Abstract
The brewer's yeast Saccharomyces cerevisiae has emerged as a versatile and robust model system for laboratory use to study toxic effects of various substances. In this study, toxicant-induced stresses of pure compounds were investigated in Saccharomyces cerevisiae utilizing a destabilized version of the green fluorescent protein optimized for expression in yeast (yEGFP3) under control of the promoter of the housekeeping plasma membrane ATPase gene PMA1. The responses of the biomarker upon increasing test compound concentrations were monitored by determining the decrease in fluorescence. The reporter assay deployed a simple and robust protocol for the rapid detection of toxic effects within a 96-well microplate format. Fluorescence emissions were normalized to cell growth determined by absorption and were correlated to internal reference standards. The results were expressed as effective concentrations (EC20). Dose-response experiments were conducted in which yeast cells were exposed in minimal medium and in the presence of 20% fetal calf serum to sublethal concentrations of an array of heavy metals, salt, and a number of stress-inducing compounds (Diclofenac, Lindane, methyl-N-nitro-N-nitrosoguanidine [MNNG], hydroxyurea, and caffeine). Long-term exposure (7 h) played a considerable role in the adaptive response to intoxication compared to early responses at 4 h exposure. The data obtained after 4 h of exposure and expressed as EC20 were compared to 50% inhibitory concentration values derived from cell line and ecotoxicological tests. This study demonstrates the versatility of the novel biomarker to complement existing test batteries to assess contaminant exposure and effects.
Conventionally, toxicologists have used bioassays based on rodent models to evaluate the toxic effects of chemical compounds and to study the mechanism of action of toxicants. However, scientific developments are required to keep in line with regulatory frameworks, such as existing EU guidelines for assessment of manufactured chemicals (67/548/EEC, 93/67/EEC, and 83/571/EEC) and the EU regulatory framework for chemicals REACH 2003 (European Commission [http://europa.eu.int/comm/enterprise/reach/index.htm]) concerning in part also existing chemicals. Scientific developments include the requirement for rapid and reliable high-throughput assays to evaluate more accurately and more mechanistically the potential hazards of large numbers of chemicals.
The yeast Saccharomyces cerevisiae is a promising model for such assays because it is amenable to genetic studies and because of the vast amount of genomics knowledge, resources, and manipulative tools associated with this unicellular fungus. The high degree of homology of essential cellular organization and metabolism shared by S. cerevisiae and higher eukaryotes has enabled study of aspects of cellular toxicity and phenomena of relevance to human biology at the molecular level (5, 30). Such research has offered many insights into the complex mechanisms underlying the sensing and response to toxicant stressors (13, 14). The degree to which gene expression profiles are conserved upon toxic stress and the regulation of key pathway elements enabled the identification of human signal transduction homologues (2, 15). Although gene expression profiling is not (yet) suited for high-throughput screening, insights in terms of hierarchic clustering of genetic stress-related networks potentially provides the means to identify surrogate markers that can be used to construct detection systems and prediction models.
PMA1, one of the most prominent housekeeping genes in S. cerevisiae, encodes the major plasma membrane H+-ATPase (35) and is essential for viability. As a highly conserved member of the P-type ATPases, the H+-ATPase is a single 100-kDa polypeptide. The electrogenic proton pump is the major source of cytosolic proton extrusion and generation of the proton motive force across the cellular membrane. The proton motive force is responsible for secondary active transport mechanisms for a variety of nutrients and is also involved in pH homeostasis. In being the major protein component of the plasma membrane (15 to 20% of total plasma membrane protein [1]), expression and activity of this proton pump are precisely regulated to match its numerous requirements (4, 11, 19, 38).
We have previously described the construction and preliminary characterization of chronic toxicity and genotoxicity test systems in sensitive S. cerevisiae host strains (21, 33, 34). We report here on the extension of the assay toward the use of the PMA1 promoter (PPMA1) of S. cerevisiae as a biomarker for the rapid detection of toxicant-induced stress on basic metabolism. A decrease in membrane potential has been suggested as primary cellular stress signal triggering the intracellular response (26). The PPMA1-mediated transcriptional activation of the yeast optimized green fluorescent protein (yEGFP3 [9]) results in the production of the green fluorescent protein (GFP). To monitor dynamic fluorescent changes, we have developed an assay whereby a fluorescence emission decrease indicates dose-dependent intoxication. To that end, a destabilized version of GFP was constructed by coupling the PEST-rich C-terminal residues of the G1 cyclin CLN2 which, as universal ubiquitin targeting sequence, confers rapid degradation of yeast proteins (25). The shift of the steady-state turnover of PPMA1-driven GFP transcription and PEST-mediated degradation upon intoxication, toward less transcription/translation and thus proportionally increased degradation, led to decreased fluorescence that served as a reporter for toxicant-induced stress. To enhance the sensitivity of the assay, the promoter-reporter constructs have been expressed in a yeast strain deleted in the multidrug ABC transporter genes PDR5, SNQ2, and YOR1 (18). Toxicant exposure in medium with 20% fetal calf serum (FCS) aimed to indicate the influence of serum components (i.e., proteins) that mediate adsorption of free chemicals and thus altered bioavailability on toxicity. With this assay we could empirically demonstrate toxic effects of heavy metals, a pharmaceutical, and other substances after 4 h of exposure.
MATERIALS AND METHODS
Plasmid construction.
To generate a multicopy (2-μm DNA-based) plasmid that constitutively expressed a destabilized version of the yeast- and fluorescence-activated cell sorting-optimized GFP (yEGFP3 [9]) in S. cerevisiae, the plasmid pYEX-PPMA1-GFP/PEST was constructed by using the episomal vector backbone of the high-copy E. coli/yeast shuttle vector pYEX-BX (Clontech, Palo Alto, CA). For that purpose a DNA fragment comprising the 3′-terminal 534 nucleotides of the G1 cyclin CLN2 which encodes the PEST-rich 178 amino acid residues conferring rapid degradation of yeast proteins (25) was amplified by PCR from S. cerevisiae strain PLY 232 (3) genomic DNA using the primers 5′-ACTGAGATCTGCATCCAACTTGAACATTTCGAGAAAGC-3′ and 5′-GAGAGAATTCCTATATTACTTGGGTATTGCCCATACC-3′, with additional nucleotides (in italics) comprising BglII and EcoRI restriction sites (underlined) to facilitate cloning. The PCR product was digested with BglII and EcoRI, and the resulting 543-bp fragment was ligated with the BglII/EcoRI 8.001-kb fragment of pYEX-yEGFP3 (J. Ludwig, unpublished data), yielding pYEX-GFP/PEST. By replacing the initial pYEX-BX CUP1 promoter sequence with that of PMA1 (pYEX-PPMA1-GFP) via the PvuII and MscI restriction sites, pYEX-PPMA1-yEGFP3/PEST was generated. The resulting reporter plasmid encodes the fusion gene (yEGFP3/PEST) encompassing the destabilizing PEST rich sequence from G1 cyclin CLN2 fused to yeast enhanced GFP under the control of the PMA1 promoter (PPMA1). All plasmids carry Ampr, LEU2, and URA3 genes for selection purposes. Recombinant plasmids, recovered from transformed Escherichia coli XL1-Blue cells were mapped by restriction analysis and confirmed by sequencing (GeneART). Computer analysis of nucleotide and amino acid sequences was performed by using the Vnti software (Informax, United Kingdom). To construct the reporter strain, the plasmid was obtained by standard DNA manipulations according to the method of Sambrook et al. (32), and approximately 1 μg of purified plasmid DNA was used to transform S. cerevisiae FYAK26/8-10B1 cells (18) by the lithium acetate method to uracil and leucine prototrophy (31).
Strains and growth conditions.
The S. cerevisiae strains used in the present study and relevant genotypes are listed in Table 1. Yeast cells were grown in minimal YNB (Difco) medium (1.7 g of yeast nitrogen base [without amino acids and without (NH4)2SO4], 10.5 g of citric acid buffer [AppliChem; final pH 6.4], and 0.5 g of amino acid dropout mix [41.7 mg of adenine, 83.3 mg of tryptophan, 16.7 mg of arginine, 16.7 mg of methionine, 25 mg of tyrosine, 25 mg of lysine, 50 mg of valine, 83.3 mg of threonine, 83.3 mg of serine, 41.7 mg of phenylalanine, 16.7 mg of asparagine, and 16.7 mg of glutamic acid; Sigma Aldrich] per liter) containing 0.5% (wt/vol) glucose as the carbon source, 20 mg of histidine, and 40 mg of leucine but lacking uracil for the maintenance of plasmids. Cells were routinely grown at 30°C. For experiments with medium containing final 20% (vol/vol) serum, and FCS was procured from Sigma Aldrich. Plasmid-dependent phenotypes were regularly checked by plasmid-loss experiments.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| FY1679-28C | MATa, ura3-52, trp1Δ63, leu2Δ1, his3Δ200, GAL2+ | 18 |
| FYAK26/8-10B1 | MATa, ura3-52, trp1Δ63, leu2Δ1, his3Δ200, GAL2+pdr5-Δ1::hisG snq2::hisG yor1-1::hisG | 18 |
| PMG-22 | FYAK26/8-10B1 [pYEX-PPMA1-GFP] | This study |
| PMG-23 | FYAK26/8-10B1 [pYEX-PPMA1-GFP/PEST] | This study |
Compounds tested in the present study.
The following compounds and/or ions were assayed for their toxicity by fluorescence repression potential: methyl-N-nitro-N-nitrosoguanidine (MNNG); the heavy metal ions Cd2+, Cr(IV), Co2+, and Cu2+; the pesticide Lindane; the CNS stimulant caffeine; the pharmaceuticals diclofenac and hydroxyurea; the PCP 3,5-dichlorophenol (3,5-DCP); and sodium chloride. All compounds were dissolved in distilled water, with the exception of Lindane, which was dissolved in a 2:3 mixture of ethanol and water. All test compounds were of analytical grade (Sigma Aldrich and Fluka).
Assay conditions and fluorescence monitoring.
Reporter repression was assayed by direct fluorescence readouts using a microplate reader (TECAN Spectra Fluoro Plus). All assays were carried out in 96-well microplates (Greiner; Cellstar #655185). For precultures, cells from a solid selective medium were transferred to liquid growth medium (∼10 ml) in 50-ml Erlenmeyer-Flasks. Cells were grown overnight to early stationary phase (with 250 rpm rotary shaking), collected by centrifugation, and resuspended in fresh YNB medium to a final optical density (OD) of 0.4 (A600; Pharmacia Ultraspec 2000). These starter cultures were used to fill the microplates (200 μl per well). Each experiment consisted of a minimum of five (for Cu2+) or eight (for NaCl) different compound concentrations (determinations were performed in hexaplicate), including the following controls: (i) negative control cultures (YNB medium with inoculum and solvent) to indicate maximum proliferative capacity and unimpaired fluorescence intensity; (ii) blank controls (compounds in appropriate concentrations and YNB medium without inoculum) to indicate endogenous compound absorbance and fluorescence; (iii) YNB medium to monitor potential contamination and medium absorbance and fluorescence; and (iv) positive control cultures (control i plus reference toxicant, e.g., 3,5-DCP) to ensure that the test and/or its components produced reliable results. Plates were covered with standard plastic lids, sealed with adhesive film to prevent evaporation, and then incubated (900-rpm horizontal shaking) at 30°C for 7 h. Compound-bound toxic effects were monitored by obtaining OD600 measurements (growth) and fluorescence readouts with an excitation wavelength of 485 nm and an emission at 535 (25-nm bandwidth) after 1, 2, 3, 4, and 7 h of incubation. For each tested compound, at least 10 replicate tests were carried out on different days.
Data collection and handling.
Concentration response curves were obtained by induction ratio (IR) calculation. The IR is the ratio of the arithmetic mean fluorescence FL (n = 6, corrected for blanks) divided by growth determined as the OD600 (n = 6, corrected for blanks) at the test concentration normalized to the corresponding values for the negative control cultures. Based on these data, EC20 values were determined by interpolation regression analysis (Probitanalysis; SPIRIT program, version 4.0, 1995). The results are summarized as the quantity of compound in mg/liter determined to reduce normalized light emission by 20% (EC20) of the mean fluorescence intensity of the negative control cultures. For detection of toxic effects the experimental rationale involved three criteria: (i) the observed parameter was potential fluorescence decrease upon compound exposure; (ii) the cell proliferation should not reach full doubling within the test period to exclude chronic toxic effects; and (iii) growth-inhibitory effects should not exceed 20% during the test period so that fluorescence decrease could unambiguously be ascribed to the chemical's toxic potential.
RESULTS
PPMA1-driven expression of destabilized GFP.
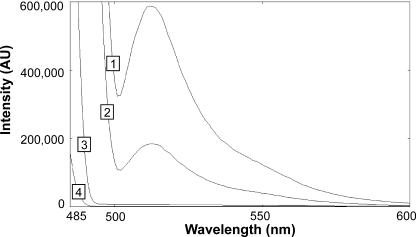
For determination of toxicant-induced stress, a plasma membrane ATPase promoter-reporter system was constructed in S. cerevisiae. The system explored PPMA1-driven transcription in conjunction with a CLN2 PEST sequence (25) that triggers rapid degradation of the yeast-enhanced version of the yEGFP chimeric construct (9) as a response to compound-mediated intoxication. According to that rationale, the entire promoter of the PMA1 gene was fused to the yEGFP gene with or without attachment of the C-terminal PEST-rich protein degradation signature of CLN2 (pPPMA1-GFP; pPPMA1-GFP/PEST). The approach deployed a S. cerevisiae strain, devoid of the pleiotropic drug transporters PDR5, SNQ2, and YOR1 (18), for increased sensitivity toward a broad spectrum of chemicals. Cells expressing the destabilized version of GFP (pPPMA1-GFP/PEST) showed a two-thirds-reduced fluorescence emission peak compared to cells transformed with the pPPMA1-GFP construct (Fig. 1).
FIG. 1.
Emission spectra (excitation at 485 nm) of 0.4 OD600 units of transformed (pPPMA1-GFP and pPPMA1-GFP/PEST, respectively) and nontransformed cells were compared to the emission spectrum of pure water. Cells were harvested by centrifugation, washed, and resuspended in sterile water. Spectrum 1 represents the light emission of cells expressing native GFP, spectrum 2 indicates the fluorescence emission of cells transformed with destabilized GFP/PEST, and spectrum 3 nontransformed cells and spectrum 4 pure water. The GFP emission maximum is at 511 nm. The steady-state fluorescence was reduced to ca. 30% by addition of the PEST degradation signature.
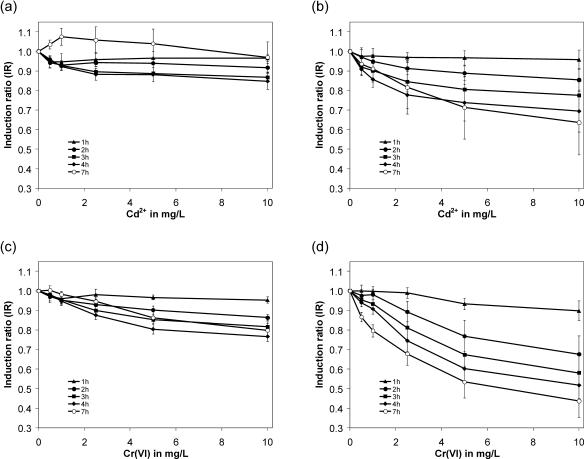
Time course experiments upon heavy metal exposure are depicted in Fig. 2. With the pPPMA1-GFP construct, cells exhibited no significant fluorescence decrease when challenged with increasing concentrations of Cr(VI) or Cd2+ (Fig. 2A and C). A reduction of the GFP signal in a dose- and time-dependent manner (Fig. 2B and D) was obtained with the modified, rapidly degradable construct. For chromium, the most prominent fluorescence decrease was obtained after 7 h of incubation. However, two criteria—cell proliferation below full doubling and growth inhibition below 20%—were not met at this time point (data not shown) so the results were regarded as not valid according to test definition. Suitable exposure times were identified as 3 and 4 h of incubation.
FIG. 2.
Time course experiments upon cadmium (a and b) and hexavalent chromium (c and d) ion exposure of native GFP and its modified, destabilized version. The diagrams are shown as concentration-response curves in mg/liter versus the calculated corresponding IRs. The data were compiled from at least 10 separate experiments with six identical wells per tested concentration. The results are expressed as means ± the standard deviation. (a and c) Cells expressing pPPMA1-GFP; (b and d) cells expressing pPPMA1-GFP/PEST. The fluorescence emission was depicted for 1-, 2-, 3-, 4-, and 7-h heavy metal incubation periods with different plot symbols (see definitions in each panel in diagrams). Only cells expressing the destabilized GFP reporter construct exhibited a concentration-dependent fluorescence emission decrease over time when challenged with cadmium or chromium ions.
Sensitivity.
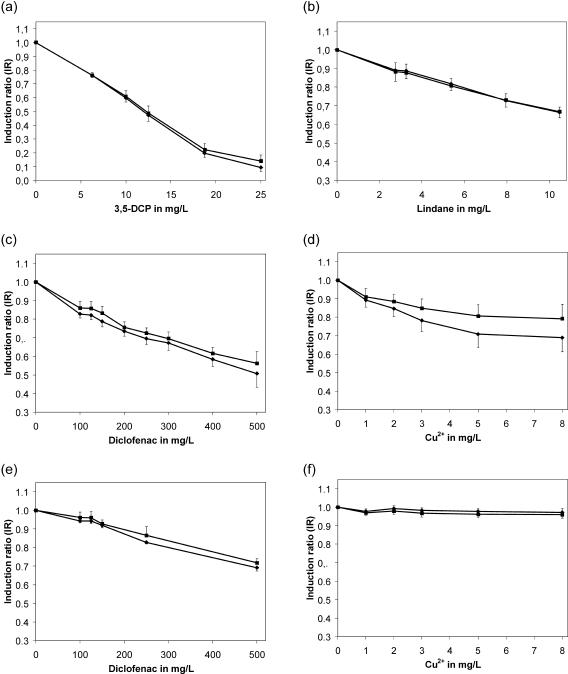
A set of chemical compounds, including DNA-damaging agents, heavy metals, commonly used chemicals, drugs, and products for domestic use, were analyzed for their toxicity by the fluorescence repression potential (Fig. 3). For toxicity assessment, EC20 values based on the fluorescence decrease were calculated for both incubation periods (3 and 4 h of exposure) and are summarized in Table 2. A rating based on 4-h EC20 values revealed the highest toxic potential for the genotoxin MNNG with 0.48 mg/liter, whereas the second analyzed genotoxin, hydroxyurea, exerted no fluorescence decrease within the tested concentration range. Hence, calculation of a corresponding EC value was not possible. With EC20 values of 6.27 mg/liter for 3,5-dichlorophenol (3,5-DCP; Fig. 3A) and 5.63 mg/liter for the insecticide Lindane (Fig. 3B), these compounds were determined to be moderately toxic compared to MNNG. The lowest EC20 value among the heavy metals, and thus the highest toxicity, was observed for hexavalent chromium with 2.0 mg/liter. Cd2+ and Cu2+ showed similarly toxic properties, with EC20 values of 2.5 mg/liter for cadmium and 2.9 mg/liter for copper ions (Fig. 3D). The absence of an observable fluorescence decrease for Co2+ ions failed to reveal an EC value. The nonsteroidal antirheumatic drug diclofenac exhibited a weak toxic potential with a calculated EC20 of 140.7 mg/liter (Fig. 3C). The central nervous stimulant caffeine exhibited a fairly weak decrease of fluorescence emission with an EC20 value of 569.5 mg/liter. The lowest toxic potential was obtained for sodium chloride with an EC20 of 7,050 mg/liter. Thus, the data represented a clear differentiation between responses to highly toxic substances such as hexavalent chromium and harmless compounds such as caffeine.
FIG. 3.
Dose response plots of pPPMA1-GFP/PEST expressing cells upon exposure of 3,5-DCP (a), Lindane (b), diclofenac (c), and copper (d) in YNB medium. Both the quantity and the quality of the diclofenac and copper responses (calculated induction ratios) were significantly altered in the presence of 20% FCS (panels e and f, respectively). The diagrams show results of 3 h (▪) and 4 h (⧫) incubation period. The data were compiled from at least 10 separate experiments, with six wells per tested concentration, and the results are expressed as means ± the standard deviation.
TABLE 2.
EC values for comparison
| Class | Compound(s) | Concn range tested (mg/liter) | Without serum
|
With serum
|
Other studies
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| EC20 in mg/liter at 3 and 4 ha | 95% CI (mg/liter)b | EC20 in mg/liter at 3 and 4 ha | 95% CI (mg/liter)b | Chronic toxicityd (EC20 and EC50 in mg/liter) | Results in mg/liter
|
|||||
| Cell line testse (IC50) | Ecotoxicological testse (IC50) | Avgf | ||||||||
| Metal ions | Cd2+ as cadmium chloride monohydrate, | 0.5-10.0 | 6.5 | 4.97-9.6 | 0.18 | |||||
| CAS (35658-65-2) | 2.5 | 1.7-3.6 | 0.80 | |||||||
| Co2+ as cobalt(II) nitrate hexahydrate, | 14.73-147.3 | NCc | NC | 19.0 | ||||||
| CAS (10026-22-9) | NC | NC | 82.9 | |||||||
| Cr(VI) as potassium dichromate, | 0.5-10.0 | 2.9 | 2.3-3.7 | 0.27 | ||||||
| CAS (7778-50-9) | 2.0 | 1.5-2.6 | 0.88 | |||||||
| Cu2+ as cupric sulfate pentahydrate, | 1.0-8.0 | 7.0 | 5.0-12.9 | NC | NC | 0.74 | 0.29-110.4 | 0.023-191.8 | 23.6 | |
| CAS (7758-98-8) | 2.9 | 2.2-3.7 | NC | NC | 2.52 | |||||
| Pharmaceuticals | Diclofenac sodium salt, | 100-500 | 170.6 | 155.3-184.9 | 350 | 310-407 | 115.7 | 11.5g | ||
| CAS (15307-79-6) | 140.7 | 122.9-156.7 | 314 | 285-352 | 246.6 | |||||
| Hydroxyurea, CAS (127-07-1) | 625-2,500 | NC | NC | 1,189 | ||||||
| NC | NC | 3,311 | ||||||||
| Pesticide | Lindane, CAS (58-89-9) | 2.75-10.48 | 5.47 | 5.16-5.79 | 3.14 | 0.43-765 | 14.6-6,363 | 349.7 | ||
| 5.63 | 5.02-6.27 | 8.28 | ||||||||
| PCPs | 3,5-Dichlorophenol, | 6.25-25.0 | 6.3 | 5.04-7.35 | 5.51 | |||||
| CAS (591-35-5) | 6.27 | 4.97-7.35 | 9.23 | |||||||
| CNS stimulant | Caffeine, CAS (58-08-2) | 150-750 | 579.0 | 431.4-1,016 | 417.1 | 368.9-473.4 | 250.2 | 119.7-49,915 | 117.0-33,746 | 40,572 |
| 569.5 | 477.5-726.3 | 373.8 | 335.7-414.5 | 640.7 | ||||||
| Genotoxic agent | MNNG, CAS (70-25-7) | 0.41-2.5 | 0.56 | 0.53-0.58 | 0.32 | |||||
| 0.48 | 0.44-0.52 | 0.92 | ||||||||
| Sodium chloride, CAS (7647-14-5) | 2,500-20,000 | 6200 | 5,480-6,860 | 4838 | 2,882-6,521 | 4,870 | 1,806-50,899 | 1,725-160,957 | 8,447 | |
| 7050 | 6,360-7,710 | 5450 | 4,033-6,728 | 11,294 | ||||||
The top value in each set is the value at 3 h; the bottom value is the value at 4 h.
95% CI, 95% confidence interval.
NC, not calculated.
Schmitt et al. (34). The top value in each set is the EC20, and the bottom value is the EC50.
That is, the average for methods with approximately the same exposure time.
Ferrari et al. (12).
Serum-bound exposure.
Experiments with medium containing 20% (vol/vol) FCS, comparable to the serum fraction of human blood, were conducted to indicate the influence of serum constituents upon the toxic properties of selected compounds. Since serum components (i.e., proteins) enable adsorption of free chemicals, bioavailability and thus toxicity could be altered. A perturbation of fluorescence readouts and/or turbidity measurements by addition of 20% (vol/vol) FCS due to its endogenous color was not observed (data not shown). Four compounds—caffeine, diclofenac, Cu2+, and sodium chloride—were analyzed in these conditions and a concentration-dependent reporter repression was observed for caffeine, diclofenac, and sodium chloride but not for copper ions (Cu2+; Fig. 3F). In the presence of FCS for diclofenac an EC20 value of 314.0 mg/liter (Fig. 3E) was calculated, for caffeine of 373.8 mg/liter and for sodium chloride of 5,450 mg/liter.
DISCUSSION
In this study, a S. cerevisiae-based biosensor utilized the housekeeping PMA1 promoter (PPMA1) for detection of early toxic effects of a variety of chemically unrelated compounds with different modes of action. PMA1 expression is adjusted according to the metabolic state of the cell, i.e., upregulated during exponential growth (38) and on respiration (11). Under nonlimiting glucose conditions (fermentative metabolism), expression is potentially regulated by the TUF/RPG/RAP1 system (two consensus sites), known to regulate expression of essential genes involved in glycolysis and active transport (4, 27), and the MCM1 protein (one consensus site [19, 27]).
To measure dynamic changes in GFP reporter fluorescence, the fused G1 cyclin CLN2 PEST-rich C-terminal genetic element, which has been reported to serve as a universal ubiquitin-targeting sequence and to reduce GFP half-life (t1/2) to approximately 30 min (25), subjected the usually very stable GFP to rapid proteolysis. The comparison of emission peaks revealed considerable lower fluorescence emission peaks for the PPMA1-GFP/PEST construct (Fig. 1). Since the PPMA1-driven transcription and/or translation rates of both GFP and GFP/PEST were not affected by compound exposure, the observed lower fluorescence intensity of GFP/PEST (∼30% of native GFP; Fig. 1) as result of rapid protein degradation was consistent with a lower steady-state fluorescence of the destabilized version. The observed reduction was lower compared to the 16-fold difference between stable GFP and GFP/PEST as reported by Mateus and Avery (25). Our GFP/PEST fluorescence profile is monitored in stationary cells with episomal multicopy expression. Background fluorescence due to the accumulation of oxidized flavines in the late growth phase did not disturb GFP emission measurements.
The balance between PPMA1-driven transcription and PEST-mediated rapid degradation of GFP was altered in a compound concentration-dependent manner toward decreased fluorescence signals upon chromium and cadmium exposure (Fig. 2B and D) in contrast to signals from native, stable GFP (Fig. 2A and C). Thus, the modified construct with destabilized chromophore was evaluated as suitable to monitor toxic effects, comprising transcription and/or translation inhibition, by measuring the decrease in fluorescence. In the presence of cadmium, 7 h of exposure even induced a fluorescence increase in cells expressing the stable variant of GFP (Fig. 2A). Such long-term exposure might invoke the cellular adaptive response, indicative of detoxification mechanisms such as chelation to glutathione and subsequent sequestration of the cadmium-glutathione conjugates into the vacuole, mediated by the Ycf1p transporter (20). According to the defined test criteria, the assay period of 4 h was determined to be suitable to meet the prerequisites of considerable fluorescence decrease, cell proliferation below full doubling, and growth inhibition below 20% (data not shown). The EC20 values of 3-h exposition were within the same order of magnitude, except for cadmium and sodium chloride.
The lowest EC values were determined for MNNG (Table 2), a genotoxic agent causing lesions such as O6-methylguanine and O4-methylthimine in DNA. DNA-damaging agents, in common with other toxicants, can induce an environmental stress response (ESR) (14). Within the ESR program, many of the repressed genes are involved in protein synthesis and metabolism. Their repression in response to stressful environments is believed to conserve energy in the cell (15). Such repression, for PPMA1 potentially mediated by the Xbp1 transcriptional repressor (24, 27), is in accord with the observed response to MNNG because the H+-ATPase is one of the most energy-consuming enzymes in yeast (25 to 40% of cellular ATP consumption [28]). In contrast, for hydroxyurea no EC value could be calculated (Table 2). This nil response might be caused by the indirect action of the antineoplastic agent that induces cell cycle arrests by a non-DNA-interactive mechanism (16), an effect beyond the 4-h observation period (test criteria, no full doubling).
Alongside MNNG, heavy metal ions (except Co2+) exhibited the highest toxic potential. Among the heavy metals, transition metals such as copper are involved in redox processes of respiratory activity, whereas others, such as cobalt, are part of complex molecules and stabilize protein and DNA structures. Cadmium, lead, mercury, and hexavalent chromium have no known biological functions and are toxic even in small doses. Accordingly, Cr(VI) and Cd2+ exhibited similar toxicity, with EC20 values of 2.0 and 2.5 mg/liter (Table 2), respectively. The intracellular reduction of Cr(VI) is believed to generate reactive metal and oxygen species (22, 36), which may confer the ESR program transcriptional repression. Copper, although essential as a micronutrient, is toxic at high concentrations, as indicated by an EC20 value of 2.9 mg/liter (Table 2) and supported by earlier observations of reduced PMA1 expression in S. cerevisiae cells cultivated in the presence of growth-inhibitory concentrations of copper (>1.5 mM) (10). Co2+ ions did not exert any toxic effect within the test concentration range.
In comparison to cell line and ecotoxicological tests with approximately similar exposure times (7, 8), the yeast EC20 assay scored among the more sensitive data. In the presence of FCS no reporter decrease, and thus loss of toxic properties, was observed (Fig. 3F) in contrast to cell line tests (7, 8). The protective effect of serum protein scavengers due to binding of metal ions to free thiol groups is well documented, involving ceruloplasmin, albumin, and transcuprein as major copper-transporting constituents in plasma (23).
Clorinated phenols (PCPs) such as 3,5-DCP are widely used by chemical industry as intermediate products for agrochemicals and have been frequently applied as wood preservatives, fungicides, bactericides, and algicides. PCPs are of ecological relevance (classified as toxic to aquatic organisms with possible long-term adverse effects), and the toxicological properties (score positive in most chronic bioassays) are based on the uncoupling of the oxidative phosphorylation. The EC20 value of 6.27 mg/liter scored 3,5-DCP as moderately toxic, although, at higher concentrations, this compound caused the strongest absolute decrease of the fluorescence signal (reduced to 10% of the corresponding control value) among all tested compounds. The moderate ranking might be due to the primarily fermentative energy gain of yeast cells in our test, a finding indicative of limitations according to the fermentative metabolic state.
For diclofenac, a 2.2-fold increase of the EC20 and thus a decrease in toxicity was observed upon serum addition (an EC20 of 350 mg/liter versus an EC20 of 170 mg/liter in serum-free medium, Table 2). As a commonly applied medication, it is known to be highly protein bound (ca. 99.5% within a concentration range of 2 to 10 mg/liter), whereby the extent of binding tends to decrease with increasing drug concentrations (6). However, the serum-free medium EC20 was 15-fold higher than those from ecotoxicological bioassays (12).
The insecticide Lindane is a neurotoxin affecting presynaptic terminals to enhance the release of neurotransmitter and is believed to interact with the GABA-A receptor-chloride channel complex (29). Corresponding to these higher eukaryote-specific modes of action, the toxicity of Lindane in yeast (EC20 of 5.47 mg/liter [Table 2]) was minor compared to certain cell line tests but twofold more sensitive than the most sensitive ecotoxicological test system (7, 8).
As a common psychoactive dietary component, caffeine was also found to be less toxic in serum-free medium with EC20 values up to 20 times above (human) blood concentrations (10 to 30 mg/liter) that cause restlessness and insomnia. Although caffeine was reported to be bound at 10 to 35% plasma proteins levels over a wide concentration range to albumin as the major binding protein (3a), the yeast reporter assay revealed a slightly increased toxicity of this compound in the presence of FCS (EC20 of 417 mg/liter versus an 569 EC20 of mg/liter in serum-free medium [Table 2]). However, both sets of data are four- to fivefold higher than the most sensitive cell line or ecotoxicological tests but 2 orders of magnitude lower than average data (Table 2) (7, 8).
Sodium is an environmentally abundant ion. However, high internal concentrations of Na+ are generally toxic for cells. S. cerevisiae cells exposed to high external Na+ encounter osmotic stress initiated ESR with corresponding expression changes (14). Na+ ions are efficiently exported by both Na+-ATPases (ENA1 to ENA4) and the Na+/H+ antiporter NHA1 (37). Corresponding to osmotic stress, increasing sodium chloride concentrations caused a substantial fluorescence decrease (EC20 of 6,200 mg/liter). Interestingly, the exposure in FCS revealed slightly increased toxicity with 1.3-fold-diminished EC20 values (EC20 4,838 mg/liter versus 6,200 mg/liter in serum-free medium, Table 2). Similar results were obtained by Kirtane et al. (17), who observed a 1.5-fold increase in the rate of Na+ entry (associated with a rise in cell Na+ content) when rat liver cells were exposed to Na+ in the presence of 10% calf serum. In both cases the data were obtained within similar exposure times.
The results of the present study indicate that the S. cerevisiae PMA1 promoter activity as a housekeeping biomarker in combination with a destabilized GFP is efficient in monitoring toxicant-induced stress by fluorescence decrease as reporter. The PPMA1 is unique in its regulatory site composition and precise response to metabolic state. This test is sufficient to enable calculation of EC values for chemically unrelated compounds, even in the presence of complex matrices such as serum. However, impairment of posttranslational regulation of the ATPase is, by nature of the test, not detected. By means of short-term concentration-response curves (4 h), differentiation of quantity and quality of toxic effects of tested chemicals can be achieved, although the EC values obtained are higher compared to long-term tests. The advantage of S. cerevisiae lies in its simple handling and cultivation, its rapid response to toxicants and indication of bioavailability, and potential extrapolation of results for the evaluation of potential risks of chemicals to human health by the high conservation of stress-sensing processes.
Acknowledgments
We thank M. Ghislain (FYSA, Belgium) for the gift of the FYAK strain, D. Sanders (University of York) for critical reading of the manuscript, and B. Kirberg for excellent technical assistance.
This study was funded in part by EC QLK3-CT-2001-00401.
REFERENCES
- 1.Ambesi, A., M. Miranda, V. V. Petrov, and C. W. Slayman. 2000. Biogenesis and function of the yeast plasma-membrane H+-ATPase. J. Exp. Biol. 203:155-160. [DOI] [PubMed] [Google Scholar]
- 2.Bergman, S., J. Ihmels, and N. Barkai. 2003. Similarities and differences in genome-wide expression data on six organisms. PLoS Biol. 2:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertl, A., J. Ramos, J. Ludwig, H. Lichtenberg-Frate, J. Reid, H. Bihler, F. Calero, P. Martinez, and P. O. Ljungdahl. 2003. Characterization of potassium transport in wild-type and isogenic yeast strains carrying all combinations of trk1, trk2, and tok1 null mutations. Mol. Microbiol. 47:767-780. [DOI] [PubMed] [Google Scholar]
- 3a.Bonati, M., R. Latini, G. Tognoni, J. F. Young, and S. Garattini. 1985. Interspecies comparison of in vivo caffeine pharmacokinetics in man, monkey, rabbit, rat, and mouse. Drug Metab. Rev. 15:1355-1383. [DOI] [PubMed] [Google Scholar]
- 4.Capieaux, E., M. L. Vignais, A. Sentenac, and A. Goffeau. 1989. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J. Biol. Chem. 264:7437-7446. [PubMed] [Google Scholar]
- 5.Castrillo, J. I., and S. G. Oliver. 2004. Yeast as touchstone in post-genomic research: strategies for integrative analysis in functional genomics. Biochem. Mol. Biol. 37:93-106. [DOI] [PubMed] [Google Scholar]
- 6.Chan, K. K., K. H. Vyas, and K. D. Brandt. 1987. In vitro binding of diclofenac sodium in plasma and synovial fluid. J. Pharm. Sci. 76:105-108. [DOI] [PubMed] [Google Scholar]
- 7.Clemedson, C., E. McFarlane-Abdulla, M. Andersson, F. A. Barile, M. C. Calleja, C. Chesné, R. Clothier, M. Cottin, R. Curren, P. Dierickx, M. Ferro, G. Fiskesjö, L. Garza-Ocanas, M. J. Gómez-Lechón, M. Gülden, B. Isomaa, J. Janus, P. Judge, A. Kahru, R. B. Kemp, G. Kerszman, U. Kristen, M. Kunimoto, S. Kärenlampi, K. Lavrijsen, L. Lewan, H. Lilius, A. Malmsten, T. Ohno, G. Persoone, R. Pettersson, R. Roguet, L. Romert, M. Sandberg, T. Sawyer, H. Seibert R., Shrivastava, M. Sjöström, A. Stammati, N. Tanaka, O. Torres Alanis, J.-U. Voss, S. Wakuri, E. Walum, X. Wang, F. Zucco, and B. Ekwall. 1996. MEIC evaluation of acute systemic toxicity. II. In vitro results from 68 toxicity assays used to test the first 30 reference chemicals and a comparative cytotoxicity analysis. ATLA 24:273-311. [Google Scholar]
- 8.Clemedson, C., F. A. Barile, B. Ekwall, M. J. Gómez-Lechón, T. Hall, K. Imai, A. Kahru, P. Logemann, F. Monaco, T. Ohno, H. Segner, M. Sjöström, M. Valentino, E. Walum, X. Wang, and B. Ekwall. 1998. MEIC evaluation of acute systemic toxicity. III. In vitro results from 16 additional methods used to test the first 30 reference chemicals and a comparative cytotoxicity analysis. ATLA 26:91-129. [Google Scholar]
- 9.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes, A. R., F. P. Peixoto, and I. Sa-Correia. 1998. Activation of the H+-ATPase in the plasma membrane of cells of Saccharomyces cerevisiae grown under mild copper stress. Arch. Microbiol. 171:6-12. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes, R., and I. Sá-Correia. 2003. Transcription patterns of PMA1 and PMA2 genes and activity of plasma membrane H+-ATPase in Saccharomyces cerevisiae during diauxic growth and stationary phase. Yeast 20:207-219. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, B., N. Paxeus, R. Lo Giudice, A. Pollio, and J. Garric. 2003. Ecotoxicological impact of pharmaceuticals found in treated wastewaters: study of carbamazepine, clofibric acid, and diclofenac. Ecotoxicol. Environ. Safety 55:359-370. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, T. S., D. di Bernardo, D. Lorenz, and J. J. Collins. 2003. Inferring genetic networks and identifying compound mode of action via expression profiling. Science 301:102-105. [DOI] [PubMed] [Google Scholar]
- 14.Gasch, A. P., P. T. Spellmann, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, J. S. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR Homolog Mec1p. Mol. Biol. Cell 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khayat, A. S., A. C. Guimarães, P. C. Cardoso, P. D. L. de Lima, M. de Oliveira Bahia, L. M. G. Antunes, and R. R. Burbano. 2004. Mutagenicity of hydroxyurea in lymphocytes from patients with sickle cell disease. Genet. Mol. Biol. 27:115-117. [Google Scholar]
- 17.Kirtane, A., N. Ismail-Beigi, and F. Ismail-Beigi. 1994. Role of enhanced Na+ entry in the control of Na,K-ATPase gene expression by serum. J. Membr. Biol. 137:9-15. [DOI] [PubMed] [Google Scholar]
- 18.Kolaczkowski, M., A. Kolaczowska, J. Luczynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4:143-158. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, M. H., and E. Grayhack. 1994. A library of yeast genomic MCM1 binding sites contains genes involved in cell cycle control, cell wall and membrane structure, and metabolism. Mol. Cell. Biol. 14:348-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Z. S., Y. P. Lu, R. G. Zhen, M. Szczypka, D. J. Thiele, and P. A. Rea. 1997. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 94:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichtenberg-Fraté, H., M. Schmitt, G. Gellert, and J. Ludwig. 2003. A yeast based method for the detection of cyto- and genotoxicity. Toxicol. In Vitro 17:709-716. [DOI] [PubMed] [Google Scholar]
- 22.Liu, K. J., and X. Shi. 2001. In vivo reduction of chromium (VI) and its related free radical generation. Mol. Cell Biochem. 222:41-47. [PubMed] [Google Scholar]
- 23.Luza, S. C., and H. C. Speisky. 1996. Liver copper storage and transport during development: implications for cytotoxicity. Am. J. Clin. Nutr. 63:812S-820S. [DOI] [PubMed] [Google Scholar]
- 24.Mai, B., and L. Breeden. 1997. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol. 17:6491-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateus, C., and S. V. Avery. 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16:1313-1323. [DOI] [PubMed] [Google Scholar]
- 26.Moskvina, E., E. M. Imre, and H. Ruis. 1999. Stress factors acting at the level of the plasma membrane induce transcription via the stress response element (STRE) of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 32:1263-1272. [DOI] [PubMed] [Google Scholar]
- 27.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao, R., and C. W. Slayman. 1996. Plasma membrane and related ATPases, p. 29-56. In R. Brambl and G. Marzluf (ed.), The mycota, vol. 3. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 29.Ratra, G. S., S. G. Kamita, and J. E. Casida. 2001. Role of human GABA(A) receptor beta3 subunit in insecticide toxicity. Toxicol. Appl. Pharmacol. 172:233-240. [DOI] [PubMed] [Google Scholar]
- 30.Resnick, M. A., and B. S. Cox. 2000. Yeast as an honorary mammal. Mutat. Res. 451:1-11. [DOI] [PubMed] [Google Scholar]
- 31.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-302. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schmitt, M., G. Gellert, J. Ludwig, and H. Lichtenberg-Fraté. 2004. Phenotypic yeast growth analysis for chronic toxicity testing. Ecotoxicol. Environ. Safety 59:142-150. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt, M., G. Gellert, J. Ludwig, and H. Lichtenberg-Fraté. 2005. Assessment of cyto- and genotoxic effects of a variety of chemicals using Saccharomyces cerevisiae. Acta Hydro-chim. Hydrobiol. 33:56-63. [Google Scholar]
- 35.Serrano R., M. Kielland-Brandt, and G. R. Fink. 1986. Yeast plasma membrane H+-ATPase is essential for growth and has homology with (Na+-K+)-, K+-, and Ca2+-ATPases. Nature 319:689-693. [DOI] [PubMed] [Google Scholar]
- 36.Sugden, K. D., and D. M. Stearns. 2000. The role of chromium(V) in the mechanism of chromate-induced oxidative DNA damage and cancer. J. Environ. Pathol. Toxicol. Oncol. 19:215-230. [PubMed] [Google Scholar]
- 37.Sychrova, H. 2004. Yeast as a model organism to study transport and homeostasis of alkali metal cations. Physiol. Res. 53:S91-S98. [PubMed] [Google Scholar]
- 38.Viegas, C. A., P. Supply, E. Capieaux, L. Van Dyck, A. Goffeau, and I. Sa-Correia. 1994. Regulation of the expression of the H+-ATPase genes PMA1 and PMA2 during growth and effects of octanoic acid in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1217:74-80. [PubMed] [Google Scholar]