Abstract
Eukaryotic cells possess an exquisitely interwoven and fine-tuned series of signal transduction mechanisms with which to sense and respond to the ubiquitous fermentable carbon source glucose. The budding yeast Saccharomyces cerevisiae has proven to be a fertile model system with which to identify glucose signaling factors, determine the relevant functional and physical interrelationships, and characterize the corresponding metabolic, transcriptomic, and proteomic readouts. The early events in glucose signaling appear to require both extracellular sensing by transmembrane proteins and intracellular sensing by G proteins. Intermediate steps involve cAMP-dependent stimulation of protein kinase A (PKA) as well as one or more redundant PKA-independent pathways. The final steps are mediated by a relatively small collection of transcriptional regulators that collaborate closely to maximize the cellular rates of energy generation and growth. Understanding the nuclear events in this process may necessitate the further elaboration of a new model for eukaryotic gene regulation, called “reverse recruitment.” An essential feature of this idea is that fine-structure mapping of nuclear architecture will be required to understand the reception of regulatory signals that emanate from the plasma membrane and cytoplasm. Completion of this task should result in a much improved understanding of eukaryotic growth, differentiation, and carcinogenesis.
INTRODUCTION
Jacques Monod's seminal discovery of the diauxic growth of bacterial cultures (243) demonstrated that microbes are particularly adept at targeting and responding to glucose, the most abundant monosaccharide in nature. A wealth of subsequent work has shown that virtually all cells possess a sophisticated genetic program that responds to this rich source of carbon and energy. Study of the glucose response in eukaryotes has been greatly aided by the development of the malleable budding yeast Saccharomyces cerevisiae as a model system (see, e.g., references 361 and 417). S. cerevisiae shares with complex multicellular eukaryotes many of the signal transduction components that detect glucose, transduce the corresponding signals to the interior of the cell, and make the needed adjustments to cellular metabolism and gene expression profiles.
This review will focus on the cytoplasmic regulators that transmit the glucose signal from the plasma membrane into the yeast nucleus, as well as the nuclear apparatus that mediates the global transcriptomic response to the sugar (both glucose repression and glucose induction). The response to glucose also involves direct modification of metabolic pathways (termed glucose or catabolite inactivation) and other processes such as mRNA turnover (7, 76, 103, 121, 153, 154, 156, 161, 166, 171-173, 232, 294, 307, 329, 334, 335, 337, 391). Although the latter phenomena are outside the scope of this review, they appear to be mediated by the same two central signal transduction pathways, Ras/cyclic AMP (cAMP)/protein kinase A (PKA) and Snf3/Rgt2/Yck, which are described below in detail.
The recent application of yeast genomics and proteomics is a first step towards comprehensive characterization of the glucose response. For example, we now know that within 20 min of glucose addition, ∼20% of the roughly 6,200 genes in S. cerevisiae exhibit a greater-than-threefold change in expression (either induction or repression), and ∼40% show at least a twofold change (408). Glucose repression largely affects genes encoding enzymes involved in respiration or alternative carbon source metabolism, and glucose induction stimulates transcription of glycolytic genes and ribosomal protein (RP) genes (see below). Genomic and proteomic analyses are particularly useful when supplemented with high-throughput genetic techniques such as synthetic genetic array analysis, which allows the identification of synthetic phenotypes by simultaneous screening of the entire collection of mutants in which each of the ∼4,700 nonessential yeast genes has been deleted (373).
The continued pursuit of such large data sets, in combination with the now-facile elimination, overexpression, or point mutation of relevant yeast regulators, should eventually permit the delineation of a global wiring diagram that mirrors its biological template when virtually challenged with a wide range of concentrations of the sugar. For such a computational model to be valid, it must at a minimum be informed by (i) an accurate molecular map of cytoplasmic and nuclear substructure, (ii) a detailed diagram of the physical and functional connections that link the relevant signal transduction components to each other and to adjacent networks, and (iii) the temporal progression of the response following the appearance of glucose. Here I review our progress toward that ambitious goal and attempt to identify the most important issues that require further clarification.
NATURE OF THE SIGNAL
Two recent surprises uncovered by global analyses of Saccharomyces cerevisiae are the extent to which the glucose induction and repression mechanisms appear to overlap and the fact that yeast cells can be tricked into generating the full-blown response (both induction and repression) in the complete absence of glucose. These and other recent developments place G-protein signaling, in particular the Ras/cAMP/PKA pathway, front and center among the cellular components that participate in early events in the glucose response.
Signaling without Extracellular Sensing, Transport, or Phosphorylation of Glucose
A recent microarray experiment from the Broach laboratory has clearly demonstrated the universality of induction/repression overlap in glucose signal transduction. A unified transcriptome-wide response (both upshifted and downshifted gene expression) was observed in yeast cells by stimulating early steps in the glucose signaling pathway; a clever experimental scheme allowed the generation and detection of this comprehensive glucose response without addition of the sugar (408). The yeast transcriptome was analyzed after heterologous induction of activated alleles of Ras2 or Gpa2, signal transmitters that act in parallel early in the cAMP-dependent protein kinase (PKA) pathway (see below). Astonishingly, virtually all of the changes in transcript levels after addition of glucose (both up-regulation and down-regulation) were also observed in the absence of glucose following expression of the activated form of Ras2. Of the genes that showed at least a threefold change in expression within an hour following glucose addition, almost all (92%) changed at least twofold in the same direction after Ras activation (Fig. 1; Table 1). The result was similar with activated Gpa2, though the intensity of the transcriptional response was reduced relative to that seen with activated Ras. Those authors also demonstrated that all of the changes in transcript levels resulting from Ras2 and Gpa2 activation were mediated by PKA (408) (Fig. 1). Importantly, since much of this transcriptional reprogramming can also be accomplished in the absence of signaling through cAMP, redundant overlapping pathways (Ras2/Gpa2/PKA dependent and Ras2/Gpa2/PKA independent) appear to be fully capable of transducing the glucose signal. Little is known about the PKA-independent mechanism, although it presumably targets a similar set of downstream factors in the absence of normal PKA catalytic activity.
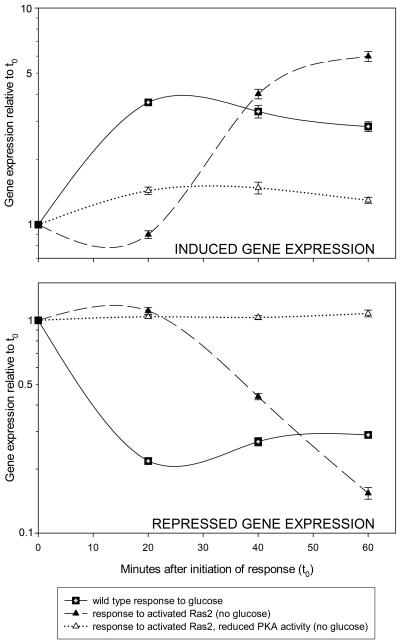
FIG. 1.
Wild-type versus Ras2 mimicry of the glucose response. Each curve represents the expression profile of genes exhibiting a twofold or greater change, both within 20 min of glucose addition and within 20 min of the appearance of activated Ras2. The average values for 250 such induced genes (upper graph) and repressed genes (lower graph) are shown (see supplemental data associated with reference 208); error bars represent standard errors of the means. Squares, expression profile of wild-type cells following glucose addition; filled triangles, expression profile of cells containing Gal-regulated Ras2* (activated Ras) following addition of galactose; open triangles, expression profile of cells containing Gal-regulated Ras2* (activated Ras) in a tpkw (weak PKA) background following addition of galactose. It is important to note that the >100-fold induction and repression observed for a small subset of glucose regulated genes (e.g., HXT genes; see reference 272) is not seen within the 60-minute time frame of this data set.
TABLE 1.
Genes that respond similarly to both glucose addition and Ras2 activationa
| Induced genes | Repressed genes
|
||||
|---|---|---|---|---|---|
| Protein biosynthesisb | Transcription | Transportc | Energy reserve metabolismd | Protein turnover | Stress response |
| RPS9A | RPA43 | NUP1 | BCY1 | PEP4 | HSP30 |
| RPS22B | RPA49 | NUP100 | SDC25 | RPN2 | SSE2 |
| RPS27A | RPB5 | NUP133 | GAL83 | RPN3 | SSA3 |
| RPL7B | RPB9 | NUP170 | PHO85 | RPN5 | SSA4 |
| RPL14B | RPC40 | MEX67 | TPS1 | RPN6 | SSD1 |
| RPL17B | RPC82 | RNA1 | CIT3 | RPN7 | STO1 |
| TIF11 | RPO26 | GSP1 | GPD1 | RPT1 | CDC55 |
| TIF3 | RPO31 | PSE1 | MDH3 | RPT3 | UBC5 |
| TIF34 | RRP43 | SXM1 | PGM2 | RPT4 | PRE1 |
| TEF4 | RRP45 | ALR21 | HXT10 | RPT5 | PRE3 |
| CDC60 | RRP5 | FET3 | MRPL23 | DOA1 | GTS1 |
| ARG8 | ARG80 | FET4 | MRPL32 | DOA4 | CRS5 |
| ARO7 | BAS1 | FKS1 | PET112 | APG13 | CTA1 |
| ARO8 | DBP2 | HNM1 | PET117 | APG14 | GRX2 |
| ASP1 | DBP3 | HXT2 | PET191 | APC9 | TRX2 |
| HIS1 | GCR2 | HXT4 | PET54 | HRD1 | MDR1 |
| HIS3 | IFH1 | LYP1 | COQ1 | HRD3 | PAU1 |
| ILS1 | HMALPHA1 | NPL3 | COQ3 | PEX14 | PAU2 |
| ILV1 | IMP4 | PHO84 | COX10 | MPD2 | PAU3 |
| LYS4 | LEU3 | ZRT1 | COX11 | SBA1 | PPZ2 |
See supplemental data associated with reference 408; a grand total of 758 genes exhibited a twofold or greater change in expression both within 20 min of glucose addition and within 20 min of the appearance of activated Ras2. Gene categories are based on Munich Information Center for Protein Sequences functional classification; a representative list from each category is shown. A total of 329 and 429 genes were in the induced and repressed categories, respectively.
Includes genes encoding translational components (including ribosomal proteins), rRNA-processing factors, and amino acid biosynthetic enzymes.
Includes genes encoding nucleocytoplasmic transport factors and plasma membrane transporters.
Includes genes encoding factors involved in carbon storage and respiration (including mitochondrial factors).
The observed transcriptome-wide up-regulation and down-regulation of gene expression in response to activated Ras was particularly surprising, since it had long been assumed (despite early hints to the contrary [see, e.g., reference 233]) that the primary glucose repression pathways in yeast are distinct from glucose induction and are also essentially Ras independent (44, 45, 118, 121, 123, 178, 315, 335, 365). Further, stimulation of the glucose response in the absence of either transport or phosphorylation of the sugar (408) is particularly significant. Transport of glucose and its conversion to glucose-6-phosphate by hexokinases had been explored as a potentially essential feature of the response (121, 314, 316) and now appears to at most participate in redundant signaling that operates through or in parallel with the cAMP-PKA pathway. Whatever extracellular or intracellular glucose sensing is needed to activate Ras or Gpa2 is therefore sufficient for both glucose induction and repression. This interpretation has the added advantage of bringing our assessment of glucose-stimulated signal transduction in yeast cells closer to what is known about the analogous response in bacterial and mammalian cells (for reviews, see references 35, 129, and 322).
Redundant but Unified Glucose Signaling Pathways
The first hint of overlap between glucose induction and repression resulted from analysis of regulators that are “downstream” in the signaling cascade, i.e., factors that for the most part function within the yeast nucleus. For example, the Rgt1 repressor blocks transcription of HXT1 through HXT4 (4 of the 18 glucose transporter genes in S. cerevisiae) (see references 118 and 272 for reviews). At high glucose concentrations, although its contact with the HXT1 promoter can no longer be detected, Rgt1 is converted into an activator through hyperphosphorylation (142, 186, 250). The hexokinase Hxk2, long known to participate in glucose repression, is also required for glucose induction of HXT1 (231, 271).
The Snf1 serine-threonine kinase represents another example of downstream regulatory overlap; Snf1 activates genes that are repressed in glucose-containing media but are derepressed and needed for growth in the presence of nonfermentable carbon sources (46). Recent transcriptomic and proteomic analyses of snf1 cells revealed a previously unnoticed upshift in expression of genes (e.g., ADH1) involved in glucose catabolism (hereafter the term “glycolytic genes” will be used for this group) (432). Thus, in addition to its well-established role in derepressing glucose-repressed genes, Snf1 also somehow represses transcription of glucose-induced genes (including HXT1) (372). Snf1 repression of glycolytic gene expression has been further substantiated by proteomic analysis of cells lacking Snf4, the γ subunit of the Snf1 kinase complex (140).
A final example of downstream regulatory overlap is the phosphoprotein Gcr1, which was first identified as an activator of glycolytic genes (13, 63, 64, 152). Transcriptomics and other analyses have confirmed that expression of numerous glucose-repressed genes is derepressed in the absence of Gcr1 at high glucose concentrations, suggesting a new role for this regulator in repression of transcription (327, 383; K. Barbara and G. M. Santangelo, unpublished data).
The roles of Rgt1, Hxk2, Snf1/Snf4, and Gcr1, as well as numerous other repressor/activator polypeptides that participate in the glucose response, are presented in greater detail in later sections of this review. In addition to these specific regulators, at least eight components of the general transcription machinery that interact with the carboxy-terminal domain of RNA polymerase II (Gal11, Sin4, Rgr1, Rox3, Srb8, Ssn2, Ssn3, and Ssn8) have been found to play both positive and negative regulatory roles; the function of many of these factors is known to be influenced by glucose signaling (43, 195). Thus, the overlapping positive and negative circuitry in the glucose response may extend from the plasma membrane through to the ultimate choice of induction or shutoff of transcription initiation of each glucose-regulated gene by RNA polymerase II.
The induction/repression duality of signal recipients such as Rgt1, Hxk2, Snf1/Snf4, Gcr1, and other transcription factors clearly requires further characterization of the precise mechanisms involved. Nonetheless, together with the transcriptomic analysis of activated Ras, it suggests the existence of a more unified and interdigitated response to glucose than had previously been appreciated. Although it now seems certain that transmission of the glucose signal is redundant (408), those signaling pathways may converge on only a relatively small group of transcription factors in the yeast nucleus (see below). Coordinated remodeling of the nuclear regulatory apparatus in response to glucose might therefore involve a parsimonious series of alterations in key regulators. Although for the sake of clarity and historical perspective the discussion of nuclear regulators is divided into separate sections of this review, in reality the mechanisms of “glucose repression” and “glucose induction” may be no more segregated than the two faces of a coin.
Early Steps in Signal Transduction
Given the sufficiency of Ras or Gpa2 activation to produce the transcriptome-wide spectrum of changes in response to glucose, it is important first to review what is known about these factors, their respective signaling pathways, and how they are activated immediately following appearance of the sugar.
The Ras/cAMP/PKA pathway.
Ras proteins are monomeric GTPases that function as switches; these so-called G proteins are inactive in the GDP-bound state and active when GTP is bound. The switch from the active to the inactive form involves hydrolysis of bound GTP by the intrinsic GTPase and is stimulated by GTPase-activating proteins (GAPs). The reverse (inactive-to-active) switch requires replacement of bound GDP with GTP, which is normally accomplished with the aid of guanine nucleotide exchange factors (GEFs). Activating mutations in Ras polypeptides (e.g., human rasVal12 or yeast Ras2Val19) increase GTP association in a GEF-independent fashion and are responsible for many human cancers (78, 419). Ras mutations are found in approximately 30% of human tumors; the frequency of oncogenic forms of Ras is as high as 50% or 90% in colorectal or pancreatic tumors, respectively (345). Although it remains dimly understood, it seems likely that there is an important connection between these recent data and a very old observation that altered glucose metabolism is a hallmark of cancerous cells in solid tumors (71, 179).
The polypeptides encoded by the two RAS genes in S. cerevisiae, Ras1 (309 residues) and Ras2 (322 residues), are >70% identical overall and approximately 90% identical over the N-terminal 180 residues (291). Growth on glucose is unaffected by the absence of either Ras1 or Ras2, whereas loss of both causes arrest in the G1 phase of the cell cycle (362, 367). These early findings were suggestive of a connection to nutrient sensing, since nutrient-deprived cells also arrest in G1 phase. Overexpression of RAS1 was also found to suppress the failure of ras2Δ cells to grow on nonfermentable carbon sources (363), suggesting that the latter defect is due to the fact that Ras1 is more weakly expressed than Ras2 (249, 361), rather than a specific function that maps to the divergent and slightly extended C terminus of Ras2.
Important effector molecules modulate Ras activity during the glucose response; posttranslational modification and membrane localization of Ras are two important features of these regulatory inputs. For example, Ras farnesylation contributes to nucleotide exchange by the GEF Cdc25 (34, 73, 134); CDC25 is an essential gene and was originally identified by a screen for mutations that cause G1 arrest (139). A second yeast RasGEF that can substitute for Cdc25 when overexpressed is encoded by the dispensable SDC25 gene (31) (note that this gene was originally named SCD25 [79]).
Ras stimulation by the RasGEFs in turn stimulates production of cAMP by the essential product of the adenylate cyclase gene (CYR1) (229). Inactivation of Ras, resulting in lower cAMP levels, is facilitated by the GAPs Ira1 and Ira2 (358, 360) (Fig. 2). Presumably due to their competing effects on Ras activation, IRA1 deletion rescues the lethality caused by deletion of the RasGEF CDC25 (359). Low-affinity and high-affinity cAMP phosphodiesterases in S. cerevisiae (Pde1 and Pde2, respectively) also antagonize the Ras signal through enzymatic removal of cAMP; in the presence of exogenous cAMP, deletion of PDE2 restores growth to ras1Δ ras2Δ strains (266, 420).
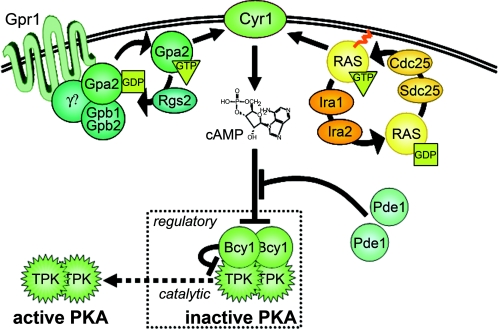
FIG. 2.
Cytoplasmic events in PKA signaling. Glucose regulation relies upon the intracellular signaling molecule (“second messenger”) cAMP. GTP-bound G proteins (Ras and Gpa2) bind independently to adenylate cyclase (Cyr1) and stimulate its production of cAMP. The monomeric Ras G proteins are anchored in the plasma membrane via a posttranslationally added palmitoyl moiety (red squiggle). RasGEFs (Cdc25 and Sdc25) and RasGAPs (Ira1 and Ira2) are also present in the Ras/Cyr1 complex and regulate adenylate cyclase by controlling the Ras switch (see text). The seven-transmembrane polypeptide Gpr1 acts upstream of Gpa2 in glucose signaling and is a member of the G protein-coupled receptor (GPCR) superfamily. Gpa2 was identified as a result of its similarity to the mammalian Gα subunits of heterotrimeric G proteins, and the putative Gβ subunits Gpb1 and Gpb2 were found via their interaction with Gpa2; a corresponding γ subunit has yet to be identified (question mark). A Gpa2GEF also awaits identification; Rgs2 is a known Gpa2GAP. Phosphodiesterases (Pde1 and Pde2) antagonize glucose signaling via enzymatic inactivation of cAMP (conversion to AMP). The PKA tetramer is the regulatory target of cAMP. While bound to the kinase subunits (TPK), the regulatory Bcy1 subunits keep PKA in an inactive state; cAMP activates the catalytic subunits by binding to Bcy1 subunits and promoting dissociation of the complex (dashed arrow).
The Ras C terminus also undergoes functionally significant posttranslational modification. Yeast Ras2 is normally palmitoylated via a thioester linkage at cysteine 318 and farnesylated via a thioether linkage at cysteine 319. Blocking both modifications with the double point mutation C318S C319S results in a nonfunctional molecule (92). Farnesylation is required for palmitoylation and all subsequent modifications, which include removal of the three C-terminal residues by proteolytic cleavage and methyl esterification of the revealed C-terminal cysteine 319 (19).
The palmitoyl moiety anchors Ras to the cytoplasmic face of the yeast plasma membrane (199); blocking addition of palmitate via the single point mutation C318S causes mislocalization of the mutated Ras2 protein to the cytoplasm (176). Ras palmitoylation is reversible and therefore may be a regulatory step. This is of particular interest because mutation of palmitoylation sites abrogates both the glucose response in yeast cells (see below) and transformation of mammalian cells by oncogenic forms of Ras (418).
The appearance of glucose thus generates one or more signals that converge on membrane-bound Ras and its modifiers (in particular the RasGEFs and RasGAPs). This glucose signaling results in a rapid but transient spike in the level of intracellular cAMP, which increases 5- to 50-fold within 1 to 2 min of glucose addition and returns to near-basal levels within 20 min. When cells express the nonpalmitoylated cytoplasmic Ras2C318S form as their only Ras protein, they fail to exhibit the glucose-induced cAMP spike and are defective in transitioning to rapid growth upon addition of glucose (160). Thus, glucose-regulated cAMP production appears to require attachment of Ras to the yeast plasma membrane. Interestingly, membrane localization of yeast Ras2 was recently shown to occur despite disruption of the classical secretory pathway that normally mediates translocation from the endoplasmic reticulum to the plasma membrane (92).
Additional signal transduction components appear to contribute to glucose signaling by contacting adenylate cyclase (Cyr1); such Cyr1 protein-protein interactions identified thus far include those with (i) an N-terminal SH3 domain in the RasGEF Cdc25 (237), (ii) the farnesylated Ras2 C terminus (73, 74, 184, 199, 338, 355), (iii) the C terminus of the putative cochaperonin Sgt1 (95), (iv) the N terminus of the ancillary factor Srv2 (CAP) via coiled-coil formation (267), and (v) the RasGAP Ira1, which mediates peripheral association of adenylate cyclase with the yeast plasma membrane (241).
It seems likely that the sole purpose of the glucose-induced spike in cAMP levels is to block the inhibitory effect of the BCY1-encoded regulatory subunit on the catalytic subunits of PKA, encoded redundantly by TPK1, TPK2, and TPK3; overexpression of any of these three genes, or deletion of BCY1, restores growth to ras1Δ ras2Δ strains (369). PKA is a heterotetramer consisting of two catalytic (Tpk) subunits and two regulatory (Bcy1) subunits (Fig. 2); addition of regulatory subunits to catalytic subunits in vitro converts protein kinase activity into a completely cAMP-dependent form (151, 177, 370).
Interestingly, the subcellular localization of PKA catalytic and regulatory subunits appears to respond to glucose signaling; Bcy1 is present in both the cytoplasm and nucleus in the absence of glucose but is exclusively nuclear in its presence (129, 130). Cross talk from the TOR (target of rapamycin) pathway, another major set of nutrient-responsive signaling components, may help to maintain PKA activity in part by regulating nuclear translocation of Tpk1 (330). Although it has yet to be demonstrated that the observed translocation of PKA subunits between compartments plays an important role in glucose signaling in yeast cells, targeting of PKA regulatory substrates would appear to undergo a qualitative and/or quantitative shift in response to the appearance or disappearance of the sugar. Further progress in this area is complicated by the fact that signaling via each of the three different catalytic subunits of PKA (Tpk1, Tpk2, and Tpk3) results in a distinct pattern of downstream gene expression readouts (309).
Although active PKA is thought to phosphorylate proteins involved in transcription, energy metabolism, and cell cycle progression (129), further study is required to identify the exact targets and sequence of protein translocation and phosphorylation events that transmit the glucose regulatory signal to the yeast nucleus. Significantly, mutations impairing specific components of the RNA polymerase II transcriptional machinery are synthetically lethal with the hyperactive Ras2Val19 allele (157), and PKA can phosphorylate a component of the Srb complex (Ssn2) both in vitro and in vivo (55). The latter may be one of but a few Ras/cAMP/PKA-mediated modifications in nuclear factors needed to accomplish the broad sweep of alterations to the yeast transcriptome upon glucose addition or depletion.
The Ras/cAMP/PKA pathway has also been implicated in aging (212, 216), thermotolerance (439), bud site selection, actin repolarization (333), glycogen accumulation (342), stress resistance (39, 407), and sporulation (39, 41); it may also regulate pseudohyphal differentiation (growth as chain-like multicellular filaments) in response to nutrient limitation. Each of these interesting topics (see reference 411 for a recent review), as well as the involvement in these processes of other nutrient-stimulated kinases (e.g., Snf1 and TOR [45, 75, 196, 197, 401, 441]), GTPases (e.g., Gpa2 [277]), and transcription factors (e.g., Msn2/Msn4 [30, 96, 112]), is outside the scope of this review.
The Gpr1/Gpa2 pathway.
Heterotrimeric G proteins are signaling molecules composed of α, β, and γ subunits. They represent a second class of factors that, like monomeric Ras polypeptides, are capable of binding guanine nucleotides (Fig. 2). The Gα subunit Gpa2 and its negatively acting regulator (GAP) Rgs2 have been shown to participate in glucose signaling in S. cerevisiae (190, 394). Gpa2 was originally identified by screening a yeast genomic library with cDNA probes encoding mammalian Gα subunits (252), and the putative β subunits Gpb1 and Gpb2 were identified as interaction partners of Gpa2 (135); no corresponding Gγ subunits involved in glucose signaling have yet been identified (but see reference 135). Whereas deletion of either GPA2 or RAS2 causes little or no impairment of growth on glucose media, deletion of both causes a severe synthetic growth defect (428). Removal of the major cAMP phosphodiesterase in the cell (Pde2) restores normal growth to gpa2Δ ras2Δ strains, suggesting that an increase in cAMP levels bypasses the loss of both Gpa2 and Ras2 (428). Most importantly, as mentioned above, the transcriptomic response to glucose is successfully mimicked in the absence of the sugar by turning on expression of either activated Gpa2 or activated Ras2; the fact that this signaling is PKA dependent confirms their redundant participation in the cAMP-dependent branch of glucose signaling (408).
A two-hybrid protein interaction screen with GPA2 as the bait led to the identification of the GPR1 gene, which encodes a member of the G protein-coupled seven-transmembrane receptor superfamily (Fig. 2). Gpr1 is located on the yeast cell surface (428). Genetic analysis strongly suggests that Gpa2 acts downstream from Gpr1 in the same signaling pathway: the low growth rates of gpr1Δ ras2Δ, gpa2Δ ras2Δ, and gpa2Δ gpr1Δ ras2Δ strains are essentially identical, and introduction of either multicopy or activated GPA2 suppresses the defect of the gpr1Δ ras2Δ strain (428). Deletion of GPR1 diminishes (although does not eliminate) genome-wide transcriptional induction and repression in response to glucose (408). The simplest explanation for these data is that Gpr1 is the only receptor coupled to Gpa2 and that it too contributes to activation of the cAMP pathway in response to glucose (190).
Although Gpa2 was originally thought to function upstream of the Sch9 kinase (368, 428), it was subsequently found that SCH9 deletion is synthetically lethal with gpr1Δ, gpa2Δ, or ras2Δ, suggesting that Sch9 acts instead in the pseudohyphal differentiation pathway that is parallel to both Gpr1/Gpa2 and Ras (217). Sch9 may nevertheless participate somehow in glucose signaling, since (i) it is the yeast ortholog of a component of the mammalian insulin response pathway (Aft/protein kinase B), (ii) it shares similarity with PKA catalytic subunits, (iii) its overexpression compensates for the loss of Ras function, and (iv) PKA activity is elevated in a sch9Δ background (72, 109).
Activation of Ras and Gpa2 by glucose.
Since artificial activation of Ras and Gpa2 is sufficient to generate the glucose response, and it seems unlikely that glucose interacts directly with the G proteins themselves, how do Ras and Gpa2 become activated in wild-type yeast cells exposed to the sugar? It is convenient to separate potential answers to this question into two categories: recognition of glucose by receptors on the outside surface of the cell versus direct signaling of Gpa2 and Ras (or their regulators) by intracellular glucose or its metabolites.
(i) Extracellular sensing.
There are 18 known hexose transporter genes in S. cerevisiae (HXT1 to HXT17 and GAL2). Deletion of seven of these genes results in the failure to transport or grow on glucose (308, 413). In this hxt1 to hxt7 null background, constitutive expression of the GAL2-encoded galactose permease restores glucose-induced cAMP synthesis (313). This suggests that the yeast hexose transporters do not participate in glucose signaling but are required only for transport.
The seven-transmembrane receptor Gpr1 may detect extracellular glucose and thereby activate Gpa2, but it is not required to signal Ras; Gpr1 or Gpa2 deletion does not prevent the glucose-induced increase in Ras2-GTP levels (66). In contrast, the Ras2-GTP increase is absent in a glucose phosphorylation-deficient (hxk1 hxk2 glk1) strain (66). Thus, Ras appears to be activated by intracellular phosphorylated glucose but not by extracellular glucose. A clear demonstration of extracellular sensing by Gpr1 will require direct evidence, for example, as suggested by recent mutagenic analysis (205), that it interacts with glucose.
(ii) Intracellular sensing.
If extracellular signaling does not participate in Ras activation (66, 313), it will be particularly important to understand how intracellular glucose triggers the glucose response. The longstanding idea that glucose phosphorylation (in particular by the glucose-induced hexokinase Hxk2; see below) and/or subsequent glycolytic reactions are required to alter yeast gene expression in response to glucose has been controversial (see references 20, 121, 244, and 314 for reviews of this topic). Since repression occurs even when the metabolic steps immediately following glucose phosphorylation are reduced to less than 2% of wild-type levels, it seems likely that nothing beyond the production of glucose-6-phosphate is required (317). Further, since cells can be tricked into the full-blown glucose response by expressing activated Ras or Gpa2, glucose phosphorylation must generate the signal through the G proteins and/or through a redundant and as-yet-obscure alternative pathway. A recent interesting suggestion is that glucose phosphorylation increases Ras2-GTP levels by inhibiting the Ira (RasGAP) proteins (66).
Although progress has been made, much more work is needed to establish the precise mechanism(s) through which Ras, Gpa2, and/or their regulatory GEFs and GAPs is signaled by glucose. Intracellular acidification (316, 365) and energy charge (the AMP/ATP ratio) (134, 244) are additional potential signaling mechanisms that require further testing. The events that immediately follow activation of either G protein are much clearer (Fig. 2). Activated Ras or Gpa2 can each generate a spike in cAMP production by adenylate cyclase; cAMP in turn releases PKA catalytic (Tpk) subunits from inhibition by its regulatory (Bcy1) subunits.
Extracellular sensing by Snf3/Rgt2.
The putative glucose sensors Snf3 and Rgt2 are 60% identical to each other and, like hexose transporters found in bacteria, plants, and mammals (228, 259, 332), contain 12 predicted transmembrane-spanning domains (272). Snf3 and Rgt2 do not appear to act as glucose transporters but instead signal the presence of the sugar (118, 178, 182, 247, 273, 314). Since the transcriptional response of Snf3/Rgt2 target genes (e.g., HXT genes encoding the glucose transporters) is generated in the absence of glucose by expression of hyperactive Ras or Gpa2, the Snf3 and Rgt2 sensors appear either to act upstream of Ras/cAMP/PKA or to play an ancillary or redundant role in the initial detection of glucose. This view is consistent with the possibility that glucose sensing by Snf3/Rgt2 is required for maintenance of glucose induction or repression for at least a subset of the transcriptome (182, 272). Further, it seems clear that the Snf3/Rgt2 regulatory pathway helps to ensure that changes in gene expression are commensurate with the glucose concentration in the environment, a mechanism that is likely to be particularly important in nature.
As mentioned above for Gpr1, a clear demonstration of extracellular sensing by Snf3 and Rgt2 will require direct evidence that they interact with glucose. Intriguing hints along these lines have already been uncovered. The V404I mutation in Rgt2 appears to abolish glucose signaling; this residue is orthologous to one in Hxt1 (F371) that is necessary for glucose transport (247). V404 in Rgt2 may therefore be required for glucose sensing because it helps to form the glucose binding domain in the receptor. Similarly, dominant mutations in both Snf3 (R229K) and Rgt2 (R231K) that cause constitutive signaling, even in the absence of glucose, map to the putative glucose binding pocket (272, 273). Progress has also been made with respect to the elaboration of a mechanism for subsequent signaling by Snf3 and Rgt2 across the plasma membrane (see below).
SIGNAL RECEPTION: GLUCOSE REPRESSION
Direct sensing of extracellular glucose by yeast cells, rather than intracellular sensing of its metabolic products, is a relatively new concept. Further study of receptor-mediated signaling in yeast may prove critical in understanding similar nutrient sensors in mammalian cells (179). How PKA, once unleashed, remodels the architecture of the yeast nucleus to produce the ultimate transcriptomic consequences of glucose induction and repression will be a central consideration of the remainder of this review.
Most of the ∼1,000 genes in S. cerevisiae that are transcriptionally downshifted during growth on glucose can be sorted into one of three groups: gluconeogenic genes, Krebs cycle/respiratory genes, and genes involved in alternate carbon source metabolism. This section will review known mechanisms through which the glucose signal is received in the yeast nucleus and modulates expression of glucose-repressed target genes.
Involvement of the Hexokinase Hxk2 in the Glucose Response
The first, irreversible step in the intracellular metabolism of glucose is C6 phosphorylation. Three enzymes (encoded by HXK1, HXK2, and GLK1) can catalyze this reaction; of these, only HXK2 is highly expressed in the presence of glucose (147). HXK2 lesions (both point and null mutations) were long ago shown to cause failures in glucose repression (99, 100, 104, 222). After much controversy, it is now deemed unlikely that Hxk2 catalytic activity plays an important role in the intracellular glucose signaling pathway (244). The surviving version ofthis model postulates that, following the onset of the phosphoryl transfer reaction, a stable transition intermediate alters Hxk2 conformation and mediates its regulatory function in altering target gene expression (20, 191). However, this model fails to explain the fact that sugars metabolized in an Hxk2-independent fashion can cause genome-wide transcriptional repression comparable to that observed after the addition of glucose to wild-type cells or upon expression of activated Ras or Gpa2 (17, 214, 312, 397, 408).
Interestingly, approximately 14% of Hxk2 is nuclear in glucose-grown cells (146). This discovery suggests an alternative explanation for the regulatory involvement of this hexokinase in the glucose response (244, 304). Hxk2 has been detected in the nucleus by enzymatic assay, immunoblotting, and fluorescent visualization of a green fluorescent protein (GFP)-tagged chimera, confirming controversial reports of nuclear hexokinase that first appeared in the early 1960s (234, 340, 390). The negatively acting DNA-bound glucose regulator Mig1 (see below) appears to mediate nuclear localization of Hxk2; Mig1 and Hxk2 have been shown to interact via two-hybrid, immunoprecipitation, and glutathione S-transferase pull-down assays (2, 245). An interaction between Hxk2 and the Mediator component Med8 has also been described, and the latter appears to bind directly to both positively and negatively acting glucose regulatory elements in DNA (57, 80, 246). A previously reported example of a metabolic enzyme that doubles as a transcriptional regulator is the galactokinase Gal1, which both phosphorylates galactose and appears to activate the transcription factor Gal4 by binding to the Gal4 inhibitor Gal80 (436).
Despite these intriguing hints, contradictory results prevent an unambiguous assignment of the Hxk2 function in glucose regulation to the nuclear form of the protein. For example, removal of a small N-terminal segment of Hxk2 (residues 7 through 16) in a putative nuclear localization sequence (Lys8-Pro-Gln-Ala-Arg12) has been reported to abolish both Hxk2 nuclear localization and glucose repression of SUC2, HXK1, and GLK1 without impairing hexokinase catalytic activity (146, 311). The exact opposite result (intact glucose signaling and low catalytic activity), however, was obtained by others upon deletion of an overlapping and slightly larger Hxk2 segment (residues 1 through 15) (231). Likewise, the phosphorylatable Ser14 residue in Hxk2 (193) is reported either to mediate (305) or not to mediate (146, 231) glucose repression. Further analysis is needed to resolve these discrepancies. Most importantly, proof of a regulatory role for nuclear Hxk2 awaits an unambiguous demonstration that its role in the glucose response relies upon the relatively small fraction of the protein that resides in the nucleus.
Snf1-Mediated Signal Transduction in Glucose Repression and Derepression
The uncertainty surrounding the role of Hxk2 subcellular localization and phosphorylation state in the glucose response is particularly important to resolve because it represents one of several potential connections between Ras/cAMP/PKA-dependent events that occur early in the signal transduction cascade and downstream modifications that have a direct impact on target gene expression. Importantly, increasing the extracellular glucose concentration, or hyperactivation of PKA via bcy1 deletion, results in reduced Hxk2 phosphorylation, whereas attenuation of PKA activity increases 32P labeling of Hxk2 (398). These data are consistent with a model in which Hxk2 dephosphorylation, which converts it from a monomer to a dimer (305), somehow transduces the glucose signal to downstream effectors, eventually resulting in repression (and perhaps also induction) of glucose-regulated genes. The nuclear form of Hxk2 might accomplish this via its direct interaction with Med8 and/or Mig1 (see above). Alternatively or additionally, such an Hxk2 signal might be received by two additional phosphoproteins that are well established as downstream components in the glucose signal transduction pathway: the Reg1-targeting subunit of the Glc7 phosphatase and the Snf1 kinase (325).
Dueling Snf1 kinase and Glc7 phosphatase mediate glucose regulation.
SNF1 was first identified in screens for regulatory factors in the glucose response (it was then named either CAT1 or CCR1) (62, 102, 440). SNF1 was also identified in a screen for mutants that could ferment glucose but not sucrose (46). Since inactivation of SNF1 also impairs utilization of galactose, maltose, and nonfermentable carbon sources, soon after its identification it was recognized as an important regulatory gene mediating glucose derepression. The SNF1 gene was cloned and found to encode a serine-threonine protein kinase (51); the existing evidence also suggested a physical and functional link to SNF4, another gene that had been identified in the “sucrose nonfermenting” screen (49, 50, 259, 261). The snf4 and snf1 phenotypes are similar, including defective utilization of the many carbon sources (e.g., sucrose, galactose, maltose, raffinose, and lactate) that require expression of glucose-repressed genes. Increased SNF1 dosage and SNF1 mutations (e.g., the SNF1-G53R allele, which produces a hyperactive kinase) can partially restore glucose derepression in the absence of SNF4 (53, 108, 204). Since Snf4 also coimmunoprecipitates with Snf1 and is required for maximal Snf1 protein kinase activity (in cells grown on a nonfermentable carbon source) (52), it is a physically associated activator of the Snf1 kinase (108, 113). Like Glc7 (and perhaps Reg1; see below), Snf1 appears to have a regulatory role in glycogen metabolism that is somehow influenced by PKA signaling (see reference 119 for a review). While this apparent connection between the signal transduction pathways governing glucose catabolism and glycogen storage is intriguing, it remains poorly defined and will not be considered further here.
Identification of a role in glucose repression for Glc7, the yeast homolog of mammalian type I protein phosphatase (PP1) (111), began with a selection for mutants defective in glucose repression of a target gene (SUC2, encoding invertase) (260). All of the recovered mutations were recessive; two of the three complementation groups found in this screen proved to be allelic with HXK2 and REG1, respectively. The third group identified a new locus (designated cid1, for constitutive invertase derepression). Complementation of one of these mutations (cid1-226, now designated glc7-T152K) (378) led to the isolation of GLC7, a locus that was also identified independently in a screen for glycogen-deficient mutants. Interestingly, in addition to SNF1 (GLC2), other glc lesions were allelic with IRA1 (GLC1), IRA2 (GLC4), and RAS2 (GLC5) (42), consistent with numerous studies connecting Ras and Gpa2 signaling with accumulation of the storage carbohydrates trehalose and glycogen (40, 41, 67, 125, 190, 284, 330, 363). Further investigation is needed to pursue this implicit regulatory linkage between glucose catabolic pathways and carbohydrate storage.
Complicating matters further, in addition to glucose repression and carbohydrate accumulation, GLC7 phosphatase is needed for a variety of other cellular functions in S. cerevisiae (14, 350, 437), including normal progression through the G2/M phase of the cell cycle (23, 24, 150, 159, 223, 288), actin organization (6, 54), translation (412), and sporulation (12, 42, 302, 357, 378, 380). Recent work has further expanded this list of functions assigned to yeast PP1; it is also required for the glucose-induced vacuolar degradation of fructose-1,6-bisphosphatase (76) and for normal Mex67-mediated export of mRNA from the nucleus to the cytoplasm (127; see also references 381, 382, and 404).
Interestingly, isolated PP1 molecules exhibit little substrate specificity and instead require regulatory subunits that alter the conformation of the phosphatase and/or target it to its substrates (65, 97). This targeting hypothesis is supported by the Glc7 localization pattern, which suggests dynamic changes throughout the yeast cell cycle (24). Glc7 binding proteins specific to the following functions have been reported: glucose repression (Reg1) (380), glycogen metabolism (Gac1 and Pig1) (59, 353, 426), cell cycle progression (Reg2 and Sds22) (120, 149, 288), actin organization (Scd5) (54), and sporulation (Gip1) (357, 380).
A short motif present in most known regulatory subunits ([RK]-X0-1-[VI]-X-[FW]) plays a critical role in the interaction with a hydrophobic groove at the interface of two β-sheets in PP1 (97, 403). Thus, multiple regulatory subunits appear to compete for Glc7 binding in vivo; for example, Gac1 overexpression affects glucose repression (426). In addition to binding regulatory subunits, the hydrophobic groove may mediate changes in the structure and/or activity of the phosphatase. It also now seems likely that many if not all targeting subunits interact with more than one site on the PP1 surface (425). In the two-hybrid assay the F468R mutation in Reg1 dramatically reduces its interaction with Glc7; the PP1 binding motif that contains this residue (RHIHF468) therefore appears to be one of the moieties required for formation of the Reg1/Glc7 complex (325). Importantly, Reg1 is the only regulatory subunit known to participate in glucose repression.
The idea that Reg1 (first identified in reference 230 and in an earlier screen as Hex2 [101]) mediates the Glc7 role in glucose repression arose because (i) reg1 and glc7 alleles were both isolated in the screen for mutants with impaired repression of SUC2 (260, 378) (see above), (b) a combination of those defective alleles exhibited no synergy in relief from glucose repression of SUC2, (iii) SNF1 deletion is epistatic to both alleles, and (iv) Reg1 overexpression suppresses the repression defect caused by glc7-T152K (379). Genetic and biochemical experiments confirmed that Reg1 and Glc7 are linked both physically and functionally (325, 379). The results of alanine-scanning mutagenesis suggested that Reg1 interacts with at least two distinct areas of the resolved PP1 structure (14). Satisfyingly, one of the Glc7 surfaces specific to its role in glucose repression is adjacent to Thr152 (14); the glc7-T152K lesion had earlier been shown to impair Reg1-Glc7 interaction (379).
A functional link between Reg1 and Snf1 was suspected long ago, primarily because REG1 deletion results in derepression of glucose-repressed genes and SNF1 deletion results in a failure to derepress many (if not all) of those same genes. The demonstration that Reg1 and Snf1 interact (218) left little doubt that the Reg1-targeted Glc7 phosphatase and Snf4-activated Snf1 kinase are indeed interconnected components of the glucose signaling pathway (106, 120, 162, 174, 378,379; see references 45, 121, and 178 for reviews) (Fig. 3). Sincethennumerous additional participants in this regulatory duel have been discovered. For example, three ancillary components of the Snf1/Snf4 kinase complex (Sip1, Sip2, and Gal83) strengthen Snf1/Snf4 association (175, 429, 430). Each of these three proteins contains an Snf1 binding internal region and an Snf4 binding C-terminal region (their N-terminal regions are divergent); they therefore appear to bridge the interaction between Snf1 and Snf4. By analogy with the heterotrimeric mammalian Snf1 homolog, the AMP-activated protein kinase, SNF1 encodes the yeast α subunit, SNF4 encodes the γ subunit, and SIP1, SIP2, and GAL83 redundantly encode the β subunit (136).
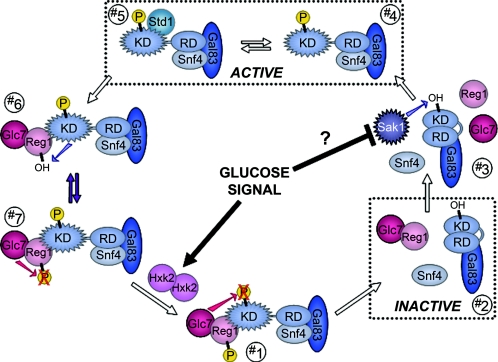
FIG. 3.
The opposing roles of Reg1/Glc7 phosphatase and the Snf1 kinase complex. The duel between Reg1/Glc7 and Snf1/Snf4/Gal83 turns the Snf1 kinase on (ACTIVE) and off (INACTIVE) by determining whether the Snf1 regulatory domain (RD) binds to and autoinhibits its kinase domain (KD). This switch plays a central role in determining the transcriptomic response to the presence or absence of glucose. The β subunit Gal83 acts as a scaffolding factor for the nuclear form of the Snf1 (α) and Snf4 (γ) subunits of the kinase. Addition of glucose to yeast cells growing on an alternative carbon source results in the dephosphorylation of active Snf1 kinase by Reg1/Glc7 (no. 1); this returns Snf1 to the autoinhibited state (no. 2) and results in export of Snf1/Gal83 to the cytoplasm (not shown). Once glucose is depleted, Sak1 phosphorylates Snf1 (no. 3), Snf1/Gal83 enters the nucleus (not shown), and Snf4 binds to the Snf1 regulatory domain, thereby releasing the catalytic domain (no. 4); Reg1 then undergoes rapid Snf1-dependent phosphorylation (no. 6), which stabilizes the interaction between Snf1 and the Reg1/Glc7 phosphatase (no. 7) and primes the latter to repress Snf1 anew should glucose reappear. Std1 interacts with the Snf1 catalytic domain and stimulates kinase activity (no. 5), although it is stoichiometrically underrepresented in Snf1 complexes. A dynamic equilibrium (purple arrows between no. 6 and 7), in which Glc7 appears to counteract Snf1 phosphorylation of Reg1 and promote its own dissociation from active kinase complexes, is at the heart of this regulatory duel. Sip5 may stabilize the Reg1/Snf1 interaction in no. 6 (not shown). Hxk2 interacts with Reg1 (not shown), and Hxk2 may respond to PKA signaling by dimerizing and mediating the switch in Glc7 phosphatase substrate selection (from Reg1 [no. 7] to Snf1 [no. 1]; see the text).
It was initially puzzling that little or no detectable phenotype resulted from mutation of all three genes that encode the β subunits of the Snf1 kinase complex (105, 106, 121, 175). However, the construction of true null alleles revealed that, as expected, the sip1Δ sip2Δ gal83Δ triple mutant does indeed display an snf1 phenotype (331). Despite the observed redundancy, the three β subunits play distinct roles in vivo, in particular with respect to Snf1 substrate specificity (143, 144, 175, 213, 254, 331, 396, 397, 430).
Besides its role in the response to glucose limitation, Snf1 has also been implicated in several other processes, including aging, thermotolerance, peroxisome biogenesis, filamentation, invasive growth, biofilm formation, meiosis, and (like Glc7) sporulation (8, 45, 137, 196, 197, 297, 401). Clearly more work is needed before we will fully understand the respective relationships between the distinct forms of the Snf1 kinase complex and each of these regulatory mechanisms. However, Gal83 is likely to be a primary contributor to glucose regulation by the Snf1 complex, since Gal83 is the most abundant β subunit, it is the only β subunit in the nucleus when cells are grown on a nonfermentable carbon source, and Snf1/Gal83 is the form responsible for most Snf1 kinase activity (144, 397).
Another trio of factors that mediate Snf1 function are the redundantly acting Snf1 kinase kinases Sak1 (YER129W, formerly known as Pak1; the new name was chosen to avoid confusion with Prk1 or p21-activated kinases), Tos1, and Elm1, each of which shares sequence similarity with the human tumor suppressor and AMP-activated protein kinase LKB1 (155, 255, 354, 422). Removal of all three yeast activities confers an snf1 phenotype. A one-to-one correspondence between these three upstream kinases and the distinct forms that contain each of the three β subunits has been ruled out (143). Since Sak1 is the most important of the trio of Snf1 kinase kinases in activating the Snf1/Gal83 form, it is likely also to be the most important regulator of Snf1-dependent glucose regulation (143). This conclusion is further supported by the observation that deletion of SAK1 suppresses many of the Snf1-dependent phenotypes observed in reg1Δ cells (255).
Like Glc7 and its regulatory subunits, the Snf1 kinase appears to be localized by its β subunits to distinct cellular subcompartments, thereby targeting it to different substrates (331, 397). In glucose-grown cells Snf1 is found exclusively in the cytoplasm; following a shift to a nonfermentable carbon source, Snf1 is targeted by Gal83 to the nucleus. The N terminus of Gal83, which is not shared by the other two β subunits Sip1 and Sip2, is required for this function; the upstream kinase Sak1 also controls Snf1 localization to the nucleus in the absence of glucose (144, 397). Upstream events in glucose signaling may also be involved, since it has been reported that PKA regulates nucleocytoplasmic shuttling of Snf1/Sip1 (144). If PKA also regulates shuttling of Snf1/Gal83, it appears to do so redundantly with another signaling pathway.
With respect to subnuclear localization, three as-yet-unconnected observations suggest the possibility that the Snf1 kinase complex operates at least in part at the nuclear periphery: Snf1 is concentrated in nuclear envelope fractions prepared from pyruvate-grown (but not glucose-grown) cells (B. Menon and G. M. Santangelo, unpublished data), Glc7 has been implicated in mRNA processing as a component of the cleavage/polyadenylation complex (127, 256, 404), and Reg1 deletion causes an RNA processing defect (381, 382). Further elaboration of this potential connection with perinuclear phenomena is needed and may provide critical information about the function and organization of the regulatory apparatus in the yeast nucleus that responds to glucose (see below).
An overview of the regulatory duel between the Reg1/Glc7 phosphatase and the Snf1 kinase complex is presented in Fig. 3. Snf1 has two domains, an N-terminal kinase domain and a C-terminal regulatory domain. In high concentrations of glucose the regulatory domain remains bound to the catalytic domain, maintaining Snf1 in an autoinhibited conformation (labeled “inactive” in Fig. 3) (325). Upon depletion of glucose, Snf4 counteracts autoinhibition of Snf1 by interacting with its regulatory domain; deletion of the latter bypasses the requirement for SNF4 in derepression of glucose-repressed genes (52, 174, 204). Snf4 binding to Snf1 therefore produces an active, open conformation of the complex (labeled “active” in Fig. 3) (174). Phosphorylation of a conserved threonine (T210) in the activation loop of Snf1 is essential for activation of the kinase; the upstream kinase Sak1 has been shown to phosphorylate this residue and is required for full Snf1 kinase activity (255). Blocking phosphorylation at this position via a T210A mutation impairs Snf4 binding to the regulatory domain and prevents the release of Snf1 autoinhibition (174, 198, 218). The T210A lesion also interferes with normal nuclear enrichment of Snf1 after a shift from high to low glucose (144). Since Snf4 localization to the nucleus is essentially carbon source independent, in glucose-grown cells nuclear Snf4 is apparently not associated with other subunits of the Snf1 kinase complex (397). Thus, Snf4 apparently does not contribute to the cytoplasmic roles of Snf1 that are distinct from its mediation of glucose derepression.
Glucose repression involves Reg1/Glc7-facilitated conversion of the kinase complex from the active to the autoinhibited state, presumably via dephosphorylation of Snf1 (325). Reg1/Glc7 appears to associate only with active kinase complexes, since the catalytic domain of Snf1 (phosphorylated at T210) is required for the interaction with Reg1 (218). Upon activation in a reg1 or glc7-T152K background, the Snf1 complex becomes trapped in the active conformation. Overexpression of Reg1, but not overexpression of the mutated form that fails to bind Glc7 (ReglF468R; see above), interferes with the Snf1/Snf4 interaction in low glucose (325). Sip5, the first protein shown to bind to both Reg1/Glc7 and the Snf1 kinase complex, may stabilize the interaction between them and thereby facilitate glucose repression, since the two-hybrid interaction between Reg1 and Snf1 is reduced threefold in a sip5Δ mutant (326).
The critical regulatory interaction between Reg1 and the Snf1 complex was presumed to occur in the nucleus, especially since that is where active Snf1 is predominantly located prior to being switched off and exported following the appearance of glucose, and Glc7 is known to be largely nuclear in growing cells (24, 437). However, in contrast with a previous report that found Reg1 in the nucleus (265), fluorescently tagged Reg1 appears to be exclusively cytoplasmic (90, 91). To resolve this discrepancy it was proposed that the activated form of Snf1 cycles rapidly between the nuclear and cytoplasmic compartments, allowing inactivation by Reg1/Glc7 to occur in the cytoplasm following the appearance of glucose (91). This hypothesis has not yet been tested, but it may explain the apparent role of Bmh1 and Bmh2 in glucose-regulated gene expression (168). Bmh1 and Bmh2 are the yeast homologs of the somewhat mysterious 14-3-3 proteins (33, 388), which may serve as mediators of Reg1 function (90) and are known to respond to Ras/PKA signaling (124, 232).
Additional information concerning the connection between upstream glucose signaling events and the Glc7/Snf1 regulatory duel will be the next topic of discussion, followed by a review of the downstream DNA-bound phosphoproteins that are known Snf1 regulatory targets and that account for much of the transcriptomic response to glucose in yeast cells.
Transmission of the Ras/cAMP-dependent glucose signal to Reg1/Glc7 and Snf1/Snf4.
Although the regulatory duel between Snf1 kinase and Glc7 phosphatase has now largely been explained at a molecular level, the input by glucose signaling that shifts the balance in favor of the closed, inactive form of the kinase remains poorly characterized. A proposal that Snf1 responds to the AMP/ATP ratio (421), which drops dramatically upon removal of glucose (7, 421), has been difficult to substantiate, since Snf1 kinase activity does not respond to these nucleotides in vitro (136). In contrast, the ample evidence that PKA and Snf1 act antagonistically in affecting a similar set of cellular functions (7, 164, 366) is reinforced by the similarity of the transcriptome-wide response of Snf1 target genes following either the addition of glucose or the expression of activated G proteins in the absence of glucose (408).
Sak1, the recently identified upstream activator of Snf1 that regulates both Snf1 kinase activity and its nuclear localization (144, 255), is an obvious potential intermediary between PKA and Snf1 (question mark in Fig. 3). If the gap between the earliest events in glucose signaling and Sak1 could be bridged, it would represent a complete chain of causality between glucose sensing at the cellular periphery and the response of DNA-bound regulators in the nucleus. Not surprisingly, however, given that its role as an Snf1 activator was only recently uncovered, little is yet known about regulation of Sak1 and whether or not the PKA pathway might be involved.
Equally mysterious is the apparent capacity of the glucose signal to change the target of Glc7 phosphatase from Reg1 to Snf1 (no. 7 and 1 in Fig. 3, respectively), which in the absence of Snf1 phosphorylation (no. 3) traps Snf1 in the autoinhibited inactive state. Interestingly, PKA signal transduction might alter Glc7 targeting through the negatively acting regulator Hxk2. As mentioned above, PKA signaling appears to cause reduced phosphorylation of Hxk2, which favors its dimeric form; the phosphatase that dephosphorylates Hxk2 in response to glucose is known to be Reg1/Glc7 (4, 305, 398). Hxk2 (presumably its underphosphorylated dimeric form) in turn interferes with dephosphorylation of Reg1 by Glc7 (325). Since only the phosphorylated form of Reg1 promotes dissociation of Snf4 from Snf1 and inactivation of the kinase, the PKA-mediated transition from monomeric to dimeric Hxk2 is potentially a direct glucose trigger that stabilizes the repressing form of Reg1/Glc7 and thereby inactivates Snf1. Reg1 and Hxk2 were reported to interact as predicted by this model (325). This is further supported by a wealth of additional data: Reg1 overexpression suppresses the glucose repression defect of an hxk2Δ mutant (325), the association between Reg1 and Glc7 is unaffected by removal of Hxk2, and the Snf1-Snf4 interaction is glucose insensitive in the absence of Hxk2 (174).
A near-complete chain of causality describing the glucose response can thus be stated as follows (Fig. 3): early events in glucose signaling stimulate PKA to inhibit phosphorylation of Hxk2, thereby targeting it to Reg1/Glc7, where Hxk2 is dephosphorylated, dimerizes, and blocks dephosphorylation of Reg1 by Glc7; Glc7 then switches targets and inactivates the Snf1 kinase, which can no longer phosphorylate its DNA-bound substrates, resulting in altered expression of the relevant target genes. Ambiguity unfortunately accompanies even this parsimonious view of the glucose regulatory cascade, since the relationship between subcellular distribution and function awaits clarification for several of the factors involved (most notably Hxk2 and Reg1).
DNA-bound repressors controlled by Snf1.
Several downstream repressors are known to be phosphorylated and inactivated by the Snf1 kinase in the absence of glucose. The first of these to be discovered was Mig1, originally identified as a multicopy inhibitor of GAL gene expression (257). Deletion of the MIG1 gene relieves glucose repression of numerous target genes (115, 131, 180, 189, 257, 258, 335, 336, 406), and its product is thought to interact with the intergenic regions of at least 90 different genes in vivo (221, 251). Mig1 is presumed to function downstream of Snf1, because deletion of MIG1 suppresses snf1 defects in glucose derepression (180, 386).
Mig1 is a Cys2His2 zinc finger protein that binds to a GC-rich core sequence (219). The consensus Mig1 binding site determined by in vitro selection is 5′-ATAAAATGCGGGGAA-3′ (221). At the time of its discovery, other zinc finger proteins were known to be capable of functioning as either activators or repressors, depending upon the chromosomal and cellular context (207). This was therefore also considered a possibility for Mig1 (257), a suggestion that later proved to be correct: both Mig1 and the related Cys2His2 zinc finger-containing repressor Mig3 (Yer028) can also function as activators (220, 221, 303, 376, 423). Although this has complicated the interpretation of Mig1 function (37, 160, 189, 376, 384, 386, 423), clearly the primary physiological role of Mig1 seems to be that of a negative regulator of glucose-repressed genes (e.g., SUC2 and GAL1).
Interestingly, the Mig1 N-terminal zinc fingers are very similar to those in the C terminus of WT1 (257), a human tumor suppressor that collaborates with p53 to repress transcription and (like p53) also activates transcription (133, 224, 235, 248, 264, 268, 306, 348). Mig1 and WT1 are both phosphoproteins, and PKA phosphorylation of WT1 appears to make an important contribution to its repressor function (323); the significance of a potential PKA target site found in the internal regulatory region of Mig1 is not yet known (257, 270).
Glucose-dependent repression by a LexA-Mig1 fusion is readily detected by a heterologous reporter gene with upstream LexA operator sites, suggesting that Mig1 can repress transcription without the assistance of other promoter-bound factors (376, 377). Analysis of LexA-Mig1 repression also led to the discovery that the Mig1 polypeptide, which has a predicted molecular mass of 55 kDa (504 residues), is phosphorylated to different extents in glucose-repressed and derepressed cells and might be a downstream target of Snf1 kinase (87, 376). Indeed, three consensus Snf1 kinase sites (Ser278, Ser311, and Ser381) map within a region of Mig1 required for glucose regulation (77, 87, 269). All three of these sites, and a fourth one that matches the Snf1 consensus imperfectly (Ser222), are phosphorylated by Snf1 in vitro (343). Mutation of these serine residues to alanine, including a Mig1 mutant in which all four residues were mutated (S222A+S278A+S311A+S381A), impaired but did not eliminate Mig1 phosphorylation and glucose repression (377). Nevertheless, in vivo phosphorylation of Mig1 is dramatically reduced in an snf1 mutant, and immunoprecipitated Snf1 (but not the kinase-dead Snf1-K84R) phosphorylates coprecipitated Mig1. Also, as expected based upon their capacity to mediate glucose repression by inhibiting Snf1 kinase activity, deletion of either REG1 or HXK2 results in Mig1 hyperphosphorylation under repressing conditions (377). Taken together, the simplest interpretation of these data is that phosphorylation of Mig1 by Snf1 is at least partly responsible for inactivation of Mig1 repressor function.
Deletions that map within an internal region containing most of the Snf1 phosphorylation sites (residues 261 to 400) converts Mig1 into a constitutive repressor that is not inactivated when glucose is depleted (270). Interestingly, deletions in Mig1 that remove all or part of the internal region from residue 261 to 400 cause it to persist in the nucleus in the absence of glucose, whereas under the same conditions wild-type Mig1 is largely exported to the cytoplasm (87). Fusion of Mig1 residues 261 to 400 to a GFP-β-galactosidase chimera confers glucose-regulated nuclear import and export upon the resulting fusion protein, and its import in the presence of glucose requires both REG1 and HXK2. In glycerol-grown cells Mig1 becomes increasingly nuclear as more Snf1 kinase sites are removed (86). Msn5, an importin β homolog originally identified due to its suppression of an snf1 mutation when overexpressed (107), interacts with Mig1 and mediates its nuclear export in response to glucose removal (86). These and other data suggest that Snf1 phosphorylation of Mig1 inactivates it by changing its subcellular localization.
According to this Mig1 nucleocytoplasmic shuttling hypothesis, Hxk2- and Reg1/Glc7-dependent inactivation of Snf1 in the presence of glucose results in hypophosphorylation of Mig1, which causes it to relocate rapidly to the nucleus; subsequent binding of Mig1 to its recognition sites upstream from glucose-regulated genes mediates transcriptional repression (86, 87). Although this model is attractive, it is difficult to reconcile with two strikingly contrasting results. First, the Mig1 target gene GAL1 is derepressed normally in an msn5 mutant grown on glycerol, despite the constitutive presence of Mig1 in the nucleus (86). Second, Mig1 remains bound to the GAL1 promoter in vivo under these same derepressing conditions (278). Both of these results suggest that Mig1 transcriptional repression of target genes is regulated via a mechanism other than nuclear export and that the key function of Snf1 phosphorylation in relieving Mig1-dependent glucose repression is to inactivate the relatively small amount of Mig1 that remains in the nucleus and is bound upstream of target genes in derepressed cells (87, 270). How this is accomplished remains obscure.
Mig3, Nrg1, and Nrg2 are three additional Snf1-regulated repressors in S. cerevisiae. Mig3 is somewhat mysterious; it was identified because it contains Cys2His2 zinc fingers that are very similar to those of Mig1 and Mig2 (26, 220). Several additional results seem to suggest that Mig3, like Mig1 but not Mig2, is an Snf1-controlled, DNA-bound glucose regulator (182, 221). It binds to the Mig1 binding sites upstream from SUC2, and a LexA-Mig3 chimera functions as a glucose-dependent repressor of reporter gene expression when bound to upstream LexA operators. When cells are shifted from glucose to galactose, Mig3 is extensively phosphorylated (at least in part by Snf1) and rapidly subjected to Snf1-dependent proteolysis (94). Nevertheless, physiologically significant regulation of target genes by Mig3 is yet to be demonstrated; in wild-type cells under standard conditions, it seems unlikely to be more than a minor regulator of glucose-responsive gene expression (220, 221). Recently the MIG3 gene was also isolated in a screen for multicopy suppressors of Rad53-GFP toxicity, but the relationship between Mig3 function and the DNA damage response awaits further clarification (94). It is hoped that transcriptomic analysis of the effects of an as-yet-untested environmental condition or genetic background will eventually reveal the Mig3 role in glucose regulation and explain the results obtained to date.
Nrg1 and Nrg2 contain Cys2His2 zinc fingers that are very similar both to each other and to those in Mig1, Mig2, and Mig3 (400). Nrg1 was first identified in two separate screens for factors mediating repression of glucose-regulated genes (1, 282), and Nrg2 was discovered because of its two-hybrid interaction with Snf1 (400). Outside of their DNA binding domains Nrg1 and Nrg2 share only 27% identity, and this is thought to account for their somewhat distinct regulatory roles in glucose repression, filamentous invasive growth, and biofilm formation (18, 196, 282, 400, 438). In the absence of glucose, neither factor is degraded, nor is their capacity to bind target sites in vivo impaired. Both Nrg1 and Nrg2 exhibit glucose-dependent repression of a heterologous reporter gene; they are also both capable of interacting with Snf1 kinase, but (unlike Mig1 and Mig3) neither appears to be phosphorylated by it (18, 400). Although Snf1 appears to modulate Nrg2 levels and is clearly required for normal Nrg1 function, the regulatory relationship between these factors is complex and in need of further study. Interestingly, like Mig1 and Mig3 (see above), Nrg1 can function as an activator in some circumstances (18).
DNA-bound activators controlled by Snf1.
The Snf1 protein kinase also regulates a number of positively acting transcription factors required for the utilization of nonfermentable carbon sources. Two of these factors, Cat8 and Sip4, both contain zinc finger DNA binding domains similar to that in Gal4 (a C6 zinc cluster) (145, 206) and bind specifically to a set of carbon source responsive elements (CSRE) under derepressing conditions (300, 395). Cat8 and Sip4 are closely related structurally and appear to have slightly different affinities for variants of the CSRE sequence (319). Although Sip4-dependent transcriptional activation has been shown to require phosphorylation of Sip4 by Snf1 kinase, removal of Sip4 has little or no effect on CSRE-regulated genes (141, 145, 206, 299). Also, unlike deletion of CAT8, which causes defective growth on nonfermentable carbon sources, deletion of SIP4 results in no obvious phenotype (206).
Cat8 regulates the expression of a wide array of genes involved in the metabolism of nonfermentable carbon sources, including genes necessary for ethanol utilization (e.g., ACS1 and ACR1/YJR095W) (28, 192), gluconeogenesis (141, 145), the glyoxylate cycle (141), lactate utilization (JEN1) (27), and isocitrate metabolism (IDP2) (27); Cat8 also regulates expression of SIP4 (141, 395). Cat8 activity is regulated on two levels: its expression is Mig1 repressed (145, 257), and its capacity to activate transcription requires a functional Snf1 kinase (303). Snf1 phosphorylates Cat8 in ethanol-grown cells (56, 303); following the addition of glucose, it is dephosphorylated in a Glc7-independent manner (303). Interestingly, constitutively expressed Cat8 fused to the Ino2 activation domain bypasses the need for a functional Snf1 in cells growing on ethanol (300).
Cat8 works together with another Snf1-regulated transcriptional activator, Adr1, for maximal expression of a small group of shared target genes (including ADH2, ACS1, and ALD6) under derepressing conditions (25, 98, 192, 356, 405, 432). Like the Mig1, Mig3, Nrg1, and Nrg2 repressors, Adr1 has a Cys2His2-type DNA binding domain (283), and its function requires Snf1 kinase (84), although the mechanism of Adr1 regulation remains unclear. It has been suggested that Snf1 and Reg1/Glc7 contribute to regulation of Adr1 by affecting its binding to promoters of target genes. Snf1 promotes Adr1 binding under derepressing conditions, while Reg1/Glc7 inhibits Adr1 binding to promoters in the presence of glucose (91, 433); it is currently unknown whether Adr1 is a direct target of Snf1 and Reg1/Glc7 or whether this effect occurs through intermediary factors. Adr1 activity has also been reported to be negatively regulated by PKA (60) in a manner independent of its DNA binding (364).
SIGNAL RECEPTION: GLUCOSE INDUCTION
The connection between PKA function and up-regulation of the ∼1,000 glucose-induced genes is even more mysterious than the mechanisms discussed above that control the ∼1,000 genes whose expression is repressed in the presence of glucose. Our clearest picture is of glucose induction by the Snf3/Rgt2 sensing mechanism, a pathway that appears to operate in a largely PKA-independent fashion and (at minimum) contributes the critical regulatory trim that allows S. cerevisiae to grow well on a broad range of glucose concentrations (from micromolar to molar concentrations) (272). The integral plasma membrane proteins Snf3 and Rgt2 mediate both transcriptional repression and derepression of four of the genes that encode glucose transporters (HXT1 to HXT4). This pathway establishes a regulatory link between glucose concentration and expression of the transporters, and it operates in a largely Snf1-independent fashion (for reviews, see references 179 and 272). Snf3 and Rgt2 are putative glucose sensors that appear to act redundantly with the Ras2/Gpa2/PKA pathway, since the latter seems sufficient for glucose signaling through both the Snf1-dependent and Snf1-independent branches (408) (see above). Interestingly, however, like activating mutations in RAS2 and GPA2, at least part of the glucose signal can be at least partially mimicked in the absence of glucose via dominant mutations in SNF3 or RGT2 (228, 272, 273). This may mean that the Snf3/Rgt2 regulatory pathway, in addition to modulating expression of key glucose transporters, signals PKA or a PKA-independent glucose response pathway and thus augments or tailors the transcriptomic response beyond its well-established effect on transporter genes.
Snf3/Rgt2 Signaling of the Rgt1 Repressor Complex
Upon transmission to the cytoplasmic side of the plasma membrane, the Snf3/Rgt2-mediated glucose signal appears to operate through YCK1 and its 77% identical paralog, YCK2, an essential gene pair encoding casein kinase (Fig. 4) (310). Like Ras (see above), Yck1 and Yck2 are tethered to the membrane via palmitate moieties attached to C-terminal Cys-Cys sequences (9, 318). Membrane anchoring of the Yck proteins is thought to facilitate an interaction with Snf3 and Rgt2 that activates the kinases following the glucose-induced conformation change in the sensors; Yck1 has been shown to interact with Rgt2 in vivo (179, 247). The targets of activated Yck kinases in this pathway are Std1 and Mth1 (247), the corepressors that interact with the Rgt1 repressor; the latter is a C6 zinc cluster protein that binds directly to DNA sites upstream from target genes (122, 163, 202, 272, 289, 332). Mutations in Rgt1 restore HXT gene expression and were originally isolated via suppression of defective glucose regulation in the snf3Δ and grr1Δ backgrounds (106, 227). Note that the Rgt1 repressor blocks transcription of glucose-induced genes, unlike the Mig and Nrg repressors, which block transcription of glucose-repressed genes (see above). Thus, the Mig and Nrg proteins act as repressors only in the presence of glucose, and Rgt1 acts as a repressor only when glucose is absent (Fig. 4).
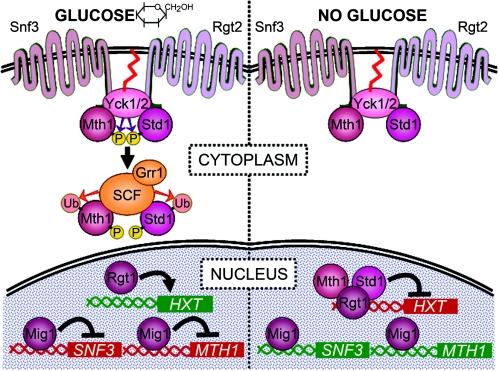
FIG. 4.
The Snf3/Rgt2 signaling pathway. Transcription of hexose transporter genes (HXT) is repressed by Rgt1 in the absence of glucose; in contrast, Mig1 represses transcription of its target genes in the presence of glucose and cross-regulates the Snf3/Rgt2 pathway as shown. Relief from the repressor function of Rgt1 occurs via glucose signaling of the Snf3/Rgt2 sensors, which stimulate phosphorylation of Mth1 and Std1 by Yck kinases; the C-terminal tails of Snf3 and Rgt2 also appear to facilitate this event by interacting with both the kinases and their substrates. The Yck polypeptides are tethered to the plasma membrane via palmitate (red squiggles). Yck-phosphorylated Mth1 and Std1 are subjected to SCFGrr1-mediated ubiquitination and degradation by the proteasome. This releases Rgt1 from its upstream DNA binding sites and results in derepression of downstream HXT target genes. Removal of Mth1 and Std1 converts Rgt1 into a transcriptional activator that may stimulate HXT expression in the absence of specific DNA binding activity.
The large (>200-residue) cytoplasmic C-terminal tails of Snf3 and Rgt2 are an important feature of this signaling pathway. Deletion of the tail domains impairs Snf3- and Rgt2-dependent gene regulation in response to glucose, and fusing the tail to Hxt1 or Hxt2 complements those defects, as does expression of the Snf3 tail domain by itself (69, 89, 228, 247, 272, 274). Although they were originally thought to play an important role in receiving the glucose signal, recent evidence suggests instead that a dynamic interaction with Mth1 and Std1 (201, 332) is the important function of the Snf3 and Rgt2 C-terminal tails in glucose signaling (247).
The membrane anchoring of Mth1 and Std1 by the cytoplasmic tails is thought to mediate phosphorylation of these Rgt1 corepressors by Snf3/Rgt2-associated Yck kinases (Fig. 4) (247). Both Std1 and Mth1 can be phosphorylated by affinity-purified Yck1 in vitro, and membrane fractions of crude extracts yield maximal Mth1 phosphorylation; mutant forms of Yck kinase were used to demonstrate that the production of phosphoforms of these regulators is Yck dependent (247). The likely site of Yck phosphorylation is a region conserved in both Std1 (residues 129 to 147) and Mth1 (residues 118 to 136), each of which contains several matches with the consensus target sequence of casein kinase I (SXXS) (247).
Once Std1 and Mth1 are phosphorylated by the Yck kinases, they are degraded via a Grr1-dependent mechanism (Fig. 4) (117, 179, 247). Mutations in GRR1 were shown long ago to impair the regulatory response to glucose (11) and have since been associated with a broad array of phenotypes, including cell elongation, decreased divalent cation transport, defects in sporulation, and slow growth or lethality in combination with defective amino acid biosynthetic pathways (21, 114, 158, 215, 427, 431).
Interestingly, the F-box motif of Grr1, which mediates its interaction with Skp1 (187), may explain the pleiotropic phenotype of grr1Δ cells (10, 209). Skp1 is a component of several structurally distinct but functionally related ubiquitin ligase complexes that target a wide variety of regulators to the 26S proteasome for degradation (for a review, see reference 414). These complexes are termed SCF to indicate their two common components, Skp1 and the cullin Cdc53, and a third exchangeable component, the F-box protein. In S. cerevisiae a total of 21 F-box proteins have been identified by sequence alignment (414). Further analysis is urgently needed, since most of these factors have neither been verified as SCF components nor characterized as to their function, and each bona fide SCF subunit is expected to confer a distinct substrate specificity upon its respective complex. Like Grr1, any one of these F-box proteins could contribute to nutrient signaling by targeting kinase substrates in response to the appearance or disappearance of glucose.
Two F-box proteins that target cell cycle regulators for degradation were first identified as a result of their important roles in cell cycle progression: Cdc4, which targets the cyclin-dependent kinase inhibitor Sic1, and Grr1, which (long after its discovery as a regulator of the glucose response) was found to target the G1 cyclins Cln1 and Cln2 (16, 110, 158, 209, 341, 393, 414). Grr1 was shown subsequently to also target Gic2, a Cdc42 effector that mediates actin polarization at bud emergence (170, 414). A breakthrough in understanding the initially puzzling GRR1 contribution to the glucose response (11, 209, 385, 387) came with the discovery that SCFGrr1, in addition to targeting the aforementioned cell cycle regulators, mediates the ubiquitination and degradation of the Rgt1 corepressors Std1 and Mth1 (117, 209, 347).
Grr1-mediated ubiquitination and degradation of Mth1 and Std1 appear to require, in addition to the Skp1 interaction with the Grr1 F box, an LRR domain comprised of 12 leucine-rich repeats that form the substrate-targeting domain in Grr1 (158, 187). This is consistent with our knowledge of structure-function relationships in other F-box proteins, which invariably possess a second motif that mediates substrate targeting (414). For example, in Cdc4 a WD40 repeat domain is sufficient for high-affinity interaction with its substrates (253).
Degradation of Mth1 in glucose-grown cells has been shown to require both phosphorylation by Yck kinase (247) and the LRR domain of Grr1, although Grr1 residues identified as essential for Cln2 interaction appear to be distinct from those required for interaction with Mth1 (117, 158, 347). No information is yet available regarding the structural requirements of the Grr1-Std1 interaction; this will likely be more difficult to determine, since Std1 apparently does not disappear as completely as Mth1 in the presence of glucose (182).
In the absence of glucose Rgt1 binds to a consensus sequence (5′-CGGANNA-3′) (186) found in multiple copies upstream of the HXT target genes (250, 275). Transcriptional repression by Rgt1 requires Mth1, which causes a conformation change that allows Rgt1 to bind to its recognition sites in DNA (117, 202, 289). Std1 also interacts with Rgt1 in the absence of glucose (289, 372), although it does not regulate the Rgt1 DNA binding activity (179). More work is needed to determine the contribution of Std1 to Rgt1-mediated repression in the pres-ence of glucose; an interesting possibility is that its interaction with Rgt1 shifts the activator/repressor equilibrium (see below) in favor of repressor function (289). Elimination of both Mth1 and Std1 appears to be required for maximal derepression of Rgt1 target genes (202).
The central events in the Snf3/Rgt2 pathway, from glucose signal generation at the plasma membrane to signal reception by a DNA-bound repressor (179, 247), are depicted in Fig. 4. As identified by global profiling (182), Snf3/Rgt2 signaling also initiates feedback and feedforward regulatory loops that are intertwined with the Snf1-dependent pathway. The Snf3/Rgt2 contribution to these regulatory loops involve three zinc finger-containing repressors, Mig2, Mig3, and Rgt1, as well as the Rgt1 corepressors Std1 and Mth1. For example, glucose signaling through Snf3/Rgt2 induces expression of the MIG2-encoded repressor, and in the absence of glucose Std1 binds to and activates Snf1 kinase (Fig. 3) (see above). The Snf1-Mig1 pathway in turn represses expression of Snf3 and Mth1. Importantly, despite these links to Snf1 kinase activity, signaling by Snf3/Rgt2 does appear to operate as an independent glucose response pathway (Fig. 4) (179, 182, 228, 272, 273). Thus, Snf3 and Rgt2 initiate a component of the glucose response that overlaps at least partially with both the PKA pathway and its Snf1 kinase branch but that can also operate independently of either.
An important remaining aspect of this glucose regulatory pathway that awaits clarification is the apparent capacity of Rgt1, under conditions (high glucose concentration) in which it no longer interacts with its recognition sites in DNA, to reverse roles and function as an activator of HXT transcription and of a heterologous reporter gene (186, 250, 271, 275) (however, see also reference 117). Although its functional significance remains unclear, transcriptional activation by Rgt1 requires the glucose sensors Snf3 and Rgt2 and the F-box protein Grr1 (275).
The Cell Cycle Connection
Eukaryotic cells grow and divide at maximal rates only when glucose is freely available. This requires sophisticated regulatory coordination between the metabolic engine of growth and the wide variety of structural events required for cellular reduplication. With respect to glucose signaling it has long seemed likely that there is a connection with the essential cyclin-dependent kinase Cdc28; the latter acts in concert with the G1 cyclins Cln1 to -3 at the primary gating event (Start) in the cell division cycle (3, 116, 132, 165, 262, 263, 276, 285, 286, 321, 371, 416, 424; see references 83, 365, and 389 for reviews). Unfortunately many of the critical relationships between the growth and cell cycles have resisted elucidation. The slow progress on this front is evident when one considers that Hartwell first codified the basic questions over 30 years ago (138): how does macromolecular synthesis communicate with the CDC28 gene product, and how are all of these diverse inputs (e.g., the pathways for carbon, nitrogen, sulfur, phosphorus, and potassium assimilation) integrated at Start?
Two mechanisms of apparent coordination between glucose signaling and cell cycle progression seem particularly worthy of further attention. The first involves Grr1, the aforementioned F-box protein that both relieves glucose repression as a component of the Snf3/Rgt2 pathway and regulates Cln-dependent Cdc28 function (209). Little is yet known about how Grr1 might integrate glucose signaling with cell cycle regulation, but it now seems clear that it responds directly to glucose sensing by Snf3 and Rgt2 at the plasma membrane. Second is the PKA pathway, which has long been considered a general regulator of cell growth in any carbon source and as such is thought to participate in the dramatic increase in the rate of cell division upon the appearance of fermentable sugars, including glucose. As discussed above, many components of the PKA pathway that were originally identified in connection with their role in glucose signaling have now been thoroughly characterized.
Progress has also been made in understanding the ultimate recipients of the glucose response in the yeast nucleus; PKA signaling increases transcription of Rap1 target genes and appears to operate through the newly identified Rap1 coregulators Fhl1/Ifh1 as well as the Rap1/Gcr1/Gcr2 transcriptional activation assemblage (81, 83, 188, 320, 328, 402, 416). Target genes activated by Rap1 fall into two main groups, glycolytic and ribosomal protein (RP) genes (211). Not surprisingly, these are the genes most abundantly transcribed by RNA polymerase II in S. cerevisiae cultures growing rapidly on glucose; 26 of the 30 most highly expressed transcription units are in one of these two groups (392). Overall the Rap1 activation mechanism(s) accounts for 60 to 75% of RNA polymerase II transcription in rapidly growing cells (324, 410).
Transcriptional Induction of Glycolytic and Ribosomal Protein Genes
Rap1 is an essential DNA binding protein required for both transcriptional repression and activation of target genes (128, 339); interestingly, it also binds to telomeres and negatively regulates telomere length (for a review, see reference 344). The promoters of most genes encoding glycolytic enzymes or ribosomal proteins contain sites occupied by Rap1 in vivo (211). The Rap1 site appears to mediate the response of these genes to the appearance or disappearance of glucose; for example, their expression is coordinately down-regulated upon depletion of glucose at the diauxic shift (85). The PKA and TOR signaling pathways are thought to regulate this response of Rap1-activated genes to nutritional status (328, 402, 410, 441).
Recent reports suggest that the PKA and TOR pathways intersect with respect to their shared role in stimulating RP expression. The transcription factors Sfp1, Fhl1, Ifh1, and Crf1 appear to be key downstream targets of this signaling in the yeast nucleus, and various unified models suggest how PKA and TOR signaling might intersect to regulate cell cycle progression by controlling the rate of ribosome production (181, 226, 292). Although additional work is needed to clarify the roles of these newly identified transcriptional regulators, together they appear to comprise the downstream targets of nutrient sensing through the TOR pathway and thus are the most likely recipients of signaling by the anticancer agent rapamycin. In contrast, the Rap1 coactivator Gcr1 appears to respond only to PKA; gcr1Δ cells exhibit no alteration in rapamycin sensitivity or rapamycin-induced down-regulation of RP gene expression (S. J. Deminoff, K. A. Willis, and G. M. Santangelo, unpublished data). Like Fhl1/Ifh1, Gcr1 makes an important contribution to nutrient sensing, since its removal causes a long delay in the glucose response (as measured by upshift of RP gene transcription, polysome formation, translation rate, and growth rate [416; see below]).
Transcriptional activation by Rap1 was shown long ago to exhibit sensitivity to disruption of GCR1 (81, 324, 435). Genome-wide analysis has now demonstrated for the entire genome that genes with a Rap1 binding site in their promoter are also Gcr1 target genes (81, 211, 324, 375, 435). A wealth of additional evidence suggests that the Rap1/Gcr1 assemblage participates directly in an activation mechanism shared by this common set of target genes. First, Gcr1 activation of glycolytic and RP genes is eliminated by point mutations or small deletions that eliminate the Rap1 binding site(s) in the corresponding promoters (374, 375). Thus, the capacity of Gcr1 to stimulate transcription of most if not all of its target genes is entirely dependent upon Rap1 binding upstream of those genes. Second, mutational inactivation of leucine zipper-mediated Gcr1 homodimerization eliminates both the GCR1 contribution to activation of Rap1 target genes and complementation of the gcr1Δ phenotype (81, 82). Indeed, loss of Gcr1 function and reduced transcriptional activation of Rap1 target genes exhibit a tight correlation across an entire spectrum of lesions that cause mild to severe impairment. Finally, fluorescently tagged Rap1 and Gcr1 colocalize to foci in the yeast nucleus in live cells (236), and epitope-tagged Rap1 and Gcr1 coimmunoprecipitate (375) and can be detected in a large (>1-MDa) complex by size affinity chromatography; this Rap1 complex exhibits faster migration in the absence of Gcr1, indicating that it is not an aggregation artifact (H. Zhou and G. M. Santangelo, unpublished data).
Despite these data and other experimental support for the hypothesis that Rap1 and Gcr1 collaborate directly to stimulate transcription of both glycolytic and RP genes, it has been difficult to rule out a competing idea, that the effect of GCR1 deletion on RP gene expression is indirect. This alternative hypothesis is based on the observation of coordinate regulation of the 138 RP genes in S. cerevisiae. Coordinate regulation links transcription of RP and other translational component genes to the growth rate of yeast cells (93, 185, 409). The idea that removal of Gcr1 indirectly causes a drop in RP transcription involves the following reasoning: the reduction in glycolytic gene transcription, which is a direct result of GCR1 deletion, causes a severe defect (slow growth relative to wild-type cells); this low growth rate in turn causes reduced RP gene transcription via a Gcr1-independent mechanism of coordinate regulation. According to this hypothesis, the rationale for assigning the “sick phenotype” label to the gcr1Δ mutant is the supposition that a bottleneck in energy generation takes place: reduced glycolytic flux in glucose-grown gcr1Δ cells impairs anaerobic metabolism, while glucose repression of aerobic metabolism remains intact. This unfortunate combination would leave cells with no good way to generate energy, thereby explaining the slow growth and indirect reduction in RP transcription.
Three independent lines of evidence demonstrate that this alternative hypothesis is inviable. First, it is now clear that glucose repression is not intact in gcr1Δ cells (327, 383). Second, analysis of strains that harbor partially functional GCR1 alleles identified several that grow and ferment glucose almost as well as the wild type but nevertheless exhibit RP transcription as defective as that observed in gcr1Δ cells (81). Third, the gcr1Δ defect in RP transcription is equivalent in all fermentable carbon sources tested thus far (glucose, sucrose, raffinose, and galactose), which, significantly (see above), includes nonrepressing sugars and pyruvate, a nonfermentable carbon source (Fig. 5). Together these data eliminate the key underlying assumption of the alternative hypothesis, i.e., that in the absence of Gcr1, glucose repression and/or slow growth causes an indirect effect on RP expression.
FIG. 5.
Transcriptomic analysis of defective gene expression in gcr1Δ cells. Microarray hybridization was done with RNA from isogenic gcr1Δ versus wild-type cultures grown to mid-logarithmic phase (a density of 1 × 106 cells/ml) on each of the carbon sources indicated (2% glucose, 3% sucrose, or 3% pyruvate). Total RNA was extracted (408) and labeled with either Cy3-CTP or Cy5-CTP by using a low-RNA-input fluorescent linear amplification kit (Agilent Technologies, Palo Alto, CA). Labeled RNA was purified with the RNeasy MinElute kit (QIAGEN) and hybridized to yeast 60-mer oligonucleotide arrays (Agilent Technologies, Palo Alto, CA) according to the manufacturer's recommendations. Array slides were scanned at 10-μm resolution with two-line averaging by using an Axon GenePix 4200A scanner and GenePix 6.0 software. Hierarchical clustering was done with Acuity 4.0, with a centered Pearson similarity metric. The data shown here represent an average of two replicate arrays, including a dye-swap control. The scale shows color intensity proportional to the log ratio of expression in gcr1Δ cells (relative to the corresponding wild-type values). The cluster diagram shows the results for the 192 genes (11 glycolytic, 110 ribosomal protein, 4 translational component, and 67 other genes) that, in all three carbon sources, exhibit at least a twofold decrease in expression in gcr1Δ cells.
Therefore, especially when combined with new insights concerning the shared participation of Rap1, Gcr1, and Gcr2 in perinuclear gene regulation (see below), it is now apparent that these global regulators work together to stimulate transcription of both glycolytic and RP genes, the two largest and most abundantly transcribed classes of glucose-induced target genes in yeast cells. Much work remains in deciphering the physical organization and relative contributions of the Rap1/Fhl1/Ifh1 and Rap1/Gcr1/Gcr2 assemblages and how they are signaled by the PKA and TOR pathways in response to the appearance of glucose. Significantly in this regard, the consequences of removing Gcr1 include a dramatic drop in translation rate, a decrease in CLN transcription, a cln3Δ-like increase in cell size, and conditional cell cycle arrest and elongated morphology in the cln3Δ and cln1Δ cln2Δ backgrounds (83, 416). An important topic for future investigation is whether (and to what extent) the respective Sfp1/Fhl1/Ifh1 and Gcr1/Gcr2 contributions to Rap1 transcriptional activation and cell cycle control are coordinated.
The Repressor/Activator Theme in Glucose Regulation
Although some information has been gleaned regarding the all-important events downstream of the transcriptional regulatory factors that mediate repression and derepression of glucose-responsive target genes, a more than rudimentary understanding of the central mechanism(s) of gene regulation in S. cerevisiae and other eukaryotes has thus far eluded our grasp. Like Mig1, Mig3, Nrg1, and Rgt1, many of the factors that bind DNA either directly or indirectly are capable of both activating and repressing transcription. This puzzling property of glucose repressors is also true of positively acting regulators such as Rap1 and Gcr1, as well as RNA polymerase II holoenzyme components such as Med1, Ssn8, Sin4, and Rgr1 (15, 58, 210, 242, 346).
How a distinction is made between these diametrically opposite activities of activation and repression based on chromosomal and/or environmental context, as well as the molecular mechanisms that execute this decision, awaits clarification. For example, an unambiguous finding regarding the mechanism of glucose repression in yeast cells is that the Ssn6/Tup1 corepressor complex plays an important role downstream of most of the factors that bind DNA directly, such as the MIG-, NRG-, and RGT1-encoded repressors. However, upon derepression both Mig1 and Ssn6/Tup1 remain associated with glucose-repressed promoters (29, 278, 279). Further, even Ssn6 appears to be capable of stimulating transcription under some circumstances (68). Thus, most if not all of the relevant negatively acting regulators remain associated with target promoters but are somehow inactivated (or their activities are reversed) under derepressing conditions. This is a much more subtle regulatory mechanism, and it operates on a much more static protein-DNA complex, than was previously believed to exist. The final sections of this review will describe a new paradigm for eukaryotic gene activation and regulation that may provide a superior context in which to understand these and other interesting new observations about glucose regulation in the yeast nucleus.
Reverse Recruitment: Rap1/Gcr1 Activation at the Nuclear Periphery
Beyond its interesting regulatory role in yeast cell physiology, a second mystery has shrouded the Rap1/Gcr1 mechanism of transcriptional activation: it conforms poorly with the recruitment paradigm of gene regulation (36, 70, 295, 296, 352, 434). For example, when a Rap1 binding site is placed upstream from heterologous reporter genes, or when the activity of Rap1-Gal4 or Rap1-LexA fusion proteins is measured, Rap1 activation is surprisingly weak (in some contexts it is undetectable) (38, 88, 349, 415). In contrast, mutational inactivation of Rap1 binding sites in the very strong glycolytic promoters typically results in a 10- to 20-fold decrease in transcription, suggesting that Rap1 is a powerful activator (see, e.g., reference 374). This discrepancy, with identical Rap1 binding elements yielding very different results depending on whether they are in their native location or upstream from a reporter, is paradoxical because no yeast protein other than Rap1 binds to its recognition motif in DNA. Moreover, weak Rap1 activation of reporter genes is not due to a structural perturbation resulting from fusion to a heterologous DNA binding domain; Rap1-LexA fusion proteins that can fully complement a deletion of endogenous RAP1 show little or no heterologous activity as transcriptional activators. This was originally thought to indicate a requirement for additional DNA-bound proteins (such as chromatin-remodeling factors) for full activation by Rap1 and its coactivators, but to date no arrangement of DNA motifs has been described that allows an isolated Rap1 site to promote transcription as efficiently as its native counterparts in glycolytic promoters.
An alternative explanation, which remains untested, is that cooperativity resulting from coupling of the various steps in transcription (i.e., activation, elongation, termination, processing, and export of the final products to the cytoplasm [225]) explains the impressive strength of Rap1 activation when the protein is bound at its native sites. Inclusion of mRNA export in such a holistic mechanism of gene expression implies a functional and/or physical connection with nuclear pores. This “gene expression machine” (GEM) explanation for the discrepancy in Rap1 activation efficiency is plausible because a pas de deux between chromatin and GEMs, an arrangement that could have evolved to optimize expression of naturally occurring promoter-open reading frame combinations whose products are in high demand, would be difficult to reproduce with a heterologous gene. (A similar discrepancy has been observed for transcriptional repression by Ssn6/Tup1, which is much stronger in natural promoters than in artificial constructs [194, 203]). Since most GEMs in glucose-grown cells appear tobe occupied with Rap1-regulated transcription units (see above) (392, 416), this would explain why Rap1 activation (and perhaps all glucose-regulated transcription) is unusually difficult to reproduce via the ubiquitous heterologous reporter assay. An important corollary of this idea is that an accurate working model for gene regulation will require detailed mapping of nuclear architecture (169, 200, 238-240, 280, 281). Interestingly, the “gene gating” hypothesis proposed by Günther Blobel in 1985 (22) may prove to have been a particularly perceptive early suggestion for how three-dimensional structure in the nucleus is used to mediate (and regulate) gene expression.
A second prominent aspect of the Rap1 activation mechanism that conforms poorly with the recruitment model of gene activation is the nature of the Gcr1 activation domain. This domain is irreducibly large (280 residues) and can be subdivided into at least four distinct hypomutable regions (82, 83); two of these hypomutable regions coincide with predicted transmembrane domains (236), and transmembrane-disrupting lesions in these domains are known to inactivate both GCR1 function and Gcr1-LexA activation of a heterologous reporter gene (82). If, as predicted by the recruitment model, these transmembrane motifs are required for incorporation of Gcr1 into its activation complex, it should have been possible to make compensatory extragenic lesions that would suppress at least one of the inactivating mutations. Exhaustive screens for such suppressing lesions by using either irradiation or chemical mutagens have failed. Further, every mutation in this region that inactivates GCR1 function also eliminates Gcr1-LexA activation of a LexA-driven reporter gene. Since Gcr1-dependent transcriptional activation of target genes is eliminated by removal of the Rap1 binding site in the corresponding promoters, this was originally interpreted to mean that contact between Rap1 and Gcr1 is a requirement for the latter to recruit the transcriptional machinery. However, targeted mutagenesis of RAP1 has been equally unsuccessful in generating detectable suppression of lesions in Gcr1 that impair activation. Furthermore, activation of a heterologous reporter by Gcr1 persists long after the disappearance of Rap1 in RAP1ts strains. Together these data suggest that when Gcr1 is bound upstream from a reporter gene, its Rap1 interaction domain is required but (paradoxically) Rap1 itself is not. The large body of existing data regarding the Gcr2 contribution to the Rap1/Gcr1 activation mechanism has likewise been difficult to reconcile with the recruitment paradigm (81, 435). Thus, all attempts to resist the straightforward interpretation of an activation domain that overlaps with transmembrane domains, i.e., that Gcr1 association with the nuclear membrane is an important feature of its transcriptional activation function, have failed.
A recent genome-wide genetic screen (synthetic genetic array analysis; see above) of gcr1Δ and gcr2Δ query strains yielded a surprising result that has finally provided an improved context for resolving these disparities and elucidating the mechanism of Rap1/Gcr1 transcriptional activation. Both GCR1 and GCR2 were found to be members of a large genetic network of nucleoporin genes (236). More specifically, deletion of each nonessential gene encoding a component of the Nup84 subcomplex exhibits a synthetic genetic defect in combination with GCR1 deletion. Follow-up biochemical, cell biological, and gene expression analyses confirmed that Rap1/Gcr1 activation occurs at the nuclear periphery and is impaired by disruption of genes encoding components of the Nup84 subcomplex. For example, Rap1 and Gcr1 are 100% and 71% as enriched in nuclear envelope fractions as the integral nuclear membrane protein Pom152, unlike control proteins such as the nucleolar factor Nop1 (<1% enriched) (236). Importantly, perinuclear localization of Rap1/Gcr1 target genes has been observed via chromatin immunoprecipitation of nuclear pore-associated genes (48), which provides independent support for the conclusion that there is a physical and functional link between the Rap1/Gcr1 assemblage and the nuclear rim.
Most surprisingly, not only do Rap1 and its coactivators Gcr1 and Gcr2 activate transcription at the nuclear periphery, but many yeast nucleoporins function as transcriptional activators; two of these were shown to do so at their normal perinuclear location (236). Transcriptional activation by nu-cleoporins is severely impaired by GCR1 deletion, suggesting that disruption of the Rap1 activation assemblage interferes with the normal organization of GEMs at the yeast nuclear periphery. This result may recapitulate a phenomenon relevant to human cancer. Perinuclear gene activation appears to be oncogenic in acute myeloid leukemia and related syndromes in humans, in which chromosomal rearrangements fuse Hox DNA binding domains to the nucleoporin hNup98 (126, 183). Interestingly, human Nup98 also activates transcription in yeast cells, and its ability to do so is impaired in the absence of Gcr1 (236). Integral GEM components, including several Mediator subunits (Ssn8, Sin4, and Med1) and Rpo21, also associate with the nuclear periphery and participate in perinuclear activation.
These data indicate the existence of an unusually intimate relationship between the Rap1/Gcr1 assemblage and GEM components, a conclusion that is further suggested by protein sequence similarity searches. For example, PSI-BLAST analysis of Gcr1 identified homology with the RNA polymerase II large subunit in the free-living protist Mastigamoeba invertens (Fig. 6) (351). The highly significant E value of this pairing is 2.0 × 10−30 (marginal significance is attained at an E value of 0.05 [5]). M. invertens and other primitively amitochondriate members of the order Pelobiontida may represent the earliest eukaryotic lineages (351). Thus, the RNA polymerase II large-subunit gene in M. invertens could represent a composite precursor of both Rpo21 and Gcr1 (Fig. 6). This potential primordial connection between nuclear pores and Rap1/Gcr1/Gcr2 function may help to explain the lack of evolutionary conservation of the latter; divergence of the nucleoporin Nup96 has been shown to cause hybrid inviability in Drosophila and may therefore contribute to speciation (293).
FIG. 6.
Both Gcr1 and Rpo21 are related to the RNA polymerase II large-subunit gene from M. invertens. The N terminus and carboxy-terminal domain (CTD) of Rpo21 exhibit strong homology with their M. invertens counterparts, whereas the internal region of the M. invertens protein exhibits weaker (although highly significant) homology with both Rpo21 and Gcr1. The transmembrane domains of Gcr1 (TM1 and TM2), as well as its primary homodimerization domain (1LZ), all of which make a critical contribution to Gcr1 function (see the text), exhibit >80% similarity with the M. invertens RNA polymerase large subunit, as indicated.
These new data begin to place the Rap1/Gcr1 activation assemblage within its proper architectural context in the yeast nucleus and (at least where this unusual regulatory complex is concerned) suggest an interesting alternative to the recruitment model of transcriptional activation. We have termed this alternative paradigm “reverse recruitment” to highlight its central feature, that Rap1/Gcr1 target genes are recruited to perinuclear GEMs; the term references the opposite presumption of the recruitment model, that the requisite factors are drawn to (or assembled on) the relevant promoters. Importantly, although this mechanism was uncovered through thediscovery of a functional connection between the Rap1/Gcr1/Gcr2 activation assemblage and GEMs at the nuclear rim, the reverse recruitment model does not imply that all activation occurs at the nuclear periphery. Indeed, promyelocytic leukemia protein bodies in the nuclear lumina of mammalian cells (61, 290, 399) could represent nodes of GEM organization that parallel what we have observed to occur at nuclear pores in yeast cells. In this regard, the reverse recruitment idea is consistent with an earlier proposal by Hutchison and Weintraub that active eukaryotic genes are localized along interchromatin channels that communicate with the nuclear periphery (167). Thus, the central feature of the reverse recruitment idea is that genes become transcriptionally active by docking with compartmentalized GEMs. Although they are relatively abundant at the periphery, it seems likely that at least some GEMs are luminal; these internal GEMs may nevertheless maintain functional and/or physical communication with their perinuclear counterparts.
The reverse recruitment idea explains much or all of the data that seemed paradoxical when attempting to fit them within the recruitment model. It presumes that Rap1 and Gcr1 are separate cogs in a large and multifaceted GEM and that the site of Gcr1 attachment to this megalith is an extensive hydrophobic interaction with either the Nup84 subcomplex, an adjacent site on the nuclear envelope itself, or both (Fig. 7). Thus, there may be two types of reverse recruitment in S. cerevisiae, i.e., transient occupation of GEMs by tightly regulated (and often weakly expressed) genes and constitutive occupation by strongly expressed genes that bind directly to a GEM component; the Rap1/Gcr1 mechanism is in the latter category. According to this idea, regulation of Rap1/Gcr1 target genes, which clearly does occur (e.g., upshift upon the appearance of glucose), might involve a novel type of regulatory control: alteration of the functional or physical relationship between perinuclear GEMs and the Rap1/Gcr1 assemblage.
FIG. 7.
Perinuclear transcriptional activation of Rap1/Gcr1 target genes via reverse recruitment. Known components of the Rap1 activation (Gcr1/Gcr2) or silencing (Sir complex) assemblages are shown. Essential nucleoporins are indicated with an asterisk. Dashed lines highlight presumptive perinuclear tethering interactions; nuclear pore complex-associated factors shown to interact with Gcr1 are underlined. The representation shown here is not intended to rule out the existence of a unified complex that can switch between activation and repression of transcription. (Reprinted from reference 236 with permission of the publisher. Copyright 2005 National Academy of Sciences, U.S.A.)
GENE REGULATION VIA REVERSE RECRUITMENT
A New Model for Eukaryotic Transcriptional Activation and Repression
As described above, the existing data suggest that reverse recruitment accounts for the bulk of transcriptional activation in growing yeast cells. What about more conventional regulatory systems: does reverse recruitment explain all types of transcriptional activation and regulation in eukaryotic nuclei? Data favoring this conclusion are already available for two conventional regulatory mechanisms, INO and GAL (32, 48) (Fig. 8). With respect to the well-studied GAL system, transcriptional activation in galactose-grown cells is accompanied by association between GAL genes and the nuclear periphery, which is dramatically reduced in glucose-grown cells (48). This result is particularly significant because analyses of galactose-regulated gene expression and the Gal4 activator were used initially to establish the recruitment paradigm. The link between transcriptional activation and perinuclear association has also been reported for INO1 (32). Thus, a growing body of evidence supports the ideas that a link exists between the nuclear periphery and transcriptional activation and that reversible association with the nuclear rim may be an integral feature of all eukaryotic gene regulation (see also a recent analysis of var genes in Plasmodium falciparum [301]).
FIG. 8.
Perinuclear transcriptional activation of GAL genes via reverse recruitment. The glucose-regulated transcriptional activator Gal4 associates with its DNA binding sites whether the downstream target genes are inactive or active, as shown. The GAL1, GAL7, and GAL10 genes are not detectably associated with the nuclear periphery under conditions (in glucose-grown cells) in which their transcription is repressed (UNINDUCED, left side). When cells are grown in the presence of galactose, these same GAL genes associate with the nuclear periphery (INDUCED, right side).
Universal application of the reverse recruitment paradigm may provide a context in which to understand numerous perplexing phenomena that have heretofore defied explanation, most importantly the capacity of regulators to exert influence on gene transcription under circumstances where either direct or indirect contact with the DNA is thought to be unlikely. The best-characterized examples of such “ghost regulation” are Rap1 repression of target genes upon disruption of the secretory pathway (208) and Rgt1 activation of HXT1 (see above). Ghost regulation cannot be reconciled with a competitor of the reverse recruitment idea that postulates an alternative explanation for the perinuclear location of activated GAL and INO genes, i.e., that transcription initiation complexes form as predicted by the recruitment model and move to the periphery only during the final stages of gene expression (i.e., during mRNA processing and export).
Glucose May Signal a Highly Networked and Structurally Stable Perinuclear Machine
Another recent development favoring the reverse recruitment idea is that proteins capable of both induction and repression of transcription, including many involved in glucose regulation (e.g., Rap1, Gcr1, Snf1, Mig1, Mig3, Nrg1, Rgt1, Ssn6, Med1, Ssn8, Sin4, Rgr1, and Fhl1), appear to be the rule rather than the exception. We have now shown that many of these factors are concentrated at the periphery of the yeast nucleus (236; B. B. Menon and G. M. Santangelo, unpublished data). It is increasingly clear that subtle allosteric alteration of bifunctional activator/repressor-type regulators is a common mechanism responsible for major regulatory shifts in the transcriptome (e.g., at Snf1/Mig1/Ssn6 targets). Reverse recruitment has the advantage of allowing a straightforward and parsimonious depiction of this type of subtle gene regulation mechanism (Fig. 9), a task that has become more and more difficult and convoluted when one adheres to the recruitment paradigm. This is in part because the latter requires so many factors to assemble in the correct configuration at each promoter. It is especially so when also considering that the recruitment model requires them to assemble in the proper sequence (i.e., in a temporally flawless fashion) at each promoter, or to preassemble and then bind as a complex or series of complexes, in order to obtain the proper transcriptional readout for each corresponding gene. Indeed, a recent genome-wide analysis of quiescence revealed that, in contrast with the recruitment model, RNA polymerase II is already positioned upstream of many inactive genes prior to their induction upon exit from stationary phase (298).
FIG. 9.
A model for glucose regulation at the nuclear periphery of S. cerevisiae. Numerous regulators involved in the transcriptomic response to glucose are components of a large nuclear assemblage (gene expression machine) that is tethered to nuclear pores (dashed lines) via multiple interactions with the Nup84 subcomplex. Red arrows depict repressed target genes, and green arrows depict activated targets. Inactivation of DNA-bound regulators (Mig1, Mig3, and Nrg1) by the Snf1 kinase is predicted to involve a subtle conformational alteration that remains obscure. Many of these glucose regulatory factors, including components of the RNA polymerase II Mediator, are bifunctional factors that can either repress or activate transcription in a context-dependent fashion (shown here for Rap1). This facile switching between transcriptional readouts underscores the mechanistic subtlety of regulatory partnering and/or conformational specificity. Association between nuclear pores and the Gcr1/Gcr2 complex is required for transcriptional activation by both Rap1 and Nup84 subcomplex components (229). The perinuclear Rap1/Gcr1/Gcr2 assemblage thus allows productive access to GEMs by glucose-induced (glycolytic and RP) genes tethered at the nuclear periphery.
With respect to glucose regulation in particular, it is also now apparent that DNA-bound transcriptional activators, readily identified in some systems (e.g., Gal4), may be curiously missing from others. Given that SUC2 has been a model target gene in the study of glucose repression, it is reasonable to have expected by now the identification of a DNA-bound SUC2 activator. (Although it seems unlikely, it remains a formal possible that the gene encoding the SUC2 activator protein has simply eluded detection.) The very successful screens for non-sucrose-fermenting mutants (46, 259, 385) have instead identified the critically important kinase subunits Snf1 and Snf4, a putative glucose sensor localized to the plasma membrane (Snf3), endosomal factors (Snf7 and Snf8 [148, 287]), and components of the Swi/Snf chromatin remodeling complex (Snf2, Snf5, Snf6, Snf11, and Snf12). None of these proteins are thought to bind DNA directly. Subsequent screens for suppressors of Snf1 (47, 386) identified two critical downstream factors in glucose repression, Mig1 (Ssn1) and Ssn6; the products of the other six SSN alleles associate biochemically with RNA polymerase II and participate in the Mediator-based mechanism of glucose repression by the Ssn6/Tup1 corepressor: Ssn2, Ssn3, Sin4 (Ssn4), Srb8 (Ssn5), Rox3 (Ssn7), andSsn8.
Thus, of all these Snf and Ssn loci, the only protein identified to date that exhibits specific DNA binding activity is the zinc finger protein Mig1. Mig1 (and its paralogs Mig2 and Mig3) bind to a GC core element upstream from SUC2 and other glucose-regulated genes. Mig1 and Mig3, like most of the other transcription factors identified in Snf and Ssn screens, are known to function as both repressors and activators. Nevertheless, Mig1 is presumed not to participate in activation of SUC2, which it binds to and represses in the presence of glucose. This conclusion is based on three assumptions that are well worth revisiting. First, SUC2 is derepressed normally in the absence of MIG1 (and/or MIG2). However, the redundancy of factors that bind to similar GC recognition sites in DNA makes it difficult to construct a strain in which all regulators that could bind and activate transcription are known to be eliminated. Thus, it is quite possible that MIG deletion uncovers a cryptic activator function in the SUC2 promoter. Second, Mig1 relocates to the cytoplasm in the absence of glucose. However, the Mig1 molecules remaining in the nucleus are sufficient to occupy binding sites upstream of Mig1 target genes (see above). Third, removal of the Mig1 recognition motif in the promoter does not impair SUC2 derepression. However, this argument rests on the assumption that the recruitment model of transcriptional activation is correct, and that once binding of those factors to their respective DNA recognition sites is blocked, they no longer remain in the vicinity of (i.e., tethered to) their respective target genes. In contrast, the reverse recruitment hypothesis postulates that a given regulator may remain in the vicinity of a “tethered” target gene and continue to influence its expression by altering the behavior of associated GEMs. This leaves open the possibility that Mig1 and other factors originally identified as repressors (e.g., Rgt1) might, in contrast to previous expectations, undergo a repressor-to-activator switch and activate their target genes under derepressing conditions (Fig. 9).
Reverse recruitment thus provides a viable alternative to an increasingly untenable prediction of the diffusion-based recruitment model that a disparate collection of activators, repressors, coactivators, corepressors, and GEM components are brought to each promoter as needed to provide the appropriate regulatory input. An improved understanding of nuclear substructure and its reception of regulatory signals from the plasma membrane and cytoplasm is therefore a critical prerequisite to constructing an accurate global wiring diagram for glucose signaling in S. cerevisiae and other eukaryotes. Reassessment of the regulatory relationships in other systems will likewise become necessary if reverse recruitment is as widespread an explanation for gene induction and repression as now seems plausible.
Acknowledgments
I am extremely grateful to Kristine Willis for figure design and preparation, to Youping Deng and Venkata Thodima for the graphical depiction of the ClustalW alignment of Gcr1 and Rpo21 with their M. invertens homolog, to Kellie Barbara for graciously contributing the microarray data shown in Fig. 5, to Kellie Barbara and Nicole Thompson for help with literature searches, and to Kristine Willis, Jeong-Ho Kim, Kelly Tatchell, Lucy Robinson, and Brenda Andrews for reviewing the manuscript, as well as for their keen insight and tolerance. Most importantly I thank the current and past members of the Santangelo laboratory for their perspiration, inspiration, and helpful suggestions. Finally, kudos and thanks to the many fine biomedical researchers in Mississippi with whom I have collaborated during my tenure as Director of the Mississippi Functional Genomics Network (mfgn.usm.edu/mfgn).
Work in my laboratory is supported by National Institutes of Health award RR16476.
REFERENCES
- 1.Ahn, J. H., S. H. Park, and H. S. Kang. 1995. Inactivation of the UAS1 of STA1 by glucose and STA10 and identification of two loci, SNS1 and MSS1, involved in STA10-dependent repression in Saccharomyces cerevisiae. Mol. Gen. Genet. 246:529-537. [DOI] [PubMed] [Google Scholar]
- 2.Ahuatzi, D., P. Herrero, T. de la Cera, and F. Moreno. 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279:14440-14446. [DOI] [PubMed] [Google Scholar]
- 3.Alberghina, L., R. L. Rossi, L. Querin, V. Wanke, and M. Vanoni. 2004. A cell sizer network involving Cln3 and Far1 controls entrance into S phase in the mitotic cycle of budding yeast. J. Cell Biol. 167:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alms, G. R., P. Sanz, M. Carlson, and T. A. Haystead. 1999. Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J. 18:4157-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews, P. D., and M. J. Stark. 2000. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J. Cell Sci. 113:507-520. [DOI] [PubMed] [Google Scholar]
- 7.Ashe, M. P., S. K. De Long, and A. B. Sachs. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashrafi, K., S. S. Lin, J. K. Manchester, and J. I. Gordon. 2000. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14:1872-1885. [PMC free article] [PubMed] [Google Scholar]
- 9.Babu, P., J. D. Bryan, H. R. Panek, S. L. Jordan, B. M. Forbrich, S. C. Kelley, R. T. Colvin, and L. C. Robinson. 2002. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 115:4957-4968. [DOI] [PubMed] [Google Scholar]
- 10.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 11.Bailey, R. B., and A. Woodword. 1984. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol. Gen. Genet. 193:507-512. [DOI] [PubMed] [Google Scholar]
- 12.Bailis, J. M., and G. S. Roeder. 2000. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell 14:211-221. [DOI] [PubMed] [Google Scholar]
- 13.Baker, H. V. 1986. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol. Cell. Biol. 6:3774-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker, S. H., D. L. Frederick, A. Bloecher, and K. Tatchell. 1997. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics 145:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balciunas, D., C. Galman, H. Ronne, and S. Bjorklund. 1999. The Med1 subunit of the yeast mediator complex is involved in both transcriptional activation and repression. Proc. Natl. Acad. Sci. USA 96:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barral, Y., S. Jentsch, and C. Mann. 1995. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9:399-409. [DOI] [PubMed] [Google Scholar]
- 17.Belinchon, M. M., and J. M. Gancedo. 2003. Xylose and some non-sugar carbon sources cause catabolite repression in Saccharomyces cerevisiae. Arch. Microbiol. 180:293-297. [DOI] [PubMed] [Google Scholar]
- 18.Berkey, C. D., V. K. Vyas, and M. Carlson. 2004. Nrg1 and Nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot. Cell 3:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya, S., L. Chen, J. R. Broach, and S. Powers. 1995. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc. Natl. Acad. Sci. USA 92:2984-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisson, L. F., and V. Kunathigan. 2003. On the trail of an elusive flux sensor. Res. Microbiol. 154:603-610. [DOI] [PubMed] [Google Scholar]
- 21.Blacketer, M. J., P. Madaule, and A. M. Myers. 1995. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics 140:1259-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blobel, G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA 82:8527-8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloecher, A., and K. Tatchell. 1999. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 13:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloecher, A., and K. Tatchell. 2000. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 149:125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumberg, H., A. Eisen, A. Sledziewski, D. Bader, and E. T. Young. 1987. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. Nature 328:443-445. [DOI] [PubMed] [Google Scholar]
- 26.Bohm, S., D. Frishman, and H. W. Mewes. 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bojunga, N., and K. D. Entian. 1999. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulated carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol. Gen. Genet. 262:869-875. [DOI] [PubMed] [Google Scholar]
- 28.Bojunga, N., P. Kotter, and K. D. Entian. 1998. The succinate/fumarate transporter Acr1p of Saccharomyces cerevisiae is part of the gluconeogenic pathway and its expression is regulated by Cat8p. Mol. Gen. Genet. 260:453-461. [DOI] [PubMed] [Google Scholar]
- 29.Boukaba, A., E. I. Georgieva, F. A. Myers, A. W. Thorne, G. Lopez-Rodas, C. Crane-Robinson, and L. Franco. 2004. A short-range gradient of histone H3 acetylation and Tup1p redistribution at the promoter of the Saccharomyces cerevisiae SUC2 gene. J. Biol. Chem. 279:7678-7684. [DOI] [PubMed] [Google Scholar]
- 30.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boy-Marcotte, E., P. Ikonomi, and M. Jacquet. 1996. SDC25, a dispensable Ras guanine nucleotide exchange factor of Saccharomyces cerevisiae differs from CDC25 by its regulation. Mol. Biol. Cell 7:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brickner, J. H., and P. Walter. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:1843-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bridges, D., and G. B. Moorhead. 2004. 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE 2004:re10. [DOI] [PubMed] [Google Scholar]
- 34.Broek, D., T. Toda, T. Michaeli, L. Levin, C. Birchmeier, M. Zoller, S. Powers, and M. Wigler. 1987. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell 48:789-799. [DOI] [PubMed] [Google Scholar]
- 35.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 36.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11:1301-1309. [DOI] [PubMed] [Google Scholar]
- 37.Bu, Y., and M. C. Schmidt. 1998. Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 26:1002-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchman, A. R., N. F. Lue, and R. D. Kornberg. 1988. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol. Cell. Biol. 8:5086-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S.cerevisiae. Cell 53:555-566. [DOI] [PubMed] [Google Scholar]
- 40.Cannon, J. F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cannon, J. F., J. B. Gibbs, and K. Tatchell. 1986. Suppressors of the ras2 mutation of Saccharomyces cerevisiae. Genetics 113:247-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon, J. F., J. R. Pringle, A. Fiechter, and M. Khalil. 1994. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics 136:485-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlson, M. 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13:1-23. [DOI] [PubMed] [Google Scholar]
- 44.Carlson, M. 1998. Regulation of glucose utilization in yeast. Curr. Opin. Genet. Dev. 8:560-564. [DOI] [PubMed] [Google Scholar]
- 45.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 46.Carlson, M., B. C. Osmond, and D. Botstein. 1981. Mutants of yeast defective in sucrose utilization. Genetics 98:25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson, M., B. C. Osmond, L. Neigeborn, and D. Botstein. 1984. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 107:19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 49.Celenza, J. L., and M. Carlson. 1984. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Celenza, J. L., and M. Carlson. 1984. Structure and expression of the SNF1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Celenza, J. L., and M. Carlson. 1986. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233:1175-1180. [DOI] [PubMed] [Google Scholar]
- 52.Celenza, J. L., and M. Carlson. 1989. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 9:5034-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celenza, J. L., F. J. Eng, and M. Carlson. 1989. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol. Cell. Biol. 9:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang, J. S., K. Henry, B. L. Wolf, M. Geli, and S. K. Lemmon. 2002. Protein phosphatase-1 binding to Scd5p is important for regulation of actin organization and endocytosis in yeast. J. Biol. Chem. 277:48002-48008. [DOI] [PubMed] [Google Scholar]
- 55.Chang, Y. W., S. C. Howard, and P. K. Herman. 2004. The Ras/PKA signaling pathway directly targets the Srb9p protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 15:107-116. [DOI] [PubMed] [Google Scholar]
- 56.Charbon, G., K. D. Breunig, R. Wattiez, J. Vandenhaute, and I. Noel-Georis. 2004. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell. Biol. 24:4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaves, R. S., P. Herrero, and F. Moreno. 1999. Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem. Biophys. Res. Commun. 254:345-350. [DOI] [PubMed] [Google Scholar]
- 58.Chen, S., R. W. West Jr., S. L. Johnson, H. Gans, B. Kruger, and J. Ma. 1993. TSF3, a global regulatory protein that silences transcription of yeast GAL genes, also mediates repression by α2 repressor and is identical to SIN4. Mol. Cell. Biol. 13:831-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng, C., D. Huang, P. J. Roach. 1997. Yeast PIG genes: PIG1 encodes a putative type 1 phosphatase subunit that interacts with the yeast glycogen synthase Gsy2p. Yeast 13:1-8. [DOI] [PubMed] [Google Scholar]
- 60.Cherry, J. R., T. R. Johnson, C. Dollard, J. R. Shuster, and C. L. Denis. 1989. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell 56:409-419. [DOI] [PubMed] [Google Scholar]
- 61.Ching, R. W., G. Dellaire, C. H. Eskiw, and D. P. Bazett-Jones. 2005. PML bodies: a meeting place for genomic loci? J. Cell Sci. 118:847-854. [DOI] [PubMed] [Google Scholar]
- 62.Ciriacy, M. 1977. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 154:213-220. [DOI] [PubMed] [Google Scholar]
- 63.Clifton, D., and D. G. Fraenkel. 1981. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J. Biol. Chem. 256:13074-13078. [PubMed] [Google Scholar]
- 64.Clifton, D., S. B. Weinstock, and D. B. Fraenkel. 1978. Glycolysis mutants in Saccharomyces cerevisiae. Genetics 88:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen, P., and P. T. Cohen. 1989. Protein phosphatases come of age. J. Biol. Chem. 264:21435-21438. [PubMed] [Google Scholar]
- 66.Colombo, S., D. Ronchetti, J. M. Thevelein, J. Winderickx, and E. Martegani. 2004. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J. Biol. Chem. 279:46715-46722. [DOI] [PubMed] [Google Scholar]
- 67.Colombo, S., P. Ma, L. Cauwenberg, J. Winderickx, M. Crauwels, A. Teunissen, D. Nauwelaers, J. H. de Winde, M. F. Gorwa, D. Colavizza, and J. M. Thevelein. 1998. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17:3326-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conlan, R. S., N. Gounalaki, P. Hatzis, and D. Tzamarias. 1999. The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274:205-210. [DOI] [PubMed] [Google Scholar]
- 69.Coons, D. M., P. Vagnoli, and L. F. Bisson. 1997. The C-terminal domain of Snf3p is sufficient to complement the growth defect of snf3 null mutations in Saccharomyces cerevisiae: SNF3 functions in glucose recognition. Yeast 13:9-20. [DOI] [PubMed] [Google Scholar]
- 70.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 71.Crabtree, H. G. 1929. Observation of the carbohydrate metabolism of tumors. Biochem. J. 23:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crauwels, M., M. C. Donaton, M. B. Pernambuco, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1997. The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway. Microbiology 143:2627-2637. [DOI] [PubMed] [Google Scholar]
- 73.Crechet, J. B., E. Jacquet, A. Bernardi, and A. Parmeggiani. 2000. Analysis of the role of the hypervariable region of yeast Ras2p and its farnesylation in the interaction with exchange factors and adenylyl cyclase. J. Biol. Chem. 275:17754-17761. [DOI] [PubMed] [Google Scholar]
- 74.Crechet, J. B., R. H. Cool, E. Jacquet, and J. Y. Lallemand. 2003. Characterization of Saccharomyces cerevisiae Ras1p and chimaeric constructs of Ras proteins reveals the hypervariable region and farnesylation as critical elements in the adenylyl cylase signaling pathway. Biochemistry 42:14903-14912. [DOI] [PubMed] [Google Scholar]
- 75.Crespo, J. L., and M. N. Hall. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66:579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui, D. Y., C. R. Brown, and H. L. Chiang. 2004. The type 1 phosphatase Reg1p-Glc7p is required for the glucose-induced degradation of fructose-1,6-biphosphatase in the vacuole. J. Biol. Chem. 279:9713-9724. [DOI] [PubMed] [Google Scholar]
- 77.Dale, S., W. A. Wilson, A. M. Edelman, and D. G. Hardie. 1995. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361:191-195. [DOI] [PubMed] [Google Scholar]
- 78.Dalley, B. K., and J. F. Cannon. 1996. Novel, activated RAS mutations alter protein-protein interactions. Oncogene 13:1209-1220. [PubMed] [Google Scholar]
- 79.Damak, F., E. Boy-Marcotte, D. Le-Roscouet, R. Guilbaud, and M. Jacquet. 1991. SDC25, a CDC25-like gene which contains a RAS-activating domain and is a dispensable gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de la Cera, T., P. Herrero, F. Moreno-Herrero, R. S. Chaves, and F. Moreno. 2002. Mediator factor Med8p interacts with the hexokinase 2: implication in the glucose signalling pathway of Saccharomyces cerevisiae. J. Mol. Biol. 319:703-714. [DOI] [PubMed] [Google Scholar]
- 81.Deminoff, S. J., and G. M. Santangelo. 2001. Rap1p requires Gcr1p and Gcr2p homodimers to activate ribosomal protein and glycolytic genes, respectively. Genetics 158:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deminoff, S. J., J. Tornow, and G. M. Santangelo. 1995. Unigenic evolution: a novel genetic method localizes a putative leucine zipper that mediates dimerization of the Saccharomyces cerevisiae regulator Gcr1p. Genetics 141:1263-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deminoff, S. J., K. Willis, and G. M. Santangelo. 2003. Coordination between eukaryotic growth and cell cycle progression: RAP1/GCR1 transcriptional activation mediates glucose-dependent CLN function. Recent Res. Dev. Genet. 3:1-16. [Google Scholar]
- 84.Denis, C. L. 1984. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 108:833-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 86.DeVit, M. J., and M. Johnston. 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9:1231-1241. [DOI] [PubMed] [Google Scholar]
- 87.DeVit, M. J., J. A. Waddle, and M. Johnston. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8:1603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Devlin, C., K. Tice-Baldwin, D. Shore, and K. T. Arndt. 1991. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol. Cell. Biol. 11:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dlugai, S., S. Hippler, R. Wieczorke, and E. Boles. 2001. Glucose-dependent and -independent signalling functions of the yeast glucose sensor Snf3. FEBS Lett. 505:389-392. [DOI] [PubMed] [Google Scholar]
- 90.Dombek, K. M., N. Kacherovsky, and E. T. Young. 2004. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 279:39165-39174. [DOI] [PubMed] [Google Scholar]
- 91.Dombek, K. M., V. Voronkova, A. Raney, and E. T. Young. 1999. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol. Cell. Biol. 19:6029-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dong, X., D. A. Mitchell, S. Lobo, L. Zhao, D. J. Bartels, and R. J. Deschenes. 2003. Palmitoylation and plasma membrane localization of Ras2pby a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol. Cell.Biol. 23:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Donovan, D. M., and N. J. Pearson. 1986. Transcriptional regulation of ribosomal proteins during a nutritional upshift in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:2429-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dubacq, C., A. Chevalier, and C. Mann. 2004. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol. Cell. Biol. 24:2560-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dubacq, C., R. Guerois, R. Courbeyrette, K. Kitagawa, and C. Mann. 2002. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1:568-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Durchschlag, E., W. Reiter, G. Ammerer, and C. Schuller. 2004. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J. Biol. Chem. 279:55425-55432. [DOI] [PubMed] [Google Scholar]
- 97.Egloff, M. P., D. F. Johnson, G. Moorhead, P. T. Cohen, P. Cohen, and D. Barford. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16:1876-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eisen, A., W. E. Taylor, H. Blumberg, and E. T. Young. 1988. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol. Cell. Biol. 8:4552-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Entian, K. D. 1980. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol. Gen. Genet. 178:633-637. [DOI] [PubMed] [Google Scholar]
- 100.Entian, K. D., and D. Mecke. 1982. Genetic evidence for a role of hexokinase isozyme PII in carbon catabolite repression in Saccharomyces cerevisiae. J. Biol. Chem. 257:870-874. [PubMed] [Google Scholar]
- 101.Entian, K. D., and F. K. Zimmermann. 1980. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 177:345-350. [DOI] [PubMed] [Google Scholar]
- 102.Entian, K. D., and F. K. Zimmermann. 1982. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J. Bacteriol. 151:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Entian, K. D., and J. A. Barnett. 1992. Regulation of sugar utilization by Saccharomyces cerevisiae. Trends Biochem. Sci. 17:506-510. [DOI] [PubMed] [Google Scholar]
- 104.Entian, K. D., F. K. Zimmermann, and I. Scheel. 1977. A partial defect in carbon catabolite repression in mutants of Saccharomyces cerevisiae with reduced hexose phosphorylation. Mol. Gen. Genet. 156:99-105. [DOI] [PubMed] [Google Scholar]
- 105.Erickson, J. R., and M. Johnston. 1993. Genetic and molecular characterization of GAL83: its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics 135:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Erickson, J. R., and M. Johnston. 1994. Suppressors reveal two classes of glucose repression genes in the yeast Saccharomyces cerevisiae. Genetics 136:1271-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Estruch, F., and M. Carlson. 1990. Increased dosage of the MSN1 gene restores invertase expression in yeast mutants defective in the SNF1 protein kinase. Nucleic Acids Res. 18:6959-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Estruch, F., M. A. Treitel, X. Yang, and M. Carlson. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fabrizio, P., F. Pozza, S. D. Pletcher, C. M. Gendron, and V. D. Longo. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292:288-290. [DOI] [PubMed] [Google Scholar]
- 110.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 111.Feng, Z. H., S. E. Wilson, Z. Y. Peng, K. K. Schlender, E. M. Reimann, and R. J. Trumbly. 1991. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J. Biol. Chem. 266:23796-23801. [PubMed] [Google Scholar]
- 112.Ferguson, S. B., E. S. Anderson, R. B. Harshaw, T. Thate, N. L. Craig, and H. C. Nelson. 2005. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 169:1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 114.Flick, J. S., and M. Johnston. 1991. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol. Biol. Cell 11:5101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Flick, J. S., and M. Johnston. 1992. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics 130:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Flick, K., D. Chapman-Shimshoni, D. Stuart, M. Guaderrama, and C. Wittenberg. 1998. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol. Cell. Biol. 18:2492-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Flick, K. M., N. Spielewoy, T. I. Kalashnikova, M. Guaderrama, Q. Zhu, H. C. Chang, and C. Wittenberg. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Cell. Biol. 14:3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91-109. [DOI] [PubMed] [Google Scholar]
- 119.Francois, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125-145. [DOI] [PubMed] [Google Scholar]
- 120.Frederick, D. L., and K. Tatchell. 1996. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol. Cell. Biol. 16:2922-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ganster, R. W., W. Shen, and M. C. Schmidt. 1993. Isolation of STD1, a high-copy-number suppressor of a dominant negative mutation in the yeast TATA-binding protein. Mol. Cell. Biol. 13:3650-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gelade, R., S. Van de Velde, P. Van Dijck, and J. M. Thevelein. 2003. Multi-level response of the yeast genome to glucose. Genome Biol. 4:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gelperin, D., J. Weigle, K. Nelson, P. Roseboom, K. Irie, K. Matsumoto, and S. Lemmon. 1995. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:11539-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Geymonat, M., L. Wang, H. Garreau, and M. Jacquet. 1998. Ssa1p chaperone interacts with the guanine nucleotide exchange factor of ras Cdc25p and controls the cAMP pathway in Saccharomyces cerevisiae. Mol. Microbiol. 30:855-864. [DOI] [PubMed] [Google Scholar]
- 126.Ghannam, G., A. Takeda, T. Camarata, M. A. Moore, A. Viale, and N. R. Yaseen. 2004. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J. Biol. Chem. 279:866-875. [DOI] [PubMed] [Google Scholar]
- 127.Gilbert, W., and C. Guthrie. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell 13:201-212. [DOI] [PubMed] [Google Scholar]
- 128.Gotta, M., and S. M. Gasser. 1996. Nuclear organization and transcriptional silencing in yeast. Experientia 52:1136-1147. [DOI] [PubMed] [Google Scholar]
- 129.Griffioen, G., and J. M. Thevelein. 2002. Molecular mechanisms controlling the localization of protein kinase A. Curr. Genet. 41:199-207. [DOI] [PubMed] [Google Scholar]
- 130.Griffioen, G., P. Anghileri, E. Imre, M. D. Baroni, and H. Ruis. 2000. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J. Biol. Chem. 275:1449-1456. [DOI] [PubMed] [Google Scholar]
- 131.Griggs, D. W., and M. Johnston. 1991. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. USA 88:8597-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hall, D. D., D. D. Markwardt, F. Parviz, and W. Heideman. 1998. Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 17:4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han, Y., S. San-Marina, J. Liu, and M. D. Minden. 2004. Transcriptional activation of c-myc proto-oncogene by WT1 protein. Oncogene 23:6933-6941. [DOI] [PubMed] [Google Scholar]
- 134.Haney, S. A., and J. R. Broach. 1994. Cdc25p, the guanine nucleotide exchange factor for the Ras proteins of Saccharomyces cerevisiae, promotes exchange by stabilizing Ras in a nucleotide-free state. J. Biol. Chem. 269:16541-16548. [PubMed] [Google Scholar]
- 135.Harashima, T., and J. Heitman. 2002. The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10:163-173. [DOI] [PubMed] [Google Scholar]
- 136.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 137.Harkness, T. A., K. A. Shea, C. Legrand, M. Brahmania, and G. F. Davies. 2004. A functional analysis reveals dependence on the anaphase-promoting complex for prolonged life span in yeast. Genetics 168:759-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hartwell, L. H. 1974. Saccharomyces cerevisiae cell cycle. Bacteriol. Rev. 38:164-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hartwell, L. H., R. K. Mortimer, J. Culotti, and M. Culotti. 1973. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics 74:267-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Haurie, V., F. Sagliocco, and H. Boucherie. 2004. Dissecting regulatory networks by means of two-dimensional gel electrophoresis: application to the study of the diauxic shift in the yeast Saccharomyces cerevisiae. Proteomics 4:364-373. [DOI] [PubMed] [Google Scholar]
- 141.Haurie, V., M. Perrot, T. Mini, P. Jeno, F. Sagliocco, and H. Boucherie. 2001. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 276:76-85. [DOI] [PubMed] [Google Scholar]
- 142.Hazbun, T. R., and S. Fields. 2002. A genome-wide screen for site-specific DNA-binding proteins. Mol. Cell Proteomics 1:538-543. [DOI] [PubMed] [Google Scholar]
- 143.Hedbacker, K., R. Townley, and M. Carlson. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hedbacker, K., S. P. Hong, and M. Carlson. 2004. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 24:8255-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hedges, D., M. Proft, and K. D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for depression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Herrero, P., C. Martinez-Campa, and F. Moreno. 1998. The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 434:71-76. [DOI] [PubMed] [Google Scholar]
- 147.Herrero, P., J. Galindez, N. Ruiz, C. Martinez-Campa, and F. Moreno. 1995. Transcription regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast 11:137-144. [DOI] [PubMed] [Google Scholar]
- 148.Hierro, A., J. Sun, A. S. Rusnak, J. Kim, G. Prag, S. E. Emr, and J. H. Hurley. 2004. Structure of the ESCRT-II endosomal trafficking complex. Nature 431:221-225. [DOI] [PubMed] [Google Scholar]
- 149.Hisamoto, N., D. L. Frederick, K. Sugimoto, K. Tatchell, and K. Matsumoto. 1995. The EGP1 gene may be a positive regulator of protein phosphatase type 1 in the growth control of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3767-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hisamoto, N., K. Sugimoto, and K. Matsumoto. 1994. The Glc7 type 1 protein phosphatase of Saccharomyces cerevisiae is required for cell cycle progression in G2/M. Mol. Cell. Biol. 14:3158-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hixson, C. S., and E. G. Krebs. 1980. Characterization of a cyclic AMP-binding protein from bakers' yeast. Identification as a regulatory subunit of cyclic AMP-dependent protein kinase. J. Biol. Chem. 255:2137-2145. [PubMed] [Google Scholar]
- 152.Holland, M. J., T. Yokoi, J. P. Holland, K. Myambo, and M. A. Innis. 1987. The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehydes-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Holzer, H. 1976. Catabolite inactivation in yeast. Trends Biochem. Sci. 1:178-181. [Google Scholar]
- 154.Holzer, H. 1989. Proteolytic catabolite inactivation in Saccharomyces cerevisiae. Revis Biol. Celular 21:305-319. [PubMed] [Google Scholar]
- 155.Hong, S. P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Horak, J., J. Regelmann, and D. H. Wolf. 2002. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J. Biol. Chem. 277:8248-8254. [DOI] [PubMed] [Google Scholar]
- 157.Howard, S. C., Y. V. Budovskaya, Y. W. Chang, and P. K. Herman. 2002. The C-terminal domain of the largest subunit of RNA polymerase II is required for stationary phase entry and functionally interacts with the Ras/PKA signaling pathway. J. Biol. Chem. 277:19488-19497. [DOI] [PubMed] [Google Scholar]
- 158.Hsiung, Y. G., H. C. Chang, J. Pellequer, R. La Valle, S. Lanker, and C. Wittenberg. 2001. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21:2506-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ip11/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 160.Hu, Z., J. O. Nehlin, H. Ronne, and C. A. Michels. 1995. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr. Genet. 28:258-266. [DOI] [PubMed] [Google Scholar]
- 161.Hu, Z., Y. Yue, H. Jiang, B. Zhang, P. W. Sherwood, and C. A. Michels. 2000. Analysis of the mechanism by which glucose inhibits maltose induction of MAL gene expression in Saccharomyces. Genetics 154:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang, D., K. T. Chun, M. G. Goebl, and P. J. Roach. 1996. Genetic interactions between REG1/HEX2 and GLC7, the gene encoding the protein phosphatase type 1 catalytic subunit in Saccharomyces cerevisiae. Genetics 143:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hubbard, E. J., R. Jiang, and M. Carlson. 1994. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:1972-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hubbard, E. J., X. L. Yang, and M. Carlson. 1992. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics 130:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hubler, L., J. Bradshaw-Rouse, and W. Heideman. 1993. Connections between the Ras-cyclic AMP pathway and G1 cyclin expression in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13:6274-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hung, G. C., C. R. Brown, A. B. Wolfe, J. Liu, and H. L. Chiang. 2004. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J. Biol. Chem. 279:49138-49150. [DOI] [PubMed] [Google Scholar]
- 167.Hutchison, N., and H. Weintraub. 1985. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell 43:471-482. [DOI] [PubMed] [Google Scholar]
- 168.Ichimura, T., H. Kubota, T. Goma, N. Mizushima, Y. Ohsumi, M. Iwago, K. Kakiuchi, H. U. Shekhar, T. Shinkawa, M. Taoka, T. Ito, and T. Isobe. 2004. Transcriptomic and proteomic analysis of a 14-3-3 gene-deficient yeast. Biochemistry 43:6149-6158. [DOI] [PubMed] [Google Scholar]
- 169.Isogai, Y., and R. Tjian. 2003. Targeting genes and transcription factors to segregated nuclear compartments. Curr. Opin. Cell Biol. 15:296-303. [DOI] [PubMed] [Google Scholar]
- 170.Jaquenoud, M., M. P. Gulli, K. Peter, and M. Peter. 1998. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 17:5360-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jiang, H., I. Medintz, and C. A. Michels. 1997. Two glucose sensing/signaling pathways stimulate glucose-induced inactivation of maltose permease in Saccharomyces. Mol. Biol. Cell 8:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jiang, H., I. Medintz, B. Zhang, and C. A. Michels. 2000. Metabolic signals trigger glucose-induced inactivation of maltose permease in Saccharomyces. J. Bacteriol. 182:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jiang, H., K. Tatchell, S. Liu, and C. A. Michels. 2000. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:411-422. [DOI] [PubMed] [Google Scholar]
- 174.Jiang, R., and M. Carlson. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105-3115. [DOI] [PubMed] [Google Scholar]
- 175.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jiang, Y., C. Davis, and J. R. Broach. 1998. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 17:6942-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Johnson, K. E., S. Cameron, T. Toda, M. Wigler, and M. J. Zoller. 1987. Expression in Escherichia coli of BCY1, the regulatory subunit of cyclic AMP-dependent protein kinase from Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 262:8636-8642. [PubMed] [Google Scholar]
- 178.Johnston, M. 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 15:29-33. [DOI] [PubMed] [Google Scholar]
- 179.Johnston, M., and J. H. Kim. 2005. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33:247-252. [DOI] [PubMed] [Google Scholar]
- 180.Johnston, M., J. S. Flick, and T. Pexton. 1994. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3834-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kaniak, A., Z. Xue, D. Macool, J. H. Kim, and M. Johnston. 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kasper, L. H., P. K. Brindle, C. A. Schnabel, C. E. Pritchard, M. L. Cleary, and J. M. van Deursen. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kido, M., F. Shima, T. Satoh, T. Asato, K. Kariya, and T. Kataoka. 2002. Critical function of the Ras-associating domain as a primary Ras-binding site for regulation of Saccharomyces cerevisiae adenylyl cyclase. J. Biol. Chem. 277:3117-3123. [DOI] [PubMed] [Google Scholar]
- 185.Kief, D. R., and J. R. Warner. 1981. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 1:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kim, J. H., J. Polish, and M. Johnston. 2003. Specificity and regulation of DNA binding in the yeast glucose transporter gene repressor Rgt1. Mol. Cell. Biol. 23:5208-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kishi, T., and F. Yamao. 1998. An essential function of Grr1 for the degradation of Cln2 is to act as a binding core that links Cln2 to Skp1. J. Cell Sci. 111:3655-3661. [DOI] [PubMed] [Google Scholar]
- 188.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klein, C. J., L. Olsson, and J. Nielsen. 1998. Glucose control in Saccharomyces cerevisiae: the role of Mig1 in metabolic functions. Microbiology 144:13-24. [DOI] [PubMed] [Google Scholar]
- 190.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 191.Kraakman, L. S., J. Winderickx, J. M. Thevelein, and J. H. de Winde. 1999. Structure-function analysis of yeast hexokinase: structural requirements for triggering cAMP signalling and catabolite repression. Biochem. J. 343:159-168. [PMC free article] [PubMed] [Google Scholar]
- 192.Kratzer, S., and H. J. Schuller. 1997. Transcriptional control of the yeast acetyl-CoA synthase gene, ACS1, by the positive regulators CAT8 and ADR1 and the pleiotropic repressor UME6. Mol. Microbiol. 26:631-641. [DOI] [PubMed] [Google Scholar]
- 193.Kriegel, T. M., J. Rush, A. B. Vojtek, D. Clifton, and D. G. Fraenkel. 1994. In vivo phosphorylation site of hexokinase 2 in Saccharomyces cerevisiae. Biochemistry 33:149-152. [DOI] [PubMed] [Google Scholar]
- 194.Kuchin, S., and M. Carlson. 1998. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol. 18:1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kuchin, S., V. K. Vyas, and M. Carlson. 2003. Role of the yeast Snf1 protein kinase in invasive growth. Biochem. Soc. Trans. 31:175-177. [DOI] [PubMed] [Google Scholar]
- 198.Kuchin, S., V. K. Vyas, E. Kanter, S. P. Hong, and M. Carlson. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kuroda, Y., N. Suzuki, and T. Kataoka. 1993. The effect of posttranslational modifications on the interactions of Ras2 with adenylyl cyclase. Science 259:683-686. [DOI] [PubMed] [Google Scholar]
- 200.Laemmli, U. K., and R. Tjian. 1996. A nuclear traffic jam: unraveling multicomponent machines and compartments. Curr. Opin. Cell Biol. 8:299-302. [PubMed] [Google Scholar]
- 201.Lafuente, M. J., C. Gancedo, J. C. Jauniaux, and J. M. Gancedo. 2000. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 35:161-172. [DOI] [PubMed] [Google Scholar]
- 202.Lakshmanan, J., A. L. Mosley, and S. Ozcan. 2003. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 203.Lee, M., S. Chatterjee, and K. Struhl. 2000. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8-Tup1 corepressor in yeast. Genetics 155:1535-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Leech, A., N. Nath, R. R. McCartney, and M. C. Schmidt. 2003. Isolation of mutations in the catalytic domain of the Snf1 kinase that render its activity independent of the Snf4 subunit. Eukaryot. Cell 2:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lemaire, K., S. Van de Velde, P. Van Dijck, and J. M. Thevelein. 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16:293-299. [DOI] [PubMed] [Google Scholar]
- 206.Lesage, P., X. Yang, and M. Carlson. 1996. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell. Biol. 16:1921-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Levine, M., and J. L. Manley. 1989. Transcriptional repression of eukaryotic promoters. Cell 59:405-408. [DOI] [PubMed] [Google Scholar]
- 208.Li, B., C. R. Nierras, and J. R. Warner. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol. 19:5393-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li, F. N., and M. Johnston. 1997. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16:5629-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane, D. J. Stillman, and R. D. Kornberg. 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92:10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 212.Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez, V. C. Culotta, G. R. Fink, and L. Guarente. 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418:344-348. [DOI] [PubMed] [Google Scholar]
- 213.Lin, S. S., J. K. Manchester, and J. L. Gordon. 2003. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278:13390-13397. [DOI] [PubMed] [Google Scholar]
- 214.Lodi, T., C. Donnini, and I. Ferrero. 1991. Catabolite repression by galactose in overexpressed GAL4 strains of Saccharomyces cerevisiae. J. Gen. Microbiol. 137:1039-1044. [DOI] [PubMed] [Google Scholar]
- 215.Loeb, J. D., T. A. Kerentseva, T. Pan, M. Sepulveda-Becerra, and H. Liu. 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153:1535-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Longo, V. D. 2003. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 38:807-811. [DOI] [PubMed] [Google Scholar]
- 217.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ludin, K., R. Jiang, and M. Carlson. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:6245-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lundin, M., J. O. Nehlin, and H. Ronne. 1994. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell. Biol. 14:1979-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lutfiyya, L. L., and M. Johnston. 1996. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 16:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lutfiyya, L. L., V. R. Iyer, J. DeRisi, M. J. DeVit, P. O. Brown, and M. Johnston. 1998. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150:1377-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ma, H., and D. Botstein. 1986. Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol. Cell. Biol. 6:4046-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.MacKelvie, S. H., P. D. Andrews, and M. J. Stark. 1995. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol. Cell. Biol. 15:3777-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Maheswaran, S., S. Park, A. Bernard, J. F. Morris, F. J. Rauscher III, D. E. Hill, and D. A. Haber. 1993. Physical and functional interaction between WT1 and p53 proteins. Proc. Natl. Acad. Sci. USA 90:5100-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 226.Marion, R. M., A. Regev, E. Segal, Y. Barash, D. Koller, N. Friedman, and E. K. O'Shea. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101:14315-14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Marshall-Carlson, L., J. L. Celenza, B. C. Laurent, and M. Carlson. 1990. Mutational analysis of the SNF3 glucose transporter of Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1105-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Marshall-Carlson, L., L. Neigeborn, D. Coons, L. Bisson, and M. Carlson. 1991. Dominant and recessive suppressors that restore glucose transport in a yeast Snf3 mutant. Genetics 128:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Matsumoto, K., I. Uno, Y. Oshima, and T. Ishikawa. 1982. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 79:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Matsumoto, K., T. Yoshimatsu, and Y. Oshima. 1983. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J. Bacteriol. 153:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mayordomo, I., and P. Sanz. 2001. Human pancreatic glucokinase (GlkB) complements the glucose signalling defect of Saccharomyces cerevisiae hxk2 mutants. Yeast 18:1309-1316. [DOI] [PubMed] [Google Scholar]
- 232.Mayordomo, I., J. Regelmann, J. Horak, and P. Sanz. 2003. Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 544:160-164. [DOI] [PubMed] [Google Scholar]
- 233.Mbonyi, K., L. van Aelst, J. C. Arguelles, A. W. Jans, and J. M. Thevelein. 1990. Glucose-induced hyperaccumulation of cyclic AMP and defective glucose repression in yeast strains with reduced activity of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 10:4518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.McEwen, B. S., V. G. Allfrey, and A. E. Mirsky. 1963. Studies on energy-yielding reactions in thymus nuclei. II. Pathways of aerobic carbohydrate catabolism. J. Biol. Chem. 238:2571-2578. [PubMed] [Google Scholar]
- 235.McKay, L. M., B. Carpenter, and S. G. Roberts. 1999. Regulation of the Wilms' tumour suppressor protein transcriptional activation domain. Oncogene 18:6546-6554. [DOI] [PubMed] [Google Scholar]
- 236.Menon, B. B., N. J. Sarma, S. Pasula, S. J. Deminoff, K. A. Willis, K. E. Barbara, B. Andrews, and G. M. Santangelo. 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102:5749-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mintzer, K. A., and J. Field. 1999. The SH3 domain of the S. cerevisiae Cdc25p binds adenylyl cyclase and facilitates Ras regulation of cAMP signalling. Cell Signal. 11:127-135. [DOI] [PubMed] [Google Scholar]
- 238.Misteli, T. 2004. Spatial positioning; a new dimension in genome function. Cell 119:153-156. [DOI] [PubMed] [Google Scholar]
- 239.Misteli, T. 2005. Concepts in nuclear architecture. Bioassays 27:477-487. [DOI] [PubMed] [Google Scholar]
- 240.Misteli, T., and D. L. Spector. 1998. The cellular organization of gene expression. Curr. Opin. Cell Biol. 10:323-331. [DOI] [PubMed] [Google Scholar]
- 241.Mitts, M. R., J. Bradshaw-Rouse, and W. Heideman. 1991. Interactions between adenylate cyclase and the yeast GTPase-activating protein IRA1. Mol. Cell. Biol. 11:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mizuno, T., and S. Harashima. 2000. Activation of basal transcription by a mutation in SIN4, a yeast global repressor, occurs through a mechanism different from activator-mediated transcriptional enhancement. Mol. Gen. Genet. 263:48-59. [DOI] [PubMed] [Google Scholar]
- 243.Monod, J. 1942. Recherches sur la croissance des cultures bacteriennes. Ph.D. thesis. University of Paris, Paris, France.
- 244.Moreno, F., and P. Herrero. 2002. The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:83-90. [DOI] [PubMed] [Google Scholar]
- 245.Moreno, F., D. Ahuatzi, A. Riera, C. A. Palomino, and P. Herrero. 2005. Glucose sensing through the Hxk2-dependent signalling pathway. Biochem. Soc. Trans. 33:265-268. [DOI] [PubMed] [Google Scholar]
- 246.Moreno-Herrero, F., P. Herrero, J. Colchero, A. M. Baro, and F. Moreno. 1999. Analysis by atomic force microscopy of Med8 binding to cis-acting regulatory elements of the SUC2 and HXK2 genes of Saccharomyces cerevisiae. FEBS Lett. 459:427-432. [DOI] [PubMed] [Google Scholar]
- 247.Moriya, H., and M. Johnston. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 101:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Morris, J. F., S. L. Madden, O. E. Tournay, D. M. Cook, V. P. Sukhatme, and F. J. Rauscher III. 1991. Characterization of the zinc finger protein encoded by the WT1 Wilms' tumor locus. Oncogene 6:2339-2348. [PubMed] [Google Scholar]
- 249.Mosch, H. U., E. Kubler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mosley, A. L., J. Lakshmanan, B. K. Aryal, and S. Ozcan. 2003. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278:10322-10327. [DOI] [PubMed] [Google Scholar]
- 251.Mukherjee, S., M. F. Berger, G. Jona, X. S. Wang, D. Muzzey, M. Snyder, R. A. Young, and M. L. Bulyk. 2004. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36:1331-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nakafuku, M., T. Obara, K. Kaibuchi, I. Miyajima, A. Miyajima, H. Itoh, S. Nakamura, K. Arai, K. Matsumoto, and Y. Kaziro. 1988. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc. Natl. Acad. Sci. USA 85:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 254.Nath, N., R. R. McCartney, and M. C. Schmidt. 2002. Purification and characterization of Snf1 kinase complexes containing a defined β subunit composition. J. Biol. Chem. 277:50403-50408. [DOI] [PubMed] [Google Scholar]
- 255.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nedea, E., X. He, M. Kim, J. Pootoolal, G. Zhong, V. Canadien, T. Hughes, S. Buratowski, C. L. Moore, and J. Greenblatt. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278:33000-33010. [DOI] [PubMed] [Google Scholar]
- 257.Nehlin, J. O., and H. Ronne. 1990. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 9:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nehlin, J. O., M. Carlberg, and H. Ronne. 1991. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10:3373-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Neigeborn, L., and M. Carlson. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108:845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Neigeborn, L., and M. Carlson. 1987. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Neigeborn, L., K. Rubin, and M. Carlson. 1986. Suppressors of SNF2 mutations restore invertase derepression and cause temperature-sensitive lethality in yeast. Genetics 112:741-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Newcomb, L. L., D. D. Hall, and W. Heideman. 2002. AZF1 is a glucose-dependent positive regulator of CLN3 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:1607-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Newcomb, L. L., J. A. Diderich, M. G. Slattery, and W. Heideman. 2003. Glucose regulation of Saccharomyces cerevisiae cell cycle genes. Eukaryot. Cell 2:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Nichols, K. E., G. G. Re, Y. X. Yan, A. J. Garvin, and D. A. Haber. 1995. WT1 induces expression of insulin-like growth factor 2 in Wilms' tumor cells. Cancer Res. 55:4540-4543. [PubMed] [Google Scholar]
- 265.Niederacher, D., and K. D. Entian. 1991. Characterization of Hex2 protein, a negative regulatory element necessary for glucose repression in yeast. Eur. J. Biochem. 200:311-319. [DOI] [PubMed] [Google Scholar]
- 266.Nikawa, J., P. Sass, and M. Wigler. 1987. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nishida, Y., F. Shima, H. Sen, Y. Tanada, C. Yanagihara, Y. Yamawaki-Kataoka, K. Kariya, and T. Kataoka. 1998. Coiled-coil interaction of N-terminal 36 residues of cyclase-associated protein with adenylyl cyclase is sufficient for its function in Saccharomyces cerevisiae ras pathway. J. Biol. Chem. 273:28019-28024. [DOI] [PubMed] [Google Scholar]
- 268.Oren, M. 2003. Decision making by p53: life, death and cancer. Cell Death Differ. 10:431-442. [DOI] [PubMed] [Google Scholar]
- 269.Ostling, J., and H. Ronne. 1998. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur. J. Biochem. 252:162-168. [DOI] [PubMed] [Google Scholar]
- 270.Ostling, J., M. Carlberg, and H. Ronne. 1996. Functional domains in the Mig1 repressor. Mol. Cell. Biol. 16:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ozcan, S., and M. Johnston. 1995. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ozcan, S., and M. Johnston. 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63:554-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ozcan, S., J. Dover, A. G. Rosenwald, S. Wolfi, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93:12428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Ozcan, S., J. Dover, and M. Johnston. 1998. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17:2566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ozcan, S., T. Leong, and M. Johnston. 1996. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 16:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Paalman, J. W., R. Verwaal, S. H. Slofstra, A. J. Verkleij, J. Boonstra, and C. T. Verrips. 2003. Trehalose and glycogen accumulation is related to the duration of the G1 phase of Saccharomyces cerevisiae. FEMS Yeast Res. 3:261-268. [DOI] [PubMed] [Google Scholar]
- 277.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Papamichos-Chronakis, M., T. Gligoris, and D. Tzamarias. 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Papamichos-Chronakis, M., T. Petrakis, E. Ktistaki, I. Topalidou, and D. Tzamarias. 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9:1297-1305. [DOI] [PubMed] [Google Scholar]
- 280.Parada, L. A., S. Sotiriou, and T. Misteli. 2004. Spatial genome organization. Exp. Cell Res. 296:64-70. [DOI] [PubMed] [Google Scholar]
- 281.Parada, L., and T. Misteli. 2002. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12:425-432. [DOI] [PubMed] [Google Scholar]
- 282.Park, S. H., S. S. Koh, J. H. Chun, H. J. Hwang, and H. S. Kang. 1999. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Parraga, G., S. J. Horvath, A. Eisen, W. E. Taylor, L. Hood, E. T. Young, and R. E. Klevit. 1988. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science 241:1489-1492. [DOI] [PubMed] [Google Scholar]
- 284.Parrou, J. L., B. Enjalbert, and J. Francois. 1999. STRE- and cAMP-independent transcriptional induction of Saccharomyces cerevisiae GSY2 encoding glycogen synthase during diauxic growth on glucose. Yeast 15:1471-1484. [DOI] [PubMed] [Google Scholar]
- 285.Parviz, F., and W. Heideman. 1998. Growth-independent regulation of CLN3 mRNA levels by nutrients in Saccharomyces cerevisiae. J. Bacteriol. 180:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Parviz, F., D. D. Hall, D. D. Markwardt, and W. Heideman. 1998. Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J. Bacteriol. 180:4508-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Peck, J. W., E. T. Bowden, and P. D. Burbelo. 2004. Structure and function of human Vps20 and Snf7 proteins. Biochem. J. 377:693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Peggie, M. W., S. H. MacKelvie, A. Bloecher, E. V. Knatko, K. Tatchell, and M. J. Stark. 2002. Essential functions of Sds22p in chromosome localization of PP1. J. Cell Sci. 115:195-206. [DOI] [PubMed] [Google Scholar]
- 289.Polish, J. A., J. H. Kim, and M. Johnston. 2005. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 169:583-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Pombo, A., P. Cuello, W. Schul, J. B. Yoon, R. G. Roeder, P. R. Cook, and S. Murphy. 1998. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 17:1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Powers, S., T. Kataoka, O. Fasano, M. Goldfarb, J. Strathern, J. Broach, and M. Wigler. 1984. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell 36:607-612. [DOI] [PubMed] [Google Scholar]
- 292.Powers, T. 2004. Ribosome biogenesis: giant steps for a giant problem. Cell 119:901-902. [DOI] [PubMed] [Google Scholar]
- 293.Presgraves, D. C., L. Balagopalan, S. M. Abmayr, and H. A. Orr. 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423:715-719. [DOI] [PubMed] [Google Scholar]
- 294.Prieto, S., B. J. de la Cruz, and I. E. Scheffler. 2000. Glucose-regulated turnover of mRNA and the influence of poly(A) tail length on half-life. J. Biol. Chem. 275:14155-14166. [DOI] [PubMed] [Google Scholar]
- 295.Ptashne, M. 2003. Regulated recruitment and cooperativity in the design of biological regulatory systems. Phil. Trans. R. Soc. London 361:1223-1234. [DOI] [PubMed] [Google Scholar]
- 296.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 297.Purnapatre, K., S. Piccirillo, B. L. Schneider, and S. M. Honigberg. 2002. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells 7:675-691. [DOI] [PubMed] [Google Scholar]
- 298.Radonjic, M., J.-C. Andrau, P. Lijnzaad, P. Kemmeren, T. T. J. P. Kockelkorn, D. van Leenen, N. L. van Berkum, and F. C. P. Holstege. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 18:171-183. [DOI] [PubMed] [Google Scholar]
- 299.Rahner, A., A. Scholer, E. Martens, G. Gollwitzer, and H. J. Schuller. 1996. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene. CAT8. Nucleic Acids Res. 24:2331-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Rahner, A., M. Hiesinger, and H. J. Schuller. 1999. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 34:146-156. [DOI] [PubMed] [Google Scholar]
- 301.Ralph, S. A., C. Scheidig-Benatar, and A. Scherf. 2005. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. USA 102:5414-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ramaswamy, N. T., L. Li, M. Khalil, and J. F. Cannon. 1998. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics 149:57-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Randez-Gil, F., N. Bojunga, M. Proft, and K. D. Entian. 1997. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol. Cell. Biol. 17:2502-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Randez-Gil, F., P. Herrero, P. Sanz, J. A. Prieto, and F. Moreno. 1998. Hexokinase PII has a double cytosolic-nuclear localisation in Saccharomyces cerevisiae. FEBS Lett. 425:475-478. [DOI] [PubMed] [Google Scholar]
- 305.Randez-Gil, F., P. Sanz, K. D. Entian, and J. A. Prieto. 1998. Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol. Cell. Biol. 18:2940-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Rauscher, F. J., III. 1993. The WT1 Wilms tumor gene product: a developmentally regulated transcription factor in the kidney that functions as a tumor suppressor. FASEB J. 7:896-903. [PubMed] [Google Scholar]
- 307.Regelmann, J., T. Schule, F. S. Josupeit, J. Horak, M. Rose, K. D. Entian, M. Thumm, and D. H. Wolf. 2003. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell 14:1652-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Reifenberger, E., K. Freidel, and M. Ciriacy. 1995. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 16:157-167. [DOI] [PubMed] [Google Scholar]
- 309.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Robinson, L. C., E. J. A. Hubbard, P. R. Graves, A. A. DePaoli-Roach, P. J. Roach, C. Kung, D. W. Haas, C. H. Hagedorn, M. Goebl, M. R. Culbertson, and M. Carlson. 1992. Yeast casein kinase I homologues: an essential gene pair. Proc. Natl. Acad. Sci. USA 89:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rodriguez, A., T. De La Cera, P. Herrero, and F. Moreno. 2001. The hexokinase 2 protein regulates the expression of GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J. 355:625-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Rodriguez, C., P. Sanz, and C. Gancedo. 2003. New mutations of Saccharomyces cerevisiae that partially relieve both glucose and galactose repression activate the protein kinase Snf1. FEMS Yeast Res. 3:77-84. [DOI] [PubMed] [Google Scholar]
- 313.Rolland, F., J. H. de Winde, K. Lemaire, E. Boles, J. M. Thevelein, and J. Winderickx. 2000. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38:348-358. [DOI] [PubMed] [Google Scholar]
- 314.Rolland, F., J. Winderickx, and J. M. Thevelein. 2001. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 26:310-317. [DOI] [PubMed] [Google Scholar]
- 315.Rolland, F., J. Winderickx, and J. M. Thevelein. 2002. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2:183-201. [DOI] [PubMed] [Google Scholar]
- 316.Rolland, F., V. Wanke, L. Cauwenberg, P. Ma, E. Boles, M. Vanoni, J. H. de Winde, J. M. Thevelein, and J. Winderickx. 2001. The role of hexose transport and phosphorylation in cAMP signalling in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 1:33-45. [DOI] [PubMed] [Google Scholar]
- 317.Rose, M., W. Albig, and K. D. Entian. 1991. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur. J. Biochem. 199:511-518. [DOI] [PubMed] [Google Scholar]
- 318.Roth, A. F., Y. Feng, L. Chen, and N. G. Davis. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Roth, S., J. Kumme, and H. J. Schuller. 2004. Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr. Genet. 45:121-128. [DOI] [PubMed] [Google Scholar]
- 320.Rudra, D., Y. Zhao, and J. R. Warner. 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Russell, M., J. Bradshaw-Rouse, D. Markwardt, and W. Heideman. 1993. Changes in gene expression in the Ras/adenylate cyclase system of Saccharomyces cerevisiae: correlation with cAMP levels and growth arrest. Mol. Cell. Biol. 4:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Saier, M. H., Jr. 1996. Cyclic AMP-independent catabolite repression in bacteria. FEMS Microbiol. Lett. 138:97-103. [DOI] [PubMed] [Google Scholar]
- 323.Sakamoto, Y., M. Yoshida, K. Semba, and T. Hunter. 1997. Inhibition of the DNA-binding and transcriptional repression activity of the Wilms' tumor gene product, WT1, by cAMP-dependent protein kinase-mediated phosphorylation of Ser-365 and Ser-393 in the zinc finger domain. Oncogene 15:2001-2012. [DOI] [PubMed] [Google Scholar]
- 324.Santangelo, G. M., and J. Tornow. 1990. Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites. Mol. Cell. Biol. 10:859-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Sanz, P., G. R. Alms, T. A. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Sanz, P., K. Ludin, and M. Carlson. 2000. Sip5 interacts with both the Reg1/Glc7 protein phosphatase and the Snf1 protein kinase of Saccharomyces cerevisiae. Genetics 154:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Sasaki, H., and H. Uemura. 2005. Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast 22:111-127. [DOI] [PubMed] [Google Scholar]
- 328.Schawalder, S. B., M. Kabani, I. Howard, U. Choudhury, M. Werner, and D. Shore. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432:1058-1061. [DOI] [PubMed] [Google Scholar]
- 329.Scheffler, I. E., B. J. de la Cruz, and S. Prieto. 1998. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Int. J. Biochem. Cell. Biol. 30:1175-1193. [DOI] [PubMed] [Google Scholar]
- 330.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24:338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Schmidt, M. C., and R. R. McCartney. 2000. β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wolfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Schneper, L., A. Krauss, R. Miyamoto, S. Fang, and J. R. Broach. 2004. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell 3:108-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Schork, S. M., G. Bee, M. Thumm, and D. H. Wolf. 1994. Site of catabolite inactivation. Nature 369:283-284. [DOI] [PubMed] [Google Scholar]
- 335.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139-160. [DOI] [PubMed] [Google Scholar]
- 336.Schuller, H. J., and K. D. Entian. 1991. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J. Bacteriol. 173:2045-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Schwartz, D., C. J. Decker, and R. Parker. 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9:239-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Shima, F., Y. Yamawaki-Kataoka, C. Yanagihara, M. Tamada, T. Okada, K. Kariya, and T. Kataoka. 1997. Effect of association with adenylyl cyclase-associated protein on the interaction of yeast adenylyl cyclase with Ras protein. Mol. Cell. Biol. 17:1057-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10:408-412. [DOI] [PubMed] [Google Scholar]
- 340.Siebert, G. 1961. Enzyme and substrate der glykolyse in isolierten zellkernen. Biochem. Z. 334:369. [Google Scholar]
- 341.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 342.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Smith, F. C., S. P. Davies, W. A. Wilson, D. Carling, and D. G. Hardie. 1999. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 453:219-223. [DOI] [PubMed] [Google Scholar]
- 344.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 345.Sobering, A. K., M. J. Romeo, H. A. Vay, and D. E. Levin. 2003. A novel Ras inhibitor, Eri1, engages yeast Ras at the endoplasmic reticulum. Mol. Cell. Biol. 23:4983-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Song, W., I. Treich, N. Qian, S. Kuchin, and M. Carlson. 1996. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol. Cell. Biol. 16:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker, and C. Wittenberg. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Stanhope-Baker, P., P. M. Kessler, W. Li, M. L. Agarwal, and B. R. Williams. 2004. The Wilms tumor suppressor-1 target gene podocalyxin is transcriptionally repressed by p53. J. Biol. Chem. 279:33575-33585. [DOI] [PubMed] [Google Scholar]
- 349.Stanway, C. A., A. Chambers, A. J. Kingsman, and S. M. Kingsman. 1989. Characterization of the transcriptional potency of sub-elements of the UAS of the yeast PGK gene in a PGK mini-promoter. Nucleic Acids Res. 17:9205-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Stark, M. J. 1996. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 12:1647-1675. [DOI] [PubMed] [Google Scholar]
- 351.Stiller, J. W., E. C. Duffield, and B. D. Hall. 1998. Amitochondriate amoebae and the evolution of DNA-dependent RNA polymerase II. Proc. Natl. Acad. Sci. USA 95:11769-11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Struhl, K., D. Kadosh, M. Keaveney, L. Kuras, and Z. Moqtaderi. 1998. Activation and repression mechanisms in yeast. Cold Spring Harbor Symp. Quant. Biol. 63:413-421. [DOI] [PubMed] [Google Scholar]
- 353.Stuart, J. S., D. L. Frederick, C. M. Varner, and K. Tatchell. 1994. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 14:896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 355.Suzuki, N., H. R. Choe, Y. Nishida, Y. Yanawaki-Kataoka, S. Ohnishi, T. Tamaoki, and T. Kataoka. 1990. Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc. Natl. Acad. Sci. USA 87:8711-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Tachibana, C., J. Y. Yoo, J. B. Tagne, N. Kacherovsky, T. I. Lee, and E. T. Young. 2005. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 25:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Tanaka, K., B. K. Lin, D. R. Wood, and F. Tamanoi. 1991. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc. Natl. Acad. Sci. USA 88:468-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Tanaka, K., K. Matsumoto, and A. Toh-e. 1989. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:757-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Tanaka, K., M. Nakafuku, T. Satoh, M. S. Marshall, J. B. Gibbs, K. Matsumoto, Y. Kaziro, and A. Toh-e. 1990. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60:803-807. [DOI] [PubMed] [Google Scholar]
- 361.Tatchell, K. 1986. RAS genes and growth control in Saccharomyces cerevisiae. J. Bacteriol. 166:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Tatchell, K., D. T. Chaleff, D. DeFeo-Jones, and E. M. Scolnick. 1984. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature 309:523-527. [DOI] [PubMed] [Google Scholar]
- 363.Tatchell, K., L. C. Robinson, and M. Breitenbach. 1985. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc. Natl. Acad. Sci. USA 82:3785-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Taylor, W. E., and E. T. Young. 1990. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc. Natl. Acad. Sci. USA 87:4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 366.Thompson-Jaeger, S., J. Francois, J. P. Gaughran, and K. Tatchell. 1991. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics 129:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 368.Toda, T., S. Cameron, P. Sass, and M. Wigler. 1988. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 2:517-527. [DOI] [PubMed] [Google Scholar]
- 369.Toda, T., S. Cameron, P. Sass, M. Zoller, and M. Wigler. 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277-287. [DOI] [PubMed] [Google Scholar]
- 370.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Tokiwa, G., M. Tyers, T. Volpe, and B. Futcher. 1994. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371:342-345. [DOI] [PubMed] [Google Scholar]
- 372.Tomas-Cobos, L., and P. Sanz. 2002. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem. J. 398:657-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 374.Tornow, J., and G. M. Santangelo. 1990. Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UASRPG) found initially in genes encoding ribosomal proteins. Gene 90:79-85. [DOI] [PubMed] [Google Scholar]
- 375.Tornow, J., X. Zeng, W. Gao, and G. M. Santangelo. 1993. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 12:2431-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Treitel, M. A., and M. Carlson. 1995. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA 92:3132-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Tu, J., and M. Carlson. 1994. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tu, J., and M. Carlson. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Tu, J., W. Song, and M. Carlson. 1996. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4199-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Tung, K. S., and A. K. Hopper. 1995. The glucose repression and RAS-cAMP signal transduction pathways of Saccharomyces cerevisiae each affect RNA processing and the synthesis of a reporter protein. Mol. Gen. Genet. 247:48-54. [DOI] [PubMed] [Google Scholar]
- 382.Tung, K. S., L. L. Norbeck, S. L. Nolan, N. S. Atkinson, and A. K. Hopper. 1992. SRN1, a yeast gene involved in RNA processing, is identical to HEX2/REG1, a negative regulator in glucose repression. Mol. Cell. Biol. 12:2673-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Turkel, S., T. Turgut, M. C. Lopez, H. Uemura, and H. V. Baker. 2003. Mutations in GCR1 affect SUC2 gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 268:825-831. [DOI] [PubMed] [Google Scholar]
- 384.Tzamarias, D., and K. Struhl. 1995. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 9:821-831. [DOI] [PubMed] [Google Scholar]
- 385.Vallier, L. G., and M. Carlson. 1991. New SNF genes, GAL11 and GRR1 affect SUC2 expression in Saccharomyces cerevisiae. Genetics 129:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Vallier, L. G., and M. Carlson. 1994. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics 137:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Vallier, L. G., D. Coons, L. F. Bisson, and M. Carlson. 1994. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics 136:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.van Hemert, M. J., G. P. van Heusden, and H. Y. Steensma. 2001. Yeast 14-3-3 proteins. Yeast 18:889-895. [DOI] [PubMed] [Google Scholar]
- 389.Vanoni, M., R. L. Rossi, L. Querin, V. Zinzalla, and L. Alberghina. 2005. Glucose modulation of cell size in yeast. Biochem. Soc. Trans. 33:294-296. [DOI] [PubMed] [Google Scholar]
- 390.van Tuinen, E., and H. Riezman. 1987. Immunolocalization of glyceraldehydes-3-phosphate dehydrogenase, hexokinase, and carboxypeptidase Y in yeast cells at the ultrastructural level. J. Histochem. Cytochem. 35:327-333. [DOI] [PubMed] [Google Scholar]
- 391.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7:1191-1200. [DOI] [PubMed] [Google Scholar]
- 392.Velculescu, V. E., L. Zhang, W. Zhou, J. Vogelstein, M. A. Basrai, D. E. Bassett Jr., P. Hieter, B. Vogelstein, and K. W. Kinzler. 1997. Characterization of the yeast transcriptome. Cell 88:243-251. [DOI] [PubMed] [Google Scholar]
- 393.Verma, R., R. M. Feldman, and R. J. Deshaies. 1997. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell 8:1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Versele, M., J. H. de Winde, and J. M. Thevelein. 1999. A novel regulator of G protein signaling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 18:5577-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Vincent, O., and M. Carlson. 1998. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17:7002-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Vincent, O., and M. Carlson. 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Vojtek, A. B., and D. G. Fraenkel. 1990. Phosphorylation of yeast hexokinases. Eur. J. Biochem. 190:371-375. [DOI] [PubMed] [Google Scholar]
- 399.von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Vyas, V. K., S. Kuchin, and M. Carlson. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Vyas, V. K., S. Kuchin, C. D. Berkey, and M. Carlson. 2003. Snf1 kinases with different β-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 23:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wade, J. T., D. B. Hall, and K. Struhl. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432:1054-1058. [DOI] [PubMed] [Google Scholar]
- 403.Wakula, P., M. Beullens, H. Ceulemans, W. Stalmans, and M. Bollen. 2003. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 278:18817-18823. [DOI] [PubMed] [Google Scholar]
- 404.Walsh, E. P., D. J. Lamont, K. A. Beattie, and M. J. Stark. 2002. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 41:2409-2420. [DOI] [PubMed] [Google Scholar]
- 405.Walther, K., and H. J. Schuller. 2001. Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology 147:2037-2044. [DOI] [PubMed] [Google Scholar]
- 406.Wang, J., and R. Needleman. 1996. Removal of Mig1p binding site converts a MAL63 constitutive mutant derived by interchromosomal gene conversion to glucose insensitivity. Genetics 142:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Wang, L., G. Renault, H. Garreau, and M. Jacquet. 2004. Stress induces depletion of Cdc25p and decreases the cAMP producing capability in Saccharomyces cerevisiae. Microbiology 150:3383-3391. [DOI] [PubMed] [Google Scholar]
- 408.Wang, Y., M. Pierce, L. Schneper, C. G. Guldal, X. Zhang, S. Tavazoie, and J. R. Broach. 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2:0610-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Warner, J. R. 1989. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev. 53:256-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 411.Weeks, G., and G. B. Spiegelman. 2003. Roles played by Ras subfamily proteins in the cell and developmental biology of microorganisms. Cell Signal. 15:901-909. [DOI] [PubMed] [Google Scholar]
- 412.Wek, R. C., J. F. Cannon, T. E. Dever, and A. G. Hinnebusch. 1992. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2 alpha kinase GCN2. Mol. Cell. Biol. 12:5700-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces Cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 414.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695:133-170. [DOI] [PubMed] [Google Scholar]
- 415.Willett, C. E., C. M. Gelfman, and M. J. Holland. 1993. A complex regulatory element from the yeast gene ENO2 modulates GCR1-dependent transcriptional activation. Mol. Cell. Biol. 13:2623-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Willis, K. A., K. E. Barbara, B. B. Menon, J. Moffat, B. Andrews, and G. M. Santangelo. 2003. The global transcriptional activator of Saccharomyces cerevisiae, Gcr1p, mediates the response to glucose by stimulating protein synthesis and CLN-dependent cell cycle progression. Genetics 165:1017-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Wills, C. 1990. Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 25:245-280. [DOI] [PubMed] [Google Scholar]
- 418.Willumsen, B. M., A. D. Cox, P. A. Solski, C. J. Der, and J. E. Buss. 1996. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene 13:1901-1909. [PubMed] [Google Scholar]
- 419.Wilson, B. A., M. Khalil, F. Tamanoi, and J. F. Cannon. 1993. New activated RAS2 mutations identified in Saccharomyces cerevisiae. Oncogene 8:3441-3445. [PubMed] [Google Scholar]
- 420.Wilson, R. B., and K. Tatchell. 1988. SRA5 encodes the low-Km cyclic AMP phosphodiesterase of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 422.Woods, A., S. R. Johnstone, K. Dickerson, F. C. Leiper, L. G. Fryer, D. Neumann, U. Schlattner, T. Wallimann, M. Carlson, and D. Carling. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13:2004-2008. [DOI] [PubMed] [Google Scholar]
- 423.Wu, J., and R. J. Trumbly. 1998. Multiple regulatory proteins mediate repression and activation by interaction with the yeast Mig1 binding site. Yeast 14:985-1000. [DOI] [PubMed] [Google Scholar]
- 424.Wu, M., L. Newcomb, and W. Heideman. 1999. Regulation of gene expression by glucose in Saccharomyces cerevisiae: a role for ADA2 and ADA3/NGG1. J. Bacteriol. 181:4755-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Wu, X., and K. Tatchell. 2001. Mutations in yeast protein phosphatase type 1 that affect targeting subunit binding. Biochemistry 40:7410-7420. [DOI] [PubMed] [Google Scholar]
- 426.Wu, X., H. Hart, C. Cheng, P. J. Roach, and K. Tatchell. 2001. Characterization of Gac1p, a regulatory subunit of protein phosphatase type I involved in glycogen accumulation in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:622-635. [DOI] [PubMed] [Google Scholar]
- 427.Xu, X., J. D. Wightman, B. L. Geller, D. Avram, and A. T. Bakalinsky. 1994. Isolation and characterization of sulfite mutants of Saccharomyces cerevisiae. Curr. Genet. 25:488-496. [DOI] [PubMed] [Google Scholar]
- 428.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Yang, X., E. J. Hubbard, and M. Carlson. 1992. A protein kinase substrate identified by the two-hybrid system. Science 257:680-682. [DOI] [PubMed] [Google Scholar]
- 430.Yang, X., R. Jiang, and M. Carlson. 1994. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 13:5878-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Yang, Z., and L. F. Bisson. 1996. The SKS1 protein kinase is a multicopy suppressor of the snf3 mutation of Saccharomyces cerevisiae. Yeast 12:1407-1419. [DOI] [PubMed] [Google Scholar]
- 432.Young, E. T., K. M. Dombek, C. Tachibana, and T. Ideker. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278:26146-26158. [DOI] [PubMed] [Google Scholar]
- 433.Young, E. T., N. Kacherovsky, and K. Van Riper. 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 277:38095-38103. [DOI] [PubMed] [Google Scholar]
- 434.Zaman, Z., A. Z. Ansari, L. Gaudreau, J. Nevado, and M. Ptashne. 1998. Gene transcription by recruitment. Cold Spring Harbor Symp. Quant. Biol. 63:167-171. [DOI] [PubMed] [Google Scholar]
- 435.Zeng, X., S. J. Deminoff, and G. M. Santangelo. 1997. Specialized Rap1p/Gcr1p transcriptional activation through Gcr1p DNA contacts requires Gcr2p, as does hyperphosphorylation of Gcr1p. Genetics 147:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Zenke, F. T., R. Engles, V. Vollenbroich, J. Meyer, C. P. Hollenberg, and K. D. Breunig. 1996. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 272:1662-1665. [DOI] [PubMed] [Google Scholar]
- 437.Zhang, S., S. Guha, and F. C. Volkert. 1995. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol. Cell. Biol. 15:2037-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Zhou, H., and F. Winston. 2001. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Zhu, X., N. Demolis, M. Jacquet, and T. Michaeli. 2000. MSI1 suppresses hyperactive RAS via the cAMP-dependent protein kinase and independently of chromatin assembly factor-1. Curr. Genet. 38:60-70. [DOI] [PubMed] [Google Scholar]
- 440.Zimmermann, F. K., I. Kaufmann, H. Rasenberger, and P. Haubetamann. 1977. Genetics of carbon catabolite repression in Saccharomycess cerevisiae: genes involved in the derepression process. Mol. Gen. Genet. 151:95-103. [DOI] [PubMed] [Google Scholar]
- 441.Zurita-Martinez, S. A., and M. E. Cardenas. 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 4:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]