Abstract
The microbial composition of acid streamers (macroscopic biofilms) in acidic, metal-rich waters in two locations (an abandoned copper mine and a chalybeate spa) in north Wales was studied using cultivation-based and biomolecular techniques. Known chemolithotrophic and heterotrophic acidophiles were readily isolated from disrupted streamers, but they accounted for only <1 to 7% of the total microorganisms present. Fluorescent in situ hybridization (FISH) revealed that 80 to 90% of the microbes in both types of streamers were β-Proteobacteria. Terminal restriction fragment length polymorphism analysis of the streamers suggested that a single bacterial species was dominant in the copper mine streamers, while two distinct bacteria (one of which was identical to the bacterium found in the copper mine streamers) accounted for about 90% of the streamers in the spa water. 16S rRNA gene clone libraries showed that the β-proteobacterium found in both locations was closely related to a clone detected previously in acid mine drainage in California and that its closest characterized relatives were neutrophilic ammonium oxidizers. Using a modified isolation technique, this bacterium was isolated from the copper mine streamers and shown to be a novel acidophilic autotrophic iron oxidizer. The β-proteobacterium found only in the spa streamers was closely related to the neutrophilic iron oxidizer Gallionella ferruginea. FISH analysis using oligonucleotide probes that targeted the two β-proteobacteria confirmed that the biodiversity of the streamers in both locations was very limited. The microbial compositions of the acid streamers found at the two north Wales sites are very different from the microbial compositions of the previously described acid streamers found at Iron Mountain, California, and the Rio Tinto, Spain.
Microorganisms that inhabit the most extreme acidic niches on our planet (acidophiles) are highly diverse in terms of their physiologies and phylogenetic relationships (4, 18). There have been many reports of macroscopic microbial growth in extremely acidic (pH <3) metal-rich environments (mostly associated with mines and mine drainage waters) following the initial description by Lackey (26). These growths may occur as gelatinous filaments in flowing mine waters (acid streamers), as stalactite-like forms hanging from pit props and underground roofs (microbial pipes and snotites), or as thick biofilms. One of the first attempts to isolate bacteria from acid streamers was the attempt of Dugan et al. (11), although all of the isolates obtained in that study were neutrophilic rather than acidophilic. In contrast, acidophilic chemolithotrophic and heterotrophic acidophilic bacteria were isolated by McGinness and Johnson from acid streamers found in an abandoned pyrite mine (Cae Coch) in north Wales (32). These researchers considered acid streamers to be mixed communities of iron- and sulfur-oxidizing acidophiles, which acted as the primary producers in the mine, and heterotrophic bacteria, which utilized lysates and exudates produced by the autotrophs. Interestingly, one of the isolates (designated CCH7) was a ferrous iron-oxidizing filamentous isolate that formed streamer-like growths in laboratory cultures (24). In contrast, Wakao et al. (37) isolated only iron- and sulfur-oxidizing bacteria from acid streamers draining an iron sulfide mine in Japan (pH 1.8 to 2.2) and concluded that the streamers were composed (predominantly) of the chemolithotroph Acidithiobacillus ferrooxidans, which was embedded in a gelatinous matrix.
The advent of biomolecular techniques has eliminated the need to use culture-dependent methods to elucidate the microbial diversity of extremely acidic environments, as well as other environments. Bond et al. (7) studied a 1-cm-thick slime biofilm that developed on the surface of finely disseminated pyrite ore in Iron Mountain, California (water pH 0.77 to 1.31; temperature, 31.5 to 36.8°C) initially by performing a phylogenetic analysis of 16S rRNA genes and later (6) by performing a fluorescent in situ hybridization (FISH) analysis using a variety of gene probes designed from clone library data. The dominant sequences identified in clone libraries were sequences of bacteria related to the autotrophic iron oxidizer Leptospirillum ferriphilum. Archaeal (Thermoplasmales lineage), Acidimicrobium/“Ferrimicrobium,” and δ-proteobacterial gene sequences were also detected. Analysis of slimes (biofilms) and snotites from different locations within Iron Mountain showed that although there were variations in the relative numbers of the different prokaryotes present, overall the samples analyzed showed limited biodiversity, with Ferroplasma, Leptospirillum, Sulfobacillus, and Acidimicrobium-related species dominating the microbial communities. Very different results were reported by Lopez-Archilla et al. (28) for “large dendritic filaments” (up to 1.5 m long and 5 cm thick) found in the Rio Tinto, Spain (pH 1.67 to 2.19; temperature, 21°C). They also used a combination of 16S rRNA gene clone library construction and analysis and FISH. Attempts to amplify archaeal genes from the filaments were unsuccessful. A bacterial clone library obtained from the filaments included γ-Proteobacteria (50%), α-Proteobacteria (36%), Firmicutes (8%), Actinobacteria (3%), and β-Proteobacteria (3%). Although most of the phylotypes were closely related to previously characterized bacteria, none of the sequences obtained was related to any known acidophile. Analysis of the filaments using FISH was hampered by problems with disrupting the gelatinous growths. However, as observed with the clone libraries, the most abundant bacteria appeared to be γ-proteobacteria, and no archaea were detected. The results of the analysis of the macroscopic filaments contrasted markedly with the results of Gonzalez-Toril et al. (15), who examined the diversity of planktonic bacteria in the Rio Tinto and found that the dominant bacteria were Acidithiobacillus ferrooxidans, Leptospirillum ferrooxidans, and Acidiphilium spp., all of which are known acidophilic prokaryotes.
In this paper we describe the microbial diversity of acid streamers that occur in mine water draining an abandoned copper mine site (Mynydd Parys) and in a chalybeate spa adjacent to the Cae Coch pyrite mine (both located in north Wales). Using a combination of cultivation-dependent and cultivation-independent approaches, we showed that the streamer communities have limited biodiversity and that their compositions are very different from the compositions of streamers at other sites that have been described.
MATERIALS AND METHODS
Mine sites and water analysis.
Acid streamer samples were obtained at two sites in north Wales: the abandoned Mynydd Parys copper mines at Anglesey and the Trefriw Wells Spa, which is adjacent to an abandoned pyrite mine (Cae Coch). Descriptions of the geochemistry of the Mynydd Parys and Cae Coch mines have been given elsewhere (22). On-site analyses of the pH, redox potential, temperature, and conductivity were performed with a Hanna water tester (VWR, United Kingdom), and dissolved oxygen was measured using a D400 dissolved oxygen meter (Whatman, United Kingdom). Ferrous iron concentrations were measured by using filtered (<0.2 μm; cellulose nitrate; Whatman, United Kingdom) samples and the ferrozine assay (29). The concentrations of other dissolved metals were determined by atomic absorption spectrometry, and sulfate concentrations were measured turbidimetrically as barium sulfate (Hydrocheck, Cambridge, United Kingdom). Dissolved organic carbon concentrations in filtered water samples were measured using a Protoc analyzer (Pollution & Process Monitoring Ltd., United Kingdom).
Sampling of acid streamers.
Acid streamer growths (ca. 1 to 10 cm3) were collected and taken to the laboratory within 1 h of sampling. For the Mynydd Parys site, streamer samples were taken from a discharge portal (the “Mona adit”) and 1 m and 10 m downstream of this portal. At the Trefriw Wells Spa, streamer samples were obtained within a small cave excavation at the site. In this cave, acidic, iron-rich water percolating from the hillside above the spa was fed into a small pool that was about 1.5 m in diameter and 1 m deep. The overflow water from the pool was channeled through a shallow open drain in the cave and then into a second open drain outside the cave. Acid streamer samples were obtained from the end of the pipe that delivered water to the pool, from the bottom and the main body of the pool (“benthic” and “planktonic” zones), and from the drainage channels inside and outside the cave (“drain 1” and “drain 2,” respectively).
Total counts and plate counts of bacteria and phylogenetic analysis of isolates.
Acid streamer samples were dissected into ca. 1-g (wet weight) subsamples and put into sterile centrifuge tubes containing 5 ml basal salts solution (23), adjusted to pH 3.5, and vortexed for 5 min. The homogenized suspensions were centrifuged (5,000 × g, 30 s) to remove any undispersed streamer material. The supernatant (“streamer cell suspension”) was either used for plating or fixed for molecular analysis. For total cell counting, fixed cell suspensions were filtered through 25-mm black polycarbonate membranes (pore size, 0.2 μm; Whatman) and stained with 4′,6-diamidino-2-phenylindole (DAPI) (10 ml of a 1-μg/ml solution). Following several washes with deionized water, the membranes were placed onto glass slides and viewed using an ECLIPSE E600 fluorescence microscope (Nikon, Japan). At least 100 bacteria per membrane were counted.
A sample of a streamer cell suspension was serially diluted in the acidified basal salts solution and spread onto a variety of solid media, including acidic overlay media for autotrophic and heterotrophic acidophiles (19, 23) and R2 agar for neutrophilic microorganisms (34). Inoculated plates were incubated for up to 6 weeks at 20°C aerobically and in anaerobic jars (using the AnaeroGen and CampyGen systems to create anaerobic and microaerobic environments; Oxoid, United Kingdom). The morphological characteristics of colonies that grew on solid media were used to differentiate organisms and to facilitate initial identification of isolates as iron oxidizers, sulfur oxidizers, or heterotrophs, using criteria described elsewhere (25). Isolates were purified by repeated single-colony isolation and then transferred to and cultured in appropriate liquid media (23).
Cells from liquid media were harvested by centrifugation, and the DNA was released by resuspension of the cell pellets in lysis solution (0.05 M NaOH, 0.25% sodium dodecyl sulfate), followed by incubation at 95°C for 15 min. Cell lysates were diluted 10-fold in MilliQ-grade water (Millipore Inc., United States) adjusted to pH 7.5 with Tris. The 16S rRNA genes of isolates were amplified from diluted lysates using touchdown PCR (10) with primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) (27) and conditions described previously (33). PCR products were purified using a QIAquick PCR purification kit (QIAGEN, United Kingdom) and were sequenced by capillary electrophoresis using a Beckman Coulter dye terminator cycle sequencing kit and a CEQ8000 genetic analysis system (Beckman Coulter, United Kingdom). The resulting gene sequences (for GenBank accession numbers see Table 3) were compared with the sequences available in the GenBank database using BLAST (2).
TABLE 3.
Identification of acid streamer isolates based on analysis of 16S rRNA gene sequences
| Isolate | GenBank accession no. | Physiological classa | Closest cultivated relative (GenBank accession no.) | % Identity |
|---|---|---|---|---|
| Mynydd Parys streamer isolates | ||||
| PK11 | NSb | Iron-oxidizing autotroph | Acidithiobacillus ferrooxidans NO-37 (AF376020) | 99.9 |
| KP1 | AY765991 | Iron-oxidizing heterotroph | “Ferrimicrobium acidiphilum” (AF251436) | 93.1 |
| KP3 | AY765992 | Heterotroph | Acidobacterium capsulatum (D26171) | 91.8 |
| PK35 | AY765993 | Heterotroph | Wheal Jane isolate WJ7 (AY096034) | 99.7 |
| PK40 | AY765994 | Heterotroph | Acidiphilium sp. strain C-1 (D30769) | 99.7 |
| PK44 | AY455806 | Heterotroph | Thiomonas sp. strain NO115 (AY455807) | 99.5 |
| PK46 | AY765995 | Heterotroph | Acidiphilium cryptum ATCC 33463T (D30773) | 99.9 |
| PK48 | AY765996 | Heterotroph | Acidiphilium acidophilum ATCC 27807T (D86511) | 97.5 |
| PK51 | AY765997 | Heterotroph | Wheal Jane isolate WJ2 (AY096032) | 100 |
| M21 | AY765998 | Heterotroph | Acidocella sp. strain NO-12 (AF376021) | 99.1 |
| Trefriw Spa streamer isolates | ||||
| CCW10 | NS | Iron-oxidizing autotroph | Acidithiobacillus ferrooxidans NO-37 (AF376020) | 99.7 |
| CCW68 | NS | Iron-oxidizing autotroph | Acidithiobacillus ferrooxidans ATCC 23270T (AF465604) | 99.5 |
| CS11 | AY765999 | Iron-oxidizing heterotroph | “Ferrimicrobium acidiphilum” (AF251436) | 93.1 |
| CCP3 | AY766000 | Heterotroph | Acidiphilium sp. strain NO-17 (AF376026) | 99.5 |
| CCW30 | AY766001 | Heterotroph | Acidocella sp. strain NO-12 (AF376021) | 99.4 |
Physiological class refers to the phenotype initially used to discriminate isolates.
NS, not submitted to GenBank.
FISH.
To fix microorganisms, the dispersed streamer cell suspensions were harvested by centrifugation at 10,000 × g for 10 min, washed by resuspension in phosphate-buffered saline (PBS) prior to mixing (1:3, vol/vol) with 4% paraformaldehyde in PBS, and incubated at 4°C overnight. Fixed cell suspensions were washed with PBS, resuspended in equal volumes of PBS and absolute ethanol, and stored at −20°C. FISH analysis was performed using a modification of the method described by Bond and Banfield (5) and a suite of rRNA-targeting probes (Table 1). Initially, the probes used were broad-range probes (e.g., probes targeting α-, β-, or γ-proteobacteria, Firmicutes, etc.). In the second phase of the FISH analysis we used probes that targeted acidophiles previously detected in acidic mine waters (e.g., Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans) and other probes that targeted bacteria detected as isolates or clones obtained from acid streamers from the two north Wales sites. When the probe used targeted gram-positive bacteria, fixed cells were first incubated (4°C for 10 min) with lysozyme (1,027.5 U/ml) prior to dehydration with ethanol. Hybridization was carried out simultaneously using 25 ng of fluorescein-labeled eubacterium-specific probe EUB338 and 25 ng of a more specific, Cy3-labeled probe (MWG Biotech, Ebersberg, Germany). Mounting medium (70% glycerol in 100 mM sodium tetraborate [pH 9.2] containing 3 mg N-propyll gallate ml−1) was used to reduce fading of the labeled probe signals. Various concentrations of formamide (0 to 50%, vol/vol) were tested to optimize specificity and to maximize the signal response for each newly designed probe, using pure cultures of target and related microorganisms (Table 1). For some probes it was necessary to use additional helper probes at the same concentration in order to enhance the fluorescence signal. These unlabeled oligonucleotides were designed to target the 16S rRNA immediately upstream and downstream of the labeled probe and to improve the access of the probe to the desired site on the rRNA molecule (14). To enumerate different groups of bacteria, Cy3-labeled cells were counted relative to the cells stained with the EUB338 probe. To assess relative eubacterial abundance and archaeal abundance, targeted cells were counted relative to the cells stained by DAPI.
TABLE 1.
16S rRNA and other oligonucleotide probes used for FISH analysis and formamide concentrations required for optimum specificity
| Probea | Target | Probe sequence (5′-3′) | Formamide concn (%) | Reference |
|---|---|---|---|---|
| EUB338 | Eubacteria | GCTGCCTCCCGTAGGAGT | 0-50 | 3 |
| ARCH915 | Archaea | GTGCTCCCCCGCCAATTCCT | 40 | 36 |
| ALF1B | α-Proteobacteria, some δ-Proteobacteria, and most spirochetes | CGTTCGYTCTGAGCCAGb | 20 | 31 |
| BET42ac | 23S rRNA of most β-Proteobacteria | GCCTTCCCACTTCGTTT | 35 | 31 |
| GAM41ac | 23S rRNA of most γ-Proteobacteria | GCCTTCCCACATCGTTT | 35 | 31 |
| LGC0355 | Low-G+C-content gram-positive bacteria | GGAAGATTCCCTACTGCTG | 20 | This study |
| TF539 | Acidithiobacillus ferrooxidans | CAGACCTAACGTACCGCC | 20 | 35 |
| ATT0223 | Acidithiobacillus thiooxidans | AGACGTAGGCTCCTCTTC | 40 | This study |
| LF655 | Leptospirillum groups 1, 2, and 3 | CGCTTCCCTCTCCCAGCCT | 35 | 5 |
| ACM732 | Acidimicrobium and “Ferrimicrobium” | GTACCGGCCCAGATCGCTG | 35 | 5 |
| ACM995 | Acidimicrobium ferrooxidans | CTCTGCGGCTTTTCCCTCCATG | 10 | Norrisd |
| FMR0732 | Streamer isolates KP1 and CS11 | GTGTCGGCCCAGATTGCTG | 30 | This study |
| ABI0199 | Streamer isolate PK35 | CTCCTCAAGTGGATTACTC | 10 | This study |
| ABI1002e | Streamer isolate KP3 | CTATTTCTAGGGGTGTCC | 10 | This study |
| ABI1002 h1f | Upstream of ABI1002 | ATACAGACCCATTGCTGG | 10 | This study |
| ABI1002 h2f | Downstream of ABI1002 | TGTACATTTCGAGCCCAG | 10 | This study |
| TM1G0138 | Thiomonas group 1 | GCAGTTATCCCCCATCAAT | 40 | This study |
| TM2G0138 | Thiomonas group 2 | GTAGTTATCCCCCATCACA | 40 | This study |
| WJ20646e | γ-Proteobacteria WJ2 and streamer isolate PK51 | TACCGTACTCCAGCAAGC | 40 | This study |
| WJ20646 h1f | Upstream of WJ20646 | GGGAATTCCACCTTTCTC | 40 | This study |
| WJ20646 h2f | Downstream of WJ20646 | CAGTATCCACCGCCATT | 40 | This study |
| GALTS0084e | Streamer clone TrefC4 | CCACTAACCTGGGAGCAA | 40 | This study |
| GALTS h1f | Upstream of GALTS0084 | GATATATTACTCACCCGTTCG | 40 | This study |
| GALTS h2f | Downstream of GALTS0084 | GCCCCCAGGCCCGTTCGA | 40 | This study |
| BSC0459 | Streamer clones TrefC11 and MPKCSC9 | TCCAGGTTATTCGCCTGA | 30 | This study |
Most probes were labeled with the fluorochrome Cy3; the only exception was the EUB338 probe, which was labeled with the fluorescein derivative Alexa-fluor 488.
Y = C or T.
Hybridization with probes BET42a and GAM41a was carried out in the presence of a 10-fold excess of the other unlabeled oligonucleotide.
P. Norris (University of Warwick, United Kingdom), unpublished data.
Equal concentrations of unlabeled helper oligonucleotide probes were used with probes ABI1002, WJ20646, and GALTS0084.
Unlabeled helper probe used in conjunction with a labeled partner to enhance fluorescence.
Terminal restriction fragment length polymorphism (T-RFLP) analysis of extracted DNA.
DNA was extracted from intact acid streamer samples using UltraClean soil DNA kits (MO BIO Laboratories, Inc., United States). Eubacterial 16S rRNA genes were amplified as described above in three separate 25-μl PCR mixtures, using fluorescently labeled (D4-phosphoramadite; ResGen, United Kingdom) primer 27f and unlabeled primer 1492r. Pooled PCR products were purified (QIAquick), and 5 μl of each resulting mixture was subjected to separate digestion reactions using restriction enzymes HhaI, MspI, and AluI. After 2 h, the digestion reactions were stopped by addition of 5 μl of a freshly prepared stop solution (2 μl of 5 M sodium acetate, 2 μl of 100 mM EDTA, 1 μl of a solution containing 20 mg glycogen ml−1) and 60 μl of absolute ethanol. The digested DNA was collected by centrifugation (16,100 × g, 20 min) and washed twice with cold 70% (vol/vol) ethanol. After drying, DNA pellets were resuspended in 40 μl of molecular biology-grade formamide (Fisher Scientific, United Kingdom), and 4 μl of the resulting solution was mixed with 36 μl of formamide containing 0.5 μl of a 640-nucleotide (nt) size standard labeled with fluorochrome D1 (ResGen, United Kingdom). The sizes of the resulting terminal restriction enzyme fragments (T-RFs) were determined by capillary electrophoresis using a CEQ8000 genetic analysis system (Beckman Coulter, United Kingdom) and by comparison of their mobilities with the mobilities of the size standard fragments. The peak areas for each T-RF (which were directly related to fluorescence intensity) relative to the total peak areas were used to determine the relative abundance of individual microorganisms (considered to be represented by a single T-RF with the same relative abundance following digestion with the three individual enzymes) within the acid streamer communities.
Clone libraries of amplified 16S rRNA genes from acid streamer DNA.
16S rRNA gene clone libraries were constructed using acid streamers taken from the Mona adit at Mynydd Parys and from the pipe feeding the pool at the Trefriw Wells Spa. Community 16S rRNA genes were amplified and purified from extracted streamer DNA using the PCR method described above, except that nonlabeled primer 27f-G (primer 27f modified by addition of a G to the 5′ end during synthesis) was used to facilitate TA cloning (8). PCR products were ligated into the pGEM-T Easy vector (Promega, United States), and the resulting plasmids were transformed into Escherichia coli strain DH5-α according to the manufacturer's instructions. Plasmids from successful transformants were purified using the Concert rapid plasmid miniprep system (Life Technologies). Prior to sequencing, DNA inserts were subjected to restriction fragment length polymorphism analysis, using HhaI to determine the number of different 16S rRNA genes that had been cloned. A representative plasmid for each of the distinct restriction enzyme banding patterns was chosen for sequencing. The novel 16S rRNA gene sequences were compared to the sequences deposited in the GenBank database using BLAST (2).
Isolation of streamer-forming bacteria.
Following the analysis of the biomolecular data obtained for the acid streamer bacterial communities, a variety of liquid cultures were set up to enrich for the dominant streamer-forming bacteria at the two north Wales sites, using ferrous iron, ammonium, urea, and yeast extract as potential substrates or electron donors. In addition, a novel overlay solid medium (23), in which tryptone soya broth (TSB) was eliminated and potassium phosphate was added at a concentration equivalent to the concentration present in TSB, was used to plate streamer bacteria.
Nucleotide sequence accession number.
The Trefriw Wells Spa and Mynydd Parys clone sequences have been deposited in the GenBank database under accession numbers AY766002 (TrefC4), AY766003 (TrefC11), and AY766004 (MPKCSC9).
RESULTS
Mine water analysis.
Physicochemical characteristics of waters from the areas where acid streamer samples were obtained are shown in Table 2. The spa water and mine water were both characterized by low concentrations of dissolved oxygen and low redox potentials (as the dominant form of soluble iron was ferrous iron). At Mynydd Parys, there was very little detectable change in water chemistry along the 10-m drainage stream where acid streamer samples were obtained. In contrast, at the Trefriw Wells Spa site, most of the iron in the feed water was oxidized from ferrous iron to ferric iron by the time that it left the cave, and there was a corresponding large increase (ca. 150 mV) in the redox potential. Many of the physicochemical parameters recorded at the two sites were fairly similar, with the notable exception of the concentrations of copper and zinc, which were far higher in the water from the former Mynydd Parys copper mine. Although the sulfate concentrations were not determined in Trefriw Wells Spa waters at the times referred to in Table 2, they were generally ca. 1,000 mg liter−1 on other occasions that the waters were analyzed.
TABLE 2.
Physicochemical data for the two acid mine drainage sites from which streamer biofilms were obtained
| Parameter | Mynydd Parysa | Trefriw Wells Spab
|
|||
|---|---|---|---|---|---|
| Feed pipe | Pool | Drain 1 | Drain 2 | ||
| pH | 2.7/2.4 | 2.7 | 2.6 | 2.9 | 2.5 |
| Temp (°C) | 8.8/12.4 | 10.1 | 9.8 | 9.5 | 13.3 |
| Eh (mV) | 420/526 | 657 | 670 | 813 | 712 |
| Conductivity (μS/cm) | 2,994/3,160 | 1,550 | 1,520 | 1,640 | 1,520 |
| Dissolved oxygen concn (mg liter−1) | 0.55/1.4 | 0.6 | 0.9 (0.5) | 1.6 | 3.9 |
| Dissolved organic carbon concn (mg liter−1) | 6.0/6.9 | 4.5 | 4.3 | 12.2 | 8.0 |
| Fe2+ concn (mg liter−1) | 473/520 | 218 | 193 | 22 | 14 |
| Fetotal concn (mg liter−1) | 514/NDc | 223 | 259 | 100 | 41 |
| SO42− concn (mg liter−1) | 1,552/1,846 | ND | ND | ND | ND |
| Mn concn (mg liter−1) | 5/ND | 1.1 | 1.1 | ND | 1.5 |
| Cu concn (mg liter−1) | 35/ND | <1 | <1 | ND | <1 |
| Zn concn (mg liter−1) | 50/ND | <1 | <1 | ND | <1 |
Data for 1 February 2002/data for 24 July 2002.
The feed pipe and drain 1 data are data for 12 March 2004; the pool and drain 1 data are data for 3 April 2003. Most of the pool data are data for water taken from the top 1 m of the pool; the only exception is the dissolved oxygen value in parentheses, which is the value for water at the base (“benthic zone”) of the pool.
ND, not determined.
Plate counts and DAPI counts.
Due to the difficulty in quantitatively dispersing the streamers, relative rather than absolute numbers of cultivatable bacteria in dispersed biofilms (streamer cell suspensions) were obtained by plating on solid media. Maximum counts of iron oxidizers were obtained on aerobically incubated plates for the Mynydd Parys streamers and on plates incubated microaerobically for the Trefriw Wells Spa streamers; far fewer colonies were obtained on solid media incubated under anaerobic conditions in both cases (data not shown). The vast majority of isolates were acidophiles; very few colonies grew on R2 medium. A comparison of plate counts and total (DAPI-stained) cell counts indicated that the plating efficiency was low (0.07 to 7.2%) in all cases.
Phylogenetic analysis of streamer isolates.
Representative streamer isolates, differentiated on the basis of colony morphology, were identified from an analysis of amplified 16S rRNA genes (Table 3). All of the iron-oxidizing autotrophic isolates from acid streamers at both sites were Acidithiobacillus ferrooxidans rather than Leptospirillum spp. In addition, “Ferrimicrobium”-like isolates (iron-oxidizing heterotrophs belonging to the class Actinobacteria) were isolated from both sites; two isolates, KP1 from Mynydd Parys and CS11 from Trefriw Wells Spa, exhibited 100% 16S rRNA gene sequence identity (for ca. 1,400 bp). These isolates formed streamer-like growths in liquid media, similar to an isolate (CCH7) obtained previously from the Cae Coch mine (24). Like CCH7, both KP1 and CS11 oxidized iron and required yeast extract for growth. Both isolates were very closely related (99.9% sequence identity for the 16S rRNA genes) to a clone (TRA2-10) detected in pH 2.5 water at the Iron Mountain mine (13).
Non-iron-oxidizing heterotrophic acidophiles isolated from acid streamers at both sites were Acidiphilium and Acidocella spp. (α-Proteobacteria). In addition, the heterotrophic isolates from the Mynydd Parys streamers included bacteria distantly related to Acidobacterium capsulatum, one of which (PK35) was closely related (99.7% 16S rRNA gene identity) to an isolate obtained from a passive acid mine drainage treatment system (19), and a γ-proteobacterium (PK51) that was also related (100% identity) to isolate WJ2 from the same treatment site. Isolate KP3 from Mynydd Parys was more closely related (96.7% identity) to a clone from Iron Mountain (TRB82) than to any other previously characterized bacterium. One other bacterium (PK44) isolated from Mynydd Parys streamers on a “heterotrophic” solid medium was found to be a Thiomonas-like β-proteobacterium. Subsequent work showed that this isolate is able to oxidize thiosulfate, ferrous iron, and arsenite (9).
In the case of the Mynydd Parys streamers, Acidithiobacillus ferrooxidans accounted for about 94% of the plate isolates, the γ-proteobacterium PK51 accounted for about 4.5%, and Acidiphilium-like bacteria accounted for about 2%. Other isolates accounted for <1% of the colonies obtained. Similarly, Acidithiobacillus ferrooxidans colonies were the dominant plate isolates from the Trefriw Wells Spa streamers, followed by “Ferrimicrobium”-like bacteria.
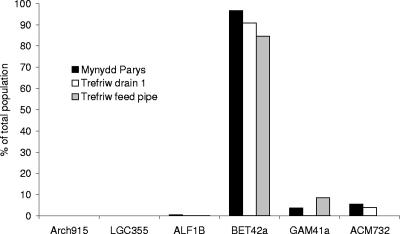
Preliminary FISH analysis of dispersed acid streamers.
The initial analysis of acid streamers using FISH supported the plate isolation data to some extent, although in general, FISH highlighted major deficiencies in the cultivation approach. The prokaryotes in the streamers were shown to be exclusively (>99.9%) bacteria; no cells were detected using the archaeal probe (Fig. 1). Using domain-specific probes, we found that the majority of cells in both types of acid streamers were proteobacteria. While α-, β-, and γ-Proteobacteria were all detected by FISH, the dominant group in both Mynydd Parys and Trefriw Wells Spa streamers was the β-Proteobacteria (Fig. 1). No Firmicutes were found in the acid streamer samples using the low-G+C-content gram-positive probe LGC355, although other gram-positive prokaryotes (Actinobacteria) were detected using a probe (ACM732) that targeted bacteria belonging to the related genera Acidimicrobium and “Ferrimicrobium.”
FIG. 1.
Composition of the acid streamers from Mynydd Parys and Trefriw Wells Spa, as determined by FISH analysis using probes targeting the 16S rRNA of major groups of bacteria and archaea (Arch915, archaea; LGC355, low-G+C-content gram-positive bacteria; ALF1B, α-Proteobacteria; BET42a, β-Proteobacteria; GAM41a, γ-Proteobacteria; ACM732, Actinobacteria).
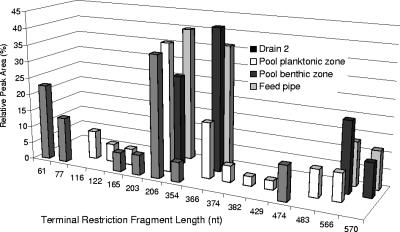
Analysis of microbial diversity in acid streamers by T-RFLP analysis.
Fragment analysis of eubacterial 16S rRNA genes that were amplified from Trefriw Wells Spa acid streamers and digested with HhaI revealed two dominant microbes, each of which was represented by a unique T-RF. Each of these organisms accounted for between 25% and 45% of the total bacterial community, as determined from a comparison of the relative peak areas of the corresponding T-RFs to the total peak area of all T-RFs (Fig. 2). The Mynydd Parys (Mona adit) streamer DNA was dominated (>80%) by one microbe that had an HhaI T-RF whose size was identical (206 nt) to the size of one of the two T-RFs from the Trefriw Wells Spa streamers; two other T-RFs (158 and 570 nt) each accounted for <10% of the total T-RF peak area (data not shown). Analysis of acid streamers at Mynydd Parys, sampled approximately 6 months later, showed that the same T-RF (206 nt, HhaI digest) still dominated the acid streamers at the Mona adit and also the streamer samples obtained 1 m and 10 m downstream of the adit, accounting for >90% of the total T-RFs (data not shown).
FIG. 2.
T-RFLP analysis of acid streamers from Trefriw Wells Spa, showing the relative abundance of each terminal restriction fragment of amplified 16S rRNA genes digested with HhaI (as a percentage of the total peak area of all terminal restriction fragments).
Analysis of 16S rRNA gene libraries.
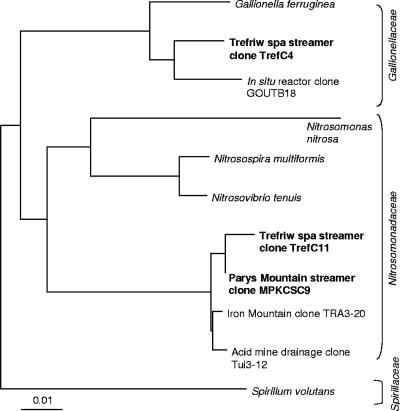
Due to the apparent limited biodiversity of the streamers suggested by the T-RFLP analysis, only 10 clones were selected randomly from each library, and digestion profiles were analyzed. Only two different RFLP banding patterns were found for the Trefriw Wells Spa clones, and one of these was identical to the pattern for 9 of the 10 Mynydd Parys clones. The length of the T-RF of the common clones (TrefC11 and MPKCSC9) was identical to the length of the dominant fragment found by T-RFLP analysis of the Mynydd Parys acid streamers and one of the two T-RFs from the Trefriw Wells Spa streamers when preparations were digested with HhaI, MspI, or AluI. The gene inserts (ca. 1,400 bp) of these clones were found to exhibit 99.6% sequence similarity to each other, and the closest matches (99% gene similarity) in the GenBank database were found to be a clone (TRA3-20) identified previously at the Iron Mountain site and another clone (Tui3-12) found in acid mine drainage in New Zealand. The closest relatives (ca. 90% gene identity) among cultivated organisms are a group of ammonium-oxidizing β-Proteobacteria (the Nitrosomonadaceae). The other clone obtained from the Trefriw Wells Spa (TrefC4) corresponded to the second dominant T-RF (366-nt HhaI fragment) found in acid streamers from this site, although corresponding T-RFs were not detected in Mynydd Parys streamers. The 16S rRNA gene sequence of this clone was 96.9% identical to the sequence of GOUTB18, an uncultured bacterial clone obtained from an in situ bioreactor treating monochlorobenzene-contaminated groundwater (1), and 95.4% identical to the sequence of Gallionella ferruginea (a neutrophilic iron-oxidizing β-proteobacterium). A phylogenetic tree showing the relationships of the major clones from the Trefriw Wells Spa and Mynydd Parys streamers to other environmental clones and previously characterized bacteria is shown in Fig. 3.
FIG. 3.
Phylogenetic tree showing the relationship between the major clones identified in the Trefriw Wells Spa and Mynydd Parys acid streamers and other environmental clones and classified bacteria. Scale bar = 0.01 nucleotide substitution per site.
Secondary FISH analysis of dispersed acid streamers.
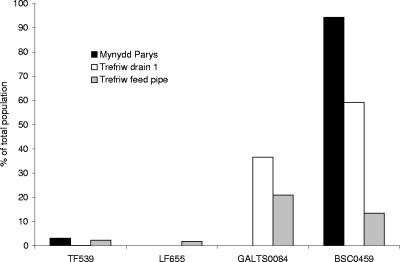
In the second phase of our FISH analysis of the acid streamers we used probes that targeted known chemolithotrophic acidophiles and plate isolates (Table 3) and also probes that targeted bacteria detected by the clone libraries. Acidithiobacillus ferrooxidans was detected in acid streamers from both sites, but the numbers were relatively low (<0.1 to 3% of the total bacteria); also, Leptospirillum was found to be present in only one of the Trefriw Wells Spa streamers, in relatively small numbers (1.8% of the total bacteria) (Fig. 4). While the preliminary FISH analysis had confirmed that the organisms in the microbial communities of streamers from both sites were predominantly β-Proteobacteria, FISH analysis using probe TM1G138 designed to target the only β-proteobacterium isolated (a Thiomonas sp.) showed that this bacterium accounted for <0.1% of the cells in the Mynydd Parys streamers and was not detectable (<0.01% of the total cells) in the Trefriw Wells Spa streamers. A second probe (TM2G138), targeting another group of Thiomonas species (9), also failed to detect cells (<0.01%) in acid streamers from either site. Similar results were obtained with most other probes that targeted bacteria that had been isolated on solid media.
FIG. 4.
Relative counts of bacteria in acid streamer samples targeted by a range of Cy3-labeled oligonucleotide probes, expressed as percentages of the fluorescein-labeled eubacterial probe (EUB338) counts.
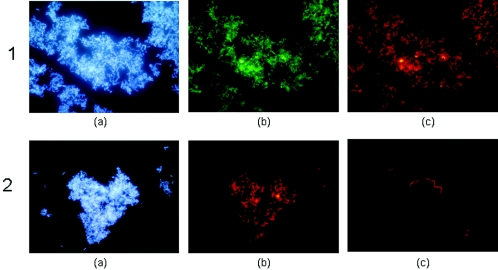
Two new probes were designed using gene sequences obtained from the clone libraries. The probe used to target the Gallionella-like microbe (GALTS0084) from Trefriw Wells Spa was a modification of Gallprobe 1, which was previously designed to target G. ferruginea (17). This probe stained 36.5% and 21.9% of the bacteria in the dispersed acid streamers from the feed pipe and drain 1, respectively, at the Trefriw Wells Spa, but no cells in the Mynydd Parys streamers were detected using this probe. A second probe, BSC0459, which was based on consensus cloned gene sequences from this study, targeted the TrefC11 and MPKCSC9 clones. This probe hybridized with 13.5% of the bacteria in streamers from the feed pipe and 59% of the bacteria in drain 1 streamers from the Trefriw Wells Spa and >90% of the viable eubacteria in streamers from the Mona adit at Mynydd Parys (Fig. 4). Images of acid streamers from the Trefriw Wells Spa stained with the BSC0459 and GALTS0084 probes are shown in Fig. 5.
FIG. 5.
DAPI-stained and FISH images of acid streamers from the Trefriw Wells Spa, obtained by using gene probes that targeted the two major clones identified in minilibraries. Image 1a, streamer from drain 1, stained with DAPI; image 1b, same image stained with fluorescein-labeled bacterial probe EUB338; image 1c, same image stained with Cy3-labeled probe BSC0459 (which targets clone TrefC11); image 2a, streamer from the pool benthic zone, stained with DAPI; image 2b, same image stained with Cy3-labeled probe GALTS0084 (which targets clone TrefC4); image 2c, another image of GALTS0084-stained streamer, showing the spirillum-like morphology which is typical of Gallionella.
Isolation of a streamer-forming bacterium.
The liquid media used did not enrich for the previously uncultivated streamer-forming bacteria represented by the TrefC4, TrefC11, and MPKCSC9 clones. Ammonium and urea enrichments (used because of the phylogenetic affiliation of the TrefC11 and MPKCSC9 clones) were negative, ferrous iron enrichments were dominated by Acidithiobacillus ferrooxidans, and yeast extract enrichments were dominated by Acidiphilium-like heterotrophic acidophiles or else overrun with fungi. In contrast, the modified TSB-free overlay solid medium was successfully used to isolate large numbers of bacteria that grew as small ferric iron-stained colonies and (in the case of Mynydd Parys streamers) increased the plating efficiency from <2% to >70% of the total cells in disrupted streamers. A representative isolate was purified by repeated single-colony isolation and was cultivated in ferrous sulfate-basal salts liquid medium (pH 2.2 to 2.5), in which it grew autotrophically by ferrous iron oxidation, forming small streamer-like growths. Sequence analysis of its 16S rRNA gene confirmed that this isolate was the β-proteobacterium represented by the TrefC11 and MPKCSC9 clones.
DISCUSSION
The use of molecular techniques for studying microbial ecology is now well established. The power of these techniques lies in their ability to identify and enumerate microorganisms without reliance on cultivation, which, when used in isolation, can lead to erroneous conclusions about the structure and dynamics of microbial communities. In this study, we used a cultivation approach in tandem with cultivation-independent techniques to characterize the microbial populations of streamer biofilms that dominate acidic ferruginous waters at two sites in north Wales.
Acidophilic (although not neutrophilic) bacteria were readily isolated from dispersed streamers from both sites using selective overlay media, and the dominant isolates were iron-oxidizing eubacteria, most of which were identified as Acidithiobacillus ferrooxidans. In contrast, although the total numbers of heterotrophic bacteria were lower, these organisms were more diverse and included iron-oxidizing isolates related to the proposed actinobacterial genus “Ferrimicrobium” (93% 16S rRNA gene similarity to “Ferrimicrobium acidophilum” [18]) and (in the case of Mynydd Parys streamers) bacteria distantly related to Acidobacterium capsulatum. Although few microbes belonging to the phylogenetic group that includes Acidobacterium capsulatum have been cultivated so far, molecular studies have shown that they are the dominant microbes in many habitats (21). Other iron-oxidizing isolates (found only in Mynydd Parys streamers) were moderate acidophiles, and they grew on ferrous iron-thiosulfate overlay medium (pH 4 to 4.5) but not on ferrous iron or ferrous iron-tetrathionate overlay medium (pH ∼2.6). These isolates were two distinct Proteobacteria, one related to Thiomonas and the other related to a previous isolate (WJ2) obtained from a constructed passive wetland ecosystem treating acid mine drainage in Cornwall, England (19). Like the “Ferrimicrobium”-like iron oxidizers, these proteobacterial isolates grew heterotrophically.
The low plating efficiency showed that the bacteria that were isolated accounted for only a small proportion of the streamer microorganisms at both sites. As a first step in determining the identities of the noncultivated microorganisms, FISH analysis using domain- and group- or class-specific probes was carried out with dispersed streamers, and the results showed that the streamer communities were essentially entirely bacterial (and dominated by β-Proteobacteria), a fact confirmed by the negative results obtained when attempts were made to amplify 16S rRNA genes from the streamers using archaeon-specific PCR primers.
T-RFLP analysis of amplified 16S rRNA genes was used to obtain an overview of the biodiversity of the streamer communities. While this technique suffers from differential cell lysis, DNA extraction, and PCR bias problems (20), it can provide a nonsubjective semiquantitative analysis of the diversity of microorganisms in an environmental sample, as it did in the present study. The results indicated that there was very limited biodiversity in the Mynydd Parys streamers, in which a single T-RF accounted for >80% of the T-RFs obtained. The streamer samples from the Trefriw Wells Spa appeared to be more heterogeneous and showed greater variation in different sampling locations, but again the T-RFLP data obtained indicated that there was limited overall biodiversity, with two T-RFs dominating the profiles obtained.
The limited biodiversity of acid streamers from the two sites facilitated construction and analysis of clone libraries prepared from amplified 16S rRNA genes, since it was necessary to screen only a relatively small number of clones to identify the 16S rRNA genes that contained the T-RFs detected by T-RFLP analysis. Analysis of the nearly complete gene sequences confirmed that the dominant bacterium obtained from the Mynydd Parys streamers was identical to one of the dominant bacteria obtained from Trefriw Wells Spa and that this organism was an uncultivated β-proteobacterium. The other dominant microbe in Trefriw Wells Spa streamers was also an uncultivated, but distinct, β-proteobacterium. The final confirmation that the PCR-dependent analyses (T-RFLP and clone library analyses) provided accurate data came from FISH analysis of the streamers using specific probes that were designed from sequences of these two dominant microbes. The species represented by one of these organisms accounted for >90% of the bacteria in the Mynydd Parys streamer sample, while the two β-Proteobacteria species together accounted for >90% and >30% of the bacteria in the “drain 1” and “feed pipe” streamers from Trefriw Wells Spa, respectively.
The identities of the two β-Proteobacteria detected in the streamers in the north Wales sites could not be established readily. The closest cultivated relative of the bacterium represented by clone TrefC4 (which has not been isolated in pure culture yet) is the neutrophilic (pH range, pH 5.0 to 6.5) iron oxidizer G. ferruginea. G. ferruginea can grow autotrophically using ferrous iron, sulfide, or thiosulfate as an electron donor and can also utilize glucose, fructose, and sucrose, and it is most frequently encountered in waters containing relatively small amounts of dissolved oxygen (16, 17, 30). Clone TrefC4 is, however, only distantly related to G. ferruginea (95% 16S rRNA gene sequence similarity) and certainly represents a new species. The fact that the bacterium represented by clone TrefC4 can grow in very acidic waters (pH as low as pH 2.6 in the present study) suggests that it differs from G. ferruginea in at least one important physiological trait. A phylogenetic analysis suggested that the bacterium represented by clone TrefC4 may be an iron oxidizer, which is consistent with its occurrence in the iron-rich waters in the chalybeate spa, although this should be confirmed when this bacterium is isolated in pure culture. In contrast, the closest characterized relatives of the bacterium represented by clones TrefC11 and MPKCSC9 are ammonium-oxidizing neutrophiles. Attempts to enrich ammonium oxidizers using either ammonia or urea from the two sites were unsuccessful (data not shown), and the concentrations of ammonium in the waters at the sites (and mine waters in general) were very low, in contrast to the concentrations of ferrous iron. The overlay solid media that were used initially to isolate acidophilic prokaryotes (which support the growth of all known iron- and sulfur-oxidizing acidophiles, such as Acidithiobacillus and Leptospirillum spp.) were ineffective for isolating the dominant acid streamer bacteria. However, a modified “organic-free” variant proved to be very useful for isolating large numbers of ferric iron-stained colonies from the Mynydd Parys streamers, which were shown to be the bacterium represented by the TrefC11 and MPKCSC9 clones. The limited amount of physiological characterization carried out so far for this isolate confirmed that it is an autotrophic iron-oxidizing acidophile. In this regard, it is interesting that another acidophilic iron-oxidizing genus, Leptospirillum, occurs in the phylum Nitrospirae, which also includes nitrifying bacteria.
The results of the present study show that the microbial communities that comprise macroscopic streamer growths found in acidic iron-rich mine waters can vary widely from site to site. Although there was some similarity between the streamers in the two north Wales locations, these streamers differed markedly from those that have been studied using cultivation-independent approaches at the Richmond mine (Iron Mountain, California) (6, 7, 12) and the Rio Tinto (Spain) (28). The Richmond mine is extremely acidic (pH 0.6 to 1.2), and the temperatures within the mine are much higher (up to 43°C) than those at the two north Wales sites. Acid streamer and slime communities in the Richmond mine are dominated by Leptospirillum spp., the archaeon “Ferroplasma acidarmanus” (particularly in the more extreme niches), and Sulfobacillus spp.; acidophilic and acid-tolerant fungi have also been detected. In general, the physicochemistry of the Rio Tinto (pH ∼2.0; variable temperature) is more similar to the physicochemistry of the water at Mynydd Parys and the Trefriw Wells Spa, although the concentrations of dissolved oxygen are much greater in the Rio Tinto. Long, filamentous streamer growths in the Rio Tinto were found to be mainly composed of γ-Proteobacteria (Pseudomonas) and α-Proteobacteria (Sphingomonas), none of which were related to known iron oxidizers or any other acidophilic prokaryotes. While the factors that control the microbial compositions of acid streamer communities have not been determined, it seems likely that the physicochemical characteristics (especially pH, temperature, and dissolved oxygen and metal concentrations) of the mine waters in which these macroscopic growths occur have a major effect on the diversity of the indigenous microbial communities.
Acknowledgments
K.C. is grateful to the NERC (United Kingdom) (grant NER/S/C/2001/06450) and Rio Tinto Technology for providing a research studentship. K.B.H. and D.B.J. thank the DTI/BBSRC (United Kingdom) (grant 5/BRM18412) for partial support of this work.
REFERENCES
- 1.Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 5.Bond, P. L., and J. F. Banfield. 2001. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149-161. [DOI] [PubMed] [Google Scholar]
- 6.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownstein, M. J., J. D. Carpten, and J. R. Smith. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20:1004-1010. [DOI] [PubMed] [Google Scholar]
- 9.Coupland, K., F. Battaglia-Brunet, K. B. Hallberg, M. C. Dictor, F. Garrido, and D. B. Johnson. 2004. Oxidation of iron, sulfur and arsenic in mine waters and mine wastes: an important role for novel Thiomonas spp., p. 639-646. In M. Tsezos, A. Hatzikioseyian, and E. Remoudaki (ed.), Biohydrometallurgy: a sustainable technology in evolution. National Technical University of Athens, Zografou, Greece.
- 10.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dugan, P. R., C. B. Macmillan, and R. M. Pfister. 1970. Aerobic heterotrophic bacteria indigenous to pH 2.8 mine water: predominant slime-producing bacteria in acid streamers. J. Bacteriol. 101:982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, K. J., P. L. Bond, T. M. Gihring, and J. F. Banfield. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796-1799. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, K. J., B. M. Goebel, T. M. Rodgers, M. O. Schrenk, T. M. Gihring, M. M. Cardona, B. Hu, M. M. McGuire, R. J. Hamers, N. R. Pace, and J. F. Banfield. 1999. Geomicrobiology of pyrite (FeS2) dissolution: case study at Iron Mountain, California. Geomicrobiol. J. 16:155-179. [Google Scholar]
- 14.Fuchs, B. M., F. O. Glockner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Toril, E., E. Llobet-Brossa, E. O. Casamayor, R. Amann, and R. Amils. 2003. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69:4853-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallbeck, L., and K. Pedersen. 1991. Autotrophic and mixotrophic growth of Gallionella ferruginea. J. Gen. Microbiol. 137:2657-2661. [Google Scholar]
- 17.Hallbeck, L., F. Stahl, and K. Pedersen. 1993. Phylogeny and phenotypic characterization of the stalk-forming and iron-oxidizing bacterium Gallionella ferruginea. J. Gen. Microbiol. 139:1531-1535. [DOI] [PubMed] [Google Scholar]
- 18.Hallberg, K. B., and D. B. Johnson. 2001. Biodiversity of acidophilic microorganisms. Adv. Appl. Microbiol. 49:37-84. [DOI] [PubMed] [Google Scholar]
- 19.Hallberg, K. B., and D. B. Johnson. 2003. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 71:139-148. [Google Scholar]
- 20.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 21.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins, D. A., and D. B. Johnson. 1993. Abandoned metal mines: a unique mineralogical and microbiological resource. J. Russell Soc. 5:40-44. [Google Scholar]
- 23.Johnson, D. B. 1995. Selective solid media for isolating and enumerating acidophilic bacteria. J. Microbiol. Methods 23:205-218. [Google Scholar]
- 24.Johnson, D. B., M. A. Ghauri, and M. F. Said. 1992. Isolation and characterization of an acidophilic, heterotrophic bacterium capable of oxidizing ferrous iron. Appl. Environ. Microbiol. 58:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, D. B., N. Okibe, and K. B. Hallberg. 2005. Differentiation and identification of iron-oxidizing acidophilic bacteria using cultivation techniques and amplified ribosomal DNA restriction enzyme analysis (ARDREA). J. Microbiol. Methods 60:299-313. [DOI] [PubMed] [Google Scholar]
- 26.Lackey, J. B. 1938. The flora and fauna of surface waters polluted by acid mine drainage. Public Health Rep. 53:1499-1507. [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 28.Lopez-Archilla, A. I., E. Gerard, D. Moreira, and P. Lopez-Garcia. 2004. Macrofilamentous microbial communities in the metal-rich and acidic River Tinto, Spain. FEMS Microbiol. Lett. 235:221-228. [DOI] [PubMed] [Google Scholar]
- 29.Lovley, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reduced ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lütters-Czekalla, S. 1990. Lithoautotrophic growth of the iron bacterium Gallionella ferruginea with thiosulfate or sulfide as energy-source. Arch. Microbiol. 154:417-421. [Google Scholar]
- 31.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 32.McGinness, S., and D. B. Johnson. 1993. Seasonal variations in the microbiology and chemistry of an acid mine drainage stream. Sci. Tot. Environ. 132:27-41. [Google Scholar]
- 33.Okibe, N., M. Gericke, K. B. Hallberg, and D. B. Johnson. 2003. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred tank bioleaching operation. Appl. Environ. Microbiol. 69:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrenk, M. O., K. J. Edwards, R. M. Goodman, R. J. Hamers, and J. F. Banfield. 1998. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science 279:1519-1522. [DOI] [PubMed] [Google Scholar]
- 36.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 37.Wakao, N., H. Tachibana, Y. Tanaka, Y. Sakurai, and H. Shiota. 1985. Morphological and physiological characteristics of streamers in acid mine drainage water from a pyritic mine. J. Gen. Appl. Microbiol. 31:17-28. [Google Scholar]