Short abstract
The major native forms of 14-3-3s are homo- and hetero-dimers, the biological functions of which are to interact physically with specific client proteins and thereby effect a change in the client. As a result, 14-3-3s are involved in a vast array of processes such as the response to stress, cell-cycle control, and apoptosis, serving as adapters, activators, and repressors. There are currently 133 full-length sequences available.
Abstract
Multiple members of the 14-3-3 protein family have been found in all eukaryotes so far investigated, yet they are apparently absent from prokaryotes. The major native forms of 14-3-3s are homo- and hetero-dimers, the biological functions of which are to interact physically with specific client proteins and thereby effect a change in the client. As a result, 14-3-3s are involved in a vast array of processes such as the response to stress, cell-cycle control, and apoptosis, serving as adapters, activators, and repressors. There are currently 133 full-length sequences available in GenBank for this highly conserved protein family. A phylogenetic tree based on the conserved middle core region of the protein sequences shows that, in plants, the 14-3-3 family can be divided into two clearly defined groups. The core region encodes an amphipathic groove that binds the multitude of client proteins that have conserved 14-3-3-recognition sequences. The amino and carboxyl termini of 14-3-3 proteins are much more divergent than the core region and may interact with isoform-specific client proteins and/or confer specialized subcellular and tissue localization.
The 14-3-3 protein family is highly conserved and is represented throughout the eukaryotic branch of life. The proteins were discovered in 1967 during a study of the soluble acidic proteins of the mammalian brain [1] and were named on the basis of fraction number during DEAE-cellulose chromatography and location after starch gel electrophoresis. For 25 years, 14-3-3s were generally thought to reside exclusively in animal brain tissue and to be involved in the function of neurons. During this early period of research, 14-3-3s were characterized as a heterogeneous family of dimeric proteins with a monomer mass of 25-32 kDa and multiple isoelectric points. One of the first biochemical functions of the 14-3-3s to be identified was the activation of the neurotransmitter pathway enzymes tyrosine hydroxylase and tryptophan hydroxylase, in a reaction requiring calcium and the cAMP-dependent kinase or calmodulin-dependent protein kinase II [2]. Once 14-3-3s were found in Arabidopsis thaliana [3], maize [4] and other plants, and in tissues other than the brain, however, perspectives on their presence and roles broadened, and now 14-3-3s have been found in every eukaryote that has been screened for their presence. The vast number of organisms containing 14-3-3s suggests that this family of proteins is involved in many important biological processes [5].
Gene organization and evolutionary history
Each 14-3-3 protein sequence can be roughly divided into three sections: a divergent amino terminus, the conserved core region and a divergent carboxyl terminus. The high level of conservation in the core region is demonstrated in Figure 1a, which shows a similarity plot derived from an alignment of selected 14-3-3s. A phylogenetic tree generated from the full alignment of the core region is shown in Figure 2 and demonstrates that plant 14-3-3s fall into two groups, an epsilon (ε) group and a non-epsilon group [6,7]. The plant non-epsilon group is very different from plant ε isoforms and animal isoforms. Additional complex groupings and subgroupings are apparent, but the tree must be evaluated carefully because not all species are represented by a complete genome sequence [8]. The presence of the divergent termini and the few amino-acid changes that do occur within the conserved region result in multiple isoforms in most organisms and present the potential for client-specific interactions occurring in distinct cellular locations [9].
Figure 1.
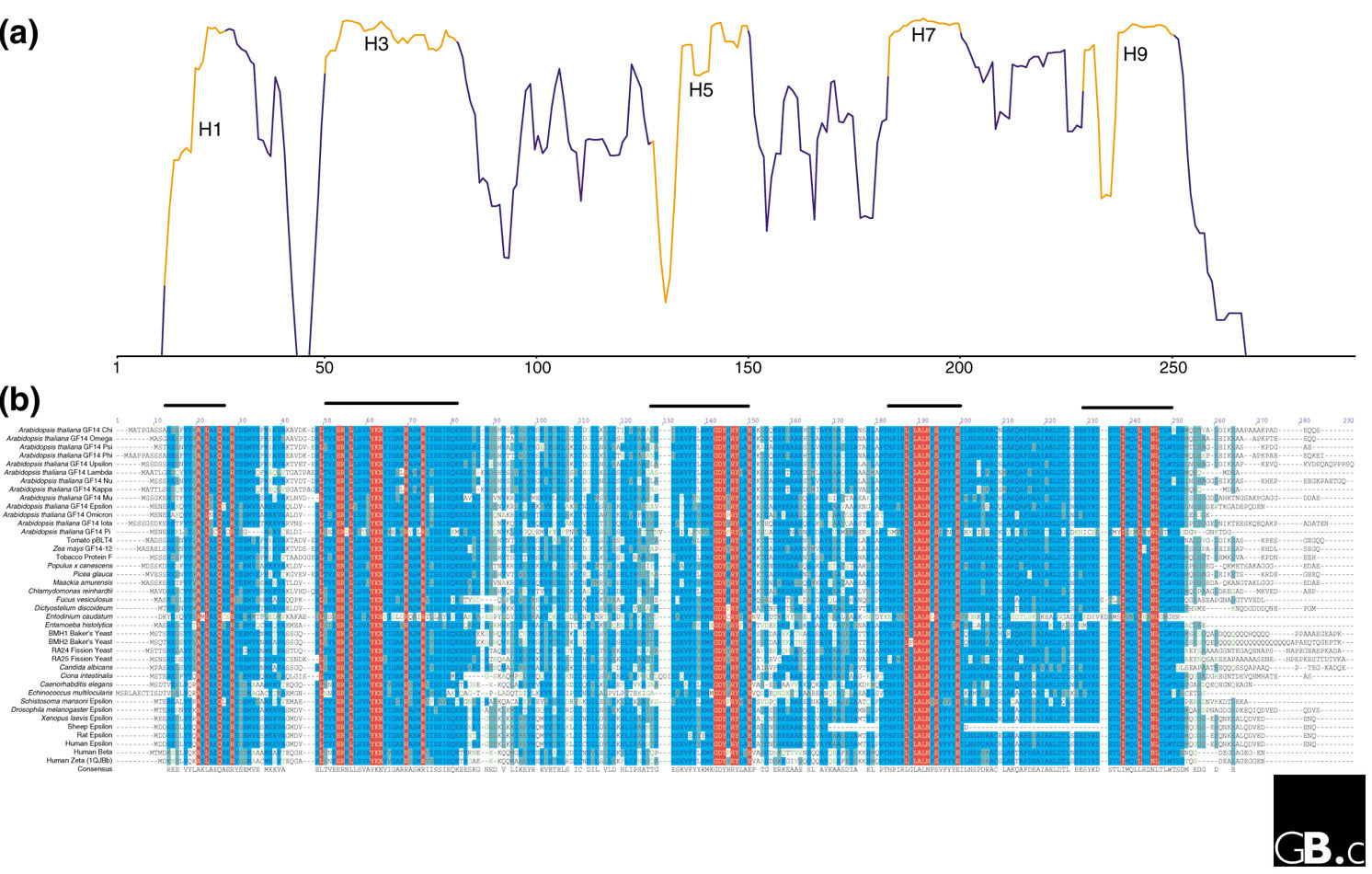
Conservation of 14-3-3 proteins. (a) A graph of percentage similarity derived from (b), which shows a multiple sequence alignment of 40 selected 14-3-3 isoforms. Highly conserved helices in (a) are in red and are indicated by numbers that correspond to helices in the crystal structure of Figure 3. The alignment in (b) was created using Clustal W [39]. Residues in red are 100% conserved across all isoforms; residues in blue are highly conserved. These red and blue colors correspond to the red and blue regions of the crystal structure shown in Figure 3.
Figure 2.
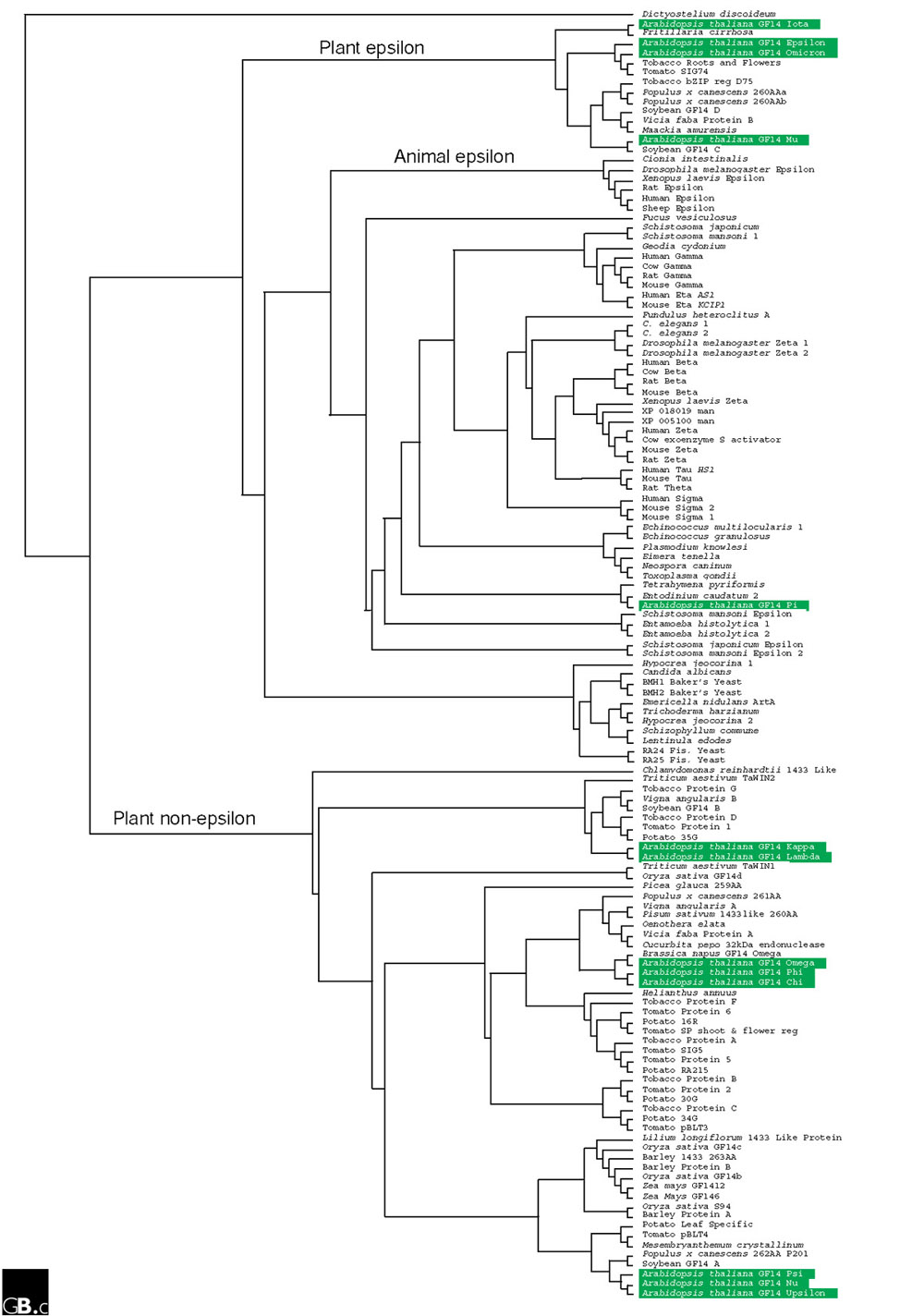
Phylogenetic relationships of 14-3-3s. This tree is a rooted cladogram from a neighbor-joining analysis [40] of the 133 different full-length 14-3-3 isoforms that are currently available in the GenBank database [37], with Dictyostelium discoideum 14-3-3 as the outgroup. Arabidopsis isoforms are highlighted in green. The separation of plant epsilon and non-epsilon 14-3-3 proteins is clearly visible.
Although 14-3-3s are found throughout the eukaryotes, Arabidopsis is an excellent model system for studying 14-3-3s for two reasons: it is a higher eukaryote with a fully sequenced genome and it has a large family of thirteen 14-3-3 isoforms [7]; there is also a wealth of knowledge on key biological pathways. In Arabidopsis, the 14-3-3 epsilon group has five members (μ (mu), ε, π (pi), ι (iota), and ο (omicron) and the non-epsilon group has eight members (κ (kappa), λ (lambda), ψ (psi), ν (nu), υ (upsilon), ω (omega),  (phi), and χ (chi)). The presence and chromosomal location of all Arabidopsis 14-3-3 isoforms is known (Table 1); there is at least one 14-3-3 on each of the five chromosomes. Similar diversity in chromosomal distribution occurs in other species where multiple 14-3-3s have been found.
(phi), and χ (chi)). The presence and chromosomal location of all Arabidopsis 14-3-3 isoforms is known (Table 1); there is at least one 14-3-3 on each of the five chromosomes. Similar diversity in chromosomal distribution occurs in other species where multiple 14-3-3s have been found.
Table 1.
Genetic, cellular and functional information on Arabidopsis 14-3-3s
| Gene name | Isoform | Locus* | Cellular localization† | Tissue distribution | Gene accession number [37] | |||
| GRF1 | Chi (χ) | At4g09000 | N | Pollen, stigma papillar cells | U09377 | |||
| GRF2 | Omega (ω) | At1g78300 | ? | ? | U09376 | |||
| GRF3 | Psi (ψ) | At5g38480 | ? | Stems, leaves, flowers | U09375 | |||
| GRF4 | Phi ( |
At1g35160 | Nm, Pm, Ct | ? | AF001414 | |||
| GRF5 | Upsilon (υ) | At5g16050 | Nm, Pm, Ct | ? | AF001415 | |||
| GRF6 | Lambda (λ) | At5g10450 | N, Pm, Ct | Stems, leaves, flowers | AF145298 | |||
| GRF7 | Nu (ν) | At3g02520 | Nm, Pm, Ct | ? | AF145299 | |||
| GRF8 | Kappa (κ) | At5g65430 | N, Pm/Cw, Ct | ? | AF145300 | |||
| GRF9 | Mu (μ) | At2g42590 | Sg | Leaves | AF145301 | |||
| GRF10 | Epsilon (ε) | At1g22300 | Ne, Pm, Ct, Sg | Leaves | AF145302 | |||
| GRF11 | Omicron (o) | At1g34760 | ? | Stems, roots, flowers | AF323920 | |||
| GRF12 | Iota (ι) | At1g26480 | ? | Flowers | AF335544 | |||
| GRF13 | Pi (π) | At1g78220 | ? | ? | AAF18556.1 | |||
*The number after 'At' † N, nucleus; Nm, nuclear membrane; Pm, plasma membrane; Ct, cytoplasm; Cw, cell wall; denotes the chromosome. Sg, leaf starch grain; Ne, nuclear envelope [7,14,30]. For further information on 14-3-3s, please see the Ferl lab website [38].
Characteristic structural features
The conserved middle core region of the 14-3-3s encodes an amphipathic groove that forms the main functional domain, a cradle for interacting with client proteins. Extensive investigation of the 14-3-3 binding site of the mammalian serine/threonine kinase Raf-1 has produced a consensus sequence for 14-3-3-binding, RSxpSxP (in the single-letter amino-acid code, where x denotes any amino acid and p indicates that the next residue is phosphorylated) [10] which has been verified through peptide library screening [11]. A common, but not exclusive, requirement of 14-3-3 ligands is the phosphorylation of a serine or threonine residue in the target sequence. The phosphorylated consensus sequence does not, however, fully represent every ligand that 14-3-3s can bind: they are also known to bind other non-phosphorylated sequences such as GHSL [12], and WLDLE [13]. A common, but not exclusive, requirement of 14-3-3 ligands is the phosphorylation of a serine residue in the target sequence. For those client proteins whose target sequences undergo phosphorylation, the binding of 14-3-3s to the target is the major step of a signal-transduction event. Despite the simplicity of the binding-site requirements, a diverse array of proteins potentially interact with 14-3-3s; some reports suggest that as many as 20% of Arabidopsis proteins are clients for 14-3-3s [14].
Our knowledge of the three-dimensional structure of 14-3-3s is based on the model derived from X-ray diffraction of the crystals of the ζ and τ mammalian isoforms [15,16]. The high level of conservation of 14-3-3 amino acid sequence in the conserved core allows the general features of this structural model to be applied to all known 14-3-3s. The monomer consists of nine α helices organized in an antiparallel manner, forming an L-shaped structure (Figure 3). The interior of the L-structure is composed of four helices: H3 and H5, which contain many charged and polar amino acids, and H7 and H9, which contain hydrophobic amino acids. These four helices form the concave amphipathic groove that interacts with target peptides. An alignment of all currently known full-length isoforms provides evidence that this groove is over 70% conserved (Figures 1 and 3). Five of the most highly conserved regions correspond to helices H1, H3, H5, H7, and h9 (Figure 1). The conserved amphipathic groove is the site for ligand binding; amino acids Lys49, Arg56, and Arg127 in mammalian 14-3-3 ζ sequences have been demonstrated to interact with the phosphorylated amino acids of ligands (serine/threonine kinases Raf-1 and Bcr) by mutational [17,18,19] and co-crystallization [11,13] experiments; the latter have also shown that both phosphorylated and non-phosphorylated ligands bind in the amphipathic groove [13]. The peptides bound by 14-3-3s adopt an extended conformation, which is thought to reduce steric hindrance between neighboring amino acids of the ligand [20].
Figure 3.
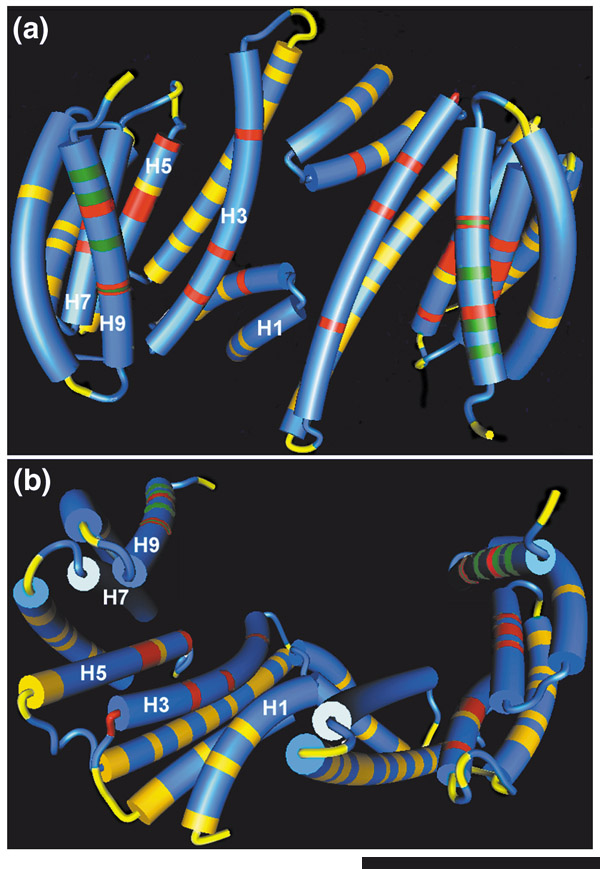
The crystal structure of 14-3-3s. The model shown is derived from the human 14-3-3 ζ isoform (PDB1QJB [22]) and is shown from (a) the top and (b) one side as visualized by the 3Dmol software found in the Vector NTI 7.0 Suite. Helix numbers are denoted from H1 near the amino terminus to H9 near the carboxyl terminus. Red and blue areas correspond to residues of 100% identity and high conservation, respectively, and correspond to colors on the alignment (Figure 1). Yellow areas correspond to regions of reduced similarity and green areas indicate the nuclear export signal [22].
The 14-3-3 dimerization interaction occurs between the amino-terminal helix H1 of one monomer and helices H3 and H4 of the opposing monomer; the high conservation of amino-acid sequence along helices H1 and H3 among various isoforms allows 14-3-3s to heterodimerize (Figure 3) [21,22]. Two identical or different client proteins can be bound simultaneously by the dimer. This dual interaction means that possible roles of 14-3-3s include acting as adapters capable of bringing disparate client proteins together or moving or rearranging two different regions of the same protein. An example of the potential role of 14-3-3 dimers as adapters comes from studies of 14-3-3s interacting with the plant plasma-membrane proton ATPase and the plant toxin fusicoccin [6]. It has been suggested that the 14-3-3 dimer binds the carboxy-terminal autoinhibitory (C-TA) domain of the ATPase in the presence of magnesium, creating a binding site where fusicoccin can then interact [6]. Once fusicoccin is bound, the complex of 14-3-3 and the ATPase is stabilized and the C-TA domain is displaced, allowing the ATPase to become fully active.
Localization and function
In general, 14-3-3s are distributed widely throughout the cell, supporting the argument that they are involved in multiple protein-protein interactions in a plethora of biological roles. There is, however, some differential subcellular localization, suggesting an element of specialization among specific isoforms. Localization data have been collected for eight of the Arabidopsis isoforms using isoform-specific antibodies and fusions of 14-3-3 to green fluorescent protein (GFP; Table 1). These data do not necessarily exhaust all possible locations for each isoform; instead, they support the idea that certain isoforms are recruited to distinct subcellular locations. The localization of 14-3-3 κ and υ was studied using carboxy-terminal GFP fusions in transgenic Arabidopsis [14]. Fusions of κ with GFP tended to localize to the plasma membrane, whereas υ-GFP fusions tended to be found in the cytosol [14]. Additional data collected using microscopy and immuno-cytochemistry of total nuclear extracts showed that at least five different forms of 14-3-3s are found in the nucleus [23]. Chloroplast stromal extracts screened with isoform-specific antibodies showed that 14-3-3 μ and ε (members of the epsilon group) and 14-3-3 υ and ν (members of the non-epsilon group) were the only 14-3-3s prominently located in the chloroplast [24]. The presence of the two non-epsilon members 14-3-3 ν and υ in the chloroplast suggests that these proteins, although located on distinct branches of the phylogenetic tree, may share similar roles and cellular locations [25]. 14-3-3 ε and μ were also found in starch grains [26]; ε has also been found at the nuclear envelope during 14-3-3-GFP studies. GFP-ω fusion studies showed that 14-3-3 nuclear localization is regulated by the cell-cycle [27]; generally, 14-3-3-ω-GFP fusions were excluded from the nucleus, but they accumulated in the nucleus just after nuclear division and then relocated back out of the nucleus just before completion of cytokinesis [28]. In addition, a nuclear export signal was identified in the 14-3-3s of the fission yeast Schizosaccharomyces pombe that is required (in concert with the Crm1 nuclear export machinery) for the shuttling of the mitosis-inducing protein Cdc25 out of the nucleus following DNA damage [29]. Nuclear shuttling has emerged as an important biological role for 14-3-3s [29], and the nuclear export signal (I/LxxxLxxxLxL) is highly conserved in the 133 full-length 14-3-3 sequences currently available (Figures 1 and 3).
The 14-3-3s are also differentially expressed among tissues and organs (Table 1). Arabidopsis 1ψ, λ, μ, and ε are found in the leaves; ψ and λ are also expressed in the stems and flowers [30], and χ is expressed in pollen grains and stigma papillar cells [25]. The fact that 14-3-3s are differentially expressed in various tissues and differentially localized in subcellular compartments adds a layer of complexity to the examination and determination of biological roles for 14-3-3s, a complexity that must be reconciled with the highly conserved nature of 14-3-3-target interactions.
The identification of mutants and the use of transgenic organisms have provided some insight into some of the biological roles and locations of some 14-3-3s. For instance, a mutation in the RAD24 protein, one of the two 14-3-3s in S. pombe, reduces the yeast's ability to keep DNA damage in check [28]. In Saccharomyces cerevisiae, disruption of the 14-3-3 genes BMH1 and BMH2 creates a lethal phenotype that can be rescued by introducing 14-3-3 isoforms from Arabidopsis, Dictyostelium discoideum, or Homo sapiens [31]. The Leonardo (14-3-3 ζ) protein of Drosophila melanogaster is known to regulate presynaptic function, and its mutation results in the death of mature embryos [32]. Two transgenic Arabidopsis lines, one carrying an antisense construct against 14-3-3 μ and another against 14-3-3 ε show dramatic increases in starch production in leaves [26].
Various 14-3-3s in a variety of species have been found to interact with proteins involved in signal transduction (such as Raf-1 [33]), apoptosis (such as the Bcl2-related protein Bad [34]), cell-cycle control (such as Cdc25 [35]), starch synthesis [26], nitrogen metabolism [36], and ATP regulation (reviewed in [6]).
Frontiers
Initially, discoveries of 14-3-3s were almost coincidental in nature; in many cases their identification was serendipitous after investigating other biochemical questions. Once it became established that these proteins were ubiquitous, research was directed toward identifying the number and sequences of isoforms present in different species as well as determining their functional diversity. As more genomes are sequenced, experimental tasks will move towards elucidation of general roles as well as investigation of isoform-specific roles in order to address the implications of 14-3-3 family diversity within organisms. Such studies are key to understanding the current conundrum: the conservation of 14-3-3s throughout eukaryotes suggests that some central biological roles might be served by any 14-3-3 protein, yet the diversity of 14-3-3 isoforms argues for a multitude of specific roles. Indeed, some combination of both concepts might be the case, with some roles being served by any isoform, and other roles requiring isoform-specific interactions. In any case, all the current data suggest that interactions involving 14-3-3 proteins are critical for the correct function of higher-order biological systems.
The presence of 14-3-3 proteins in most, if not all, eukaryotic cells, but not in any prokaryotic cells, offers an interesting opportunity to study the early evolutionary history of this protein family and the concomitant development of eukaryotic regulatory processes.
References
- Moore B, Perez V. Specific acidic proteins of the nervous system. In: Carlson FD, editor. In Physiological and Biochemical Aspects of Nervous Integration. Englewood Cliffs, NJ: Prentice-Hall; 1967. pp. 343–359. The first identification of 14-3-3 proteins in mammal brains. [Google Scholar]
- Ichimura T, Isobe T, Okuyama T, Yamauchi T, Fujisawa H. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. This study was the first to assign a role to 14-3-3 proteins. It was shown that 14-3-3s activate tyrosine and tryptophan hydroxylases in a calcium- and kinase-dependent manner. [DOI] [PubMed] [Google Scholar]
- Lu G, DeLisle AJ, de Vetten NC, Ferl RJ. Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc Natl Acad Sci USA. 1992;89:11490–11494. doi: 10.1073/pnas.89.23.11490. The first identification of 14-3-3 proteins in Arabidopsis thaliana. They were found to be associated with G-box-binding factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten NC, Lu G, Ferl RJ. A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. Plant Cell. 1992;4:1295–1307. doi: 10.1105/tpc.4.10.1295. The first identification of 14-3-3 proteins in maize. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays. 2001;23:936–946. doi: 10.1002/bies.1134. A review discussing the roles of 14-3-3s in cell-cycle control, signal transduction, and apoptosis. [DOI] [PubMed] [Google Scholar]
- Chung HJ, Sehnke PC, Ferl RJ. The 14-3-3 proteins: cellular regulators of plant metabolism. Trends Plant Sci. 1999;4:367–371. doi: 10.1016/s1360-1385(99)01462-4. A review of the participation of 14-3-3s in plant regulatory events, including the regulation of plasma membrane H1-ATPase, nitrate reductase and sucrose phosphate synthase. [DOI] [PubMed] [Google Scholar]
- DeLille JM, Sehnke PC, Ferl RJ. The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001;126:35–38. doi: 10.1104/pp.126.1.35. A review of Arabidopsis 14-3-3 gene structure and phylogeny. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist M, Sehnke P, Ferl RJ, Sommarin M, Larsson C. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J Mol Evol. 2000;51:446–458. doi: 10.1007/s002390010107. This study describes the binding affinity of Arabidopsis 14-3-3 isoforms to a unique peptide and discusses the potential for functional specificity for these proteins across all multicellular species. A substantial family tree is also presented. [DOI] [PubMed] [Google Scholar]
- Ferl RJ. 14-3-3 proteins and signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:49–73. doi: 10.1146/annurev.arplant.47.1.49. A comprehensive review of the history, functions, cell-specific expression, evolution and structure of 14-3-3 proteins. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. This study was the first to show that 14-3-3 is a specific phosphoserine-binding protein. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. Two different binding motifs were identified after screening mammalian and yeast peptide libraries with 14-3-3 proteins. Additionally, the crystal structure of 14-3-3 ζ bound to a phosphoserine target peptide was solved. [DOI] [PubMed] [Google Scholar]
- Andrews RK, Harris SJ, McNally T, Berndt MC. Binding of purified 14-3-3 zeta signaling protein to discrete amino acid sequences within the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX-V complex. Biochemistry. 1998;37:638–647. doi: 10.1021/bi970893g. This study presents another non-phosphorylated target, GHSL, discovered via co-purification experiments. [DOI] [PubMed] [Google Scholar]
- Petosa C, Masters SC, Bankston LA, Pohl J, Wang B, Fu H, Liddington RC. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. 14-3-3 crystal structures in complex with phosphorylated and unphosphorylated peptide were solved to determine the location of the binding site. [DOI] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. Consummating signal transduction: the role of 14-3-3 proteins in completion of signal-induced transitions in protein activity. Plant Cell. 2002;14:S339–S354. doi: 10.1105/tpc.010430. A comprehensive review of the involvement of 14-3-3 proteins in signal transduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. Crystal structure of the mammalian 14-3-3 ζ isoform. [DOI] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. Crystal structure of the mammalian 14-3-3 τ isoform. [DOI] [PubMed] [Google Scholar]
- Zhang SH, Kobayashi R, Graves PR, Piwnica-Worms H, Tonks NK. Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3beta protein. J Biol Chem. 1997;272:27281–27287. doi: 10.1074/jbc.272.43.27281. Mutation of serine to alanine severely reduced the level of 14-3-3 and PTPH1 binding. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang L, Liddington R, Fu H. Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3zeta disrupt its interaction with Raf-1 kinase. J Biol Chem. 1998;273:16297–16304. doi: 10.1074/jbc.273.26.16297. Mutation of hydrophobic residues on the 14-3-3 amphipathic groove shows that these residues are involved with binding the client protein. The mutated residues are conserved among isoforms, suggesting a general importance for ligand binding. [DOI] [PubMed] [Google Scholar]
- Thorson JA, Yu LW, Hsu AL, Shih NY, Graves PR, Tanner JW, Allen PM, Piwnica-Worms H, Shaw AS. 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol Cell Biol. 1998;18:5229–5238. doi: 10.1128/mcb.18.9.5229. Overexpression of 14-3-3s causes increased phosphorylation of Raf-1. Similarly, removal of 14-3-3 reduces the kinase activity of Raf-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. Comprehensive review of 14-3-3-ligand interaction on a structural basis, a proposal of 14-3-3 function in several pathways, and a discussion of regulatory mechanisms. [DOI] [PubMed] [Google Scholar]
- Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. Evidence that 14-3-3 proteins form both hetero- and homodimers. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. A nuclear export signal was identified in 14-3-3 proteins and shown to share topology with other signals recognized by the Crm1 nuclear export machinery. [DOI] [PubMed] [Google Scholar]
- Bihn EA, Paul AL, Wang SW, Erdos GW, Ferl RJ. Localization of 14-3-3 proteins in the nuclei of Arabidopsis and maize. Plant J. 1997;12:1439–1445. doi: 10.1046/j.1365-313x.1997.12061439.x. Scanning confocal microscopy and immunocytochemistry with mono-clonal antibodies against plant 14-3-3 proteins were used for localization studies. [DOI] [PubMed] [Google Scholar]
- Sehnke PC, Henry R, Cline K, Ferl RJ. Interaction of a plant 14-3-3 protein with the signal peptide of a thylakoid-targeted chloroplast precursor protein and the presence of 14-3-3 isoforms in the chloroplast stroma. Plant Physiol. 2000;122:235–242. doi: 10.1104/pp.122.1.235. Immuno-electron microscopy of leaf tissue and western-blotting analysis of chloroplast fractions with monoclonal anti-14-3-3 antibodies localized 14-3-3 proteins to the chloroplast stroma and the stromal side of thylakoid membranes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty CJ, Rooney MF, Miller PW, Ferl RJ. Molecular organization and tissue-specific expression of an Arabidopsis 14-3-3 gene. Plant Cell. 1996;8:1239–1248. doi: 10.1105/tpc.8.8.1239. In situ hybridization showed that expression of χ (chi) mRNA was prominent in epidermal tissue of roots, petals, and sepals of flower buds, papillae cells of flowers, siliques, and endosperm of immature seeds. These results show plant 14-3-3 gene expression exhibits cell-and tissue-specific localization rivaling that observed for 14-3-3 proteins within the mammalian brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, Chung HJ, Wu K, Ferl RJ. Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc Natl Acad Sci USA. 2001;98:765–770. doi: 10.1073/pnas.021304198. 14-3-3 proteins in Arabidopsis were shown to regulate starch synthesis through the use of two antisense constructs of 14-3-3 ε and 14-3-3 μ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. 14-3-3-GFP fusions were observed in hypocotyl cells by confocal microscopy and showed localization to nuclei undergoing cytokinesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JC, al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Carr AM. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science. 1994;265:533–535. doi: 10.1126/science.8036497. This investigation identified two 14-3-3 proteins, Rad24 and Rad25, that are involved in the DNA-damage checkpoint during mitosis. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. The Rad24 14-3-3 protein controls the intracellular distribution of Cdc25. [DOI] [PubMed] [Google Scholar]
- Chung H-J, Shanker S, Ferl RJ. Sequences of five Arabidopsis general regulatory factor (GRF) genes encoding 14-3-3 proteins. Plant Physiol. 1999;120:1206. doi: 10.1104/pp.107.1.283. An entry in the Plant Gene Register. Sequences were obtained from stem and flower tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heusden GP, van der Zanden AL, Ferl RJ, Steensma HY. Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Lett. 1996;391:252–256. doi: 10.1016/0014-5793(96)00746-6. In this exquisite study, the genes encoding two indigenous 14-3-3 iso-forms in bakers' the phenotypes were rescued by replacement of the genes with those of four of six Arabidopsis isoforms. [DOI] [PubMed] [Google Scholar]
- Broadie K, Rushton E, Skoulakis EM, Davis RL. Leonardo, a Drosophila 14-3-3 protein involved in learning, regulates presynaptic function. Neuron. 1997;19:391–402. doi: 10.1016/s0896-6273(00)80948-4. Immunolocalization studies showed that the Leonardo 14-3-3 protein was expressed at synaptic connections enriched in presynaptic boutons of the neuromuscular junction. Null leonardo mutants die as mature embryos. [DOI] [PubMed] [Google Scholar]
- Chang HC, Rubin GM. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. This study discusses how 14-3-3 ε protein appears to function in multiple receptor tyrosine kinase signal transduction pathways. [DOI] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. An example of how a 14-3-3 is involved with apoptosis. In the presence of a survival factor, Bad was phosphorylated on two serine residues embedded in 14-3-3 consensus binding sites. Only the non-phosphorylated Bad promotes cell death. [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. An example of 14-3-3s involved in cell-cycle control. A mutation in human Cdc25c prevented the phosphorylation of Ser216, thus preventing 14-3-3 binding. Conditional overexpression of this mutant perturbed mitotic timing and allowed cells to escape the G2 checkpoint arrest induced by either unreplicated DNA or radiation-induced damage. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 14-3-3 proteins associate with the regulatory phosphoryla-tion site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 1996;398:26–30. doi: 10.1016/s0014-5793(96)01188-x. 14-3-3 proteins were shown to regulate nitrate reductase by binding a synthetic phosphonitrate reductase peptide. [DOI] [PubMed] [Google Scholar]
- Entrez nucleotide view http://www.ncbi.nlm.nih.gov/entrez/query.fcgi Access to the GenBank sequence database.
- The laboratory of Robert J. Ferl http://www.hos.ufl.edu/ferllab/ Our lab's website contains information on 14-3-3s and related research.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. CLUSTAL W is a common method used to generate multiple sequence alignments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. The neighbor-joining method is a statistical method used to generate possible phylogenetic trees. [DOI] [PubMed] [Google Scholar]


