°C for 10 min was added.
The amount of PCR product was estimated on an agarose gel, and 3 μl of the product was added to 0.8 μl of 500 ROX-labeled internal size standard (Applied Biosystems, Monza, Italy) and 15 μl of deionized formamide. Samples were denatured at 95°C for 8 min and placed into an ice bath for 5 min. LH-PCR fragments were loaded on an ABI Prism 310 capillary electrophoresis system and run in denaturing conditions using POP-4 running polymer. Samples were run for 40 min at 15 kV. The injection time of each sample was 5 s at 15 kV. The LH-PCR data were analyzed with Genescan 3.1.2 software (Applied Biosystems), and a threshold of 50 fluorescent units was used, corresponding to three times the value of the highest peak detectable in the PCR-negative control run in the same conditions.
For denaturing gradient gel electrophoresis (DGGE) analysis, the primers GC357f, containing a 40-bp GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′), and 907r (5′-CCGTCAATTCCTTTGAGTTT-3′) were used to amplify the 16S rRNA gene as previously described (36). Polyacrylamide gels (7% of a 37:1 acrylamide-bisacrylamide mixture in 1× Tris-acetate-EDTA [TAE] buffer), with a gradient of 40 to 60% denaturant (100% denaturing polyacrylamide was defined as 7 M urea) and 40% formamide according to the method of Muyzer et al. (28), were made with a gradient maker (Bio-Rad, Milan, Italy) according to the manufacturer's guidelines. Gels were run for 15 h at 110 V in 1× TAE buffer at a constant temperature of 60°C in a D-Code electrophoresis system (Bio-Rad). The gels were stained for 30 min in 1× TAE buffer containing SYBR Green (Molecular Probes, Leiden, The Netherlands). Visualization and digital image recording was performed with GelDoc 2000 apparatus (Bio-Rad) by using the Diversity Database software (Bio-Rad).
Sequencing of DGGE bands.
DGGE bands were excised from the gel by using a sterile blade, transferred to 50 μl of MilliQ water, and frozen at −20°C until reamplification. Prior to amplification the bands in the Tris-EDTA were eluted at 37°C for 6 h. Reamplification was done by using the primers 357f (without GC clamp) and 907r and 9 μl of the eluted DNA fragments as a template. The following protocol was used: initial DNA denaturation at 94°C for 4 min, followed by 30 cycles at 94°C for 30 s, 64°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 7 min. PCR products were purified by using a QIAquick PCR Purification Kit (QIAGEN, Milan, Italy) according to the manufacturer's instructions. Purified products were then sequenced with the 357f primer by using a DYEnamic ET Terminator Cycle Sequencing Kit (Pharmacia) and an ABI 310 automated sequencer (Applied Biosystems). The resulting sequences were compared to the sequence database at the National Center for Biotechnology Information by using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) (1).
Additional sequence of the 16S rRNA of ST1-C endosymbiont was extended by two additional specific PCRs as follows. From the sequence of the DGGE bands, two primers were designed to amplify the regions outside the 5′ and 3′ ends of the fragment, as explained below. The specificity of the primers was evaluated by aligning the selected sequence in the National Center for Biotechnology Information database by using BLAST. Hence, the two primers named Endo F1 (5′-GTACAGGAGCAAAAAAGCC-3′) and Endo R3 (5′-AGTTAAAGACCAGTAAGCT-3′) were used in combination, respectively, with the reverse primer 1495R (5′-CTACGGCTACCTTGTTACGA-3′) and the forward universal primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) to amplify with specific reactions the flanking regions at the 5′ and 3′ end of the DGGE fragment. After amplification the fragments were sequenced and aligned with the original sequence from the DGGE fragment. The final sequence was then used for analyzing the phylogenetic position of the bacterium.
“Ca. Phytoplasma vitis” 16S rRNA gene was amplified by PCR test with the primer pair fAY-rEY (∼300 bp) (25, 27) or a specific nested PCR previously described, with primer pair R16F2n-R16SR2 for the first round of PCR and R16[V]F1/R16[V]R1 (22). The obtained fragments were hence sequenced.
Phylogenetic analysis.
16S rRNA sequences of ST1-C endosymbiont and of the putative phytoplasma were subjected to BLAST analysis and aligned with close relatives, as well as with other unrelated bacterial sequences. Alignment with the corresponding 16S rRNAs was performed by using software available at the Ribosomal Database Project website (11); secondary structure was taken into account when this was done. Phylogenetic analyses were performed by using Jukes and Cantor distance estimation with the TREECON 1.3b package (40). A 50% majority rule bootstrap consensus tree (1,000 replicates) was generated. Gaps were treated as a fifth base.
Prevalence of ST1-C endosymbiont and “Ca. Phytoplasma vitis” in S. titanus populations.
The alignment of ST1-C 16S rRNA sequence with related bacterial sequences was used to design primers that specifically target a fragment of the new sequence. During the process of designing primers, regions in which there was high variability were chosen, and the BLAST program was used to examine the number of mismatches that candidate primers had with all other known 16S rRNA sequences, in order to avoid potential amplification from other bacteria. The forward and reverse primers were Endo F1 and Endo R3, respectively. Endo F1 and Endo R3 had no matches with any bacterial or invertebrate sequences in GenBank at the time of checking and correspond to positions 443 to 465 and to positions 751 to 770, respectively, in the 16S rRNA sequence of Escherichia coli strain K-12 (accession number AE000452). They amplify a 305-bp fragment of the newly discovered sequence. For searching “Ca. Phytoplasma vitis,” the nested PCR previously described was used (22, 23). A total of 103 S. titanus individuals, including those examined by DGGE and LH-PCR, were screened by PCR. A further 15 individuals were dissected as described above, and each organ or body part was then analyzed by PCR. PCR amplifications were performed in 50 μl of buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin) containing each deoxynucleotide triphosphate at a concentration of 0.12 mM, 0.6 μM concentrations of each primer, 1 U of Taq polymerase (Perkin-Elmer), and 1 μl of DNA sample. The cycling conditions were as follows: 94°C for 4 min; five cycles of 94°C for 1 min, 50°C for 45 s, and 72°C for 1 min; 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min; and 7 min at 72°C.
Nucleotide sequence accession number.
The nucleotide sequence of ST1-C endosymbiont 16S rRNA gene was deposited in the EMBL nucleotide sequence database (GenBank/EMBL/DDBJ) under the accession number AM042540.
RESULTS
Characterization of the bacterial community associated with S. titanus.
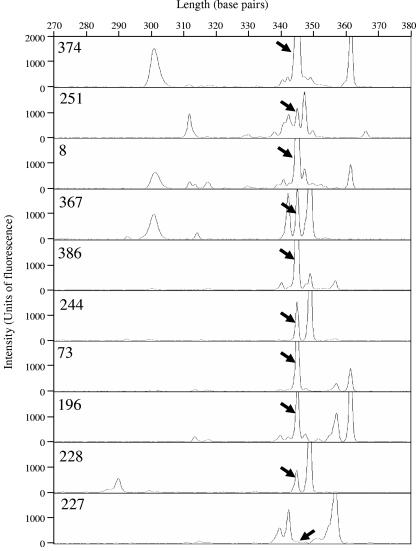
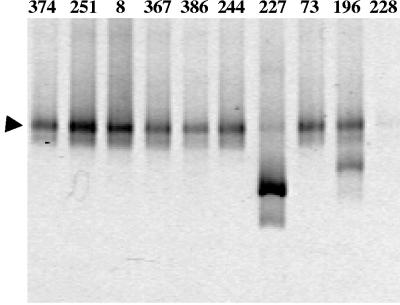
LH-PCR was used to survey the diversity of the bacterial community associated with S. titanus. This technique uses universal bacterial primers to amplify a defined portion of the 16S rRNA gene, including two variable regions, and can discriminate different bacterial species basing on length polymorphisms between different 16S rRNA genes. We initially surveyed 10 insects recovered in three different areas of Piedmont using as a template for LH-PCR the DNA extracted from the whole insect. Interestingly, we found one of the dominant peaks (345 bp) in the profiles obtained from the whole insect DNA conserved in almost all of the individuals examined, suggesting a stable bacterial association with the insect (Fig. 1). The bacterial community associated with S. titanus was then analyzed by DGGE of a longer portion (about 600 bp) of the 16S rRNA gene. Some variability was again observed in the community profiles among different individuals. A band, named ST1, with a high intensity was found to be associated with almost all of the individuals examined (Fig. 2).
FIG. 1.
Example of electropherograms revealing part of the bacterial community. LH-PCR profiles were obtained from DNA extracted from whole insects recovered from FD-affected vineyards in Piedmont, Italy. Numbers refer to the individuals reported in Table 1. The peaks with a length compatible to that of ST1-C endosymbiont are indicated with arrows.
FIG. 2.
DGGE patterns on a 7% polyacrylamide denaturing gel (40 to 60% urea and formamide) obtained from DNA extracted from whole insects recovered from FD-affected vineyards in Piedmont, Italy. Numbers over the lanes refer to the individuals reported in Table 1. The arrowhead indicates ST1 band identified by sequencing as explained in the text.
Phylogeny of bacteria associated with S. titanus.
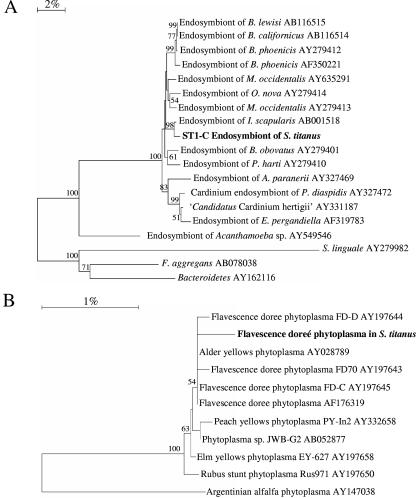
The sequence of the major band ST1 shared by almost all of the samples analyzed by DGGE (Fig. 2) was 99% identical to a bacterium in the phylum Bacteroidetes associated with the tick Ixodes scapularis. By using a combination of universal primers for the domain Bacteria at the 5′ and 3′ ends of the 16S rRNA gene and two primers specific for the sequence of the ST1 band in the DGGE profile, almost the entire sequence of this 16S rRNA gene was obtained. This sequence (named ST1-C) was used to construct a phylogenetic tree including the most-related sequences found in the public database (Fig. 3A). The nearly complete ST1-C sequence was confirmed to be closely related to that of a bacterium associated with the tick Ixodes scapularis (98% identity). The ST1-C sequence was in a branch of the neighbor-joining tree with sequences derived from endosymbionts of several species of the genus Brevipalpus, including the feminizing symbiont of B. phoenicis (41, 42), and of other acarine genera such as Metaseiulus, Oppiella, and Petrobia (17, 42). A group of sequences in another branch of the neighbor-joining tree closely related with the ST1-C sequence included endosymbionts of Encarsia pergandiella (44), Aspidiotus paranerii (42), and Plagiomerus diaspidis (46). All of these sequences were grouped with “Ca. Cardinium hertigii,” an endosymbiont of Encarsia wasps (96% identity with ST1-C sequence) (44, 45). Sequences related to these endosymbionts have never been previously associated with S. titanus or other leafhoppers (46).
FIG. 3.
Phylogenetic trees showing the phylogenetic positions of ST1-C endosymbiont (A) and “Ca. Phytoplasma vitis” (B) 16S rRNA genes. The trees were built without an outgroup sequence. The numbers at each node represent percentages of bootstrap replications calculated from 1,000 replicate trees. The scale bar represents the sequence divergence.
Few other bacteria were found from DGGE fingerprinting of S. titanus. Among these, several γ-Proteobacteria, β-Proteobacteria, and Bacteroidetes were found in many of the samples. Among γ-Proteobacteria, several sequences were associated with the genus Asaia. Bacteria of this genus have been described as epiphytic and associated with different plant species (18). Other sequences affiliated with the γ-Proteobacteria were homologous to the genus Stenotrophomonas. Species in this genus have been detected in feces or associated with insects such as Collembola (12). In this genus the species S. maltophilia has been described for its capacity to produce chitinases (19). Chitin is a common polymer in the insect body and may be used by these bacteria as growth substrate. Among Bacteroidetes, besides the ST1-C, a sequence related to the genus Chriseobacterium was detected. It was not possible to find an evident and clear correlation among specific sequences and the sex, the age, or the origin of the insect.
By using primers specifically designed for the amplification of “Ca. Phytoplasma vitis,” it was possible to obtain the expected fragment from one of the insects examined (Table 1). The amplified fragments were sequenced and showed 98% nucleotide identity with the “Ca. Phytoplasma vitis” (Fig. 3B). Restriction analysis with TacI restriction enzyme showed that it belongs to the 16SrV subgroup C (data not shown). Fragments with homology to this bacterium were not identified in PCR-DGGE profiles.
Localization of a bacterial endosymbiont and “Ca. Phytoplasma vitis” in adult S. titanus.
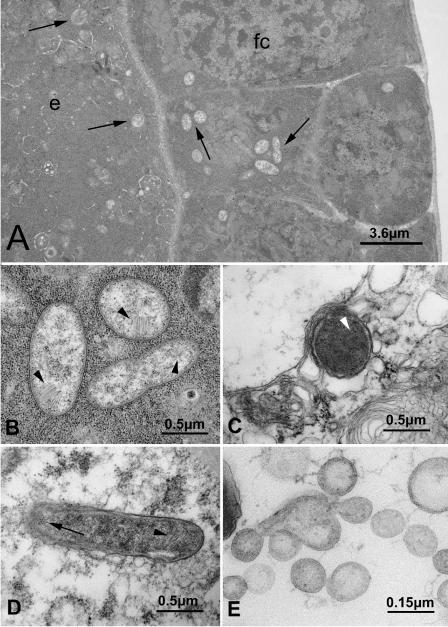
Different organs of individuals of S. titanus fed on grapevine plants with strong symptoms of FD and in which a phytoplasma was diagnosed by PCR were examined by TEM (Fig. 4). Many different bacterial morphotypes were observed in the gut of the S. titanus in contrast with the low diversity observed by LH-PCR and DGGE analysis (data not shown).
FIG. 4.
Transmission electron micrographs showing the putative ST1-C and “Ca. Phytoplasma vitis” in different organs or tissues of Scaphoideus titanus. (A) ST1-C (arrows) in follicle cells (fc) and in the egg (e). (B) Details of panel A showing three ST1-C cells with the typical brush-like structure (arrowheads). (C) ST1-C in the salivary glands (arrowhead). (D) ST1-C in the fat body showing transverse (arrow) and tangential (arrowhead) sections of the brush-like structure. (E) Cells of “Ca. Phytoplasma vitis” in the lumen of the midgut.
TEM examination of the ovaries showed numerous bacteria in both oocytes and follicle cells (Fig. 4A). Morphological analysis of these bacterial cells revealed a two-layered envelope (an outer cell wall and an inner plasma membrane) and a brush-like array of microfilament-like structures that appear to be characteristic of the symbiont. This parallel array of tubes extended from the cytoplasmatic membrane into the cytosol of the bacterium (Fig. 4B). This structure was the same as that described in the tick Ixodes scapularis symbiont (20) and in “Ca. Cardinium hertigii,” a bacterial endosymbiont of Encarsia pergandiella (44, 45). Bacterial cells with this typical morphology were found in the follicle cells and developing oocytes of both the previtellogenic and the vitellogenic phases (Fig. 4A and B). No other bacterial morphotypes were found in all of the ovarian cells examined. Cells with the same morphotypes of “Ca. Cardinium hertigii,” including the same brush-like structure in the cytoplasm (45), were also found in the salivary glands (Fig. 4C) and fat bodies of S. titanus adults (Fig. 4D).
TEM analysis also showed many small (0.25 to 0.5 μm) spherical cells with a triple-layered membrane resembling the typical morphotype of “Ca. Phytoplasma vitis” (Fig. 4E). These cells were found in the midgut (Fig. 4E), suggesting that the phytoplasma enters the intestine of S. titanus and, probably, through the hemolymph arrives to the salivary glands. Phytoplasma cells were never found associated with the ovary cells.
Prevalence of ST1-C endosymbiont and “Ca. Phytoplasma vitis” in S. titanus.
A wider collection of 103 individuals (including those analyzed by DGGE and LH-PCR) of S. titanus recovered in different years from different areas in Piedmont was screened by specific PCR assays to evaluate the prevalence of ST1-C endosymbiont and “Ca. Phytoplasma vitis” in populations of S. titanus and in the insect tissues (Table 1).
ST1-C endosymbiont was found in 97 of the 103 field-collected individuals (minimal field infection rate, 94.2%), including six field-collected individuals that were fed in the laboratory on plants with heavy symptoms of the phytoplasmosis. Of the 58 females examined, 55 (94.8%) were found to be positive. Of the 30 individual males tested, 28 were also positive (93.3%), confirming that the Bacteroidetes endosymbiont is not restricted to the ovaries but colonizes other nonsexual tissues. Fourteen of the 15 nymphs tested (93.3%) were positive. Direct sequencing of PCR products obtained from four adults (two males and two females) showed they were identical to the ST1-C 16S rRNA.
Of 103 individuals (minimal field infection rate, 21.4%), including field-collected individuals, fed in the laboratory on plants with heavy symptoms of the FD, 22 yielded positive results in the nested PCR assay on the 16S rRNA gene specific for the “Ca. Phytoplasma vitis” (Table 1). The identity of the fragment was confirmed by direct sequencing of the PCR fragment from two individuals. A total of 16 of the 58 females (27.6%) and 6 of the 30 males (20%) were positive, whereas none of the nymphs tested gave detectable amplification signals. Among the different vineyards Vignale showed the high percentage (29.5%) of insects infected by “Ca. Phytoplasma vitis.”
Fifteen adults (eleven females and four males) were dissected, and specific PCR analyses for ST1-C endosymbiont and “Ca. Phytoplasma vitis” 16S rRNA genes were performed with DNA extracted from the different organs (Table 2). ST1-C endosymbiont was detected in the fat bodies, intestine, and salivary glands of all of the four adult males analyzed. All of the 11 females were positive for the fat bodies, ovaries, and intestines, while 14 of the 15 tested (93.3%) were positive for the salivary glands. “Ca. Phytoplasma vitis” was detected in 1 of the 11 female and in 1 of the 4 male fat bodies, ovaries, and intestines analyzed, while it was not detected in the ovaries of any of the insects analyzed (neither females nor males).
TABLE 2.
Prevalence of “Ca. Phytoplasma vitis” and ST1-C endosymbionts in different organs or tissues of S. titanus as determined by specific PCR assays
| Insect IDa | Insect type and no.b | “Ca. Phytoplasma vitis”c
|
ST1-C endosymbiontc
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Salivary glands | Intestine | Fat bodies | Ovaries | Salivary glands | Intestine | Fat bodies | Ovaries | ||
| 440, 445, 447, 451, 452, 455, 464, 466, 467, 468, 469, 471, 472*, 473, 478 | 15 (F, 11; M, 4) | 2/15 (F, 1/11; M, 1/4) | 2/15 (F, 1/11; M, 1/4) | 2/15 (F, 1/11; M, 1/4) | 0/11 (F, 0/11) | 14/15 (F, 10/11; M, 4/4) | 15/15 (F, 11/11; M, 4/4) | 15/15 (F, 11/11; M, 4/4) | 11/11 (F, 11/11) |
Refer to Table 1 for sample origins. Individual marked with an asterisk was used in PCR-DGGE screening and for cloning and sequencing. ID, identification number.
Numbers indicate the individuals sampled for each category: M, males; F, females.
Number of individuals positive for the specific PCR assay with respect to the number of total individuals tested. M, males; F, females.
DISCUSSION
Our study of the bacterial diversity associated with S. titanus was initiated by using methods independent from cultivation and based on length (LH-PCR) and sequence (DGGE) polymorphisms of the 16S rRNA gene, using total DNA extracted from the whole insect body. We found a low microbial diversity with both LH-PCR and DGGE that used primers targeting different regions of the 16S rRNA gene. The diversity profiles obtained with each of the methods indicated that a single sequence predominated in the bacterial microflora. This is in contrast to the results obtained with TEM analysis of the insect intestinal lumen that showed a rather morphologically heterogeneous bacterial population (results not shown). This contradictory result has two possible explanations: (i) the method for extracting the DNA from the insect body did not efficiently lyse all of the bacteria living in the insect or (ii) it reflects the dominance of a single type of bacterium associated with S. titanus. Indeed, by using stronger cell lysis protocols, we were able to obtain a higher number of peaks in the LH-PCR profiles (data not shown). However, under these conditions we found one of the dominant peaks (345 bp) conserved among almost all of the insects examined, indicating that a specific bacterium colonizes the insect body.
BLAST and phylogenetic analyses of almost the entire 16S rRNA gene sequence initially derived from the main band present in the DGGE profiles placed it among the homologous sequences of members of phylum Bacteroidetes. Thus, there is evidence that S. titanus harbors a dominant bacterium, the ST1-C endosymbiont, belonging to the Bacteroidetes phylum. ST1-C endosymbiont showed the highest level of 16S rRNA nucleotide sequence identity with a bacterium associated with the tick I. scapularis (20) (Fig. 3A). The best-characterized bacterium affiliated with ST1-C is “Ca. Cardinium hertigii,” an endosymbiont of the parasitoid wasp Encarsia pergandiella (45). The level of 16S rRNA gene identity (96%) between ST1-C and “Ca. Cardinium hertigii” suggests that the bacterium associated with S. titanus may represent a separate species from “Ca. Cardinium hertigii.” “Ca. Cardinium hertigii” and related bacteria are characterized by a peculiar brush-like structure attached to the inner membrane (44, 45), which we also observed by TEM in the intracellular bacteria present in different tissues of S. titanus (see discussion below). Since molecular phylogenetic analysis placed ST1-C among a group of bacteria that show the peculiar brush-like structure of “Ca. Cardinium hertigii,” we can assume that the bacteria observed by TEM with the same brush-like structure putatively correspond to ST1-C.
In our study, bacteria with the typical brush-like structure of the ST1-C endosymbiont were initially observed in the ovaries of S. titanus females. However, the presence of “Cardinium-like” bacteria was not restricted to the ovaries: it was also detected in the fat body and the salivary glands of both females and males. These findings suggest that ST1-C (i) is transovarially transmitted to the insect progeny; (ii) has a complex life cycle in the insect body, colonizing different and rather separate tissues; and (iii) possibly has an important role in the biology of S. titanus. These hypotheses are supported by the high prevalence of ST1-C in the populations of S. titanus. A minimal field infection rate of 94.2% was found by PCR, with very similar values in the males and females.
The possible types of interactions between arthropods and bacterial endosymbionts are (i) parasitism (the bacteria harm the hosts), (ii) commensalism or transient mutualism (the bacteria benefit the hosts or cause no harm, but are not necessary for host survival), (iii) reproductive parasitism (the bacteria do not harm the hosts in the normal sense but cause reproductive alterations, such as skewed sex ratios), and (iv) obligatory mutualism (e.g., the bacteria provide essential compounds to the hosts) (31).
Bacteria phylogenetically related to “Ca. Cardinium hertigii” or showing its typical brush-like structure have been observed in insects and acarines (both mites and ticks) (20, 46). As is the case for Wolbachia (38), these bacteria have been shown to be associated with a variety of reproductive alterations, including parthenogenesis, feminization of genetic males, and CI (44, 45). In S. titanus, no data are currently available that indicate any bias in the sex ratio. In addition, since our survey indicates that the prevalence of ST1-C is similar in males and females, we cannot predict a role of this bacterium as a sex-determining factor. The very high prevalence of ST1-C in females and males of different populations of S. titanus could be the result of a selective sweep of the bacterium. We could thus hypothesize that ST1-C diffused into the S. titanus population through a selective sweep caused by CI, as occurs for CI-inducing Wolbachia (38). Alternatively, the interaction between the bacterium and S. titanus may be mutualistic. If the mechanism underlying the interaction between ST1-C and S. titanus could be clarified and the occurrence of CI demonstrated, a potential target for developing future control strategies against FD could arise, similar to the case of Wolbachia (see, for example, reference 43). Also, the demonstration of a mutualistic interaction between ST1-C and S. titanus would perhaps offer a new target for the development of novel control strategies. An intriguing aspect of the association between ST1-C and S. titanus is the fact that this bacterium has been detected in the salivary glands, an ideal place to interact with saliva-transmitted agents such as “Ca. Phytoplasma vitis.”
“Ca. Phytoplasma vitis” was found by a specific PCR test in more than 21% of the S. titanus individuals analyzed. The analysis of the different organs of the insects indicated that “Ca. Phytoplasma vitis” is present in the intestines, the fat body, and the salivary glands. These results support a biological cycle of the FD phytoplasma in S. titanus similar to that described in studies on the laboratory model Euscelidius variegatus (24). In this model, the phytoplasma is acquired by the insect from FD-infected plants and, after multiplication in the insect body, passes to other tissues to reach the salivary glands, from which it is reinoculated in plants. In the intestine of S. titanus, presumably the first organ where the phytoplasma grows after infection, cells with a morphology strongly resembling that of “Ca. Phytoplasma vitis” were observed by TEM. The PCR study on the prevalence of ST1-C and “Ca. Phytoplasma vitis” in S. titanus indicated that the two bacteria coexist in the same insect individuals and in the same organs: the intestine, the fat body, and the salivary glands. The colocalization of the two bacteria in the same host tissues makes it possible to study the potential interactions between these bacteria in the insect body and makes ST1-C an interesting candidate for the symbiotic control of the FD agent, e.g., through a paratransgenesis approach (5, 10, 33).
Acknowledgments
We thank Franco Faoro for helpful comments on the TEM results and Nathan Lo for critically reading and editing the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Angelini, E., D. Clair, M. Borgo, A. Bertaccini, and E. Boudon-Padieu. 2001. Flavescence Doree in France and Italy: occurrence of closely related Phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 40:79-86. [Google Scholar]
- 3.Baldridge, G. D., N. Y. Burkhardt, J. A. Simser, T. J. Kurtti, and U. G. Munderloh. 2004. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood thick Dermacentor andersoni. Appl. Environ. Microbiol. 70:6628-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batlle, A., A. Lavina, C. Kuszala, D. Clair, J. Larrue, and E. Boudon-Padieu. 1997. Detection of Flavescence Doree phytoplasma in grapevine in northern Spain. Vitis 36:211-212. [Google Scholar]
- 5.Beard, C. B., R. V. Durvasula, and F. F. Richards. 1998. Bacterial symbiosis in arthropods and the control of disease transmission. Emerg. Infect. Dis. 4:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beard, C. G., C. Cordon-Rosales, and R. V. Durvasula. 2002. Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu. Rev. Entomol. 47:123-141. [DOI] [PubMed] [Google Scholar]
- 7.Beninati, T., N. Lo, L. Sacchi, C. Genchi, H. Noda, and C. Bandi. 2004. A novel alpha-Proteobacterium resides in the mitochondria of ovarian cells of the tick Ixodes ricinus. Appl. Environ. Microbiol. 70:2596-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianco, P., A. Alma, P. Casati, G. Scattini, and A. Arzone. 2001. Transmission of 16Srv phytoplasmas by Scaphoideus titanus Ball in northern Italy. Plant Prot. Sci. 37:49-56. [Google Scholar]
- 9.Bianco, P. A., P. Casati, R. E. Davis, and A. Fortusini. 1996. Prevalence of aster yellows (AY) and elm yellows (EY) group phytoplasmas in symptomatic grapevines in three areas of northern Italy. Vitis 35:195-199. [Google Scholar]
- 10.Bextine, B., C. Lauzon, S. Potter, D. Lampe, and T. A. Miller. 2004. Delivery of a genetically marked Alcaligenes sp. to the glassy-winged sharpshooter for use in a paratransgenic control strategy. Curr. Microbiol. 48:327-331. [DOI] [PubMed] [Google Scholar]
- 11.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czarnetzki, A. B., and C. C. Tebbe. 2004. Diversity of bacteria associated with Collembola: a cultivation-independent survey based on PCR-amplified 16S rRNA genes. FEMS Microbiol. Ecol. 49:217-227. [DOI] [PubMed] [Google Scholar]
- 13.Doi, Y. M., M. Teranaka, K. Yora, and H. Asuyama. 1967. Mycoplasma or PLT-group-like microorganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches' broom, aster yellows, or paulonia witches' broom. Ann. Phytopatol. Soc. Jpn. 33:259-266. [Google Scholar]
- 14.Doyle, J. J., and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissues. Focus 12:13-15. [Google Scholar]
- 15.IRPCM Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group. 2004. “Candidatus Phytoplasma,” a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54:1243-1255. [DOI] [PubMed] [Google Scholar]
- 16.Jagoueix-Eveillard, S., F. Tarendeau, K. Guolter, J. L. Danet, J. M. Bove, and M. Garnier. 2001. Catharanthus roseus genes regulated differentially by mollicute infections. Mol. Plant-Microbe Interact. 14:225-233. [DOI] [PubMed] [Google Scholar]
- 17.Jeyaprakash, A., and M. A. Hoy. 2004. Multiple displacement amplification in combination with high-fidelity PCR improves detection of bacteria from single females or eggs of Metaseiulus occidentalis (Nesbitt) (Acari: Phytoseiidae). J. Invertebr. Pathol. 86:111-116. [DOI] [PubMed] [Google Scholar]
- 18.Katsura, K., H. Kawasaki, W. Potacharoen, S. Saono, T. Seki, Y. Yamada, T. Uchimura, and K. Komagata. 2001. Asaia siamensis sp. nov., an acetic acid bacterium in the alpha-proteobacteria. Int. J. Syst. Evol. Microbiol. 51:559-563. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, D. Y., R. M. Reedy, J. Bick, and P. V. Oudemans. 2002. Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl. Environ. Microbiol. 68:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtti, T. J., U. G. Munderloh, T. G. Andreadis, L. A. Magnarelli, and T. N. Mather. 1996. Tick cell culture isolation of an intracellular prokaryote from the tick Ixodes scapularis. J. Invertebr. Pathol. 67:318-321. [DOI] [PubMed] [Google Scholar]
- 21.Lee, I. M., R. E. Davis, and D. E. Gundersen. 2000. Phytoplasma: phytopathogenic mollicutes. Annual Rev. Microbiol. 54:221-255. [DOI] [PubMed] [Google Scholar]
- 22.Lee, I. M., D. E. Gundersen, R. D. Hammond, and R. E. Davis. 1994. Use of mycoplasma-like organism (MLOs) group specific oligonucleotide primers for nested-PCR assay to detect mixed-MLO infection in a single host plant. Phytopathology 84:559-566. [Google Scholar]
- 23.Lee, I. M., R. D. Hammond, R. E. Davis, and D. E. Gundersen. 1993. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasma-like organism. Phytopathology 83:834-842. [Google Scholar]
- 24.Lefol, C., J. Lherminier, E. Boudon-Padieu, J. Larrue, C. Louis, and A. Caudwell. 1994. Propagation of Flavescence Dorèe MLO (mycoplasma-like organisms) in the leafhopper vector Euscelidius Variegatus. Kbm. J. Invertebr. Pathol. 63:285-293. [Google Scholar]
- 25.Marcone, C., A. Ragozzino, B. Schneider, U. Lauer, D. Smart, and E. Seemuller. 1996. Genetic characterization and classification of two phytoplasmas associated with spartium witches'-broom disease. Plant Dis. 80:365-371. [Google Scholar]
- 26.Martini, M., S. Botti, C. Marcone, C. Marzachi, P. Casati, P. A. Bianco, R. Benedetti, and A. Bertaccini. 2002. Genetic variability among flavescence dorée phytoplasmas from different origins in Italy and France. Mol. Cell Probes 16:197-208. [DOI] [PubMed] [Google Scholar]
- 27.Marzachì, C., S. Palermo, A. Boarino, F. Veratti, M. D'Aquilio, A. Loria, and G. Boccardo. 2001. Optimization of an one-step PCR assay for the diagnosis of the Flavescence Doreé-related phytoplasmas in field-grown grapevines and vector populations. Vitis 40:213-217. [Google Scholar]
- 28.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Namba, S. 2002. Molecular biological studies on phytoplasmas. J. Gen. Plant Pathol. 68:257-259. [Google Scholar]
- 30.Newman, K. L., R. P. Almeida, A. H. Purcell, and S. E. Lindow. 2004. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA 101:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neill, S. L., A. A. Hoffmann, and J. H. Werren. 1997. Influential passengers. Oxford University Press, New York, N.Y.
- 32.Oshima, K., S. Kakizawa, H. Nishigawa, H. Y. Jung, W. Wei, S. Suzuki, R. Arashida, D. Nakata, S. Miyata, M. Ugaki, and S. Namba. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic Phytoplasma. Nat. Genet. 36:27-29. [DOI] [PubMed] [Google Scholar]
- 33.Rio, R. V. M., Y. Hu, and S. Aksoy. 2004. Strategies for the home team: symbioses exploited for vector-borne disease control. Trends Microbiol. 12:325-336. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacchi, L., E. Bigliardi, S. Corona, T. Beninati, N. Lo, and A. Franceschi. 2004. A symbiont of the tick Ixodes ricinus invades and consumes mitochondria in a mode similar to that of the parasitic bacterium Bdellovibrio bacteriovorus. Tissue Cell 31:442-444. [DOI] [PubMed] [Google Scholar]
- 36.Sass, A. M., H. Sass, M. J. Coolen, H. Cypionka, and J. Overmann. 2001. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea). Appl. Environ. Microbiol. 67:5392-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stouthamer, R., J. A. Breeuwer, and G. D. Hurst. 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53:71-102. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 41.Weeks, A. R., F. Marec, and J. A. J. Breeuwer. 2001. A mite species that consists entirely of haploid females. Science 292:2479-2482. [DOI] [PubMed] [Google Scholar]
- 42.Weeks, A. R., R. Velten, and R. Stouthamer. 2003. Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proc. R. Soc. Lond. B Biol. Sci. 270:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zabalou, S., M. Riegler, M. Theodorakopoulou, C. Stauffer, C. Savakis, and K. Bourtzis. 2004. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. USA 101:15042-15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zchori-Fein, E., Y. Gottlieb, S. E. Kelly, J. K. Brown, J. M. Wilson, T. L. Karr, and M. S. Hunter. 2001. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc. Natl. Acad. Sci. USA 98:12555-12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zchori-Fein, E., S. J. Perlman, S. E. Kelly, N. Katzir, and M. S. Hunter. 2004. Characterization of a “Bacteroidetes” symbiont in Encarsia wasps (Hymenoptera: Aphelinidae): proposal of “Candidatus Cardinium hertigii.” Int. J. Syst. Evol. Microbiol. 54:961-968. [DOI] [PubMed] [Google Scholar]
- 46.Zchori-Fein, E., and S. J. Perlman. 2004. Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 13:2009-2016. [DOI] [PubMed] [Google Scholar]