Abstract
Arabidopsis possesses several genes related to the multidrug resistance (MDR) genes of animals, one of which, AtMDR1, was shown to be induced by the hormone auxin. Plants having mutations in AtMDR1 or its closest relative, AtPGP1, were isolated by a reverse genetic strategy. Auxin transport activity was greatly impaired in atmdr1 and atmdr1 atpgp1 double mutant plants. Epinastic cotyledons and reduced apical dominance were mutant phenotypes consistent with the disrupted basipetal flow of auxin. The auxin transport inhibitor 1-naphthylphthalamic acid was shown to bind tightly and specifically to AtMDR1 and AtPGP1 proteins. The results indicate that these two MDR-like genes of Arabidopsis encode 1-naphthylphthalamic acid binding proteins that are required for normal auxin distribution and auxin-mediated development.
INTRODUCTION
The transporters encoded by mammalian multidrug resistance (MDR) genes, often referred to as P-glycoproteins, enhance the efflux of hydrophobic drugs from the cytoplasm in an energy-dependent fashion (Gottesman et al., 1995; Ferté, 2000). The MDR transporters belong to a natural subgroup of ATP binding cassette (ABC) transporters designated cluster II by Decottignies and Goffeau (1997) in their analysis of the 29 ABC transporters present in the yeast genome. This classification system has since been adopted for the ABC transporters present in plants as well (Davies and Coleman, 2000; Sánchez-Fernández et al., 2001). Molecules in cluster II are topologically similar in having five or six membrane-spanning domains followed by a cytoplasmic ATP binding domain, which is followed by another five or six membrane-spanning domains and another ATP binding domain. Thus, MDR-like molecules are large membrane proteins composed of two related, repeated halves. Other cluster II molecules, well known from a variety of organisms, include CFTR, STE6, YCF1, and the multidrug-related proteins (MRPs). Their substrates include inorganic anions, peptides, heavy metal chelates, and organic glutathione conjugates. Especially relevant to the present work is the recent finding that an Arabidopsis MRP, AtMRP5, may pump auxin conjugates into the vacuole (Gaedeke et al., 2001).
Mammalian MDR transporters clearly enhance the efflux of cytotoxic drugs from cells, but it is still unclear whether they pump drug substrates directly across plasma membranes (Ambudkar et al., 1999) or alter transmembrane drug distribution indirectly as a consequence of other MDR-mediated transport processes (Roepe, 2000). Another uncertainty is the physiological role of MDR proteins in normal, pharmacologically unchallenged cells. Studies have indicated that human MDR1 and mouse MDR1a can modulate volume-activated chloride channel activity (Hardy et al., 1995; Higgins, 1995). Lipid translocating activity also has been demonstrated for mouse and human MDRs (Ruetz and Gros, 1994; van Helvoort et al., 1996; Borst et al., 2000; Ferté, 2000). Moreover, a large body of recent work indicates that MDR proteins participate in the immune system and in apoptosis. Johnstone et al. (2000) recently reviewed these and other MDR functions. Rea et al. (1998) and Theodoulou (2000) have written comprehensive reviews of the comparatively new field of plant ABC transporters.
With so many diverse activities attributed to the products of MDR genes, it is not possible to know a priori the function of the MDR-like genes present in the Arabidopsis genome. The only hint of their function comes from a study in which one MDR-like gene, Arabidopsis thaliana P-glycoprotein1 (AtPGP1), was overexpressed in sense and antisense orientations (Sidler et al., 1998). Compared with that of the wild type, overexpression of AtPGP1 was associated with longer hypocotyls when seedlings were grown in dim light but not in bright light. Antisense seedlings developed shorter hypocotyls in the same light conditions. The authors suggested that AtPGP1 may transport a growth-regulating molecule from the shoot apex. One candidate for such a growth-regulating molecule would be the auxin indoleacetic acid (IAA) because its synthesis is believed to occur in the shoot apex, where AtPGP1 expression was greatest. Brassinolides and gibberellins are other growth-promoting molecules that may possess the general structural features shared by MDR substrates (Seelig, 1998). In this article, we describe genetic, biochemical, and physiological investigations of AtMDR1, a previously unstudied MDR-like gene from Arabidopsis. AtMDR1 and AtPGP1 are shown to play an important role in controlling the growth and form of Arabidopsis in both juvenile and adult stages of development, probably by influencing the distribution of the hormone auxin.
RESULTS
Isolation of AtMDR1
Previous electrophysiological studies demonstrated that 5-nitro-2-(3-phenylpropylamino)-benzoic acid (NPPB) potently and reversibly inhibited an anion channel being studied because of its role in blue light signal transduction (Cho and Spalding, 1996; Parks et al., 1998). A screen for genes differentially expressed in response to NPPB treatment was conducted to identify proteins related to anion channel function, the rationale being that plants may alter the expression of the anion channel or a regulator of an anion channel to compensate for the pharmacologically imposed suppression of channel activity. An NPPB-upregulated gene was identified in Brassica napus seedlings, and its sequence was used to isolate a nearly identical gene from an Arabidopsis cDNA library. The Arabidopsis gene was named AtMDR1 because it encoded a cluster II ABC transporter with a predicted amino acid sequence that was 36% identical to human MDR1 (Chen et al., 1990). AtMDR1 is one of several MDR-like genes in the Arabidopsis genome, and it is 52% identical to its closest relative in the genome, which was named AtPGP1 by Dudler and Hertig (1992). (In the phylogenetic analysis of Arabidopsis ABC molecules recently performed by Sánchez-Fernández et al. [2001], the AtMDR1 gene is referred to as AtMDR11, and AtPGP1 is called AtMDR1.) The general topology of the predicted AtMDR1 protein is shown in Figure 1A. The RNA gel blot in Figure 1B shows that NPPB treatment increased AtMDR1 transcript abundance similar to the originally isolated Brassica paralog.
Figure 1.
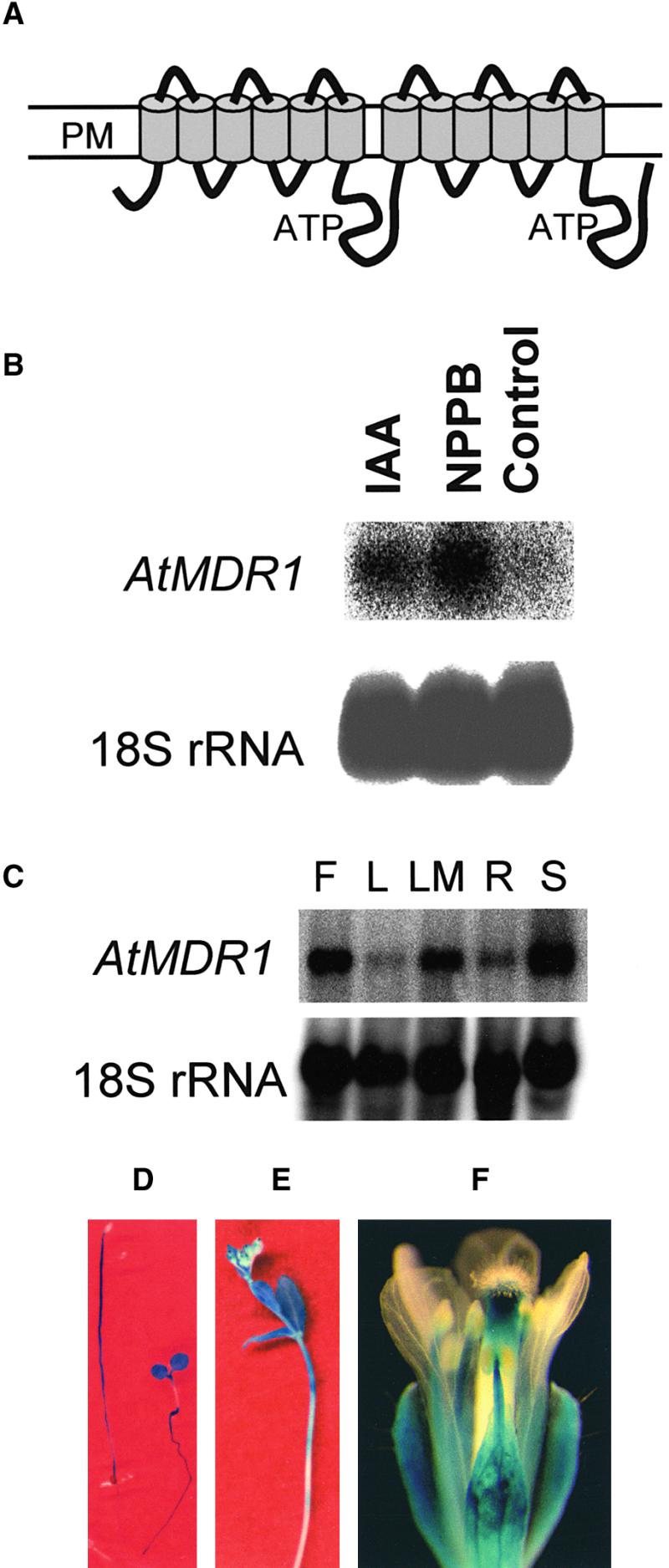
Topology and Expression Pattern of AtMDR1.
(A) Two groups of six transmembrane helices are predicted, each followed by cytoplasmic domains containing ATP binding domains. An alternate model having seven membrane-spanning helices in the first group also is supported. PM, plasma membrane.
(B) NPPB- and IAA-induced expression of AtMDR1 mRNA. Each lane of the gel contained 30 μg of total RNA extracted from Arabidopsis seedlings grown for 5 days in the presence or absence of 20 μM NPPB or 1 μM IAA, and the blot was probed with 32P-labeled AtMDR1 genomic DNA and also with 18S rRNA to confirm equal RNA loading.
(C) Tissue-specific expression pattern of AtMDR1 mRNA. Each lane contained 30 μg of total RNA from flowers (F), rosette leaves (L), rosette leaves plus shoot apical meristem (LM), roots (R), and seedlings (S). A blot of the RNA gel was probed with AtMDR1 genomic DNA and also with 18S rRNA to confirm equal RNA loading.
(D) GUS activity patterns in etiolated (left) and light-grown (right) AtMDR1::GUS seedlings. Light increased AtMDR1 promoter activity in cotyledons and roots but suppressed it in hypocotyls of these 5-day-old seedlings.
(E) GUS activity in the primary inflorescence of an AtMDR1::GUS plant. Staining was strongest in the apical portions of the stem and the youngest cauline leaves.
(F) GUS activity pattern in the flower of an AtMDR1::GUS plant. All floral whorls except petals displayed staining.
Expression Pattern of AtMDR1 and Induction by IAA
The RNA gel blot in Figure 1C shows that AtMDR1 was expressed in seedlings, roots, rosette leaves, flowers, and aboveground tissues before bolting (rosette leaves plus shoot apical meristem). Evidence that AtMDR1 mRNA was relatively more abundant in the vegetative shoot meristem was obtained by comparing the signal in lanes containing leaf RNA with that in lanes containing leaf plus meristem RNA. To better resolve the spatial expression pattern of AtMDR1, transgenic plants carrying 4 kb of upstream AtMDR1 sequence fused to the β-glucuronidase (GUS) reporter gene were histochemically stained at different stages of development. The hypocotyls but not the cotyledons of dark-grown seedlings were strongly stained (Figure 1D). Seedlings grown in white light (80 μmol·m−2·sec−1) expressed GUS in the cotyledons, petioles, shoot apex region, and roots but not in the hypocotyls (Figure 1D). The time course or wavelength dependence of the light-induced decrease in expression in the hypocotyl has not been investigated. In adult plants, GUS activity was detected in the apical region of the inflorescence and stem nodes (Figure 1E). Floral parts, especially the stamen filaments and the apical portion of the style, showed GUS staining, but the petals did not (Figure 1F).
This GUS staining pattern was reminiscent of expression driven by auxin-inducible promoters (Wyatt et al., 1993; Wong et al., 1996; Gil and Green, 1997), suggesting that AtMDR1 expression may be under the control of IAA. The RNA gel blot in Figure 1B demonstrated that exogenous IAA induced AtMDR1 expression. Induction by IAA and greater expression in tissues expected to have higher auxin concentrations (apical meristem and youngest leaves of seedlings) raised the possibility that AtMDR1 function could be related to auxin.
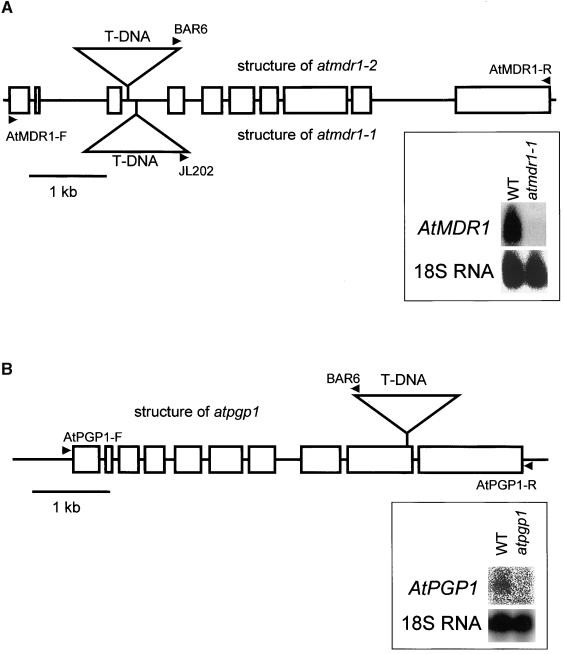
Isolation of atmdr1 and atpgp1 Mutant Alleles
Mutants lacking functional AtMDR1 were sought to help study the in planta function of this ABC transporter. Two separate populations of T-DNA insertional mutants from the University of Wisconsin Arabidopsis Knockout Facility were screened for the presence of T-DNA insertions in the AtMDR1 gene, as described by Krysan et al. (1999). After a series of polymerase chain reaction (PCR) amplifications using the AtMDR1-R primer, which is specific to the AtMDR1 coding sequence, and either of two T-DNA–specific primers (Figure 2A), individual plants homozygous for a T-DNA insertion in the AtMDR1 gene were isolated from each of the two mutant populations. Both atmdr1-1 and atmdr1-2 contained a T-DNA insertion in the third intron but at different locations (Figure 2A). No AtMDR1 mRNA was detected in atmdr1-1 seedlings (Figure 2A). The lack of signal in the mutant lane indicates that the probe used here and in Figures 1B and 1C does not hybridize to other MDR-like genes.
Figure 2.
Genomic Structure of T-DNA Insertion Alleles.
(A) atmdr1-1 and atmdr1-2.
(B) atpgp1.
Rectangles represent the exons and lines represent the untranslated regions, including introns of the two MDR-like genes. T-DNAs, represented by triangles, are not drawn to scale. Arrowheads indicate primers used to screen for the mutants. RNA gel blots provide evidence that the mutations severely or completely block expression. Fifty micrograms (A) or 30 μg (B) of total RNA from either Wassilewskija or mutant seedlings was subjected to electrophoresis, blotted, and probed as indicated. WT, wild type.
This screen was redesigned and repeated to isolate a T-DNA insertion mutation in AtPGP1, the gene most closely related to AtMDR1 in the Arabidopsis genome database. Figure 2B shows the location of the T-DNA insertion and the PCR primers used to isolate the atpgp1-1 allele, which probably is a null allele because no mRNA was detected on a blot (Figure 2B).
Phenotypic Characterization of atmdr1
Figure 3A shows that at the seedling stage, both alleles of atmdr1 displayed downward-folded, or epinastic, cotyledons and first true leaves. The angle formed by the shorter cotyledon petioles was smaller in atmdr1 seedlings. The epinastic phenotype was more visible in dim light than in bright light (data not shown). The rosette leaves of adult plants were somewhat curled and wrinkled along the margin (Figure 3B). Bolting of the inflorescence stem was delayed by 2.8 days on average relative to that of the wild type, and the growth rate of the bolt was also slower than in the wild type. These two effects of the mutation caused atmdr1-1 and atmdr1-2 plants to be shorter than wild-type plants after 27 days of growth (Figure 3B). The mutant ultimately achieved the same height as wild-type plants (data not shown). These phenotypes coincided for the most part with the sites of AtMDR1 expression indicated by GUS staining.
Figure 3.
Morphology of atmdr1, atpgp1, and atmdr1 atpgp1 Plants.
(A) Seedlings grown for 6 days in white light. From left: wild type, atmdr1-1, atmdr1-2, and atmdr1-1 molecularly complemented with a genomic copy of AtMDR1.
(B) Plants grown for 27 days. From left: wild type, atmdr1-1, atmdr1-2, and atmdr1-1 molecularly complemented with a genomic copy of AtMDR1.
(C) Phenocopy of atmdr1 by applying the auxin 2,4-D to the wild type. From left: untreated wild type, wild type treated with 1 μM 2,4-D for 6 hr before the photograph was taken, and atmdr1-1. All seedlings were 4 days old.
(D) Lack of obvious phenotype in atpgp1 mutants. Wild-type (left) and atpgp1 (right) plants grown for 27 days.
(E) The atpgp1 mutation does not affect hypocotyl elongation. Wild-type (WT) and mutant seedlings were grown in darkness or dim white light (photon fluence rate of 2 μmol·m−2·sec−1, 16-hr-light/8-hr-dark cycle) for 11 days. Hypocotyl lengths were determined with software from digital images of the seedlings. Error bars indicate ±sem.
(F) Phenotypes of single and double mutant seedlings. From left: wild type, atmdr1-1, atpgp1, and atmdr1-1 atpgp1 (two example seedlings). Cotyledon epinasty of atmdr1 seedlings is extreme in the double mutant but absent in atpgp1 seedlings.
(G) Dark-grown wild-type and atmdr1 atpgp1 seedlings. Etiolated wild-type seedlings (left two seedlings) have longer, straight hypocotyls compared with the shorter and curved hypocotyls of the double mutants (right two seedlings).
(H) Curled leaf phenotype of double mutants. Abaxial view of a rosette leaf from a atmdr1-1 atpgp1 seedling (left) and adaxial view of a wild-type leaf of the same age (right).
(I) Dwarf phenotype of double mutant plant (30-day-old atmdr1-1 atpgp1) beginning to bolt. Bar = 1 cm.
(J) Reduced apical dominance in adult double mutant plants. Left, a wild-type plant (64 days old) ceased flowering; right, a mutant plant still produced flowers after 109 days and produced many more secondary inflorescences.
(K) Morphology of double mutant flowers. Petals and sepals were removed to show stamens and pistils of wild-type (left) and atmdr1-2 atpgp1 flowers (right). Filaments of the double mutant did not elongate sufficiently to self-pollinate normally.
Epinasty of the cotyledons and first leaves was the most obvious aspect of the atmdr1 mutants, and this could be phenocopied in wild-type plants by the application of auxin. Figure 3C shows that transferring wild-type seedlings to agar plates containing 1 μM 2,4-D phenocopied the atmdr1 mutants within 6 hr of treatment. This result may be evidence that the atmdr1 mutation caused an increase in auxin concentration in the cotyledons, leading to the abnormal growth differentials that produce leaf epinasty. Some etiolated atmdr1-1 and atmdr1-2 seedlings could be distinguished from wild-type seedlings by a slight waviness in the hypocotyl that, like epinasty, results from abnormal growth differentials.
Molecular Complementation of the atmdr1 Mutant
To determine if these phenotypes were attributable exclusively to the loss of functional AtMDR1, atmdr1-1 was transformed with a full-length genomic copy of AtMDR1, including ∼4 kb of the upstream promoter region and ∼1 kb of the 3′ untranslated region. All kanamycin-resistant T1 seedlings displayed wild-type morphology (Figures 3A and 3B), and all T2 seedlings from independent transgenic lines segregated wild-type and mutant morphologies at a ratio of approximately 3:1. These results confirm that the lesion in AtMDR1 is responsible for the atmdr1-1 phenotype.
Phenotypic Characterization of atpgp1
No morphological differences between atpgp1-1 and wild-type plants were detected at the seedling or adult stages of development (Figure 3D). The light-dependent difference in hypocotyl length one would expect based on the phenotype of antisense plants reported by Sidler et al. (1998) was not observed (Figure 3E). It is possible that the antisense phenotype they observed resulted from the reduced expression of more than one family member. In any event, the proposal that AtPGP1 exerts significant control over hypocotyl growth rate in dim light is not supported by this mutant analysis.
Phenotypic Characterization of atmdr1 atpgp1
Creating mutants that lack more than one member of a gene family enables tests of genetic interaction and redundancy between members. A double mutant lacking both AtMDR1 and AtPGP1 was constructed by allowing F1 individuals from a cross of atpgp1-1 with atmdr1-1 or atmdr1-2 to self. Approximately one in 16 of the F2 seedlings from both crosses displayed extremely downward curved cotyledons and curled true leaves when grown in the light (Figure 3F) and short, distinctly wavy hypocotyls when grown in the dark (Figure 3G). PCR analysis confirmed that only seedlings displaying the more severe atmdr1 phenotype were doubly homozygous for the two mutations.
The rosette leaves of double mutant plants were irregular in shape, wrinkled, and severely curled (Figure 3H). At the flowering stage, double mutant plants were very stunted (Figure 3I). In addition to growing slowly, the primary inflorescence stems and floral pedicels also were wavy, indicating that the direction of growth changed periodically during the elongation phase of these organs (data not shown). A photograph taken after 109 days of growth (Figure 3J) shows that the double mutant produced abundant secondary inflorescence stems, indicating a large reduction in apical dominance. The fertility of the flowers was poor because the filaments did not elongate sufficiently to position the anthers above the stigma at anthesis (Figure 3K). Hand-pollinated flowers of the doubly homozygous mutant plants produced viable double mutant seed having a normal appearance.
Polar Auxin Transport in atmdr1 and atpgp1 Plants
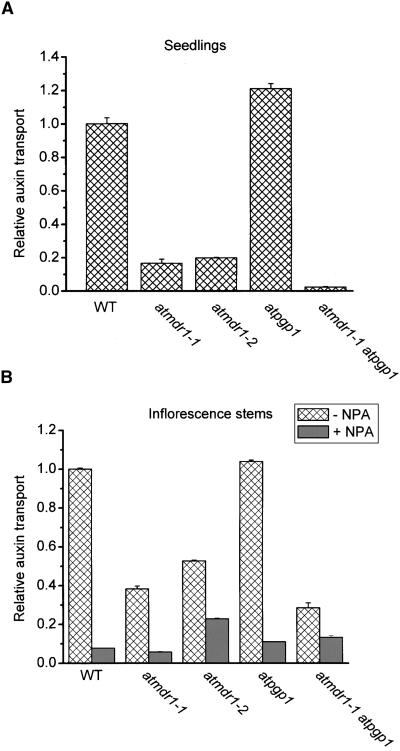
One interpretation of the various single and double mutant phenotypes is that AtMDR1 and AtPGP1 function together to transport auxin from apical regions of auxin synthesis to more basal tissues. This hypothesis was tested by comparing rates of polar auxin transport in atmdr1, atpgp1, atmdr1 atpgp1, and wild-type plants.
Three different auxin transport assays were performed using tissues wherein AtMDR1 is strongly expressed. The first measured the basipetal movement of 14C-IAA applied as a single microdroplet to the apex of light-grown seedlings, as described by Murphy et al. (2000). Auxin transport measured this way was reduced severely in both alleles of atmdr1 and especially in the double mutant, but not in the atpgp1 seedlings (Figure 4A). The second assay measured the basipetal transport of IAA through the apical portion of etiolated seedlings inverted in a reservoir containing 14C-IAA (Garbers et al., 1996). The results (data not shown) were similar to those in Figure 4A. Importantly, there were no significant differences between atmdr1, atpgp1, and wild-type seedlings in the small amounts of acropetal auxin transport. Third, the basipetal transport of auxin in excised segments of the upper inflorescence stem was measured using another standard assay (Ruegger et al., 1997; Brown et al., 2001). As shown in Figure 4B, atmdr1 mutants and the atmdr1 atpgp1 double mutant, but not atpgp1, were clearly defective in basipetal auxin transport. Again, as in the case of the seedling assay, there were no significant differences in acropetal transport between the mutant and wild-type inflorescences. The majority of the residual amount of basipetal auxin transport in the mutants was blocked by 1-naphthylphthalamic acid (NPA; Figure 4B). Thus, under these specific conditions, the activities of AtMDR1 and AtPGP1 are required for normal NPA-sensitive polar auxin transport in the apical regions of inflorescence stems.
Figure 4.
Auxin Transport Activity in the Wild Type and Mutants.
(A) Auxin transport activity in seedlings. The atmdr1 mutation dramatically impaired auxin transport in seedlings, but the atpgp1 mutation did not. Each assay used 10 seedlings. Values shown are means ±sd of three independent experiments.
(B) Polar auxin transport activity in apical sections of inflorescence. As in seedlings, the atmdr1 mutation dramatically impaired polar auxin transport but the atpgp1 mutation did not. The double mutant was more impaired. WT, wild type. Values shown are means of three independent experiments ±sd.
AtMDR1 Binds NPA
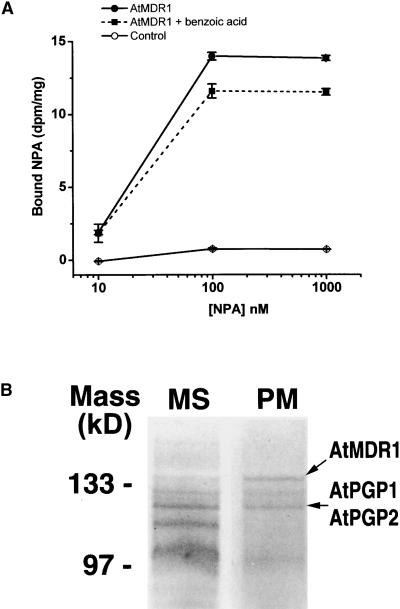
The effect of the atmdr1 mutations on IAA transport resembled the effect of NPA, a chemical inhibitor of polar auxin transport. AtMDR1 was expressed in yeast cells to test the hypothesis that it could be involved in the inhibition of auxin transport by NPA. As shown in Figure 5A, NPA bound specifically to AtMDR1-expressing yeast but not to control yeast carrying the empty vector. The binding was saturable and was outcompeted by excess unlabeled NPA (see Methods). Importantly, bound NPA was not displaced by benzoic acid, a similar hydrophobic organic acid. The transformed yeast also were used to determine whether AtMDR1 enhanced the rate of IAA efflux from cells preloaded with 14C-IAA, but the results were negative (data not shown). AtMDR1 may not function as an auxin transporter by itself.
Figure 5.
NPA Binding by MDR-like Molecules.
(A) AtMDR1 expressed in yeast specifically binds NPA. Values are means of three independent experiments ±sd. Open circles, specific NPA binding to untransformed yeast strain; closed circles, specific NPA binding to yeast expressing AtMDR1; closed squares, NPA binding in the presence of a 1000-fold concentration of benzoic acid to yeast expressing AtMDR1.
(B) Arabidopsis membrane proteins identified by high-affinity NPA binding. Shown are Coomassie blue–stained, 10% SDS–polyacrylamide gels of high molecular mass proteins purified by gel filtration and NPA affinity chromatography. Proteins were first solubilized from microsomal (MS) or plasma membrane (PM) vesicles derived from seedlings.
To determine if the NPA binding by AtMDR1 in yeast also occurs in plants, we analyzed specific NPA binding activity in microsomal vesicles prepared from 5-day-old wild-type, atmdr1, and atpgp1 seedlings. At 20 nM 14C-NPA, the average NPA binding activity of microsomes from atmdr1 (0.082 ± 0.022 pmol/mg) was 36% lower than that of microsomes from the wild type (0.128 ± 0.016 pmol/mg). Student's t test revealed that this difference was statistically significant (t = 2.929, P < 0.05). The average NPA binding activity of microsomes from atpgp1 (0.098 ± 0.025 pmol/mg) also was reduced compared with that of wild-type microsomes, but not to a statistically significant extent (t = 1.751, P > 0.1). These results suggest that mutation of AtMDR1 significantly decreased the NPA binding capacity of membranes. The loss of binding may have been considerably more severe than the 36% average in the cells in which AtMDR1 is the predominantly expressed family member.
Remarkably, AtMDR1 and AtPGP1 were purified independently from Arabidopsis membrane preparations by NPA affinity chromatography in an effort to purify NPA binding aminopeptidases from the plasma membrane (Murphy et al., 2000, 2001). Figure 5B shows that four major high molecular mass protein bands eluted from the NPA column by >300 mM KCl were relatively more abundant in the plasma membrane–enriched preparations (see Methods). The actual amino acid sequences obtained from three of these four proteins were (K)ILLLDEATSALDA(K)GVSTFS, (K)FLSW(R)VVMAFV-GESRA, and (K)LFSFADFYD(K)VGNFLHYIS. (K) and (R) indicate inferred lysine and arginine residues cleaved in trypsin digests. Comparison of these sequences with Arabidopsis genomic, cDNA, and predicted protein databases established that the three proteins were the products of AtMDR1, AtPGP1, and AtPGP2, respectively.
An interpretation consistent with the results presented here is that AtMDR1 and AtPGP1 are NPA binding molecules required for the proper transport and distribution of auxin from sites of synthesis.
DISCUSSION
The loss of AtMDR1 activity affects auxin transport (Figure 4) either directly because AtMDR1 is an auxin transporter or indirectly by an unspecified mechanism. The induction of AtMDR1 by auxin (Figure 1B) and the auxin-treated appearance of atmdr1 seedlings (Figure 3C) raise the possibility that AtMDR1 contributes to auxin homeostasis by mediating efflux. Genetically impairing efflux from apical sources would be expected to result in greater than normal concentrations of auxin in the cotyledons, petioles, and young leaves of atmdr1 and atmdr1 atpgp1 seedlings. Accumulation of auxin in these organs may be the cause of the epinasty displayed by these mutants. However, morphology is a secondary indicator of hormone levels, and more direct measurements of auxin, or molecular markers of auxin concentration, are needed to test such an auxin-distribution explanation.
In general, there was good agreement between the sites of strong expression in wild-type plants and the organs that displayed morphological abnormalities in the mutants. For example, AtMDR1 was expressed strongly in the cotyledons and petioles of light-grown seedlings, the same organs that developed epinastic curvatures in the mutants. Also, expression of AtMDR1 was obvious in the apical portions of the inflorescence stem, which grew slower when AtMDR1 was absent. AtPGP1 expression was not detected in the inflorescence stem or seedling hypocotyl (Sidler et al., 1998), and those organs were not noticeably affected by the atpgp1 mutation (Figure 3). AtMDR1 was highly expressed in hypocotyls of etiolated seedlings, and atmdr1 seedlings often displayed a slightly S-shaped hypocotyl, unlike the straight hypocotyls of wild-type seedlings (data not shown). Expression of AtPGP1 was not detected in the hypocotyls of etiolated seedlings (Sidler et al., 1998), in accordance with the lack of phenotype in atpgp1 seedlings.
When grown in the light, the epinastic attitude of the cotyledons and first leaves was the most obvious phenotype of atmdr1 seedlings (Figure 3A). The loss of AtPGP1 did not contribute to this epinasty unless AtMDR1 also was absent (Figure 3F). This nonreciprocal redundancy also occurred in etiolated seedlings, as indicated by the strongly curved hypocotyl of double mutants (Figure 3G). Apparently, AtMDR1 activity was able to compensate for a deficiency in AtPGP1 in both etiolated and light-grown seedlings, but AtPGP1 was not able to compensate for a lack of AtMDR1. One possible explanation for this nonreciprocal redundancy is that both proteins perform similar functions but AtMDR1 is expressed at greater levels than AtPGP1, so the loss of the latter is of less consequence.
The inflorescence stem of the double mutant also was curved and grew more slowly than that of the wild type. It appears that mutating both of these MDR-like transporters impairs the rate and coordination of growth in tissues in which the mutations also affect auxin transport (Figure 4). Thus, the mutants produced here may prove useful in studies of auxin-mediated growth control, such as the differential growth underlying tropic curvatures.
The mutant analysis indicates that both AtMDR1 and AtPGP1 function in floral development. Although neither single mutant displayed an obvious floral phenotype, flowers of the double mutant displayed low fertility. The fertility defect appears unrelated to gametophyte formation or function. Instead, it can be explained by inefficient delivery of pollen to the stigma caused by improper elongation of the stamens (Figure 3K), consistent with previous reports of a role for auxin in filament elongation (Fei and Sawhney, 1999; Lobello et al., 2000). Improper stamen elongation also was responsible for reduced fertility in the auxin-resistant mutant axr1 (Lincoln et al., 1990). Again, the floral phenotype appeared only in the atmdr1 atpgp1 double mutant, indicating functional redundancy between the two genes, even though AtPGP1 expression apparently is not detectable in filaments (Sidler et al., 1998).
The enhanced proliferation of secondary inflorescences is another informative phenotype that developed only when both AtMDR1 and AtPGP1 were mutated (Figure 3J). This finding can be interpreted as a classic loss of apical dominance, the phenomenon in which basipetally transported auxin suppresses the growth of basal lateral buds. Although this aspect of the phenotype would appear to reflect reduced auxin concentrations and is similar to the phenotype of strong axr1 alleles (Lincoln et al., 1990), other aspects, such as epinasty, are evidence of excessive auxin and are reminiscent of auxin-overproducing mutants (Boerjan et al., 1995; Zhao et al., 2001). Thus, it appears that mutations in AtMDR1 and AtPGP1 create areas of auxin accumulation and deficit, each of which is associated with altered morphologies. These phenotypes may be the result of a disruption in the basipetal distribution of auxin either by inhibiting polar transport or by limiting auxin movement from sites of synthesis into the polar transport stream, or both.
MDR-like transporters are best known for their ATP-driven active transport of hydrophobic substrates. Auxin efflux, on the other hand, should be energetically passive because the electrochemical potential of anionic IAA, the form that predominates at the neutral pH of cytoplasm, is much greater inside than outside. Indeed, substantial genetic and biochemical evidence indicates that auxin efflux during polar auxin transport occurs by passive transport through PIN-type channels (Chen et al., 1998; Gälweiler et al., 1998; Luschnig et al., 1998; Müller et al., 1998). Thus, active pumping of IAA by an MDR transporter would seem unnecessary. But it is possible that protonated IAA (IAAH) is pumped out of sites of synthesis when cytoplasmic concentrations exceed a threshold level. IAAH is much more abundant in the apoplast for the same reasons that the anionic form predominates in the cytoplasm, and it is electrically neutral, so its efflux would require active transport. AtMDR1 could perform this active transport provided that its Km for IAAH is reasonably well matched with the nanomolar concentrations expected for IAAH in the cytoplasm. After export from sites of synthesis and subsequent uptake by the appropriate cells, polar transport of this auxin through stems would occur via PIN-mediated passive transport. This mechanism could explain the inferred accumulation of auxin in the apices of atmdr1 seedlings, but it would not explain the reduced auxin transport in the inflorescence stems of atmdr1. Perhaps AtMDR1 pumps IAAH out of auxin-generating apices and also regulates PIN-mediated efflux of anionic IAA in the specific stem cells that perform polar auxin transport (Gälweiler et al., 1998). Another possibility is that conjugated IAA, formed in tissues containing high levels of auxin, is in fact the natural MDR substrate, as suggested by Gaedeke et al. (2001) for AtMRP5. If so, auxin hydrolases such as ILR1, IAR1, and IAR3 (Davies et al., 1999; Lasswell et al., 2000) would be required at some subsequent step because free IAA rather than a conjugated form is transported through Arabidopsis hypocotyls (Brown et al., 2001).
Two results, neither inconsistent with a role for AtMDR1 in the movement of auxin from sites of synthesis, argue in favor of AtMDR1 encoding an important component of the polar auxin transport mechanism: the impaired IAA transport in segments of atmdr1 inflorescences (Figure 4B), and the binding of NPA by native and heterologously expressed AtMDR1 (Figure 5). High-affinity, specific binding of 3H-NPA by a fraction containing purified AtMDR1, AtPGP1, and AtPGP2 has been demonstrated (Murphy et al., 2001). Binding of NPA by AtMDR1 probably occurs in planta as well, because atmdr1 microsomal membranes displayed a 36% decrease in NPA binding capacity despite being derived from whole seedlings that included tissues not expressing AtMDR1. Together, this evidence supports the conclusion that AtMDR1 and, to a lesser extent, AtPGP1 are responsible for a significant fraction of NPA binding sites in seedling membranes. More work is needed to establish what biological effects of NPA are caused by this binding.
Although the results presented here indicate that MDR-like molecules are essential for the normal polar transport of IAA, other molecules also may be essential. NPA binds to yeast expressing AtMDR1 (Figure 5A), but compared with control cells, those same yeast did not display higher rates of auxin efflux (data not shown). A reasonable conjecture is that other proteins, such as PIN-type channels, function together with MDR-like proteins to create an efficient, NPA-sensitive auxin efflux mechanism. Such a mechanism is in agreement with the evidence that NPA modulates auxin efflux channels by binding to a site distinct from the anion (PIN) channel (reviewed by Lomax et al., 1995) and is consistent with the impaired IAA transport in atmdr1 plants (Figure 4). There is considerable precedent in the animal literature for cluster II ABC transporters acting as regulators of passive channels with which they interact physically (Higgins, 1995; Vanoye et al., 1997; Bryan and Aguilar-Bryan, 1999; Schreiber et al., 1999; Ji et al., 2000). Genetic and biochemical studies of the interaction between the MDR-like and PIN-like molecules are presently under way.
Specific flavonoids, particularly quercetin and kaempferol, have been shown to inhibit auxin efflux in zucchini hypocotyls and to displace NPA bound to plasma membrane vesicles (Jacobs and Rubery, 1988; Faulkner and Rubery, 1992). More recent studies took advantage of mutations in the general phenylpropanoid biosynthetic pathway (Murphy et al., 2000; Brown et al., 2001) to demonstrate that endogenous flavonoids modulate auxin transport in a tissue-specific manner in Arabidopsis (Murphy et al., 2000; Peer et al., 2001). These results relate to the present results because flavonols and isoflavones strongly inhibit P-glycoprotein (MDR1) activity in a variety of human cell types. Kaempferol and quercetin were found to be the most effective inhibitors (Chieli et al., 1995; Castro and Altenberg, 1997; Conseil et al., 1998; Ferté et al., 1999; Perez-Victoria et al., 1999). Perhaps flavonoids modulate auxin distribution in plants by binding to MDR-like transporters, which may be considered essential for normal auxin transport and the development of plant form.
METHODS
Plant Material and Growth Conditions
Brassica napus seedlings were grown in continuous white light for 5 days on minimal salt medium (1 mM KCl and 1 mM CaCl2) in the presence or absence of 20 μM 5-nitro-2-(3-phenylpropylamino)-benzoic acid (NPPB) before the roots were harvested to isolate sufficient mRNA for subtractive cloning of differentially expressed genes. Arabidopsis thaliana (Wassilewskija ecotype) was used in all experiments except for the blot in Figure 2A, which was made with RNA isolated from the Columbia ecotype. Seedlings generally were grown on 0.5 × Murashige and Skoog (1962) medium containing 1.5% sucrose. For induction experiments, 20 μM NPPB (Calbiochem, San Diego, CA) or 1 μM indoleacetic acid (IAA; Sigma, St. Louis, MO) was added to the medium. Adult plant material was obtained by growing plants in soil under white light with a 16-hr-light/8-hr-dark cycle at 23°C. For protein purification and auxin transport experiments, seedlings were grown as described previously (Murphy and Taiz, 1995).
Cloning of AtMDR1
Root mRNA, extracted from B. napus seedlings treated either with or without 20 μM NPPB, was the starting material. Reverse transcription, subtractive hybridization, and amplification of subtracted cDNA fragments were performed reciprocally with the two mRNA populations as specified by the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA). Polymerase chain reaction (PCR) fragments representing differentially expressed mRNAs were cloned into pGEM-T vector (Promega, Madison, WI) and introduced into Escherichia coli, and their induction or repression by NPPB was confirmed by analyses of RNA gel blots. One of the few clones that displayed particularly strong differential expression was selected for further analysis.
A full-length Arabidopsis ortholog (AtMDR1) of the selected B. napus clone was isolated by probing a 3- to 6-kb size-selected cDNA library (Arabidopsis Biological Resource Center, Ohio State University, Columbus). The cDNA was subcloned into pBluescript SK+ (Stratagene, La Jolla, CA), and its sequence was determined. Sequence comparisons were performed using the BLAST program. The secondary structure of AtMDR1 was predicted using the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html; Hofmann and Stoffel, 1993).
AtMDR1 Expression Analysis
For the purposes of measuring mRNA levels, plant tissues were frozen and ground in liquid N2. Total RNA was isolated using RNA ISOLATOR (Genosys Biotechnologies, The Woodlands, TX). RNA was separated on an agarose gel by electrophoresis, transferred to a nylon membrane, and probed with 32P-labeled AtMDR1 genomic DNA generated by PCR using the gene-specific primers shown in Figure 2. The same blot was probed with 32P-labeled Arabidopsis 18S rDNA to confirm that equal amounts of RNA had been loaded in each lane.
To create the AtMDR1 promoter::reporter construct, two PCR primers flanking the regions from −1 to −4008 were made (5′-gcgggcccGGTTTTTTGTAGATCCGAGTTAAAG-3′ and 5′-cgctgcagG-TAAGGGACATATTTATGAATATACAAC-3′). The underlined portions are nonnative sequences representing SmaI and PstI restriction sites, respectively. PCR reactions were performed using genomic DNA as a template and Ex-Taq polymerase (PanVera, Madison, WI). The PCR product was inserted upstream of the β-glucuronidase (GUS) coding sequence present in the pPZP 211G binary vector, which was formed by engineering the GUS followed by the nopaline synthase terminator of PBI 101.1 (Jefferson et al., 1987) into pPZP 211 (Hajdukiewicz et al., 1994). Agrobacterium tumefaciens was transformed with the binary vector and introduced into Arabidopsis Wassilewskija wild-type plants by vacuum infiltration (Bechtold et al., 1993). Kanamycin-resistant plants in the T1 and T2 generations were histochemically stained to detect GUS activity by immersing seedlings or tissues in 50 mM sodium phosphate buffer, pH 7.0, containing 0.5 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (Research Organics, Cleveland, OH) overnight at 37°C followed by dechlorophylation in 70% ethanol.
Molecular Complementation of atmdr1
Bacterial artificial chromosome DNA from Institut für Genbiologische Forschung (IGF) clone F3J22 (Arabidopsis Biological Resource Center) was digested with EaeI, and the 12-kb EaeI fragment containing AtMDR1 was cloned into pBluescript SK+ (Stratagene) and recloned into the binary vector pPZP 221B, which differs from pPZP 221 (Hajdukiewicz et al., 1994) by the addition of a Basta-resistant selectable marker. The resulting construct was transferred to Agrobacterium strain ABI, which was used to transform atmdr1-1 plants by vacuum infiltration (Bechtold et al., 1993).
Isolation of Mutants
T-DNA–mutagenized Arabidopsis lines were screened as described by Krysan et al. (1999). The gene-specific PCR primer sets used for screening were as follows: AtMDR-5′, 5′-CATTTTATAATAACGCTG-CGGACATAC-3′; AtMDR-3′, 5′-CTTGAATCACACCAATGCAATCAA-CACCTC-3′; AtPGP1-5′, 5′-GTCCGAAATGCAAGGCCTTGAGCT-TCTCCC-3′; AtPGP1-3′, 5′-CTGTATCATTCGCGCGTAGATTCCATC-AGG-3′. These primers and Ex-Taq polymerase (PanVera) were used in PCR reactions to determine which of the nested subpools of DNA contained the insertion of interest. At each step, the PCR products were separated on an agarose gel by electrophoresis, blotted onto a nylon membrane, and probed with the gene of interest to determine if the amplified fragments were genuine. The genotypes of isolated plants were determined by PCR with two gene-specific primers and T-DNA border–specific primer in one tube. In the case of homozygous plants, only one band representing insertion was detectable, whereas in the case of heterozygous plants, the insertion and another band amplified with gene-specific primers only were detected together on the agarose gel.
Auxin Transport Assays
Auxin transport assays of intact light-grown seedlings were performed as described previously (Murphy et al., 2000) with the exception that 0.1 μL of 50 nM 14C-IAA with a specific activity of 5 mCi/mmol (Sigma) was applied to seedlings. Auxin transport in etiolated hypocotyls was measured by the method of Garbers et al. (1996) with the exception that 1.45 μM 14C-IAA was used instead of 3H-IAA. Auxin transport in the inflorescence stem was measured using the method described previously (Ruegger et al., 1997; Brown et al., 2001), except, again, 1.45 μM 14C-IAA was used in place of 3H-IAA. Because the lengths of mutant and wild-type inflorescences differed at any given time, inflorescences were selected for assays when they reached 6 cm. Upper inflorescence segments were excised 0.5 cm below the apical tip.
Generation and Analysis of Yeast Overexpressing AtMDR1
To create the pAtMDR-y yeast expression vector, a 3.9-kb BstYI and SalI fragment of AtMDR1 cDNA was inserted at a BamHI-SalI site between the alcohol dehydrogenase (ADH) promoter and the Cyc1 terminator of the p426ADH yeast overexpression vector (Mumberg et al., 1995). Yeast strain AD12345678u− (matα, pdr1-3, his1, ura3, pdr5Δ, yor1Δ, snq2Δ, ycf1Δ, pdr10Δ, pdr11Δ, pdr15Δ, pdr3Δ) (Decottignies et al., 1998) was transformed with pAtMDR-y or p426ADH by the lithium acetate method. Expression of AtMDR1 in yeast carrying the pAtMDR-y (YAtMDR) but not in the control yeast was confirmed by RNA gel blot analysis (data not shown).
To measure 1-naphthylphthalamic acid (NPA) binding, yeast were grown in complete medium minus uracil with aeration until the OD600 of cell culture reached 0.5. Cells were spun down in tared microcentrifuge tubes, and the resulting cell pellets were weighed. Optimum pellet weight was determined to be 50 mg for assay in 1 mL of medium incubated in 2-mL tubes with shaking for aeration. The medium was amended with 10 nM 3H-NPA (57 Ci/mmol) or 10 nM 3H-NPA and 10 μM cold NPA (±10 μM benzoic acid for competition experiments). To determine the saturability of binding, separate series of experiments were conducted with all concentrations increased 10-fold times and 100-fold (3H-NPA at 100 nM and 1 μM, respectively). Samples were incubated at 30°C for 1 hr, spun down, and washed three times for 5 min with complete medium minus uracil. The pellet was resuspended with scintillation fluid and counted in a scintillation counter (Beckman LS 5810) calibrated to 14C standard to convert counts per minute to disintegrations per minute. For each experiment, three replicates were used. The reported values are means of specific binding (3H-NPA bound minus 3H-NPA bound in the presence of 1000-fold cold NPA) for each experiment. The stock solution of NPA had an activity of 30 Ci/mmol and a concentration of 28 μM.
Isolation and Identification of NPA Binding Proteins
Plasma membrane–enriched microsomal preparations of 5-day-old Arabidopsis seedlings were prepared as described previously (Murphy and Taiz, 1999a, 1999b). The purification of NPA binding proteins by affinity purification has been described in detail elsewhere (Murphy et al., 2001). Briefly, membrane proteins were detergent solubilized in a buffer containing 0.05% 3-([3-cholamidopropyl]dimethylammonio)-1-propanesulfonic acid/0.1% Brij 35. NPA was immobilized on amine-terminated magnetic beads (Sigma) using the manufacturer's protocol, with the exceptions that NPA was used in 5× molar excess and coupling reactions were conducted at >pH 6.5 at 0°C for 24 hr to prevent NPA hydrolysis. Coupled beads were washed extensively and used immediately. After further purification by gel permeation and anion exchange chromatography, fractions with Tyr aminopeptidase activity were mixed with NPA-conjugated magnetic beads, incubated at 4°C for 2 hr with gentle shaking, and then collected with a magnet. The beads were washed three times for 10 min at 4°C with a 0.05% Triton X-100 buffer containing 50 mM KCl. Fractions then were eluted in a 50 mM step gradient beginning at 100 mM and ending at 500 mM. Eluted fractions were dialyzed against 50 mM KCl buffer and then assayed for Tyr aminopeptidase and NPA binding activity as described previously (Murphy et al., 2000).
Proteins purified as described above were separated by SDS-PAGE and visualized with Coomassie Brilliant Blue R 250 staining. Coomassie blue–stained bands were excised, digested with trypsin, and separated by HPLC. Amino acid sequences of at least two peak fractions from each band then were analyzed by Edman degradation as described previously (Murphy et al., 2001). Molecular masses were confirmed in some cases by time of flight mass spectrometry (TOFL-MS). Sequences were compared with Arabidopsis genomic, cDNA, and predicted protein databases for unique matches. The presence of predicted cDNA then was confirmed by reverse transcriptase–mediated PCR.
Microsomal membrane preparation of 5-day-old seedlings was performed as described previously (Brown et al., 2001). 3H-NPA binding assays were performed in a 200-μL total volume of NPA-binding buffer (NBB) with protein at a final concentration of 0.2 mg/mL, as described by Brown et al. (2001). Microsomes were incubated for 1 hr at 4°C with 20 nM 3H-NPA in the presence or absence of 20 μM NPA. The addition of unlabeled NPA allowed measurement of specific binding (bound 3H-NPA minus bound 3H-NPA in the presence of 20 μM NPA). After incubation, samples were filtered over 0.3% (v/v) polyethyleneimine-treated GF/B filters and washed with 5 mL of cold NBB. The filters were counted in 2.5 mL of scintillation fluid using a liquid scintillation counter.
Accession Numbers
The GenBank accession numbers for the AtMDR1 and AtPGP1 genes are BAB02129 and A42150.
Acknowledgments
This work was supported by a National Science Foundation grant (Career Award IBN-9734478) to E.P.S.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010350.
References
- Ambudkar, S.V., Dey, S., Hrycyna, C.A., Ramachandra, M., Pastan, I., and Gottesman, M.M. (1999). Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39 361–398. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III Sci. Vie 316 1194–1199. [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inzé, D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst, P., Zelcer, N., and van Helvoort, A. (2000). ABC transporters in lipid transport. Biochim. Biophys. Acta 1486 128–144. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis thaliana. Plant Physiol. 126 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, J., and Aguilar-Bryan, Z. (1999). Sulfonylurea receptors: ABC transporters that regulate ATP-sensitive K+ channels. Biochim. Biophys. Acta 1461 285–303. [DOI] [PubMed] [Google Scholar]
- Castro, A.F., and Altenberg, G.A. (1997). Inhibition of drug transport by genistein in multidrug-resistant cells expressing P-glycoprotein. Biochem. Pharmacol. 53 89–93. [DOI] [PubMed] [Google Scholar]
- Chen, C., Clark, D., Ueda, K., Pastan, I., Gottesman, M., and Roninson, I. (1990). Genomic organization of the human multidrug resistance (MDR1) gene and origin of P-glycoproteins. J. Biol. Chem. 265 506–514. [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieli, E., Romiti, N., Cervelli, F., and Tongiani, R. (1995). Effects of flavonols on P-glycoprotein activity in cultured rat hepatocytes. Life Sci. 57 1741–1751. [DOI] [PubMed] [Google Scholar]
- Cho, M.H., and Spalding, E.P. (1996). An anion channel in Arabidopsis hypocotyls activated by blue light. Proc. Natl. Acad. Sci. USA 93 8134–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conseil, G., Baubichon-Cortay, H., Dayan, G., Jault, J.M., Barron, D., and Di Pietro, A. (1998). Flavonoids: A class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proc. Natl. Acad. Sci. USA 95 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, T.G.E., and Coleman, J.O.D. (2000). The Arabidopsis thaliana ATP-binding cassette proteins: An emerging superfamily. Plant Cell Environ. 23 431–433. [Google Scholar]
- Decottignies, A., and Goffeau, A. (1997). Complete inventory of the yeast ABC proteins. Nat. Genet. 15 137–145. [DOI] [PubMed] [Google Scholar]
- Decottignies, A., Grant, A.M., Nichols, J.W., de Wet, H., McIntosh, D.B., and Goffeau, A. (1998). ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273 12612–12622. [DOI] [PubMed] [Google Scholar]
- Dudler, R., and Hertig, C. (1992). Structure of an mdr-like gene from Arabidopsis thaliana: Evolutionary implications. J. Biol. Chem. 267 5882–5888. [PubMed] [Google Scholar]
- Faulkner, I.J., and Rubery, P.H. (1992). Flavonoids and flavonoid sulphates as probes of auxin-transport regulation in Cucurbita pepo hypocotyl segments and vesicles. Planta 186 618–625. [DOI] [PubMed] [Google Scholar]
- Fei, H., and Sawhney, V.K. (1999). Role of plant growth substances in MS33-controlled stamen filament growth in Arabidopsis. Physiol. Plant. 105 165–170. [Google Scholar]
- Ferté, J. (2000). Analysis of the tangled relationship between P-glycoprotein-mediated multidrug resistance and the lipid phase of the cell membrane. Eur. J. Biochem. 267 277–294. [DOI] [PubMed] [Google Scholar]
- Ferté, J., Kuhnel, J.M., Chapius, G., Rolland, Y., Lewin, G., and Schwaller, M.A. (1999). Flavonoid-related modulators of multidrug resistance: Synthesis, pharmacological activity, and structure-activity relationships. J. Med. Chem. 42 478–489. [DOI] [PubMed] [Google Scholar]
- Gaedeke, N., Klein, M., Kolukisaoglu, U., Forestier, C., Müller, A., Ansorge, M., Becker, D., Mamnun, Y., Kuchler, K., Schulz, B., Mueller-Roeber, B., and Martinoia, E. (2001). The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 20 1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Garbers, C., Delong, A., Deruere, J., Bernasconi, P., and Söll, D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15 2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Gil, P., and Green, P.J. (1997). Regulatory activity exerted by the SAUR-AC1 promoter region in transgenic plants. Plant Mol. Biol. 34 803–808. [DOI] [PubMed] [Google Scholar]
- Gottesman, M.M., Hrycyna, C.A., Schoenlein, P.V., Germann, U.A., and Pastan, I. (1995). Genetic analysis of the multidrug transporter. Annu. Rev. Genet. 29 607–649. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hardy, S.P., Goodfellow, H.R., Valverde, M.A., Gill, D.R., Sepulveda, V., and Higgins, C.F. (1995). Protein kinase C-mediated phosphorylation of the human multidrug resistance P-glycoprotein regulates cell volume-activated chloride channels. EMBO J. 14 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, C.F. (1995). The ABC of channel regulation. Cell 82 693–696. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). TMbase: A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347 166. [Google Scholar]
- Jacobs, M., and Rubery, P.H. (1988). Naturally occurring auxin transport regulators. Science 241 346–349. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H.L., Chalfant, M.L., Jovov, B., Lockhart, J.P., Parker, S.B., Fuller, C.M., Stanton, B.A., and Benos, D.J. (2000). The cytosolic termini of the β- and γ-ENaC subunits are involved in the functional interactions between cystic fibrosis transmembrane con-ductance regulator and epithelial sodium channel. J. Biol. Chem. 275 27947–27956. [DOI] [PubMed] [Google Scholar]
- Johnstone, R.W., Ruefli, A.A., and Smyth, M.J. (2000). Multiple physiological functions for multidrug transporter P-glycoprotein? Trends Biochem. Sci. 25 1–6. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasswell, J., Rogg, L.E., Nelson, D.C., Rongey, C., and Bartel, B. (2000). Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12 2395–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobello, G., Fambrini, R., Baraldi, R., Lercari, B., and Pugliesi, C. (2000). Hormonal influence on photocontrol of the protandry in the genus Helianthus. J. Exp. Bot. 51 1403–1412. [PubMed] [Google Scholar]
- Lomax, T.L., Muday, G.K., and Rubery, P.H. (1995). Auxin transport. In Plant Hormones: Physiology, Biochemistry, and Molecular Biology, 2nd ed, P.J. Davies, ed (Norwell, MA: Kluwer Academic Publishers), pp. 509–530.
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A., Guan, C., Gälweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119–122. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Murphy, A., and Taiz, L. (1995). A new vertical mesh transfer technique for metal-tolerance studies in Arabidopsis: Ecotype variation and copper-sensitive mutants. Plant Physiol. 108 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A., and Taiz, L. (1999. a). Naphthylphthalamic acid is enzymatically hydrolyzed at the hypocotyl-root transition zone and other tissues of Arabidopsis thaliana seedlings. Plant Physiol. Biochem. 37 413–430. [Google Scholar]
- Murphy, A., and Taiz, L. (1999. b). Localization and characterization of soluble and plasma membrane aminopeptidase activities in Arabidopsis seedlings. Plant Physiol. Biochem. 37 431–443. [Google Scholar]
- Murphy, A., Peer, W.A., and Taiz, L. (2000). Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211 315–324. [DOI] [PubMed] [Google Scholar]
- Murphy, A.S., Hoogner, K., Peer, W.A., and Taiz, L. (2001). Identification, purification, and molecular cloning of N-1-naphthylphthalamic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Parks, B.M., Cho, M.H., and Spalding, E.P. (1998). Two genetically separable phases of growth inhibition induced by blue light in Arabidopsis seedlings. Plant Physiol. 118 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Brown, D., Taiz, L., Muday, G.K., and Murphy, A.S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis thaliana. Plant Physiol. 126 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria, J.M., Chiquero, M.J., Conseil, G., Dayan, G., Di Pietro, A., Barron, D., Castanys, S., and Gamarro, F. (1999). Correlation between the affinity of flavonoids binding to the cytosolic site of Leishmania tropica multidrug transporter and their efficiency to revert parasite resistance to daunomycin. Biochemistry 38 1736–1743. [DOI] [PubMed] [Google Scholar]
- Rea, P.A., Li, Z.-S., Lu, Y.-P., Drozdowicz, Y.M., and Martinoia, E. (1998). From vacuolar GS-X pumps to multispecific ABC transporters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 727–760. [DOI] [PubMed] [Google Scholar]
- Roepe, P.D. (2000). What is the precise role of human MDR1 protein in chemotherapeutic drug resistance? Curr. Pharm. Design 6 241–260. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., Muday, G., and Estelle, M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruetz, S., and Gros, P. (1994). Phosphatidylcholine translocase: A physiological role for the mdr2 gene. Cell 77 1071–1081. [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández, R., Davies, T.G.E., Coleman, J.O.D., and Rea, P.A. (2001). The Arabidopsis thaliana ABC protein superfamily: A complete inventory. J. Biol. Chem. 276 30231–30244. [DOI] [PubMed] [Google Scholar]
- Schreiber, R., Hopf, A., Mall, M., Greger, R., and Kunzelmann, K. (1999). The first-nucleotide binding domain of the cystic-fibrosis transmembrane conductance regulator is important for inhibition of the epithelial Na+ channel. Proc. Natl. Acad. Sci. USA 96 5310–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig, A. (1998). A general pattern for substrate recognition by P-gly-coprotein. Eur. J. Biochem. 251 252–261. [DOI] [PubMed] [Google Scholar]
- Sidler, M., Hassa, P., Hasan, S., Ringli, C., and Dudler, R. (1998). Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10 1623–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou, F.L. (2000). Plant ABC transporters. Biochim. Biophys. Acta 1465 79–103. [DOI] [PubMed] [Google Scholar]
- van Helvoort, A., Smith, A.J., Sprong, H., Fritzsche, I., Schinkel, A.H., Borst, P., and van Meer, G. (1996). MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 87 507–517. [DOI] [PubMed] [Google Scholar]
- Vanoye, C.G., Altenberg, G.A., and Reuss, L. (1997). P-glycoprotein is not a swelling-activated Cl− channel: Possible role as a Cl− channel regulator. J. Physiol. Lond. 502 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L.M., Abel, S., Shen, N., de la Foata, M., Mall, Y., and Theologis, A. (1996). Differential activation of the primary auxin response genes, PS-IAA4/5 and PS-IAA6, during early development. Plant J. 9 587–599. [DOI] [PubMed] [Google Scholar]
- Wyatt, R.E., Ainley, W.M., Nagao, R.T., Conner, T.W., and Key, J.L. (1993). Expression of the Arabidopsis AtAUX2-11 auxin-responsive gene in transgenic plants. Plant Mol. Biol. 22 731–749. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309. [DOI] [PubMed] [Google Scholar]