Abstract
Until recently, methicillin-resistant Staphylococcus aureus (MRSA) was considered the prototype of a hospital-acquired bacterial pathogen. However, recent reports have shown that MRSA has now emerged in the community. Characterization of specific markers for distinguishing the origin of isolates could contribute to improved knowledge of MRSA epidemiology. The release of whole-genome sequences of hospital- and community-acquired S. aureus strains allowed the development of whole-genome content analysis techniques, including microarrays. We developed a microarray composed of 8,191 open reading frame-specific oligonucleotides covering >99% of the four sequenced S. aureus genomes (N315, Mu50, MW2, and COL) to evaluate gene contents of hospital- and community-onset S. aureus strains. In parallel, pulsed-field gel electrophoresis, variable number of tandem repeats, antibiogram, staphylococcal cassette chromosome-mec element typing, and presence of the Panton-Valentine leukocidin gene were evaluated in a collection of 15 clinical isolates. Clusters obtained with microarrays showed a high degree of similarity with those obtained by pulsed-field gel electrophoresis or variable number of tandem repeats. Clusters clearly segregated hospital-onset strains from community-onset strains. Moreover, the microarray approach allowed definition of novel marker genes and chromosomal regions specific for given groups of isolates, thus providing better discrimination and additional information compared to pulsed-field gel electrophoresis and variable number of tandem repeats. Finally, the comparative genome hybridization approach unraveled the occurrence of multiple horizontal transfer events leading to community-onset MRSA as well as the need for a specific genetic background in recipient strains for both the acquisition and the stability of the mec element.
Methicillin-resistant Staphylococcus aureus (MRSA) is the causative agent of a wide diversity of diseases ranging from benign skin infections to life-threatening diseases such as endocarditis, osteomyelitis, sepsis, and toxic shock syndrome. MRSA was previously described as a typical nosocomial pathogen (2, 4, 30, 37), but recently, several outbreaks in the community have been reported (9, 29). Although exportation of nosocomial MRSA lineages to the community explains some cases, epidemiologic and genetic elements suggest that the community holds specific strains harboring different genetic backgrounds (11, 20, 52).
Specific markers have been identified to distinguish community- from hospital-acquired strains, such as antibiotic susceptibility profiles (antibiograms), toxin contents (toxinograms), and staphylococcal cassette chromosome (SCC)-mec typing. Most of these markers are carried by mobile genetic elements or located in genomic islands, suggesting transmission by horizontal transfer (3, 38, 57). Phylogenic trees arising from molecular techniques such as sequencing of highly conserved genes (multilocus sequence typing [MLST]) or from enzymatic cleavage of genomic DNA (pulsed-field gel electrophoresis [PFGE]) demonstrated that community and hospital strain populations are clearly divergent (57), originating from different ancestral clones. These molecular methods contributed to the solution of central phylogenetic issues. MLST revealed a powerful macroevolutionary tool recognizing common ancestries, while PFGE displayed appropriate resolution power for distinguishing outbreak events and providing microevolutionary analysis (6, 36). However, these methods appear poorly informative for the study of specific gene contents in whole bacterial genomes. Identification of specific genes would considerably deepen our knowledge of epidemiologic markers and bacterial mechanisms implicated in Staphylococcus aureus dissemination. Clearly, such information is accessible only with a whole-genome approach.
Recent progress in high-throughput sequencing techniques yielded the publication of numerous S. aureus genomes, thus facilitating the discovery of sequences that are variably repeated in the different sequenced strains. Those variable numbers of tandem repeats (VNTR) facilitate a new type of multilocus analysis. VNTR was successfully applied to the molecular study of numerous bacteria (40, 45, 46), including MRSA (22, 27, 47).
Genome sequence data also permitted the development of DNA oligoarrays for evaluating the presence and/or expression of genes in whole genomes of several bacterial pathogens (5, 28, 49). The use of microarrays for studying MRSA genomic contents showed that 22% of the genome is dispensable, containing mainly virulence and resistance factors. Moreover, these regions contain mediators of lateral gene transfer such as transposase and integrase genes (21). Recently, Saunders and colleagues used a microarray only composed of virulence-associated factors and core genes (used for MLST) to study the evolution and pathogenic potential of S. aureus isolates. This array yielded a coherent phylogenetic tree (using core genes) yet provided interesting epidemiological information on the potential evaluation of strain virulence (50).
To further analyze S. aureus gene contents and provide a more detailed molecular map, we developed an oligoarray (10) based on the genome of four fully sequenced S. aureus strains: MW2 (3), N315 (38), Mu50 (38), and COL (http://www.tigr.org/tdb/mdb/mdbinprogress.html). This array is composed of 8,191 open reading frame (ORF)-specific oligonucleotides allowing >99% coverage of these four genomes. To find molecular signatures specific for hospital-acquired or community-onset strains, we examined the genomic contents of 15 MRSA isolates. Strains are classified as community-onset MRSA (CO-MRSA; 13 strains) or hospital-acquired MRSA (HA-MRSA; 2 strains) according to genetic markers previously described; we also included clinical data (age, sex, sampling date). We compared clusters obtained by hybridizing genomic DNA on microarrays (genomotyping (34), PFGE, and VNTR. The microarray approach provided better discrimination. Furthermore, it allowed definition of novel marker genes and chromosomal regions specific for given groups of isolates, thus enriching our molecular arsenal for epidemiological monitoring.
MATERIALS AND METHODS
Strain collection.
CO-MRSA strains were selected from a collection recovered during a screening study performed at hospital admission between February and August 2003 (26) and from ongoing surveillance of MRSA in the Geneva community. Thirteen strains were randomly selected as potential CO-MRSA according to their antimicrobial susceptibility profiles, SCC-mec element type, and presence of the Panton-Valentine leukocidin (PVL) gene. Moreover, 2 MRSA isolates (strains I and II) were added as controls, representative of our predominant HA-MRSA clone (Table 1). Note that strains IX through XII and strain XV originated from three members of the same family.
TABLE 1.
Antimicrobial susceptibility profiles
| Strain No. | Sampling date (day.mo.yr) | Age (yr) | Sexb | PVLc | SCC-mec type | ST | Result fora:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | OXA | AMI | GEN | NOR | CLI | ERY | FUS | MUP | SXT | TET | FOS | |||||||
| I | 19.06.2003 | 66 | M | − | I | 228 | R | R | R | R | R | R | R | R | S | S | S | S |
| II | 16.05.2003 | 65 | M | − | I | 228 | R | R | R | R | R | R | R | S | R | S | S | S |
| III | 14.04.2003 | 74 | M | − | III | 239 | R | R | R | R | R | S | R | S | S | R | R | R |
| IV | 22.05.2003 | 59 | M | − | IV | 8 | R | R | S | S | R | S | S | S | S | S | S | S |
| V | 10.04.2003 | 26 | F | − | IV | 8 | R | R | S | S | S | S | S | I | S | S | S | S |
| VI | 08.05.2003 | 36 | M | + | V | 152 | R | R | R | R | S | S | S | S | S | S | S | S |
| VII | 20.05.2003 | 37 | M | + | V | 152 | R | R | R | R | S | S | S | S | S | S | S | S |
| VIII | 24.08.2003 | 29 | M | + | V | 152 | R | R | R | R | S | S | S | S | S | S | S | S |
| IX | 24.07.2003 | 40 | M | + | IV | 80 | R | R | R | S | S | R | R | R | R | S | S | S |
| X | 24.07.2003 | 9 | F | + | IV | 80 | R | R | R | S | S | R | R | R | R | S | S | S |
| XI | 26.02.2003 | 40 | M | + | IV | 80 | R | R | R | S | S | S | S | R | S | S | S | S |
| XII | 26.02.2003 | 40 | M | + | IV | 80 | R | R | R | S | S | S | S | R | S | S | S | S |
| XIII | 17.02.2003 | 50 | M | + | IV | 80 | R | R | R | S | S | S | S | R | S | S | S | S |
| XIV | 02.08.2003 | 39 | F | + | IV | NTd | R | R | R | S | S | S | S | R | S | S | S | S |
| XV | 07.08.2003 | 73 | M | + | IV | 80 | R | R | R | S | S | S | S | R | R | S | S | S |
Abbreviations: PEN, penicillin; OXA, oxacillin; AMI, amikacin; GEN, gentamicin; NOR, norfloxacin; CLI, clindamycin; ERY, erythromycin; FUS, fusidic acid; MUP, mupirocin; SXT, cotrimoxazole; TET, tetracycline; FOS, fosfomycin; R, resistant; S, susceptible; I, intermediate.
M, male; F, female.
PVL, presence (+) or absence (−) of the Panton-Valentine leukocidin gene.
NT, nontypeable. Allelic profile is 1.3.1.14.11.51.NT, which matches to 6/7 alleles of ST80. The sequence of the seventh gene, corresponding to allele “4,” suggested that this strain is probably a new ST.
Microbiologic methods.
Identification of MRSA was performed on oxacillin-resistant S. aureus plates (Oxoid, Basingstoke, United Kingdom). Further identification of MRSA was based on Pastorex agglutination (Bio-Rad, Reinach, Switzerland), DNase reaction on agar, and growth on Mueller-Hinton oxacillin plates (6 mg of oxacillin per ml). MRSA identification was confirmed with the Vitek 2 identification and susceptibility testing cards for gram-positive bacteria (bioMérieux, Marcy l'Etoile, France).
DNA extraction and purification.
Genomic DNA (gDNA) was prepared from isolated colonies grown overnight on Mueller-Hinton agar at 37°C. Briefly, 109 cells were lysed in 100 μL Tris-EDTA buffer (10 mM Tris-1 mM EDTA, pH 8) containing 50 μg/ml lysostaphin (Ambicin; Applied Microbiology, Tarrytown, NY) for 10 min at 37°C. DNA was then isolated and purified using a DNeasy kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions, including RNase treatment. DNA quantification and protein contaminations were assessed by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Rockland, DE).
PVL, mecA detection, and SCC-mec typing.
PCR assays were conducted to evaluate the presence of the mecA gene (23), the Panton-Valentine leukocidin gene, and the SCC-mec type element (24).
DNA preparation for pulsed-field gel electrophoresis.
Epidemiological typing of MRSA isolates was performed by PFGE of chromosomal DNA digested with SmaI (Bio-Rad) using a CHEF MAPPER system (Bio-Rad, Hercules, CA), according to established protocols (8). The banding patterns of the different gels were analyzed with GelCompar software (v. 4.1; Applied Maths, Belgium). Interpretation of the fragment patterns was based on published criteria (7). Patterns differing by one to six DNA fragments were considered subtypes, and those distinguished by seven or more DNA fragments were considered distinct types (14, 53).
Multiple-locus VNTR typing.
The VNTR typing assay was performed as previously described (22) but with the addition of the following two primer pairs for assessment of a total of 10 target genes: SAS-F (5′-TTG-GAA-CAT-TCG-AAT-ATA-CAG-AGT) and SAS-R (5′-TCG-ATG-TAC-TGT-CAC-TTA-ATG-ATG); plsR2-F (5′-AAT-TAC-AAC-GCC-TCA-AGC-TG) and plsR2-R (5′-GCA-CCA-TGG-ATG-ATT-ACT-TC).
SCC-mec sequencing.
Genomic DNA of strains III, VI, VII, and VIII was extracted as previously described (24). The amplification reaction was performed in a PTC 200 Peltier thermal cycler (MJ Research, Inc., Watertown, Mass.) in a 20-μl reaction volume. Amplification conditions and sequencing primers for the cassette chromosome recombinase (ccr) genes were selected according to the method of a previously published study (43). DNA sequencing was performed with an ABI Prism 3100 sequencer (Applied Biosystems). Homologies were searched using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Multilocus sequence typing.
MLST was performed on all isolates by PCR amplification of internal fragments of seven housekeeping genes by using previously described procedure and primers (17). PCR products were sequenced with an ABI Prism 3100 DNA sequencer (Applied Biosystems, Foster City, CA, USA). Allele numbers were assigned according to the program available from the MLST website (http://www.mlst.net) (Table 1).
Microarray hybridization and scanning.
Our whole-genome Staphylococcus aureus DNA microarray was designed and validated as described by Charbonnier et al. (10). Test and reference gDNAs (1 μg) were labeled with cyanine-3 or cyanine-5 dCTP (NEN, Perkin Elmer) using the BioPrime DNA labeling kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Unincorporated fluorescent nucleotides were removed using Centrisep columns (Princeton separations, EMP Biotech, Berlin, Germany). Cy-3-labeled gDNAs from the four reference strains used to design the microarray: 0.125 μg from each strain (54) were mixed with 0.5 μg of Cy-5-labeled test gDNA in hybridization buffer (Agilent Technologies, CA), for a total volume of 250 μl. The hybridization mixture was heated to 95°C for 2 min, and then hybridization was performed for 17 h at 60°C with rotation in a dedicated hybridization oven (Robbins Scientific, Sunnyvale, CA). Stringent washings were then performed according to the manufacturer's instructions. Slides were dried under nitrogen flow and scanned (Agilent Technologies, CA) using 100% photon multiplier tube power for both wavelengths using the Agilent scanner.
Microarray analysis.
Fluorescence intensities were extracted using Feature extraction software (version 6.1.1; Agilent). Local background-subtracted signals were corrected for unequal dye incorporation or unequal load of the labeled product. The algorithm consisted of a rank consistency filter and a curve fit using the default LOWESS (locally weighted linear regression) method. Additional software was developed in-house to analyze the processed data. This software filtered the data to exclude irrelevant values, as flagged by the extraction software. The background noise of each experiment was evaluated by computing the standard deviation of negative-control intensities. Features whose intensities were smaller than the standard deviation value of the negative controls were considered inefficient hybridization and discarded from further analysis. The software calculated for each spot the logarithm of the ratio between the test channel and the control channel (log ratio). Since the control signal is present in each spot, this log ratio corresponds to a per feature normalization. Computed log ratio values were further sorted into 150 bin categories and fitted with a Gaussian distribution curve, using the Levenberg-Marquardt algorithm. The software estimated the presence probability of each oligonucleotide probe (EPP), as previously described (34). As clearly documented in the work of Kim et al., we used the most stringent EPP value, as our study was focused on “strict divergent gene analysis parameter.” EPP values of ≤1% (each oligonucleotide probe) were extracted and considered absent features in the test channel; we concentrated on the subset of probes predicted to reliably detect MW2 gene targets (10). The list of absent features from each experiment was then clustered by the software using Dice distance and the group average linkage algorithm to construct a hierarchical cluster tree (15). The extensive list of these genes, absent in at least one of the tested strains is shown in a supplemental table (http://www.genomic.ch/sup3.php).
Verification of divergent genes.
Projection of absent features onto the genome map of MW2 (Genome Viewer Software) (33) identified regions of difference between our collection and MW2. Flanking primers were designed with Jellyfish software (LabVelocity) to control the size of the amplicons spanning these regions of difference.
We also designed primers to assess a selection of 11 cluster-specific genes. As each gene is covered by one to eight oligonucleotide probes on our microarray, selected gene targets ought to have every covering oligonucleotide probe present (or absent) to be selected. Target genes and regions within different clusters were amplified using 13 PCRs after protocol optimization using MW2 genomic DNA as a target control (primer sequences are shown in Table 2). PCR assay was performed in a PTC 200 Peltier thermal cycler in a 20-μl reaction volume. Reaction mixtures contained 0.2 mM concentrations of each deoxynucleoside triphosphate, 0.3 μM concentrations of each primer, 1 mM MgSO4, 0.8 U of KOD hot-start DNA polymerase (Novagen, Madison, Wis.). Cycling conditions were as follows: denaturation for 2 min at 94°C; 35 cycles of 15 s at 94°C, 20 s at 60°C, and x s (20 s/kbp, depending on the size of genes and regions) at 72°C; and a postextension of 10 min at 72°C. PCR amplification results were evaluated using the Bioanalyzer 2100 with the DNA 7500 chip kit (Agilent Technologies, CA).
TABLE 2.
Primer sequences for amplification of 11 genes and 2 regions of interesta
| Gene or region | Accession no. | Primer | Primer sequence (5′-3′) | Amplicon size or length (bp) | % GC |
|---|---|---|---|---|---|
| Genes | |||||
| I | MW0329 | F | GCATTAACACCAAAACATTTAGCT | 1,089 | 30.1 |
| R | ATAAATCAGCATGATGCAGAARGT | ||||
| II | MW1040 | F | ACTACAACTACAATTGCGTCAACA | 367 | 30.7 |
| R | AAACTAAGTTGACTGCCTTTTGTG | ||||
| III | MW1753 | F | CCTCAGTAACTGGAATAAATGCTG | 648 | 32.77 |
| R | TAAAATCTTTGATTTGAGGCGTAA | ||||
| IV | MW0305 | F | CATGCGAATTATTTCACGATTATT | 946 | 35.38 |
| R | TGCTAAAATTGCTTCTTYTTGTGT | ||||
| V | MW1327 | F | CAAGCATTAAACCATTTATTCGTC | 929 | 35.16 |
| R | ATGTTCAATGACACCWGAAACTCT | ||||
| VI | MW0760 | F | ACAGTCTTATCTAACGGCGATGTA | 646 | 28.21 |
| R | CGACATCTAGATGAAATTGTGTTG | ||||
| VII | MW0447 | F | AACGTTTATTTGAAGAGTCGAATG | 359 | 35.19 |
| R | CATATCCATTGATAGCGTTTCTCT | ||||
| VIII | MW0622 | F | GCATGAACTGGATATTTTGGATAT | 963 | 30.64 |
| R | CAATTTCATTTTGTAATGGGAAAA | ||||
| IX | MW2515 | F | AGTTAGTGACATAGCACGTGTGAA | 414 | 32.83 |
| R | GCCATTATTGCTGTATTACTTTCG | ||||
| X | MW0105 | F | GTTACTTATCGGTTTAGCGGTTTT | 1,189 | 30.1 |
| R | ATCCCTTTCCATCTTTTCATATTG | ||||
| XI | MW1864 | F | CCCTCAAAATGATATTTCRCGATA | 125 | 31.03 |
| R | TGATTTTTAACATCATTTTTGGATG | ||||
| Regions | |||||
| A | MW1204-MW1211 | F | GAAAGAACATTCCCAAATAATGAA | 5,987 | 28.77 |
| R | TATTTGCCATTTGGTGAAAAATAC | ||||
| B | MW2308-MW2313 | F | TACATCCAAATACCGCTAAGAAAA | 3,957 | 31.38 |
| R | TCAAAATGATATGGAAGTTGTTGC |
Theoretical size and GC content of amplicons were determined based on the genome sequence of MW2.
RESULTS
Strain selection and characterization.
Among 13 strains defined as potential CO-MRSA by their antimicrobial susceptibility profiles, only 10 (77%) possessed the gene of the PVL toxin, contrasting with previous reports suggesting that it was an efficient marker of CO-MRSA (57); however, further study confirmed our local prevalence (39, 42). The two other selected strains were negative for the PVL toxin yet displayed a highly resistant antibiotic profile (Table 1). The type of SCC-mec element has also been reported as an important marker for distinguishing community acquisition from hospital acquisition (22, 48). Indeed, among our PVL-positive strains, 7/10 carried a type IV cassette and 3/10 carried a type V cassette. Among the remaining 3 strains, two additional were type IV (15%) and one revealed type III (7%). Antibiotic susceptibility profiles (Table 1) showed that the two nosocomial strains displayed a broad resistance spectrum (strains I and II), as opposed to the CO-MRSA, which showed more susceptible and variable resistance patterns.
Genomotyping and hierarchical clustering by microarray.
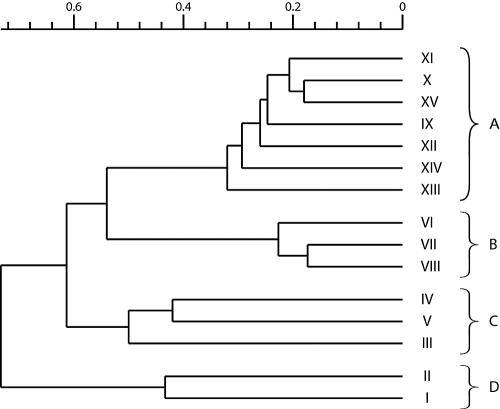
The listing of gene targets reported to be absent by genomotyping was employed for cluster analysis. We arbitrarily defined a cutoff of 52% that segregated our strain collection into four clusters (Fig. 1). Cluster A was composed of seven strains containing the PVL toxin and a SCC-mec IV element. These isolates were resistant to penicillin and oxacillin but susceptible to the majority of antibiotics tested (Table 1). We found in this cluster all isolates collected from patients of the same family (strains IX, XI, and XII were isolates from the father, XV was an isolate from the grandfather, and X was from the daughter). This cluster contained also two isolates (strains XIII and XIV) epidemiologically unrelated to the described family outbreak. We determined a similarity of >65% between strains that composed this cluster and of >70% between isolates of the same family (included in cluster A). Cluster B was composed of PVL-positive strains with a SCC-mec V element but was epidemiologically unrelated to cluster A. As shown in Table 1, these strains were resistant to gentamicin and susceptible to fusidic acid, as opposed to strains from cluster A. In this cluster, we noted a >75% similarity to and a <55% difference from cluster A. Cluster C was composed of three PVL-negative strains with variable antibiograms, two of which possessed a SCC-mec IV cassette and the remaining one possessed a type III cassette. Finally, cluster D contained the 2 HA-MRSA control strains, displaying a SCC-mec I element. This final cluster showed the least similarity (only about 30%) to the other ones.
FIG. 1.
Cluster analysis by microarrays, using an EPP value of ≤1%. Roman numerals represent strain numbers. The scale above the dendrogram shows the percentage of similarity among different strains.
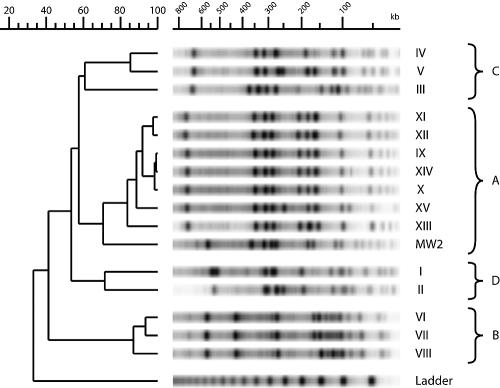
Pulsed-field gel electrophoresis.
Figure 2 shows the 16 PFGE lanes that segregated into four major clusters. These four clusters showed the same strain composition as those previously assessed by microarray analysis. Cluster A included all five strains from the family outbreak, two unrelated isolates (strains XIII and XIV), and strain MW2, considered a model of community-acquired MRSA (3). Note that strains IX, X, and XIV appeared clonally related by PFGE. Cluster C and cluster D were composed of PVL-negative CO-MRSA strains and control HA-MRSA, respectively. The most distantly related cluster, cluster B, included three PVL-positive strains with a SCC-mec V cassette.
FIG. 2.
Cluster analysis by PFGE. The scale above the dendrogram shows the percentage of similarity among different strains.
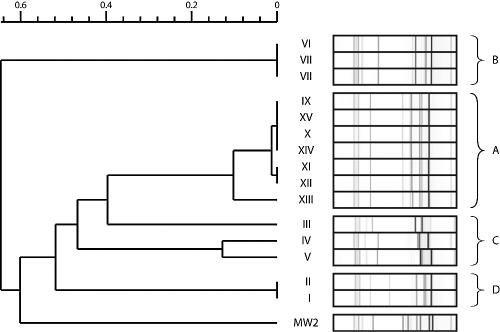
Multiple-locus VNTR typing.
VNTR results were analyzed with in-house software, as described elsewhere (22). Again, cluster composition was similar to that previously determined by microarray analysis or PFGE (Fig. 3). A high degree of similarity was observed within cluster A, except for strain XIII, which displayed a 10% divergence from the other strains composing cluster A. This represents the lowest discriminatory power among the three methods describing this cluster. As for PFGE, clusters C and D were closest to cluster A. Strains constituting cluster D appeared to be clonally related by VNTR. MW2, which coclustered with cluster A by PFGE, appeared very distantly related using VNTR. These data and clustering analysis were confirmed by another independent determination (not shown).
FIG. 3.
Cluster analysis by VNTR. Roman numerals represent strain numbers. The scale above the dendrogram shows the percentage of similarity among different strains.
MLST analysis.
All 15 MRSA isolates were further analyzed by MLST. Strains I and II belong to ST228. Strains III and IV and strain V from cluster C revealed ST239 and ST8, respectively, both being major clones belonging to CC8. Strains VI, VII, and VIII from cluster B belong to ST152, while six of seven strains from cluster A belong to ST80. The last one, strain XIV, was nontypeable (Table 1).
Circular genome view.
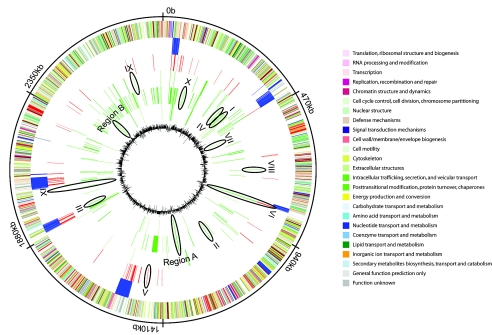
The previous identification of four different clusters was used to identify and map oligonucleotide probes into cluster-specific groups. A cluster-specific oligonucleotide probe is defined as failing to detect its target gene in all strains of a given cluster. Analyses were performed with an EPP of ≤1%, corresponding to the most stringent available condition. Of 6,201 MW2-specific oligonucleotide probes covering 2,632 ORFs, analysis revealed that 93 (3.5%) to 520 (19.7%) probes failed to detect their cognate targets. Fifty-seven to 335 of these probes were cluster specific, i.e., absent in all strains that compose a given cluster. Surprisingly, strains of the B cluster show the largest number of divergence from MW2 (335 absent oligonucleotide probes). Cluster-specific oligonucleotide probes were displayed on the genome of MW2 (Fig. 4), thus revealing that genome regions appeared discriminatory between different clusters. To confirm this observation, cluster-specific regions and selected genes were further analyzed by PCR amplification.
FIG. 4.
Functional genomic organization of the MW2 chromosome compared with 15 clinical strains. From the outside inward, the first circle represents nucleotide position in Mbp. The second circle shows ORFs on the plus and minus (37) strands, with colors according to their COG functional categories. The third circle shows genetic mobile elements and virulence factors from MW2 (3). The fourth circle highlights gene targets that are absent from cluster D compared to MW2. The fifth, sixth, and seventh circles depict gene targets that are absent in clusters C, B, and A, respectively. The eighth circle represents G/C content of MW2. Roman numerals represent gene numbers that are further analyzed.
Validation of microarray results.
We analyzed by PCR the presence of 11 genes showing a divergent presence between the four observed clusters. To select a gene, we required that all probes covering that gene revealed its absence in all strains of a given cluster. We also studied two chromosomal regions consisting of more than three contiguous ORFs that were absent in one or more clusters compared with the community-acquired reference strain MW2. Those targets appeared widely distributed on a full-genome scale (Fig. 4). Among our selection, genes I, II, III, and IV were present in every cluster except cluster B. They encode a hypothetical protein similar to low-temperature-requirement A protein, a fibrinogen-binding protein, a serine protease, and a hypothetical protein similar tothedihydroflavone-4-reductase, respectively (corresponding tothefollowing ORF numbers on the genome sequence of MW2:MW0329, MW1040, MW1753, and MW0305). Gene X (MW0105), encoding a capsular polysaccharide synthesis protein, was expected to be absent in all the strains composing cluster B. However, an amplification band was observed in all strains composing this cluster (figure not shown), suggesting a too-stringent selection of the cutoff. Two genes were selected to characterize cluster C: a hypothetical protein similar to the two-component sensor histidine kinase (gene VIII, MW0622) and a hypothetical protein (gene IX, MW2515). PCR confirmed their absence in strains from cluster C only. Gene V (MW1327) was absent in cluster D only, as determined by microarray and PCR; it encodes for a threonine deaminase homolog. Gene VI (extracellular enterotoxine L, MW0760) was absent in all the strains excepted in MW2 and strain XIII. Gene XI (MW1864) encodes for a truncated transposase present in all strains and was thus considered a positive control for genomotyping. The last gene (gene VII, MW0447) was absent from strains of cluster A; it encodes a conserved hypothetical protein.
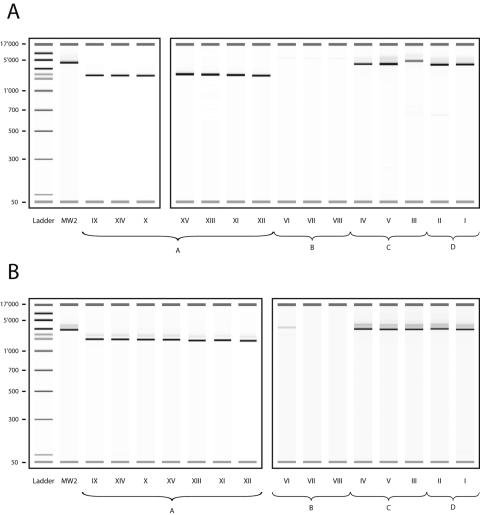
Two regions of difference were selected. As expected, regions A and B displayed 28.77 and 31.38% GC content, respectively, a lower value than the average 32.80% GC content measured in the MW2 genome (3). Primers were designed flanking the zone of interest to generate amplicons of different sizes. Jellyfish software (LabVelocity) was used for primer selection and amplicon size determination. As PCR primers were designed in conserved regions, flanking the region of divergence, we were able to calculate the theoretical size of the PCR product. Region A (MW1206 to MW1209) is composed of ABC transporters (MW1206 to MW1207) and a two-component histidine kinase sensor and regulator (MW1208 to MW1209). Based on the MW2 genome, amplification of region A (Fig. 5a) was expected to yield a 5,987-bp fragment, in agreement with our MW2 control strain that yielded a 5,585-bp amplicon. Strains constituting cluster A yielded an amplicon size of 2,761 bp (minimum, 2,600 bp; maximum, 2,944 bp) (Fig. 5A), in agreement with the predicted size of that conserved region. However, no amplification signal was recorded from strains of cluster B. In cluster C, strains IV and V displayed a shorter band than MW2, but their coclustering strain III yielded a band of 6,333 bp, with a probable insertion of approximately 300 bp.
FIG. 5.
Size variation of amplicons spanning regions A (A) and B (B). Roman numerals represent strain numbers. Each box represents a new assay.
Region B (ORFs MW2308 to MW2313) contains two hypothetical proteins and a transcriptional regulator. This second region (Fig. 5B) generated a 3,893-bp band when using strain MW2, in agreement with the predicted size of 3,957 bp. Cluster A yielded an average size of 1,929 bp (minimum, 1,821; maximum, 2,028), in agreement with the predicted conserved region. In cluster B, only strain VI yielded a 4,564-bp amplicon, thus showing a probable insertion in this region. On average, amplicons measured 4,138 bp and 4,163 bp when originating from clusters C and D, respectively. Taken together, these results suggest that our conservative genomotyping approach allowed accurate detection of two regions of difference.
DISCUSSION
Numerous recent reports about community-onset MRSA contributed to convince experts that its epidemiology represents an emerging and worldwide concern (57). The study of evolutionary relationship of MRSA in the community is controversial; while some studies report a clonal origin for CO-MRSA, others suggest a more distant relatedness between clinical isolates (16, 41). We describe here the characterization and analysis of a collection of CO-MRSA strains using whole-genome oligonucleotide microarrays together with different standardized genotyping methods.
The combined use of different genotyping methods applied on the same panel of strains yields an improvement in overall resolving power. Frequently, laboratories associate two genotyping techniques, one focusing on slow evolutionary genetic markers and another one addressing rapid evolutionary elements. MLST is considered the reference method for studying relatedness between strains (18), while PFGE is recognized as the gold standard method for outbreak analysis (32, 36). However, PFGE can display an overly discriminant power by segregating clonally related strains (6); and under special analysis criteria, PFGE and MLST have the same level of discrimination (44). Coombs et al. (13) have proved the efficiency of a combined approach using MLST and analysis of the mec element; moreover, the latter provided substantial complementary epidemiological information. One should also keep in mind that MLST can group unrelated strains under a common profile (55, 58), thus revealing insufficient discriminatory power. From our study, discriminatory power obtained by VNTR appears lower than that of PFGE and comparative genome hybridization techniques. However, strain relatedness, as established with cluster composition and arrangement, was identical using VNTR (Fig. 3) and PFGE approaches (Fig. 2). The main advantages of VNTR reside in its low cost, moderate time consumption, and high typing resolution (23). Cluster analysis by microarrays is clearly the most discriminative of the three methods. The cluster composition is identical to the two previously cited methods, although it shows a slightly different arrangement; this observation was also described by Koreen et al. (36). Moreover, the comparative genome hybridization technique, consisting of mapping the whole chromosome (at a frequency of 1 probe every 400 to 500 bp using our array), allows precise evaluation of variation between clusters and even between strains within each cluster. This global overview of the genomic composition highlights the biological signature of different clusters.
MLST studies revealed that the main clones were ST5, ST8, and ST239. Among them, some specific genetic backgrounds appear able to acquire SCC-mec elements I, II, III, and IV, while others appear capable of displaying only one of them (18, 19). This observation strongly suggests that the acquisition of the mec element occurred during several independent acquisition events, i.e., the horizontal transfer of genetic material between different strains (52). For instance, the SCC-mec type IV (associated with the presence of the PVL gene and considered a specific marker of community-onset strains) is found integrated in a wide diversity of genetic backgrounds and in different clones, including ST5 and ST8 (19). On the contrary, our study and a previous one performed in Switzerland (25) suggest that the new SCC-mec type V cassette is recovered only in a particular genetic background, namely ST152. An Australian study was recently able to link the presence of the mec V element to three different sequence types: ST152, ST8, and ST45 (13). By combining VNTR, PFGE, and microarrays, we confirmed that SCC-mec V-related strains appear extremely homogeneous and distantly related to all other strains, either CO-MRSA or HA-MRSA. This may signify that the mec V mobile element needs a particular genetic background to be acquired and stabilized.
In our study, CO-MRSA strains were found to belong to major epidemic clones: strain IV and V mec IV belong to ST8 (epidemic MRSA strains EMRSA-2 and EMRSA-6), while strain III, carrier of the mec III element, belongs to ST239. As we do not differentiate mec III from mec IIIA, we were unable to attribute this strain to the Brazilian clone (mec IIIA) (56) or to the Hungarian clone (mec III) (35, 51). All but one strain from cluster A belonged to ST80, a preponderant clone in Greece and also carrier of the Panton-Valentine toxin (1).
We were unable to attribute a sequence type to strain XIV, as sequencing results yielded a novel allelic profile. Interestingly, this profile differed from that of ST80 by one nucleotide only.
Taken together, these observations within that small strain collection suggest (i) the occurrence of multiple horizontal transfer events leading to CO-MRSA and (ii) the presence of specific genetic backgrounds for the acquisition and stability of the mec element (31).
An advantage of typing the whole bacterial genome is the possibility of detecting regions of difference between clusters of strains. In this study, we were able to identify different types of CO-MRSA strains: MRSA strains with the SCC-mec V element, CO-MRSA related to the reference strain MW2 (mec IV type), and other CO-MRSA strains (mec IV type or mec III type). Using our genomotyping analysis, our strain collection differed from MW2 by 3.5 to 19.7%. Using microarrays, Fitzgerald et al. evaluated the dispensable part of the genome of Staphylococcus aureus as 22% (21). Discrepancies can be explained by the use of different analytical methods and diversity of strain collections. For instance, we used a very conservative EPP value, whereas other authors reported the absence of genes with EPP values as high as 19% (12). Thus, we expect that our approach might have missed genes that were truly absent in different strains. Whereas Fitzgerald et al. (21) compared MRSA and methicillin-susceptible S. aureus strains of human, ovine, and bovine origin to the lab strain COL, we focused on the CO-MRSA isolated recovered in our region and compared them with the human pathogen MW2. As expected, regions of difference are mainly localized in genomic regions exhibiting low GC contents and thus likely to derive from horizontal transfer (21).
Genomotyping enables a precise overview of the genetic contents of a strain during a single experiment. Such microarray experiments highlight nonessential regions specific to some strains and permit definition of clusters based on such biological signatures. Thus, they permit not only typing and characterization of clinical isolates with high resolution capacity but also identification of genetic markers characterizing the origin of the strains. In this context, microarrays appear to be a powerful tool for comprehensive genotyping and further understanding of genetic plasticity and strain dissemination.
Acknowledgments
We thank the many staff members of the University Hospitals of Geneva in Bacteriology, Infectious Diseases, and Infection Control Program who contributed to this study.
This work was supported by grants 631-057950.99, PP00B—103002/1 (to J.S.), and 404940-106296/1 (to P.F.) from the Swiss National Science Foundation. T.K. was supported by a grant from the Department of Internal Medicine from the University Hospitals of Geneva.
REFERENCES
- 1.Aires, D. S., C. Bartzavali, I. Spiliopoulou, I. S. Sanches, M. I. Crisostomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, F. F., R. F. McGehee, Jr., and M. Finland. 1968. Methicillin-resistant Staphylococcus aureus at Boston City Hospital. Bacteriologic and epidemiologic observations. N. Engl. J. Med. 279:441-448. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc, D. S., P. Francioli, and P. M. Hauser. 2002. Poor value of pulsed-field gel electrophoresis to investigate long-term scale epidemiology of methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2:145-148. [DOI] [PubMed] [Google Scholar]
- 7.Blanc, D. S., D. Pittet, C. Ruef, A. F. Widmer, K. Muhlemann, C. Petignat, S. Harbarth, R. Auckenthaler, J. Bille, R. Frei, R. Zbinden, P. Moreillon, P. Sudre, and P. Francioli. 2002. Molecular epidemiology of predominant clones and sporadic strains of methicillin resistant Staphylococcus aureus in Switzerland and comparison with European epidemic clones. Clin. Microbiol. Infect. 8:419-426. [DOI] [PubMed] [Google Scholar]
- 8.Blanc, D. S., M. J. Struelens, A. Deplano, R. de Ryck, P. M. Hauser, C. Petignat, and P. Francioli. 2001. Epidemiological validation of pulsed-field gel electrophoresis patterns for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, K. M., A. F. Vaughn, K. L. Russell, B. Smith, D. L. Jimenez, C. P. Barrozo, J. R. Minarcik, N. F. Crum, and M. A. Ryan. 2004. Risk factors for community-associated methicillin-resistant Staphylococcus aureus infections in an outbreak of disease among military trainees in San Diego, California, in 2002. J. Clin. Microbiol. 42:4050-4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charbonnier, Y., B. M. Gettler, P. Francois, M. Bento, A. Renzoni, P. Vaudaux, W. Schlegel, and J. Schrenzel. 2005. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics 6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlebois, E. D., F. Perdreau-Remington, B. Kreiswirth, D. R. Bangsberg, D. Ciccarone, B. A. Diep, V. L. Ng, K. Chansky, and H. F. Chambers. 2004. Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 39:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, T., Y. Hosogi, K. Nishikawa, K. Abbey, R. D. Fleischmann, J. Walling, and M. J. Duncan. 2004. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 186:5473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, and P. M. Giffard. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 15.Dice, L. R. 1945. Measures of the amount of ecologic association between species. Ecology 26:297-302. [Google Scholar]
- 16.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 17.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin- resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feil, E. J., and M. C. Enright. 2004. Analyses of clonality and the evolution of bacterial pathogens. Curr. Opin. Microbiol. 7:308-313. [DOI] [PubMed] [Google Scholar]
- 20.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francois, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, B. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. Van Leeuwen, A. Van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francois, P., D. Pittet, M. Bento, B. Pepey, P. Vaudaux, D. Lew, and J. Schrenzel. 2003. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francois, P., G. Renzi, D. Pittet, M. Bento, D. Lew, S. Harbarth, P. Vaudaux, and J. Schrenzel. 2004. A novel multiplex real-time PCR assay for rapid typing of major staphylococcal cassette chromosome mec elements. J. Clin. Microbiol. 42:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harbarth, S., P. Francois, J. Schrenzel, C. Fankhauser-Rodriguez, S. Hugonnet, T. Koessler, A. Huyghe, and D. Pittet. 2005. Community-associated methicillin-resistant Staphylococcus aureus, Geneva, Switzerland. Emerg. Infect. Dis. 11:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harbarth, S., H. Sax, C. Fankhauser-Rodriguez, J. Schrenzel, A. Agostinho, and D. Pittet. Evaluating the probability of previously unknown methicillin-resistant Staphylococcus aureus carriage at hospital admission. Am. J. Med., in press. [DOI] [PubMed]
- 27.Hardy, K. J., D. W. Ussery, B. A. Oppenheim, and P. M. Hawkey. 2004. Distribution and characterization of staphylococcal interspersed repeat units (SIRUs) and potential use for strain differentiation. Microbiology 150:4045-4052. [DOI] [PubMed] [Google Scholar]
- 28.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, M. E., D. C. Mayfield, C. Thornsberry, J. A. Karlowsky, D. F. Sahm, and D. Peterson. 2002. Prevalence of oxacillin resistance in Staphylococcus aureus among inpatients and outpatients in the United States during 2000. Antimicrob. Agents Chemother. 46:3104-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen, J. H. 1986. Laboratory and epidemiologic experience with methicillin-resistant Staphylococcus aureus in the USA. Eur. J. Clin. Microbiol. 5:693-696. [DOI] [PubMed] [Google Scholar]
- 31.Katayama, Y., D. A. Robinson, M. C. Enright, and H. F. Chambers. 2005. Genetic background affects stability of mecA in Staphylococcus aureus. J. Clin. Microbiol. 43:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468-475. [DOI] [PubMed] [Google Scholar]
- 33.Kerkhoven, R., F. H. van Enckevort, J. Boekhorst, D. Molenaar, and R. J. Siezen. 2004. Visualization for genomics: the Microbial Genome Viewer. Bioinformatics 20:1812-1814. [DOI] [PubMed] [Google Scholar]
- 34.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065.1-0065.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 38.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 39.Liassine, N., R. Auckenthaler, M. C. Descombes, M. Bes, F. Vandenesch, and J. Etienne. 2004. Community-acquired methicillin-resistant Staphylococcus aureus isolated in Switzerland contains the Panton-Valentine leukocidin or exfoliative toxin genes. J. Clin. Microbiol. 42:825-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindstedt, B. A., T. Vardund, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213-222. [DOI] [PubMed] [Google Scholar]
- 41.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 42.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 43.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peacock, S. J., G. D. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pourcel, C., F. Andre-Mazeaud, H. Neubauer, F. Ramisse, and G. Vergnaud. 2004. Tandem repeats analysis for the high resolution phylogenetic analysis of Yersinia pestis. BMC Microbiol. 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramisse, V., P. Houssu, E. Hernandez, F. Denoeud, V. Hilaire, O. Lisanti, F. Ramisse, J. D. Cavallo, and G. Vergnaud. 2004. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J. Clin. Microbiol. 42:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Said-Salim, B., B. Mathema, and B. N. Kreiswirth. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect. Control Hosp. Epidemiol. 24:451-455. [DOI] [PubMed] [Google Scholar]
- 49.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saunders, N. A., A. Underwood, A. M. Kearns, and G. Hallas. 2004. A virulence-associated gene microarray: a tool for investigation of the evolution and pathogenic potential of Staphylococcus aureus. Microbiology 150:3763-3771. [DOI] [PubMed] [Google Scholar]
- 51.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49:2070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shukla, S. K., M. E. Stemper, S. V. Ramaswamy, J. M. Conradt, R. Reich, E. A. Graviss, and K. D. Reed. 2004. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J. Clin. Microbiol. 42:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 54.Talaat, A. M., S. T. Howard, W. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, Y. W., G. W. Procop, and D. H. Persing. 1997. Molecular diagnostics of infectious diseases. Clin. Chem. 43:2021-2038. [PubMed] [Google Scholar]
- 56.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaidi, N., K. Konstantinou, and M. Zervos. 2003. The role of molecular biology and nucleic acid technology in the study of human infection and epidemiology. Arch. Pathol. Lab. Med. 127:1098-1105. [DOI] [PubMed] [Google Scholar]