Abstract
Zoothamnium niveum is a giant, colonial marine ciliate from sulfide-rich habitats obligatorily covered with chemoautotrophic, sulfide-oxidizing bacteria which appear as coccoid rods and rods with a series of intermediate shapes. Comparative 16S rRNA gene sequence analysis and fluorescence in situ hybridization showed that the ectosymbiont of Z. niveum belongs to only one pleomorphic phylotype. The Z. niveum ectosymbiont is only moderately related to previously identified groups of thiotrophic symbionts within the Gammaproteobacteria, and shows highest 16S rRNA sequence similarity with the free-living sulfur-oxidizing bacterial strain ODIII6 from shallow-water hydrothermal vents of the Mediterranean Sea (94.5%) and an endosymbiont from a deep-sea hydrothermal vent gastropod of the Indian Ocean Ridge (93.1%). A replacement of this specific ectosymbiont by a variety of other bacteria was observed only for senescent basal parts of the host colonies. The taxonomic status “Candidatus Thiobios zoothamnicoli” is proposed for the ectosymbiont of Z. niveum based on its ultrastructure, its 16S rRNA gene, the intergenic spacer region, and its partial 23S rRNA gene sequence.
Symbioses of eukaryotes with chemolithoautotrophic sulfur-oxidizing (thiotrophic) bacteria are widespread and known from diverse marine sulfidic habitats, ranging from shallow subtidal sands and macrophyte debris to deep sea cold seeps and hydrothermal vents (14, 24, 25, 45). The giant peritrich ciliate Zoothamnium niveum (Ciliophora, Oligohymenophora) is host of one of the most remarkable of such symbioses. This sessile, colonial ciliate was originally described more than 150 years ago from the Red Sea (20, 30); later it was also found in the Lower Keys of Florida (Gulf of Mexico) and at the Belize Barrier Reef (Caribbean Sea) (4). Typically Z. niveum occurs on mangrove peat walls in groups of more than 100 colonies, with an estimated group life of approximately 20 days, predominantly at sites where the microbial surface mat has been destroyed (44).
The feather-like Z. niveum colonies, which reach sizes up to 1.5 cm, consist of a central stalk with branches occurring alternately on the stalk and three cellular morphotypes on the branches: (i) microzooids are the feeding stages; (ii) macrozooids are the dispersal stages, capable of leaving the colony as large swarmers to build a new colony after settlement; and (iii) terminal zooids are responsible for asexual reproduction by longitudinal fission (4, 5) (Fig. 1).
FIG. 1.
Zoothamnium niveum colony. (A) General view of the entire colony by the central stalk (st) with alternate branches (br) bearing cells. Light microscopy; scale bar, 0.5 mm. (B) Detail of the middle part of the colony, demonstrating the three different Z. niveum cell types: macrozooids (ma), microzooids (mi), and terminal zooids (t) occurring on the branches. Light microscopy, contrast enhanced; scale bar, 50 μm.
Z. niveum has been found exclusively in obligatory association with ectosymbiotic bacteria that are responsible for the ciliate's brilliant white color (4, 5). Z. niveum is virtually completely covered by these bacteria appearing as rods and coccoid rods arranged in a highly specific and permanently occurring pattern (Fig. 2). Macrozooids, terminal zooids, and branches are occupied by a monolayer of rod-shaped bacteria (average length, 1.4 μm; average width, 0.4 μm), whereas bacteria on the oral side of microzooids are coccoid to rod shaped (average length, 1.9 μm; average width, 1.0 μm) (5). The series of transitional morphological stages from rods to coccoid rods found on the microzooids suggests the presence of a single ectosymbiotic bacterial species on Z. niveum (5, 7). This regular coat, however, appears to be disturbed and replaced by a variety of other bacterial morphotypes on the basal parts of large colonies only (44).
FIG. 2.
SEM of the Zoothamnium niveum symbiosis. (A) Z. niveum microzooid with bacterial coat, consisting of coccoid rods (co) on the oral part of the microzooid, rods (ro) on the aboral part, and transitional stages of intermediate shapes (in) in between. (B) Basal part of the Z. niveum colony exhibiting large attached filaments on branch (br). Note that the ectosymbiotic coat on the microzooids is partly lost. Bar lengths, 10 μm.
Culture and incubation experiments showed that Z. niveum is unable to survive without its symbionts (4), and physiological studies applying 14C-labeled bicarbonate incubations (48) and Cartesian diver experiments (44) indicated that the Z. niveum ectosymbionts are thiotrophic bacteria. Since none of the known thiotrophic symbionts have yet been cultivated without their respective hosts, culture-independent approaches have to be employed to reveal the identity of these elusive bacteria (3).
Here we investigated by scanning electron microscopy (SEM), 16S rRNA gene sequencing, comparative sequence analysis, and specific fluorescence in situ hybridization (FISH) the thiotrophic ectosymbionts of Z. niveum. We identified a single bacterial phylotype representing the Z. niveum ectosymbiont, which is only moderately related to known symbiotic and free-living bacteria within the Gammaproteobacteria. Only on basal, senescent parts of large colonies was the symbiont replaced by a variety of other bacteria. For classification of the Z. niveum ectosymbiont we propose the tentative taxonomic status “Candidatus Thiobios zoothamnicoli.”
MATERIALS AND METHODS
Sample collection and fixation.
Samples of Zoothamnium niveum colonies were cut from peat surfaces in 1 to 3 m depth at different locations in the main channel (extending 1 km) of the mangrove island Twin Cays (16°48′N, 88°05′W; Belize Barrier Reef, Caribbean Sea) by scuba diving in July 2003. Samples (20 ciliates each) were preserved for DNA analysis (96% ethanol), for FISH (4% paraformaldehyde in phosphate-buffered saline [PBS] 0.1 M, pH 7.4, or 1% osmium tetroxide in natural seawater), and for scanning electron microscopy (2% osmium tetroxide in natural seawater). All solutions were sterile filtered. Applying osmium tetroxide fixation, natural seawater was evaporated by boiling to half of its volume and mixed 1:1 with 2% (for FISH) or 4% (for SEM) aqueous osmium tetroxide. All specimens preserved for FISH and SEM were rinsed in PBS three times for 5 min each, dehydrated in 30 and 50% ethanol for 5 min each, and stored in 70% ethanol until further treatment up to a year later.
Scanning electron microscopy.
Specimens fixed for SEM and stored in 70% ethanol were further dehydrated, critical point dried, and sputter coated (4). A Philips XL 20 scanning electron microscope was used to view the colonies.
PCR, cloning, and DNA sequencing.
Specimens stored in 96% ethanol were transferred to PBS, followed by short pulses of vortexing to mechanically disintegrate the large Z. niveum colonies; 5 μl of this PBS solution containing a mix of host tissues and symbionts was directly used as the template for PCR without prior DNA extraction. PCRs were performed in a temperature gradient thermocycler with a standard PCR cycling program and an annealing temperature of 55°C. A typical PCR mixture contained 2 mM of MgCl2, 0.2 mM of deoxynucleoside triphosphate, 1.5 U of Taq DNA polymerase (Promega GmbH, Mannheim, Germany), 50 pM of forward and reverse primer, and 5 μl of the Z. niveum mix in a total volume of 50 μl.
The forward primer 616V (5′-AGA GTT TGA TYM TGG CTC-3′) (33) targeting the 5′ end of the 16S rRNA gene of all bacteria was used in combination with the unlabeled probe GAM42a (5′-GCC TTC CCA CAT CGT TT-3′) targeting the 23S rRNA of all Gammaproteobacteria at Escherichia coli position 1027 as the reverse primer (38). PCR products were separated by electrophoresis using a 2% agarose gel. Gels were stained with ethidium bromide and visualized by UV transillumination. Amplification products in the desired size (approximately 3,000 bp) were cut out and purified with GELase (Epicenter, Madison, WI). The TopoXL cloning kit (Invitrogen Life Technologies, Karlsruhe, Germany) was used for all cloning reactions. The nucleotide sequences of cloned DNA fragments were determined by cycle sequencing of purified plasmid DNA with the Thermo Sequenase cycle sequencing kit (Amersham Life Science, Little Chalfont, United Kingdom) with dye-labeled, vector-specific primers using an automated DNA sequencer (LI-COR 4200; LI-COR Inc., Lincoln, NE) under conditions recommended by the manufacturers.
Phylogenetic analysis and probe design.
The nucleotide sequences obtained were added to a 16S rRNA database containing more than 14,000 16S rRNA sequences using the ARB program package (37). Automatic alignment was performed with the tool of the ARB package and subsequently corrected by visual inspection. The phylogenetic relationships of the obtained 16S rRNA sequences were inferred applying the following methods in the ARB package: neighbor joining (with the Jukes-Cantor correction), maximum parsimony (Phylip DNAPARS 3.573, with 100 resamplings), maximum likelihood (AxML), and Treepuzzle 5.0. The outgroup was composed of 32 phylotypes covering the main groups of the domain Bacteria. A filter considering only those alignment positions that were conserved in at least 50% of all Gammaproteobacteria sequences was applied, and only sequences with a length of more than 1,300 nucleotides were used for tree calculations.
Probe design was performed with the tool of the ARB software package (37). To demonstrate that the 16S rRNA sequence obtained is not chimeric, two differently labeled Z. niveum ectosymbiont-specific probes (targeting near the 5′ and the 3′ ends of the retrieved 16S rRNA sequence) (Table 1) were designed. The probe sequences obtained were checked with the Probe Match tool integrated in the ARB, as well as against all 16S rRNA sequences in the Ribosomal Database Project by the implemented tool Probe Match (15). The nucleotide sequences of the newly designed probes ZNS196 and ZNS1439 have been deposited at probeBase (36; www.microbial-ecology.net/probebase).
TABLE 1.
Probes used for fluorescence in situ hybridization
| Probe | Standard probe namea | Specificity | Sequence | Target sites | Reference |
|---|---|---|---|---|---|
| EUB338 | S-D-Bact-0338-a-A-18 | Most Bacteria | 5′-GCT GCC TCC CGT AGG AGT-3′ | 338-355b | Amann et al. (2) |
| EUB338II | S-*-BactP-0338-a-A-18 | Bacteria not covered by probe EUB338, e.g., many Planctomycetes | 5′-GCA GCC ACC CGT AGG TGT-3′ | 338-355b | Daims et al. (16) |
| EUB338III | S-*-BactV-0338-a-A-18 | Bacteria not covered by probe EUB338, e.g., many Verrucomicrobia | 5′-GCT GCC ACC CGT AGG TGT-3′ | 338-355b | Daims et al. (16) |
| NON338 | Not named | Negative control | 5′-ACT CCT ACG GGA GGC AGC-3′ | 338-355 | Wallner et al. (57) |
| GAM42a | L-C-gProt-1027-a-A-17 | Gammaproteobacteria | 5′-GCC TTC CCA CAT CGT TT-3′ | 1027-1043c | Manz et al. (38) |
| BET42a | L-C-bProt-1027-a-A-17 | Betaproteobacteria | 5′-GCC TTC CCA CTT CGT TT-3′ | 1027-1043c | Manz et al. (38) |
| ZNS196 | S-*-Znsym-0196-a-A-19 | Z. niveum ectosymbiont | 5′-CTG ATA GTG ACC GAA GTC T-3′ | 196-214b | This study |
| ZNS1439 | S-*-Znsym-1439-a-A-20 | Z. niveum ectosymbiont | 5′-GAG CGC CCA TTA TTA AGC TA-3′ | 1439-1458b | This study |
Fluorescence in situ hybridization.
The probes listed in Table 1 were labeled on their 5′ end with the fluorescent dyes Cy3, Cy5, or fluorescine (Fluos) (Thermo, Ulm, Germany). Specimens fixed for FISH and stored in 70% ethanol were rehydrated in 50 and 30% ethanol for 3 min each, transferred to 0.02 M Tris-HCl (pH 8), and dried on SuperFrostPlus microscope slides (Menzel-Gläser, Braunschweig, Germany) at 46°C for 10 min. FISH was carried out according to Manz (38) on whole-mount specimens. Finally, slides were washed briefly with ice-cold distilled H2O, air dried, and embedded in Vectashield (Vector Laboratories, Burlingame, CA). Optimal hybridization conditions for the newly developed probes ZNS196 and ZNS1439 were determined with Z. niveum ectosymbionts, applying a series of formamide concentrations (0 to 40%) in hybridization buffer (56).
Positive and negative hybridization controls were the EUBmix probe set (consisting of EUB338, EUB338II, and EUB338III) (2, 16), targeting all Bacteria and probe NON338, complementary to EUB338 (57), respectively. Probe GAM42a targeting all Gammaproteobacteria was always applied in combination with the unlabeled BET42a probe as a competitor, and vice versa (BET42a probe with the unlabeled GAM42a as competitor) (38). Hybridized samples were examined using a confocal laser scanning microscope (LSM 510 Meta; Carl Zeiss, Jena, Germany) equipped with two helium-neon lasers (543 and 633 nm) and an argon laser (458 to 514 nm). Image analysis was performed with the standard software package delivered with the instrument (version 3.2).
Nucleotide sequence accession number.
The 16S rRNA gene, the intergenic spacer region and the partial 23S rRNA gene sequence of the Z. niveum ectosymbiont have been deposited in the public databases GenBank/EMBL/DDBJ under accession number AJ879933.
RESULTS
Scanning electron microscopy.
The symbiotic coat of Zoothamnium niveum was found to consist of a bacterial monolayer of rods, coccoid rods, and intermediate shapes, as reported previously (45) (Fig. 2). An overgrowth of various bacteria could be observed not only for the lower stalk (44) but also for the lower branches of large and therefore senescent colonies (Fig. 2).
Amplification, cloning, and sequencing of rRNA genes, and phylogenetic inference.
Initial FISH experiments using the bacterial EUBmix (2, 16) probe GAM42a and probe BET42a, targeting all Gamma- and Betaproteobacteria, respectively, revealed that the ectosymbiotic coat of Z. niveum consists exclusively of Gammaproteobacteria. Therefore we chose a directed approach for PCR amplification of the ectosymbiont 16S rRNA gene by applying the bacterial 16S rRNA gene-targeted forward primer 616V (33) in combination with the unlabeled probe GAM42a, targeting the 23S rRNA of Gammaproteobacteria (38), as the reverse primer.
The PCR product was cloned and two clones were sequenced. The sequences obtained were found to be identical; they were 2,956 nucleotides in length and included the complete 16S rRNA gene (1,487 nucleotides), an intergenic spacer region (472 nucleotides) containing two tRNA genes (isoleucine and alanine), and the 5′ end of the 23S rRNA gene (997 nucleotides) showing a deletion at E. coli positions 136 to 160. The 16S rRNA gene sequence was analyzed in more detail and showed highest sequence similarity to two Gammaproteobacteria: 94.5% to the free-living sulfur-oxidizing bacterium ODIII6 (34), and 93.1% to the endosymbiont (named “scaly snail endosymbiont”) of a recently discovered hydrothermal vent gastropod (family Peltospiridae) from the Indian Ocean (28). Among cultivated free-living thiotrophic bacteria (and beyond strain ODIII6) Thiorhodovibrio winogradskyi and Marichromatium chilcum were each most similar (91.3%) to the recovered 16S rRNA sequence.
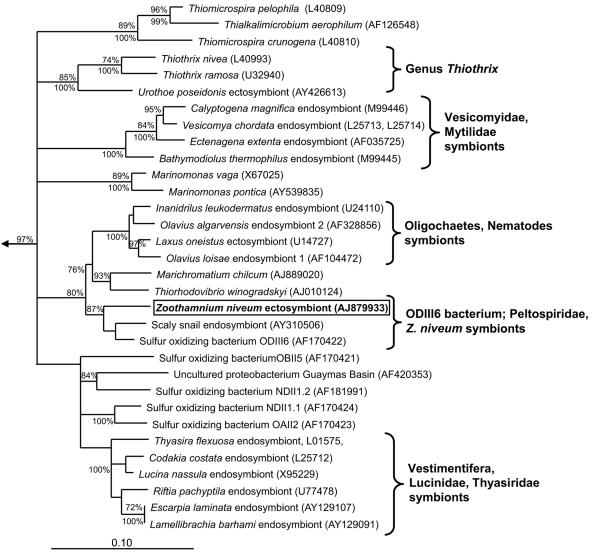
All tree methods used to resolve the phylogenetic relationships of the Z. niveum symbiont to other Gammaproteobacteria in our data set provided similar results. Within a cluster of thiotrophic symbiotic and free-living Gammaproteobacteria, the Z. niveum symbiont formed a monophyletic group together with strain ODIII6 and the scaly snail endosymbiont (Fig. 3). A characteristic deletion at E. coli 16S rRNA positions 455 to 477 was detected in all three 16S rRNA sequences within this group.
FIG. 3.
16S rRNA-based phylogenetic Treepuzzle tree (with the HKY correction/uniform model of substitution) shows the relationships of the Zoothamnium niveum ectosymbiont and chemoautotrophic symbiotic and free-living Gammaproteobacteria. For the symbiotic bacteria the respective host organisms are indicated. Treepuzzle support values are depicted above the respective branches; maximum parsimony bootstrap values are shown below the branches. Only Treepuzzle support values above 70% and parsimony bootstrap values greater than 75% are displayed. GenBank accession numbers are given in parentheses. The arrow points to the outgroup, and the bar represents 10% estimated evolutionary distance.
Fluorescence in situ hybridization.
Paraformaldehyde fixation of Z. niveum resulted in adequate FISH signal intensity of the bacterial ectosymbionts, but unfortunately caused detachment of the bacteria from their host cells. Therefore we used osmium tetroxide-fixed samples for FISH as these yielded a good signal intensity as well as an excellent morphological preservation of host cells, including an intact bacterial coat. Two specific oligonucleotide probes, ZNS196 and ZNS1439, targeting the 5′ end and the 3′ end of the obtained 16S rRNA sequences, respectively, were designed for the specific detection of the Z. niveum ectosymbiont (Table 1).
Both probes showed at least two strong mismatches to all other 16S rRNA sequences available in the public databases. However, the lowest number of mismatches was observed with 16S rRNA gene clone sequences, a pathogenic bacterium causing bacteremia in humans, and Cyanobacteria, none of which represented a suitable nontarget organism for the new probes. For both ectosymbiont-specific probes, a formamide concentration of 35% in the hybridization buffer was found to be optimal and was subsequently used for all FISH experiments.
FISH has been demonstrated previously on many occasions to be able to discriminate even single nucleotide mismatches under these highly stringent conditions (3). Thus, the conditions applied in our experiments virtually exclude the possibility of nonspecific hybridizations. Furthermore, an additional level of reliability is provided by the use of not one but two probes targeting the same symbiont specifically at two different 16S rRNA target regions. The specificity of our hybridization conditions was further demonstrated by the application of the betaproteobacterial probe BET42a, which did not hybridize to the gammaproteobacterial Z. niveum ectosymbiont (showing only one mismatch to this probe).
FISH of whole-mount Z. niveum colonies using the ectosymbiont-specific probes ZNS196 and ZNS1439 under these stringent conditions demonstrated that, except for the bacteria on the basal parts of larger colonies, all bacteria constituting the bacterial coat on the surface of Z. niveum hybridized with the symbiont-specific probes, probe GAM42a, and the EUBmix probe set. This included the rod-shaped ectosymbionts on the stalk, branches, and terminal and macro- and microzooids, as well as the coccoid rod-shaped ectosymbionts and intermediate forms between rods and coccoid rods on the microzooids (Fig. 4). Simultaneous hybridization signals of the two differently labeled Z. niveum ectosymbiotic probes showed that the retrieved 16S rRNA gene sequence was not chimeric, and the application of probe NON338 (complementary to bacterial probe EUB338) as a negative control yielded no detectable fluorescence signals (data not shown), further demonstrating that the observed fluorescence signals were specific and not caused by autofluorescence or unspecific staining of bacteria. The mostly rod-shaped or filamentous bacteria on the basal parts of large colonies were stained by the EUBmix probe set (Fig. 5). Some of these bacteria also hybridized with the Gammaproteobacteria probe GAM42a but none of them was detected with the symbiont-specific probes.
FIG. 4.
FISH micrographs of Zoothamnium niveum symbiosis. Z. niveum microzooid with ectosymbionts visualized using a hierarchical probe set: (A) EUBmix probes in Fluos (green); (B) probe GAM42a in Cy5 (blue); (C) probe ZNS196 in Cy3 (red); (D) overlay with differential interference contrast. Bar length, 10 μm.
FIG. 5.
FISH micrographs of unidentified bacteria associated with Zoothamnium niveum. Bacteria found at basal parts of large colonies visualized by differential interference contrast and probes EUBmix, GAM42a, and ZNS196. (A) Note that a few detached symbionts and symbionts on a Z. niveum microzooid in addition to eubacterial filaments and rods on stalk and braches can be seen. (B) Mainly large filamentous bacteria overgrowing a basal Z. niveum branch. Bar lengths, 10 μm.
DISCUSSION
The 16S rRNA gene sequence analysis presented in this study revealed that the thiotrophic symbiosis of Zoothamnium niveum from Twin Cays (Caribbean Sea) is a specific association involving a single, pleomorph ectosymbiont species belonging to the Gammaproteobacteria. Although our approach did not allow us to exclude microheterogeneity within this symbiont population, we were able to demonstrate the transition of the Z. niveum ectosymbiont morphology from rods to coccoid rods on the microzooids by SEM in this and in previous studies (4, 5), and by FISH using two ectosymbiont-specific probes.
Structural polymorphism is not unusual in bacteria. Molecular evidence of pleomorphic species has been reported previously, e.g., for the soil bacterium Arthrobacter globiformis (51), the chemoautotrophic bacterium Thiomicrospira thyasirae (59), certain pathogens such as Coxiella burnetii and members of the phylum Chlamydiae (29), as well as for the chemoautotrophic symbionts of bivalves and tubeworms (9, 18). Transitions from rods to coccoid shapes are usually accompanied by changes in cell size, whereby either morphotype can be larger than the other. In the growth cycle of A. globiformis, for example, irregular rods in young cultures are replaced by smaller coccoid forms in older cultures (51), whereas the chemoautotrophic thiotrophic endosymbiont of the tubeworm Riftia pachyptila is rod shaped and changes by terminal differentiation into larger cocci, showing transitional stages between the two morphotypes (8, 9), similar to the morphological stages observed for the Z. niveum ectosymbionts analyzed in this study.
In some well-studied free-living bacteria such as E. coli, the specific cell size is correlated with growth rate, which is related to nutritional and nonnutritional parameters (pH, temperature, osmotic strength, and hydrostatic pressure). Cell form modulations however are considered to be related to nutrition only (41). In the case of the thiotroph symbiont, nutrition means chemicals for sulfide oxidation and carbon fixation. Therefore, we hypothesize that the physicochemical microhabitats (54, 55) of both morphotypes, coccoid rods being restricted to the oral, feeding side of the host's microzooids and rods on all other parts of the host, differ, leading to the different morphology and size of the symbionts.
The newly identified Z. niveum ectosymbiont forms a phylogenetic group with the bacterial isolate ODIII6 and a scaly snail endosymbiont. This monophyletic group thus includes free-living and ecto- and endosymbiotic bacteria from distantly located habitats ranging from shallow waters to deep-sea environments, from macrophyte debris and sediments to bare basalt hydrothermal vents. In addition, the geographic range of this bacterial group covers the Caribbean Sea, the Mediterranean Sea, and the Indian Ocean. However, all these habitats are sulfidic and therefore provide, in a most general sense, similar ecological conditions for thiotrophic bacteria. The mangrove peat walls at Twin Cays (Caribbean Sea), where Z. niveum lives, were shown to release sulfide by diffusion in concentrations up to 250 μM (44). From shallow-water hydrothermal vents at Palaeochori Bay (Mediterranean Sea), where strain ODIII6 was isolated from sediment (49), sulfide concentrations range from 10 to 1,000 μM (52). The host of the scaly snail endosymbiont was discovered at the base of black smoker chimneys, presumably a zone of mixing between warm sulfide-rich vent fluids and cold, oxygen-rich deep-sea water (53, 58). Indeed, there is evidence that at least the free-living bacterial strain ODIII6 and the Z. niveum ectosymbiont show a similar physiological background.
Strain ODIII6 is capable of oxidizing thiosulfate in culture (34), while oxygen uptake experiments with Cartesian divers (44) indicated that the Z. niveum ectosymbiont is also a thiotroph. Furthermore, incubations with 14C-labeled bicarbonate in sulfide or thiosulfate added to seawater suggested that the Z. niveum ectosymbiont was capable of oxidizing both sulfur species to gain energy for autotrophic carbon fixation (48). Data on the physiology of the scaly snail endosymbiont are not yet available (28).
In addition, a variety of differently shaped, mostly filamentous Bacteria, some of which are Gammaproteobacteria, were found on the stalk and on some branches at the basal parts of large Z. niveum colonies. The fact that these bacteria occurred only on senescent, basal stalk and branches, and that they did not hybridize with the ectosymbiont-specific probes, suggests that they are nonspecific microbial epigrowth. Z. niveum colonies need more than 4 days to develop to a full-grown stage (43), but after 4 days, the oldest basally located host cells have already become senescent. Successively, the specific ectosymbionts are lost and appear to be replaced by other bacteria forming the nonspecific epigrowth. Eventually, Z. niveum host cells die, leaving the naked stalk and branches. These observations indicate the existence of a mechanism to ensure growth of the ectosymbiont and to prevent nonspecific microbial fouling on the host surface at the same time.
Bacterial partners in thiotrophic symbioses known so far belong to two classes of Proteobacteria. Whereas the ectosymbionts of the shrimp Rimicaris exoculata (46) and of the polychaete Alvinella pompejana (13) are Epsilonproteobacteria, the majority of chemoautotrophic sulfur-oxidizing ecto- or endosymbiotic bacteria belong to the Gammaproteobacteria. All treeing methods applied to our data set support four monophyletic groups comprising thiotrophic symbionts within a cluster of Gammaproteobacteria, in addition to the monophyletic group containing the Zoothamnium niveum ectosymbiont: (i) the endosymbionts of two bivalve families, Vesicomyidae and Mytilidae, cluster together as established previously (17); (ii) endosymbionts of vestimentiferan tubeworms (Siboglinidae, Polychaeta) (39, 42) form a group together with bivalve symbionts of the families Lucinidae and Thyasiridae (32); (iii) the gammaproteobacterial ectosymbiont of the nematode Laxus oneistus of the subfamily Stilbonematinae (47) groups together with the endosymbionts of the oligochaete subfamily Phallodrillinae (19); and (iv) an ectosymbiotic Thiothrix bacterium living on the amphipod crustacean Urothoe poseidonis was discovered recently as a novel marine phylotype within the genus Thiothrix (27).
The exact branching order between these groups of thiotrophic Gammaproteobacteria could not be resolved by comparing the results of the different treeing methods. Interestingly, a deletion at E. coli 16S rRNA positions 455 to 477 was found in the 16S rRNA gene sequence of the Z. niveum ectosymbiont, strain ODIII6, and the scaly snail endosymbiont. This deletion was thought to be unique to the genus Thiothrix within the Gammaproteobacteria and some members of the Epsilonproteobacteria (31, 35). This deletion clearly separates the monophyletic group containing the Z. niveum ectosymbiont and members of the genus Thiothrix from all other known Gammaproteobacteria. Since the occurrence of this deletion twice in separate evolutionary lineages is more unlikely than a single deletion event, it is tempting to speculate that the group containing the Z. niveum ectosymbiont and the members of the genus Thiothrix originate from a common free-living ancestor from sulfidic habitats.
According to Stackebrandt and Goebel (50) the 16S rRNA sequence similarity threshold for the unambiguous differentiation of two species is 97%. This suggests that the newly identified Z. niveum ectosymbiont and its closest relatives, isolate ODIII6 and the scaly snail endosymbiont, represent different species. We therefore propose the taxonomic status Candidatus (40) for the ectosymbiont of Z. niveum with the following description:
“Candidatus Thiobios zoothamnicoli” [thio (Gr.), sulfur; bios (Gr.), life; on (Gr.), unit, neutrum; Zoothamnium (L.), genus of the ciliate host; colo (L.), inhabit]. Phylogenetic position: Gammaproteobacteria; gram-negative-type cell wall; pleomorphic morphology, ranging from rods to coccoid rods (rods 1.4 by 0.4 μm; coccoid rods 1.9 by 1.0 μm). Basis of assignment: 16S rRNA gene, intergenic spacer region, and partial 23S rRNA gene sequence GenBank/EMBL/DDBJ accession number AJ879933, 16S rRNA-targeted oligonucleotide probes S-*-ZnSym-0196-a-A-19 (5′-CTG ATA GTG ACC GAA GTC T-3′) and S-*-ZnSym-1439-a-A-20 (5′-GAG CGC CCA TTA TTA AGC TA-3′). Not cultivated on cell-free media; ectosymbiont of Zoothamnium niveum (Ciliophora, Oligohymenophora) collected from mangrove peat walls, Barrier Reef of Belize; membrane-bound, electron-translucent vesicles resembling sulfur vesicles (4; this study).
In this study, for the first time, a chemolithoautotrophic sulfide-oxidizing ectosymbiont of a protist was identified at the molecular level. Protists with bacterial ectosymbionts are widely distributed and have also been observed for example in sulfide-rich habitats from subtidal sands (21, 22, 23, 26) and cold seeps (6, 12). Future research should thus focus on the analysis of phylogenetic relationships and ecophysiology of the bacteria involved in these associations in order to get a more complete picture of the biology of the worldwide-occurring thiotrophic protist symbioses.
Acknowledgments
The present study has been supported by Austrian Science Foundation grants P16840-BO3 (M. Bright), P13964-BIO (J. Ott), and P16566-B14 (M. Wagner and M. Horn) and the CCRE program of the National Museum of Natural History (Smithsonian Institution), Washington, D.C. (grant no. 746).
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer-Nebelsick, M., C. F. Bardele, and J. Ott. 1996. Redescription of Zoothamnium niveum (Hemprich & Ehrenberg, 1831) Ehrenberg, 1838 (Oligohymenophora, Peritrichida) a ciliate with ectosymbiotic, chemoautotrophic bacteria. Eur. J. Prostistol. 32:18-30. [Google Scholar]
- 5.Bauer-Nebelsick, M., C. F. Bardele, and J. Ott. 1996. Electron microscopic studies on Zoothamnium niveum (Hemprich & Ehrenberg, 1831) Ehrenberg 1838 (Oligohymenophora, Peritrichida) a ciliate with ectosymbiotic, chemoautotrophic bacteria. Eur. J. Prostistol. 32:202-215. [Google Scholar]
- 6.Bernhard, J. M., K. R. Buck, M. A. Farmer, and S. S. Bowser. 2000. The Santa Barbara Basin is a symbiosis oasis. Nature 403:77-80. [DOI] [PubMed] [Google Scholar]
- 7.Bright, M. 2002. Life strategies of thiotrophic ectosymbiosis, p. 19-32. In M. Bright, P. Dworschak, and M. Stachowitsch (ed.), The Vienna School of Marine Biology: a tribute to Jörg Ott. Facultas, Vienna, Austria.
- 8.Bright, M., and A. Sorgo. 2003. Ultrastructural reinvestigation of the trophosome in adult Riftia pachyptila (Vestimentifera). Invertebr. Biol. 122:347-368. [Google Scholar]
- 9.Bright, M., and O. Giere. 2005. Microbial symbiosis in Annelida. Symbiosis 38:1-45. [Google Scholar]
- 10.Brosius, J., M. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of 16S ribosomal RNA gene from E. coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 12.Buck, K. R., J. P. Barry, and A. G. B. Simpson. 2000. Monterey Bay cold seep biota: euglenozoa with chemoautotrophic bacterial epibionts. Eur. J. Protistol. 36:117-126. [Google Scholar]
- 13.Cary, S. C., M. T. Cottrell, J. L. Stein, F. Camacho, D. Desbruyeres. 1997. Molecular identification and localization of a filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl. Environ. Microbiol. 63:1124-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanaugh, C. M. 1985. Symbioses of chemoautotrophic bacteria and marine invertebrates from hydrothermal vents and reducing sediments. Biol. Soc. Wash. Bull. 6:373-388. [Google Scholar]
- 15.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 1:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 17.Distel, D. L., H. Felbeck, and C. M. Cavanaugh. 1994. Evidence for phylogenetic congruence among sulfur-oxidizing chemoautotrophic bacterial endosymbionts and their bivalve hosts. J. Mol. Evol. 38:533-542. [Google Scholar]
- 18.Distel, D. L., D. J. Lane, G. J. Olsen, S. J. Giovannoni, B. Pace, D. A. Stahl, and H. Felbeck. 1988. Sulfur-oxidizing bacterial endosymbionts; analysis of phylogeny and specificity by 16S rRNA sequences. J. Bacteriol. 170:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubilier, N., R. Amann, C. Erséus, G. Muyzer, S. Y. Park, O. Giere, and C. M. Cavanaugh. 1999. Phylogenetic diversity of bacterial endosymbionts in the gutless marine oligochete Olavius loisae (Annelida). Mar. Ecol. Prog. Ser. 178:271-280. [Google Scholar]
- 20.Ehrenberg, C. G. 1838. Die Infusionsthierchen als vollkommene Organismen. Ein Blick in das tiefere organische Leben der Natur. Leopold Voss Verlag, Leipzig, Germany.
- 21.Epstein, S. S., D. A. Bazylinski, and W. H. Fowle. 1998. Epibiotic bacteria on several ciliates from marine sediments. J. Eukaryot. Microbiol. 45:64-70. [Google Scholar]
- 22.Fenchel, T., and J. Finlay. 1989. Kentrophoros: a mouthless ciliate with a symbiotic kitchen garden. Ophelia 30:75-93. [Google Scholar]
- 23.Fenchel, T., T. Perry, and A. Thane. 1977. Anaerobiosis and symbiosis with bacteria in free-living ciliates. J. Protozool. 24:154-163. [DOI] [PubMed] [Google Scholar]
- 24.Fisher, C. R. 1990. Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev. Aquat. Sci. 2:399-436. [Google Scholar]
- 25.Fisher, C. R. 1996. Ecophysiology of primary production at deep-sea vents and seeps, p. 311-334. In F. Uiblein, J. Ott, and M. Stachowitsch (ed.), Deep-sea and extreme shallow-water habitats: affinities and adaptations. Biosystematics and Ecology Series 11. Austrian Academy of Science Press, Vienna.
- 26.Foissner, W. 1995. “Kentrophoros” (Ciliophora, Karyorelictida) has oral vestiges: a reinvestigation of “K. fistulosus” (Faure-Fremiet, 1950) using protargol impregnation. Arch. Protistenk. 146:165-179. [Google Scholar]
- 27.Gillan, D. C., and N. Dubilier. 2004. Novel epibiotic thiothrix bacterium on a marine amphipod. Appl. Environ. Microbiol. 70:3772-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goffredi, S. K., A. Waren, V. J. Orphan, C. L. Van Dover, and R. C. Vrijenhoek. 2004. Novel forms of structural integration between microbes and a hydrothermal vent gastropod from the Indian Ocean. Appl. Environ. Microbiol. 70:3082-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatch, T. P. 1999. Developmental biology, p. 29-67. In R. S. Stephens (ed.), Chlamydia. American Society for Microbiology, Washington, D.C.
- 30.Hemprich, F. W., and C. G. Ehrenberg. 1831. Symbolae physicae. Evertebrata. I. Phytozoa. Abh. Akad. Wiss. Berl.
- 31.Howarth, R., R. F. Unz, E. M. Seviour, R. J. Seviour, L. L. Blackall, R. W. Pickup, J. G. Jones, J. Yaguchi, and I. M. Head. 1999. Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater treatment plants and description of Thiothrix eikelboomii sp. nov., T. unzii sp. nov., T. fructosivorans sp. nov. and T. defluvii sp. nov. Int. J. Syst. Bacteriol. 49:1817-1827. [DOI] [PubMed] [Google Scholar]
- 32.Imhoff, J. F., H. Sahling, J. Süling, and T. Kath. 2003. 16 S rDNA based phylogeny of sulfur-oxidising bacterial endosymbionts in marine bivavles from cold-seep habitats. Mar. Ecol. Prog. Ser. 249:39-51. [Google Scholar]
- 33.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuever, J., S. M. Sievert, H. Stevens, T. Brinkhoff, and G. Muyzer. 2002. Microorganisms of the oxidative andreductive part of the sulphur cycle at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Cah. Biol. Mar. 43:413-416. [Google Scholar]
- 35.Lane, D. J., A. P. Harrison, Jr., D. Stahl, B. Pace, S. J. Giovannoni, G. J. Olsen, and N. R. Pace. 1992. Evolutionary relationships among sulfur- and iron-oxidizing eubacteria. J. Bacteriol. 174:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 1:593-600. [Google Scholar]
- 39.McMullin, E. R., S. Hourdez, S. W. Schaeffer, and C. R. Fisher. 2003. Phylogeny and biogeography of deep sea vestimentiferan tubeworms and their bacterial symbionts. Symbiosis 34:1-41. [Google Scholar]
- 40.Murray, R. G. E., and E. Stackebrandt. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int. J. Syst. Bacteriol. 45:186-187. [DOI] [PubMed] [Google Scholar]
- 41.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 42.Nelson, K., and C. R. Fisher. 2000. Speciation of the bacterial symbionts of deep-sea vestimentiferan tube worms. Symbiosis 28:1-15. [Google Scholar]
- 43.Ott, J. 1996. Sulfide ectosymbioses in shallow marine habitats, p. 369-382. In F. Uiblein, J. Ott, and M. Stachowitsch (ed.), Deep-sea and extreme shallow-water habitats: affinities and adaptations. Biosystematics and Ecology Series 11. Austrian Academy of Science Press, Austria.
- 44.Ott, J., M. Bright, and F. Schiemer. 1998. The ecology of a novel symbiosis between a marine peritrich ciliate and chemoautotrophic bacteria. Mar. Ecol. 19:229-243. [Google Scholar]
- 45.Ott, J., M. Bright, and S. Bulgheresi. 2004. Marine microbial thiotrophic ectosymbioses. Oceanogr. Mar. Biol. Annu. Rev. 42:95-118. [Google Scholar]
- 46.Polz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polz, M. F., D. L. Distel, B. Zarda, R. Amann, H. Felbeck, J. Ott, and C. M. Cavanaugh. 1994. Phylogenetic analysis of a highly specific association between ectosymbiotic, sulfur-oxidizing bacteria and a marine nematode. Appl. Environ. Microbiol. 60:4461-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rinke, C. 2002. Nutritional processes in the chemoautotrophic Zoothamnium niveum symbioses. Diploma thesis. University of Vienna, Vienna, Austria.
- 49.Sievert, S. M., T. Brinkhoff, G. Muyzer, W. Ziebis, and J. Kuever. 1999. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65:3834-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 51.Stackebrandt, E., and C. R. Woese. 1981. The evolution of the prokaryotes, p. 1-3. In M. J. Carlile, J. F. Collins, and B. E. Moseley (ed.), Molecular and cellular aspects of microbial evolution. Symposium of the Society for General Microbiology 32. Cambridge University Press, Cambridge, England.
- 52.Thiermann, F., R. Windoffer, and O. Giere. 1994. Selected meiofauna around shallow water hydrothermal vents off Milos (Greece): ecological and ultrastructural aspects. Vie Milieu 44:215-226. [Google Scholar]
- 53.Van Dover, C. L., S. E. Humphris, D. Fornari, C. M. Cavanaugh, R. Collier, S. K. Goffredi, J. Hashimoto, M. D. Lilley, A. L. Reysenbach, T. M. Shank, K. L. Von Damm, A. Banta, R. M. Gallant, D. Gotz, D. Green, J. Hall, T. L. Harmer, L. A. Hurtado, P. Johnson, Z. P. McKiness, C. Meredith, E. Olson, I. L. Pan, M. Turnipseed, Y. Won, C. R. Young III, and R. C. Vrijenhoek. 2001. Biogeography and ecological setting of Indian Ocean hydrothermal vents. Science 294:818-823. [DOI] [PubMed] [Google Scholar]
- 54.Vopel, K., M. Pöhn, A. Sorgo, and J. Ott. 2001. Ciliate-generated advective seawater transport supplies chemoautotrophic ectosymbionts. Mar. Ecol. Prog. Ser. 210:93-99. [Google Scholar]
- 55.Vopel, K., D. Thistle, J. Ott, M. Bright, and H. Roy. 2005. Wave-induced H2S flux sustains a chemoautotrophic symbiosis. Limnol. Oceanogr. 50:128-133. [Google Scholar]
- 56.Wagner, M., M. Horn, and H. Daims. 2003. Fluorescence in situ hybridisation for the identification and charcterisation of prokaryotes. Curr. Opin. Microbiol. 6:302-309. [DOI] [PubMed] [Google Scholar]
- 57.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 58.Warén, A., S. Bengtson, S. K. Goffredi, and C. L. Van Dover. 2003. A hot-vent gastropod with iron sulfide dermal sclerites. Science 302:1007. [DOI] [PubMed] [Google Scholar]
- 59.Wood, A. P., and D. P. Kelly. 1993. Reclassification of Thiobacillus thyasirisas to Thiomicrospira thyasirae comb. nov. an organism exhibiting pleomorphism in response to environmental conditions. Arch. Microbiol. 159:45-47. [Google Scholar]