Abstract
Gibberellins (GAs) are essential regulators of many aspects of plant development, including stem elongation, seed germination, and flowering. RSG is a transcriptional activator with a basic leucine zipper domain that regulates endogenous amounts of GAs through the control of a GA biosynthetic enzyme. The ubiquitous expression of RSG in plant organs suggests an involvement of post-transcriptional and/or post-translational modifications of the transcription factor. Here, we identify the 14-3-3 signaling proteins as RSG binding partners. The mutant version of RSG that could not bind to 14-3-3 proteins exhibited a higher transcriptional activity than did wild-type RSG. Consistent with this observation, the mutant RSG that could not bind to 14-3-3 proteins was localized predominantly in the nucleus, whereas wild-type RSG was distributed throughout the cell. Using the nuclear export inhibitor leptomycin B, we also showed that RSG, apparently statically localized in the cytoplasm, is capable of shuttling in and out of the nucleus. These results suggest that 14-3-3 proteins negatively modulate RSG, which is involved in the regulation of endogenous amounts of GAs, by controlling its intracellular localization.
INTRODUCTION
One of the unique features of plants is their developmental plasticity. As sessile organisms, plants have evolved highly plastic developmental programs to adapt to the changing environment. Plant hormones modulate growth and development in response to both endogenous programs and environmental stimuli. Gibberellins (GAs), which are tetracyclic diterpenoid growth factors, are essential regulators of many aspects of plant development, including seed germination, stem elongation, flower induction, and anther development. GAs are of considerable agricultural significance. World crop grain yields increased substantially in the 1960s and 1970s because farmers rapidly adopted the new varieties and cultivation methods of the so-called green revolution. The new varieties are shorter, increase grain yield at the expense of straw biomass, and are more resistant to damage by wind and rain. These characteristics are caused by mutations in a GA response modulator belonging to the GRAS family (Peng et al., 1999), which exhibit structural similarity to mammalian STAT (for signal transducers and activators of transcription) proteins (Richards et al., 2000). GA-deficient Arabidopsis mutants display characteristic phenotypes, including dark green leaves and stunted growth attributable to the inhibition of stem elongation, especially during the cell elongation phase. Recent genetic strategies using these mutants have led to the isolation of a series of genes that encode GA biosynthetic enzymes (Sun and Kamiya, 1994; Chiang et al., 1995; Xu et al., 1995; Yamaguchi et al., 1996; Helliwell et al., 1998, 2001). Both endogenous developmental programs and environmental stimuli affect the expression of these enzymes. Therefore, elucidating the transcriptional regulation of GA biosynthetic enzymes is crucial to identify the molecular mechanisms involved in plant development and to understand how these mechanisms help plants adapt to changes in their environment.
RSG (for repression of shoot growth) is a transcriptional activator with a basic leucine zipper (bZIP) domain that is involved in the regulation of endogenous amounts of GAs (Fukazawa et al., 2000). RSG bound and activated the promoter of Arabidopsis GA3, the gene encoding ent-kaurene oxidase in the GA biosynthetic pathway. The dominant negative form of RSG repressed the expression of the ent-kaurene oxidase gene in transformed tobacco plants. This downregulation reduced endogenous amounts of GAs and severely inhibited the process of cell elongation of stems, resulting in a dwarf phenotype. Thus, RSG regulates the morphology of plants by controlling endogenous amounts of GAs. Although GA biosynthesis is restricted to specific regions, including actively growing and elongating tissues (Smith, 1992), RSG is expressed ubiquitously in plant organs. This apparent inconsistency suggests an involvement of post-transcriptional and/or post-translational modifications of the transcription factor. One possible mechanism for the functional regulation of RSG is the interaction of RSG with accessory proteins, including other transcriptional activators, repressors, general transcription factors, coactivators, or chaperones.
The 14-3-3 proteins, which constitute a highly conserved isoform of homodimeric and heterodimeric molecules, associate with a number of different signaling proteins, including Raf-1, Bad, the epithelial keratins K8/K18, Cdc25, and telomerase (Liao and Omary, 1996; Muslin et al., 1996; Zha et al., 1996; Peng et al., 1997; Seimiya et al., 2000). On the basis of their interaction with various ligands, 14-3-3 proteins have been proposed to be important in controlling intracellular signaling pathways (Aitken, 1996). In addition, 14-3-3 proteins work as molecular chaperones or regulate the intracellular localization of their binding partners (Liao and Omary, 1996; Kumagai and Dunphy, 1999; Yang et al., 1999). The three-dimensional crystal structures of 14-3-3 have been solved as a dimer with a bundle of nine antiparallel α-helices in each monomer (Liu et al., 1995; Xiao et al., 1995). Many ligand proteins containing the conserved phosphorylated motifs bind into a large groove formed by amphiphilic helices of 14-3-3 (Muslin et al., 1996; Yaffe et al., 1997). In plants, 14-3-3 proteins include the regulator for the H+-ATPase in the plasma membrane (Korthout and de Boer, 1994; Marra et al., 1994; Oecking et al., 1994) and a protein that specifically inhibits nitrate reductase activity from spinach cells (Bachmann et al., 1996; Moorhead et al., 1996). The plant 14-3-3 proteins also are found as part of a transcriptional DNA binding complex. 14-3-3 proteins have been reported to associate with G-box DNA binding complexes (Lu et al., 1992; Schultz et al., 1998) and a TATA box binding protein (Pan et al., 1999). In agreement with these results, 14-3-3 proteins were found in the nucleus of plant cells (Bihn et al., 1997). These findings raise the possibility that 14-3-3 proteins may participate in the regulation of transcription factors.
Recently, molecular genetic studies using morphological mutants have revealed many proteins that exhibit homology with transcription factors involved in plant development. To understand how these transcription factors affect morphogenesis, their target genes and the functional regulation mechanism must be identified. However, information regarding target genes and post-translational regulations of transcription factors is still limited in plants. We previously found that the ent-kaurene oxidase gene is a target of RSG (Fukazawa et al., 2000). In this study, we have identified 14-3-3 family proteins as RSG binding proteins. The mutant version of RSG that could not bind to 14-3-3 proteins exhibited a higher transcriptional activity than did wild-type RSG. We also demonstrated that this mutant version of RSG was localized predominantly in the nucleus, whereas RSG was distributed throughout the cell. These observations suggest that 14-3-3 is a post-translational regulator of RSG that functions by controlling the intracellular localization of RSG.
RESULTS
Identification of 14-3-3 Proteins as RSG-Interacting Proteins
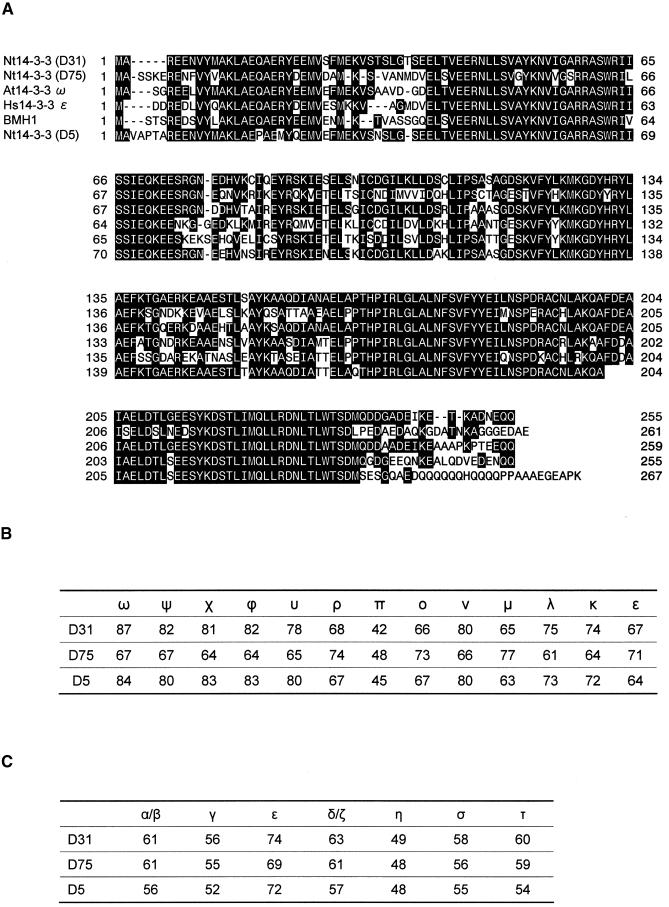
To identify proteins that interact specifically with RSG, we performed a yeast two-hybrid screen using RSG as bait on cDNA libraries made from tobacco BY-2 cells. From 2 3 106 transformants, 13 colonies expressed b-galactosidase activity and were capable of growth on a medium lacking histidine, indicating that two reporter genes were expressed. Restriction mapping and sequencing revealed that 12 clones encoded members of the 14-3-3 family of proteins. There are at least seven distinct genes for 14-3-3 in mammalian cells, giving rise to nine isotypes (a, b, g, ´, d, h, s, t, and z, with a and d being phosphorylated forms of b and z, respectively) (Aitken et al., 1995). The Arabidopsis 14-3-3 family consists of 13 members (v, c, x, f, y, r, p, o, n, m, l, k, and ´) (DeLille et al., 2001). 14-3-3s are currently designated by Greek letters, with the mammalian isoform names generally chosen from the beginning of the alphabet and the Arabidopsis isoforms chosen from the end of the alphabet. Our tobacco 14-3-3 clones represented three different genes, named D5, D31, and D75 (Figure 1A), which exhibited higher similarity to the v, v, and m types of Arabidopsis, respectively (Figure 1B). Sequence comparisons with human 14-3-3 isoforms revealed that D5, D31, and D75 have a higher similarity to the ´ type among mammalian isoforms (Figure 1C). D31 was chosen for further analysis.
Figure 1.
Comparison of Deduced Amino Acid Sequences of 14-3-3 Proteins.
(A) Alignment of amino acid sequences of 14-3-3 proteins. The sequence of the tobacco 14-3-3 protein (D31) was compared with the sequences of tobacco D5 and D75 (this work), Arabidopsis GF14ω (At14-3-3 ω; Lu et al., 1992), human 14-3-3ɛ (Hs14-3-3 ε; Chong et al., 1996), and yeast BMH1 (van Heusden et al., 1992). D5 is a partial cDNA clone. Highlighted residues indicate amino acids that are identical to those of D31.
(B) Percentage of identical amino acid residues between the tobacco 14-3-3 proteins and all of the isoforms of Arabidopsis 14-3-3 proteins.
(C) Percentage of identical amino acid residues between the tobacco 14-3-3 proteins and isoforms of human 14-3-3 proteins.
Expression of 14-3-3 in Tobacco
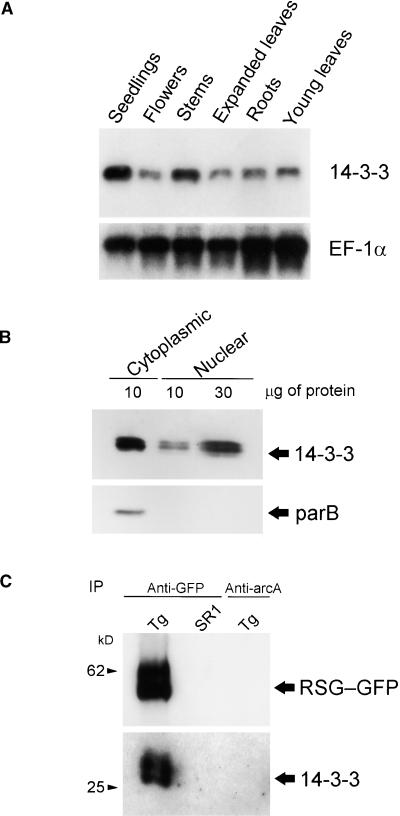
The expression of the 14-3-3 genes was investigated by RNA gel blot analysis, with total RNA isolated from tobacco plants. The mRNAs for 14-3-3 are expressed in seedlings, stems, roots, young leaves, expanded leaves, and flowers (Figure 2A). We next investigated the subcellular distribution of 14-3-3 proteins using antibodies raised against the recombinant 14-3-3 protein. Immunoblot analysis using proteins of tobacco BY-2 cells showed that 14-3-3 proteins were distributed in both cytosolic and nuclear fractions (Figure 2B). The antibodies detected at least two isoforms of 14-3-3 proteins. Tobacco parB protein (Takahashi and Nagata, 1992), which is a glutathione S-transferase, was used as a marker protein of the cytosolic fraction. Although known 14-3-3 binding proteins in plants are mainly cytosolic proteins, including nitrate reductase (Bachmann et al., 1996; Moorhead et al., 1996), sucrose-phosphate synthase (Toroser et al., 1998), and the plasma membrane H+-ATPase (Korthout and de Boer, 1994; Marra et al., 1994; Oecking et al., 1994), our result is in agreement with previous results showing that 14-3-3 proteins also were found in the nuclei of both maize and Arabidopsis cells (Bihn et al., 1997).
Figure 2.
Interaction of RSG with 14-3-3 in Vivo and Expression of 14-3-3 in Tobacco.
(A) RNA gel blot analysis of 14-3-3 mRNAs. One microgram of poly(A)+ RNA isolated from the indicated organs was used for gel blot analysis. The blot was hybridized with radiolabeled tobacco 14-3-3 cDNA (D31) and then reprobed with EF-1α cDNA as a loading control.
(B) Protein gel blot analysis of tobacco cells. Cytoplasmic and nuclear extracts prepared from tobacco BY-2 cells were separated on SDS–polyacrylamide gel electrophoresis and blotted to membranes. 14-3-3 proteins were detected with anti-14-3-3 antiserum. Tobacco parB protein was used as a cytosolic marker.
(C) Association of RSG and 14-3-3 in plant cells. The lysate of leaves expressing GFP-tagged RSG was immunoprecipitated with either anti-GFP or anti-arcA antiserum, and the bound fractions were subjected to protein gel blot analysis with anti-RSG (top) and anti-14-3-3 (bottom). Anti-RSG antibodies detected two bands in the immunoprecipitate. Although the exact nature of the proteins is unknown, double bands could be caused by proteolytic degradation and/or post-translational modifications. IP, immunoprecipitate; SR1, control SR1 tobacco plants; Tg, transgenic tobacco plants expressing RSG–GFP.
RSG Binds 14-3-3 in Vivo
To confirm the interaction between RSG and 14-3-3 proteins in plant cells, we generated transgenic tobacco plants in which the fusion gene of RSG and green fluorescent protein (GFP) was expressed under the control of the 35S promoter of the Cauliflower mosaic virus (CaMV). The RSG–GFP fusion protein was immunoprecipitated with anti-GFP antibodies, and RSG-bound materials were immunoblotted with anti–14-3-3 antibodies. Antibodies raised against tobacco arcA, a WD-40 protein (Ishida et al., 1993), were used as a negative control for immunoprecipitation. As shown in Figure 2C, 14, 14 proteins were coimmunoprecipitated with the GFP-tagged RSG. These observations indicate that RSG binds to 14-3-3 proteins in plant cells.
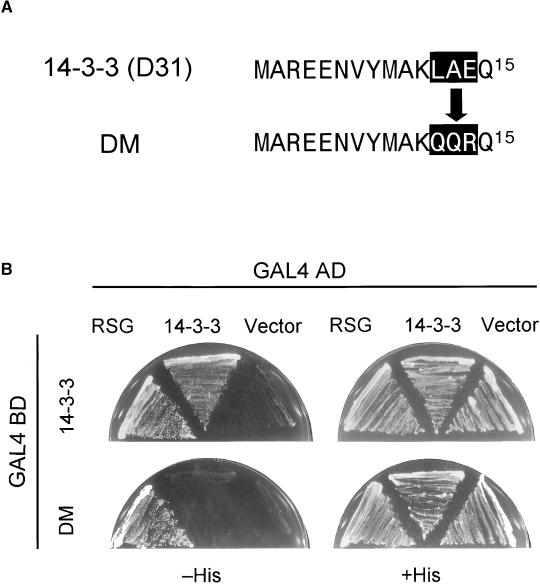
Dimerization Motif of 14-3-3
Biochemical characterization of the 14-3-3 proteins has shown that they are able to form homodimers or heterodimers through their N-terminal dimerization domains (Luo et al., 1995; Wu et al., 1997). The dimerization of 14-3-3 proteins seemed to be required for the successful involvement in signaling events of Raf-1 in mammalian cells because mutant monomeric forms of 14-3-3, although able to bind Raf-1, do not enable Raf-1 to be activated in vivo (Tzivion et al., 1998). To examine the role of the first N-terminal a-helix of tobacco 14-3-3 for dimerization, we constructed a mutant version of 14-3-3 in which the amino acid sequence 12-LAE-14 was mutated to QQR (Figure 3A). This mutation diminished the dimerization of 14-3-3 in the yeast two-hybrid assay, whereas it did not affect the binding to RSG (Figure 3B). Thus, the first N-terminal a-helix of tobacco 14-3-3 is involved in dimerization. Furthermore, this result also indicated that the ligand binding of 14-3-3 is independent of dimerization.
Figure 3.
Mutations of the N-Terminal Domain of 14-3-3 Impair Dimerization.
(A) Amino acid sequences of a 14-3-3 mutant (DM) and the wild-type protein.
(B) Yeast two-hybrid assay. Full-length tobacco 14-3-3 (D31) or its mutant version (DM) was cloned into pGBT9 as an in-frame fusion with the DNA binding domain (BD) of GAL4. Full-length RSG or tobacco 14-3-3 (D31) was cloned into pGAD424 as an in-frame fusion with the activation domain (AD) of GAL4. Yeast HF7c cells were transformed simultaneously with a combination of the indicated plasmids. Cells containing both plasmids were streaked on the plates with (+His) or without (−His) histidine but with 2 mM 3-aminotriazole.
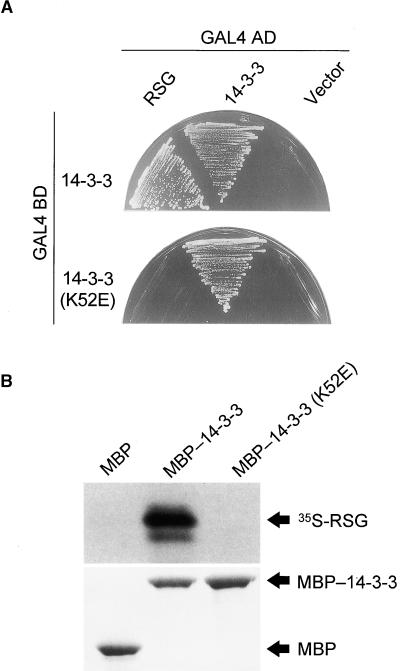
K52E Mutation Disrupts the Binding of 14-3-3 to RSG
The analysis of the crystal structure of 14-3-3 proteins revealed a conserved amphipathic groove that may allow the association of 14-3-3 proteins with the phosphoserine-containing motifs of many signaling molecules (Liu et al., 1995; Muslin et al., 1996). The Lys-49 in the amphipathic groove of mammalian 14-3-3z is indispensable for ligand binding. The point mutation (K49E) of 14-3-3z proteins disrupts the association of 14-3-3 with Raf-1 kinase without large structural changes (Zhang et al., 1997). Because Lys-49 is conserved among all 14-3-3 isoforms, Lys-49 equivalent residues in other isoforms also may participate in binding to the conventional phosphoserine-containing motif in a similar fashion. To examine whether this lysine residue is involved in binding with RSG, we made the tobacco 14-3-3 mutant K52E, which corresponds to K49E of mammalian 14-3-3z. The K52E mutation in tobacco 14-3-3 diminished the interaction with RSG in the two-hybrid assay (Figure 4A); however, this mutation did not affect the dimerization of 14-3-3, suggesting that the impaired binding to RSG is not caused by the overall structural alteration of the mutant protein. The mode of the binding of tobacco 14-3-3 to RSG seemed similar to that of mammalian 14-3-3 and the signaling molecules with the phosphoserine-containing motifs.
Figure 4.
Effect of the K52E Mutation of 14-3-3 on Its Interaction with RSG.
(A) Yeast two-hybrid assay. Full-length tobacco 14-3-3 (D31) or the K52E mutant version of 14-3-3 was cloned into pGBT9 as an in-frame fusion with the DNA binding domain (BD) of GAL4. Full-length tobacco 14-3-3 (D31) or RSG was cloned into pGAD424 as an in-frame fusion with the activation domain (AD) of GAL4. Yeast HF7c cells were transformed simultaneously with a combination of the indicated plasmids. Cells containing both plasmids were streaked on the plates without histidine but with 0.2 mM 3-aminotriazole.
(B) In vitro binding assay. MBP–14-3-3 or MBP–14-3-3(K52E) fusion protein was conjugated to amylose resin and incubated with 35S-methionine–labeled RSG. 14-3-3–associated proteins were resolved by SDS-PAGE. After electrophoresis, proteins were visualized by fluorography and Coomassie blue staining.
This observation was confirmed by in vitro binding experiments. Recombinant maltose binding protein (MBP)–14-3-3 and MBP–14-3-3(K52E) fusion proteins were absorbed to amylose resin. These immobilized 14-3-3 fusion proteins were incubated individually with in vitro–translated, 35S-methionine–labeled RSG followed by affinity chromatography. Full- length RSG bound to MBP–14-3-3 but not to the control MBP or to MBP–14-3-3(K52E) (Figure 4B). These results substantiate the importance of Lys-52 in tobacco 14-3-3 on binding to RSG, as observed in the yeast two-hybrid assay. Furthermore, they indicate that the interaction between RSG and 14-3-3 is direct.
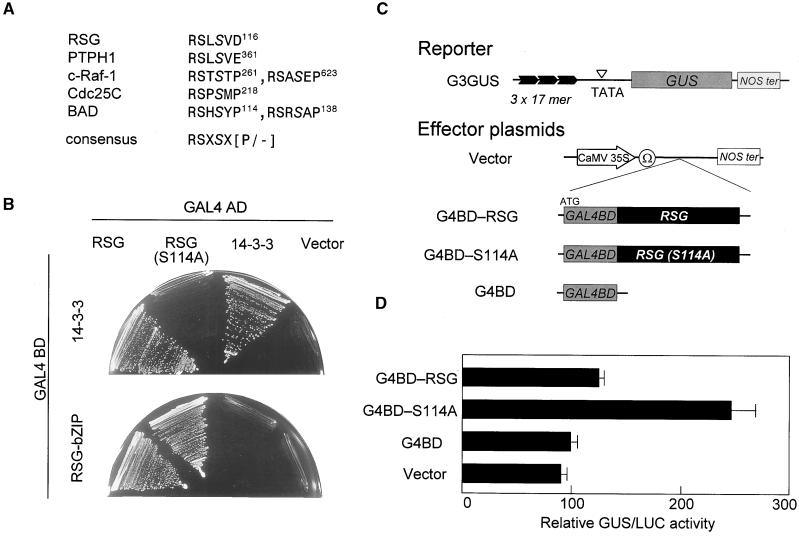
Ser-114 of RSG Is Essential for Binding to 14-3-3
It has been demonstrated that 14-3-3 proteins bind to phosphorylated motifs containing phosphoserine residues of RSXpSXP and RXY/FXpSXP (pS indicates a critical phosphoserine) in their target proteins (Muslin et al., 1996; Yaffe et al., 1997). A sequence closely related to the conventional 14-3-3 binding motif was found surrounding Ser-114 of RSG (Figure 5A). To determine whether Ser-114 of RSG was critical for the interaction between RSG and 14-3-3, we constructed a mutant version of RSG in which Ser-114 in the putative 14-3-3 binding motif was mutated to alanine. As expected, the mutation S114A of RSG diminished the ability to interact with tobacco 14-3-3 in the yeast two-hybrid assay; however, this mutation did not affect the dimerization of RSG through the bZIP domain (Figure 5B), suggesting that the impaired binding to 14-3-3 is not attributable to the gross structural alteration of the mutant protein. These results show that Ser-114 is a critical serine on binding to 14-3-3. Of note, similar 14-3-3 binding motifs are found in RSG-related proteins of Arabidopsis.
Figure 5.
Effect of the S114A Mutation of RSG on Its Function.
(A) Comparative alignment of the 14-3-3 binding motif identified in RSG with other known 14-3-3 binding sequences. Critical serine residues on 14-3-3 binding are italicized.
(B) Full-length tobacco 14-3-3 (D31) or the bZIP domain of RSG (amino acids 159 to 287) was cloned into pGBT9 as an in-frame fusion with the DNA binding domain (BD) of GAL4. Full-length RSG or the S114A mutant version of RSG was cloned into pGAD424 as an in-frame fusion with the activation domain (AD) of GAL4. Yeast HF7c cells were transformed simultaneously with a combination of the indicated plasmids. The control was the pGAD424 vector. Cells containing both plasmids were streaked on the plates without histidine but with 0.5 mM 3-aminotriazole.
(C) Scheme of the reporter and effector plasmids used in transient assays. The three tandem copies of the GAL4 binding site were fused to the TATA box of parB-driving GUS (G3GUS). The effector plasmid expressing the GAL4 DNA binding domain (G4BD) fused upstream of the full- length RSG or the S114A mutant under the control of the 35S promoter with a viral translation enhancer (Ω). NOS ter indicates the polyadenylation signal of the gene for nopaline synthetase.
(D) The S114A mutation enhances the transcriptional activity of RSG. The reporter construct G3GUS (10 μg) and the indicated effector construct (20 μg), with 35S–LUC (10 μg) as an internal control, were introduced to protoplasts of tobacco BY-2 cells. Each assay was performed in triplicate, and GUS activity was normalized by luciferase (LUC) activity. Bars indicate ±sd.
S114A Mutation Enhances Transcriptional Activity
To address the biological role of 14-3-3 binding to RSG, we conducted experiments using the GAL4 system. Transactivator proteins (effectors) were constructed as heterologous fusions between the GAL4 DNA binding domain (amino acids 1 to 147) and either the wild-type RSG or the RSG mutant S114A that cannot interact with tobacco 14-3-3 (Figure 5C). The transcriptional activity of the mutant was compared with that of the wild-type protein after transient transfection into tobacco cultured cells together with the reporter plasmid G3GUS, which contains three copies of the GAL4 target sequence element. As negative controls, the reporter plasmid G3GUS plus the empty effector plasmid or the plasmid carrying only the DNA binding domain of GAL4 were cotransfected. The transformation efficiency was monitored by introducing another reporter construct, luciferase, which was driven by the 35S promoter of CaMV (35S–LUC), together with the effector and the reporter constructs. The RSG mutant S114A that cannot interact with tobacco 14-3-3 is a more potent transcriptional activator than is wild-type RSG (Figure 5D), indicating that tobacco 14-3-3 negatively regulates RSG.
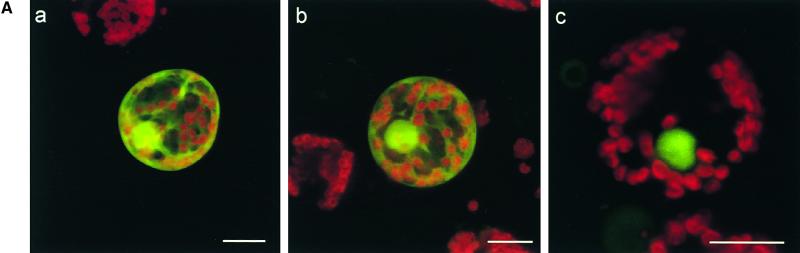
14-3-3 Directs the Intracellular Localization of RSG
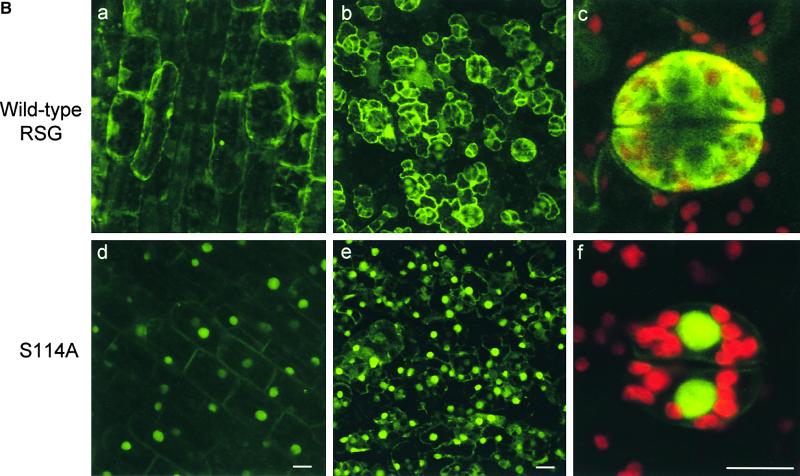
One example of a protein that has been shown to bind to 14-3-3 proteins through the conserved motif containing a phosphoserine residue is Cdc25. Cdc25 is a phosphatase that dephosphorylates and activates the cyclin-dependent kinase Cdc2, leading to entry into mitosis. The binding of 14-3-3 proteins has little or no effect on the phosphatase activity of Cdc25; however, 14-3-3 proteins sequester Cdc25 in the cytoplasm, regulating its subcellular localization to prevent access of Cdc25 to the Cdc2–cyclin B substrates (Peng et al., 1997; Kumagai and Dunphy, 1999; Yang et al., 1999). This finding prompted us to examine the possibility that 14-3-3 proteins regulate RSG by controlling its intracellular localization. To assess whether the binding of 14-3-3 proteins has a role in the localization of RSG, we investigated the intracellular localization of the GFP fusion proteins in a transient assay system with tobacco mesophyll protoplasts. RSG–GFP was distributed throughout the nucleus and cytoplasm; however, the mutant version of RSG that cannot bind 14-3-3 proteins (S114A–GFP) localized exclusively in the nucleus (Figure 6A). Furthermore, we analyzed transgenic tobacco plants in which hybrid proteins consisting of GFP and either wild-type RSG (RSG–GFP) or the S114A mutant (S114A–GFP) was expressed under the control of the 35S promoter of CaMV. In epidermal cells of young leaves and stems, RSG–GFP accumulated in both the cytoplasm and the nucleus (Figure 6B, panels a and b), whereas the S114A–GFP mutant that cannot associate with 14-3-3 was predominantly nuclear (Figure 6B, panels d and e). In guard cells, RSG–GFP was mainly cytoplasmic, whereas the S114A–GFP mutant localized exclusively in the nucleus (Figure 6B, panels c and f). These results indicated that the binding of 14-3-3 proteins to RSG might promote the cytoplasmic localization of wild-type RSG.
Figure 6.
Effects of the S114A Mutation on Intracellular Localization of RSG.
(A) Intracellular localization of GFP fusion proteins in tobacco mesophyll protoplasts. Cells were transfected with plasmids encoding GFP (a), wild-type RSG–GFP (b), or S114A–GFP (c) and observed by confocal laser scanning microscopy. More than 1000 cells were examined for each construct. Fluorescence signals of both GFP (green) and chlorophyll (red) were detected and merged. Bars = 20 μm.
(B) Intracellular localization of GFP fusion proteins in transgenic tobacco plants. For each construct, at least three independent transgenic tobacco plants expressing GFP fusion proteins were analyzed. Cells were observed by confocal laser scanning microscopy. (a) Epidermal cells of stems of transgenic tobacco plants expressing wild-type RSG–GFP. Fluorescence signals of GFP are displayed. (b) Epidermal cells of leaves of transgenic tobacco plants expressing wild-type RSG–GFP. Fluorescence signals of GFP are displayed. (c) Larger image of guard cells of leaves of transgenic tobacco plants expressing wild-type RSG–GFP. Fluorescence signals of GFP and chlorophyll are overlaid. (d) Epidermal cells of stems of transgenic tobacco plants expressing S114A–GFP. Fluorescence signals of GFP are displayed. (e) Epidermal cells of leaves of transgenic tobacco plants expressing S114A–GFP. Fluorescence signals of GFP are displayed. (f) Larger image of guard cells of leaves of transgenic tobacco plants expressing S114A–GFP. Fluorescence signals of GFP and chlorophyll are overlaid. Bars = 20 μm.
RSG Is a Nuclear-Cytoplasmic Shuttling Protein
The apparent cytoplasmic localization of RSG in plant cells suggests that RSG could be tethered to a cytoplasmic structure by a static mechanism that prevents its release and subsequent movement to the nucleus. Alternatively, the cytoplasmic localization of RSG may reflect the balance of a dynamic nuclear import/export process in which RSG is mobile and can enter the nucleus but is exported rapidly to the cytoplasm. To distinguish between these hypotheses, we examined the effect of leptomycin B (LMB), a potent inhibitor that blocks nuclear export (Nishi et al., 1994; Kudo et al., 1998, 1999). This drug inhibits CRM1/exportin 1, a receptor that mediates the nuclear export of proteins containing a nuclear export sequence (NES) (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997). Until now, the use of LMB with plant cells has been limited to a few cases; however, we confirmed that the Arabidopsis homolog of CRM1/exportin 1 contains the conserved cysteine that is alkylated by LMB (Kudo et al., 1999). As a result, plant cells would be expected to be LMB sensitive. If RSG in plant cells is tethered statically in the cytoplasm, it should remain there upon treatment with LMB. However, if RSG shuttles rapidly between the nucleus and the cytoplasm, LMB should trap it in the nucleus. The treatment of epidermal cells of leaves with LMB led to a significant redistribution of RSG–GFP within 45 min from a mainly cytoplasmic to an almost exclusively nuclear localization (Figure 7). These results indicated that RSG is exported actively to the cytoplasm and that its intracellular localization is regulated by a dynamic mechanism rather than a static retention mechanism.
Figure 7.
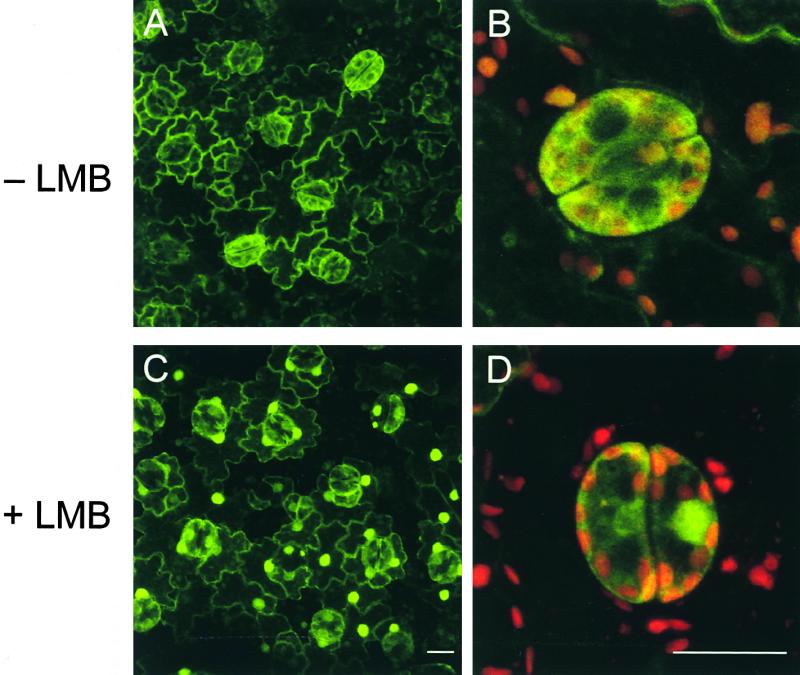
RSG Shuttles.
Epidermal cells of leaves of transgenic tobacco plants expressing wild-type RSG–GFP were incubated with 20 nM LMB for 45 min and observed by confocal laser scanning microscopy.
(A) Intracellular localization of RSG–GFP in epidermal cells of leaves before treatment with LMB. Fluorescence signals of GFP are displayed.
(B) Larger images of guard cells of leaves before treatment with LMB. Fluorescence signals of GFP and chlorophyll are overlaid.
(C) Intracellular localization of RSG–GFP in epidermal cells of leaves treated with 20 nM LMB for 45 min. Fluorescence signals of GFP are displayed.
(D) Larger images of guard cells of leaves treated with 20 nM LMB for 45 min. Fluorescence signals of GFP and chlorophyll are overlaid.
Bars in (C) and (D) = 20 μm.
DISCUSSION
Signal transduction pathways are complex networks of biochemical reactions that ultimately culminate in specific patterns of nuclear gene expression mediated by transcription factors. Intensive studies have revealed a variety of post-translational regulation mechanisms of transcription factors, including the interaction with coactivators and general transcription factors. Another possible mechanism, in which the ability of transcription factors is not necessarily affected, is the regulation of intracellular localization. In this study, we have identified 14-3-3 proteins as factors interacting with RSG that control the endogenous amounts of GAs in plants. Because mRNA for RSG is expressed in various organs (Fukazawa et al., 2000), post-transcriptional and/or post-translational modifications of RSG should be necessary for the appropriate spatial and temporal regulation of GA amounts in plants. We demonstrated an involvement of 14-3-3 proteins in the regulation of the subcellular localization of RSG, suggesting a molecular mechanism for control of its transcriptional activity. A function of 14-3-3 proteins may be to bind RSG and thereby sequester RSG in the cytoplasm so that it is unable to regulate its target genes in the nucleus. Thus, 14-3-3 proteins negatively modulate RSG through the regulation of intracellular localization.
We found that the S114A mutant of RSG that lost the ability to interact with tobacco 14-3-3 accumulates in the nuclear compartment of plant cells. This result suggested that the binding of 14-3-3 proteins might inhibit the nuclear import and/or promote the nuclear exclusion of wild-type RSG. Masking of the nuclear localization sequence and/or exposure of the NES of the complex could be a consequence of 14-3-3 binding to RSG. Alternatively, 14-3-3 binding could result in a conformational alteration of RSG that changes the interaction with nuclear import and/or export machinery. Recent investigations indicate that 14-3-3 proteins regulate the intracellular localization of their several binding partners. In the fission yeast Schizosaccharomyces pombe, a 14-3-3 protein (Rad24) functions as an attachable NES that enhances the nuclear export of Cdc25 in response to DNA damage (Lopez-Girona et al., 1999). On the other hand, in Xenopus, binding of 14-3-3 proteins to Cdc25 markedly reduces the nuclear import rate of Cdc25, allowing nuclear export mediated by CRM1 to predominate (Yang et al., 1999). Different cells use different molecular mechanisms to regulate the subcellular localization of Cdc25 by 14-3-3 proteins; however, the outcome in response to 14-3-3 binding is the same (i.e., Cdc25 is excluded from the nucleus to prevent access to the Cdc2–cyclin B substrates). Similarly, a variety of mammalian proteins involved in transcriptional control are regulated through 14-3-3 binding, including FKHRL1 (Brunet et al., 1999), histone deacetylase (Grozinger and Schreiber, 2000; McKinsey et al., 2000), TAZ (Kanai et al., 2000), and MITR (Zhang et al., 2001). Plants do not appear to have homologs of Cdc25 (Arabidopsis Genome Initiative, 2000). However, in this study, we found that 14-3-3 binding to a plant bZIP transcription factor is responsible for its cytoplasmic localization. Thus, the nuclear-cytoplasmic partitioning of regulatory factors by 14-3-3 proteins appears to be an evolutionarily ancient mechanism.
Several studies have shown that 14-3-3 proteins also associate with plant transcription factors, including the G-box binding complex (Lu et al., 1992), VP1, bZIP protein EmBP1 (Schultz et al., 1998), general transcription factor TBP, and TFIIB (Pan et al., 1999). Because 14-3-3 proteins apparently are assembled into protein–DNA complexes with these proteins, a role of 14-3-3 could be to provide the link of activator–activator and activator–general transcription factors but not the sequestration of the ligands in the cytoplasm, as in the cases of RSG and Cdc25. Another example of the involvement of plant 14-3-3 proteins in the subcellular localization of their binding partners is seen in protein import into chloroplasts (May and Soll, 2000). 14-3-3 proteins form part of a guidance complex targeting nucleus-encoded proteins to the chloroplast. Plant 14-3-3 proteins also participate in modulating activities of key metabolic enzymes, including nitrate reductase (Bachmann et al., 1996; Moorhead et al., 1996), sucrose-phosphate synthase (Toroser et al., 1998), plasma membrane H+-ATPase (Jahn et al., 1997; Oecking et al., 1997), and ATP synthases (Bunney et al., 2001), in response to environmental signals such as light and metabolite levels.
From analyses of the crystal structure of a 14-3-3 dimer and from peptide binding studies, it is predicted that the target proteins bind into a large groove formed by amphiphilic helices (Liu et al., 1995; Xiao et al., 1995). The phosphorylated peptides bind with much higher affinity to 14-3-3 proteins than do nonphosphorylated peptides, which explains the involvement of protein phosphorylation in the 14-3-3 binding to ligands (Aitken, 1996). Like the interaction of phosphotyrosine motifs with SH2 domain–containing proteins, such a phosphoserine motif may provide a general mechanism for 14-3-3 protein–target interactions. We identified a sequence in RSG closely related to the conventional 14-3-3 binding motif and demonstrated that Ser-114 in the motif is essential for 14-3-3 binding. Furthermore, lysine residue in the amphipathic groove of 14-3-3 involved in phosphoserine recognition was necessary for RSG binding (Figure 4). These results suggest that the RSG binding to 14-3-3 might be regulated by the phosphorylation of Ser-114 in RSG. When RSG dissociates from 14-3-3 proteins in response to developmental programs and environmental stimuli, probably through the dephosphorylation of Ser-114, it accumulates in the nucleus. There it may activate its target genes, including a GA biosynthetic gene.
RSG apparently is distributed statically in the cytoplasm of plant cells. However, our results using LMB demonstrated that the localization of RSG is very dynamic (Figure 7). RSG shuttles continuously between the nucleus and the cytoplasm. The cytoplasmic localization of RSG is the result of a steady state situation in which RSG enters the nucleus and is exported more rapidly back to the cytoplasm. Exclusion of RSG from the nucleus via active nuclear export suggests that the nuclear localization of RSG may be allowed only in a limited number of cells and in a restricted time frame during plant development. This intracellular compartmentalization of RSG could play a role in the strictly controlled GA biosynthesis of plants. One consequence of RSG shuttling would be to allow rapid binding of RSG with its target DNA in response to internal and external stimuli. A transcription factor usually regulates the expression of several target genes. For proper function, some target genes of RSG might require such rapid and transient expression after specific stimulation. An attractive possibility is that RSG could activate the transcription of negative regulators of RSG, including 14-3-3 proteins, to quench RSG activity, thereby establishing an autoregulatory loop like p53 (Haupt et al., 1997; Kubbutat et al., 1997) and NF-κB (Sun et al., 1993). The search for RSG's target genes is under way.
14-3-3 family proteins can dimerize via their N-terminal domains, as we demonstrated in a two-hybrid assay. Each 14-3-3 dimer can bind up to two distinct ligand molecules (Muslin et al., 1996; Yaffe et al., 1997). Thus, 14-3-3 proteins are thought to sometimes work as molecular scaffolds that allow interaction between signaling proteins that do not associate directly with each other (Braselmann and McCormick, 1995). Because 14-3-3 binding proteins include various signaling factors, such as kinases and phosphatases (Aitken, 1996), there could be other mechanisms in which RSG functions are regulated by another 14-3-3 binding protein via a 14-3-3 dimer as an adaptor. Investigation of how the interaction between RSG and 14-3-3 is controlled by both internal and external signals will help reveal the molecular mechanisms for the fine regulation of the endogenous amounts of GAs that control many aspects of plant development.
METHODS
Two-Hybrid Screen
The coding region of RSG (amino acids 164 to 351) was introduced into pGBT9 (Clontech, Palo Alto, CA) to produce bait in the two-hybrid screen. Tobacco (Nicotiana tabacum) cDNA was generated from poly(A)+ RNA isolated from 2-day-old tobacco BY-2 cells (Fukazawa et al., 2000) and cloned into yeast expression vector pGAD10. A purified plasmid library DNA was used to transform the HF7c strain. Of 2 3 106 yeast transformants, 113 colonies grew on plates without histidine but containing 3-aminotriazole (2 mM), and these were screened for LacZ activity. The library plasmids were isolated from LacZ-positive clones and transformed back into yeast. Clones that restored the activities of reporters were sequenced, and 12 clones encoding 14-3-3 proteins were isolated. The full-length cDNA clones for 14-3-3 proteins were obtained from a lgt10 cDNA library of BY-2 cells using a partial cDNA for 14-3-3 as a probe.
Site-Directed Mutagenesis
Oligonucleotide-directed mutagenesis was performed using polymerase chain reaction (PCR) with plasmid harboring full-length RSG or tobacco 14-3-3 (D31) as a template. For the substitution of Leu-12, Ala-13, and Glu-14 of 14-3-3 to Gln-12, Gln-13, and Arg-14, 59-CTCGGCTTGCcgttgttGCTTAGCCATGTAC-39 and its complementary primer were used. For the substitution of Lys-52 of 14-3-3 to Glu, 59-CCTCTCCGTGGCaTAtgAGAACGTTATCG-39 and its complementary primer were used. For the substitution of Ser-114 of RSG to Ala, 59-TTCGTAGTTTAgCAGTcGACGCCGACTT-39 and its complementary primer were used. Mutations generated with the corresponding oligonucleotides are shown in lowercase letters. After PCR, the resultant products were cloned into plasmids and sequenced.
In Vitro Binding Assay
Amylose resin (New England Biolabs, Beverly, MA) was incubated with Escherichia coli crude extracts containing maltose binding protein (MBP)–14-3-3 or MBP–14-3-3(K52E) fusion proteins to generate resin coated with 14-3-3 fusion proteins for the subsequent binding assays. Full-length RSG labeled with 35S-methionine was prepared by in vitro transcription (Epicentre Technologies, Madison, WI) and a rabbit reticulocyte lysate translation system (Amersham Pharmacia Biotech, Buckinghamshire, UK). The phosphorylation of the 14-3-3 binding motif by endogenous Ser/Thr kinase(s) within the reticulocyte lysate has been reported (Kanai et al., 2000). Portions of 35S-RSG protein were incubated with the MBP–14-3-3– or MBP–14-3-3 (K52E)–bound amylose resin for 1 hr at 4°C in a binding buffer consisting of 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, and 10 mM β-mercaptoethanol. Each resin was washed extensively with the binding buffer followed by elution with a binding buffer containing 20 mM maltose. Eluted proteins were resolved by SDS-PAGE. After electrophoresis, proteins were visualized by fluorography and Coomassie Brilliant Blue R250 staining.
Transient Assay
The 3 × 17mer oligonucleotides (containing three tandem copies of the GAL4 binding site) were cloned into the parB minimal promoter–β-glucuronidase (GUS) and named G3GUS. The 5′ end of the parB minimal promoter was 4 bp upstream of the TATA box of parB (Takahashi et al., 1995). The sequences of 3 × 17mer were 5′-AGC-TTCGGAAGACTCTCCTCCGAGCGGATTAGAAGCCGCCGACGGAAG-ACTCTCCTCCGCTGCA-3′ and 5′-AGCTTCGGAAGACTCTCCTCC-GAGCGGATTAGAAGCCGCCGACGGAAGACTCTCCTCCGCTGCA-3′. The effector plasmid 35S–GAL4BD was constructed by the insertion of a GAL4 DNA binding domain (amino acid positions 1 to 147) from the pM vector (Clontech) downstream of the Cauliflower mosaic virus (CaMV) 35S promoter with a viral translation enhancer (Ω). The cDNA fragment encoding the full-length wild-type RSG or the S114A mutant was fused downstream of the GAL4 DNA binding domain of 35S–GAL4BD. The control effector was the empty vector.
Protoplasts were prepared from BY-2 cells harvested 3 days after subculture. After collection by centrifugation, cells were treated with 1% cellulase Y-C, 0.1% pectolyase Y-23 (both enzymes from Seishin Co., Tokyo, Japan), and 0.4 M mannitol for the removal of cell walls. After incubation for 1.5 hr at 30°C, the resultant protoplasts were washed with 0.4 M mannitol. Ten micrograms of reporter plasmid, 20 μg of effector plasmid (35S–GAL4BD–RSG, 35S–GAL4BD–S114A, or 35S–GAL4BD), and 10 μg of internal control 35S–LUC (for luciferase) plasmid were used for the transient assay. Plasmid DNAs and polyethylene glycol 6000 at a final concentration of 1% (w/v) were added sequentially to a suspension of 2 × 106 protoplasts in 700 μL of an electroporation buffer consisting of 5 mM Mes-KOH, pH 5.8, 70 mM KCl, and 0.3 M mannitol to yield a final volume of 800 μL. After pulses had been delivered (voltage, 300 V; capacitance, 125 μF; Gene Pulser Transfection Apparatus; Bio-Rad, Hercules, CA) in a cuvette whose electrodes had a clearance of 0.4 cm, the suspension was incubated on ice for 10 min. Incubation was started in 12 mL of a modified Linsmaier and Skoog (1965) medium containing 0.4 M mannitol and 1% sucrose with 0.9 μM 2,4-D. After culturing at 27°C for 36 hr, cells were collected, and GUS and luciferase activities were determined.
RNA Gel Blot Hybridization
The DNA region encoding the full-length tobacco 14-3-3 (D31) was used as a probe. Hybridization conditions were 6 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS for 18 hr at 65°C. After hybridization, the membrane was washed at 65°C in 2 × SSC containing 0.1% SDS and then autoradiographed for 1 day.
Generation of Anti-RSG and Anti-14-3-3 Antibodies
The entire coding regions of RSG and tobacco 14-3-3 (D31) were cloned into a pET-30 vector (Novagen, Madison, WI) and expressed in E. coli BL21 (DE3) cells. The hexahistidine-tagged proteins were purified using nickel–nitrilotriacetic acid agarose resin (Qiagen, Hilden, Germany) as specified by the manufacturer. The eluted proteins were further resolved by SDS-PAGE. After electrophoresis, the bands corresponding to RSG and 14-3-3 were cut out of the gels. The electroeluted proteins were used to immunize rats.
Cell Fractionation
Protoplasts prepared from BY-2 cells were suspended in a nucleus isolation buffer consisting of 10 mM Hepes-KOH, pH 7.9, 10 mM MgCl2, 10% glycerol, 0.44 M sucrose, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 0.5% Triton X-100 and incubated for 30 min on ice. After centrifugation, the supernatant was used as a cytosolic extract, and the crude nuclear pellet was resuspended with nucleus isolation buffer without Triton X-100. Nuclear extracts were prepared as described by Sakai et al. (1998).
Immunoprecipitation and Protein Gel Blot Analysis
Cell lysates from leaves of transgenic tobacco plants expressing RSG–green fluorescent protein (GFP) or S114A–GFP were extracted in 50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1 mM DTT, 20% glycerol, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/mL leupeptin, antipain, and pepstatin A. Aliquots of extracts were immunoprecipitated with anti-GFP rabbit polyclonal antibody (Clontech) or rabbit antiserum raised against tobacco arcA protein followed by incubation with protein G beads. Immunoprecipitates were washed, and bound proteins were subjected to protein gel blot analysis with anti-14-3-3 (D31) serum or anti-RSG serum and visualized by enhanced chemiluminescence.
Confocal Microscopic Observation
The plant GFP expression vector pJ4–GFP was constructed by the insertion of the full-length S65T GFP (Chiu et al., 1996) cDNA into the SalI and NotI sites of the vector carrying the CaMV 35S promoter with a viral translation enhancer, the Ω sequence (Fukazawa et al., 2000). The PCR-amplified cDNA fragment encoding the full-length RSG or S114A mutant was cloned into BamHI and XbaI of pJ4–GFP.
Transfected tobacco mesophyll protoplasts were mounted in a culture medium on glass slides. The epidermal strips were peeled from the stems and leaves of transgenic tobacco plants and mounted in PBS (11.9 mM sodium phosphate, pH 7.5, 2.7 mM KCl, and 136.9 mM NaCl) on glass slides. Specimens were examined with a confocal laser scanning microscope (model TCS ST; Leica Microsystems, Heidelberg, Germany) using the 488-nm excitation line of an argon ion laser. The emission light was dispersed and recorded at 500 to 570 nm for GFP fluorescence and 600 to 670 nm for chlorophyll autofluorescence. After successive scanning for each 0.3-μm interval, the three-dimensional images of fluorescences were constructed from optical sections taken at different channels and overlaid. Images were exported as TIFF format files and further processed.
Accession Numbers
The GenBank accession numbers of the tobacco 14-3-3 cDNAs are AB071967 (D31), AB071968 (D5), and AB071969 (D75).
Acknowledgments
We are grateful to Dr. Minoru Yoshida for providing LMB and to Dr. Yasuo Niwa for providing S65T GFP. We also thank Kyoko Ohashi for technical assistance and helpful discussions. This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture (Japan) and a grant from the Ministry of Agriculture, Forestry, and Fisheries (Japan) to Y.T.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010188.
References
- Aitken, A. (1996). 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 6 341–347. [DOI] [PubMed] [Google Scholar]
- Aitken, A., Howell, S., Jones, D., Madrazo, J., and Patel, Y. (1995). 14-3-3 α and δ are the phosphorylated forms of Raf-activating 14-3-3 β and ζ. J. Biol. Chem. 270 5706–5709. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bachmann, M., Huber, J.L., Liao, P.C., Gage, D.A., and Huber, S.C. (1996). The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett. 387 127–131. [DOI] [PubMed] [Google Scholar]
- Bihn, E.A., Paul, A.L., Wang, S.W., Erdos, G.W., and Ferl, R.J. (1997). Localization of 14-3-3 proteins in the nuclei of Arabidopsis and maize. Plant J. 12 1439–1445. [DOI] [PubMed] [Google Scholar]
- Braselmann, S., and McCormick, F. (1995). BCR and RAF form a complex in vivo via 14-3-3 proteins. EMBO J. 14 4839–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet, A., Bonni, A., Zigmond, M.J., Lin, M.Z., Juo, P., Hu, L.S., Anderson, M.J., Arden, K.C., Blenis, J., and Greenberg, M.E. (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96 857–868. [DOI] [PubMed] [Google Scholar]
- Bunney, T.D., van Walraven, H.S., and de Boer, A.H. (2001). 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 98 4249–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, H.-H., Hwang, I., and Goodman, H.M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W.-L., Niwa, Y., Zheng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Chong, S.S., Tanigami, A., Roschke, A.V., and Ledbetter, D.H. (1996). 14-3-3 epsilon has no homology to LIS1 and lies telomeric to it on chromosome 17p13.3 outside the Miller-Dieker syndrome chromosome region. Genome Res. 6 735–741. [DOI] [PubMed] [Google Scholar]
- DeLille, J.M., Sehnke, P.C., and Ferl, R.J. (2001). The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126 35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I.W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukazawa, J., Sakai, T., Ishida, S., Yamaguchi, I., Kamiya, Y., and Takahashi, Y. (2000). REPRESSION OF SHOOT GROWTH, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390 308–311. [DOI] [PubMed] [Google Scholar]
- Grozinger, C.M., and Schreiber, S.L. (2000). Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, Y., Maya, R., Kazaz, A., and Oren, M. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387 296–299. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Sheldon, C.C., Olive, M.R., Walker, A.R., Zeevaart, J.A.D., Peacock, W.J., and Dennis, E.S. (1998). Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 95 9019–9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Chandler, P.M., Poole, A., Dennis, E.S., and Peacock, W.J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, S., Takahashi, Y., and Nagata, T. (1993). Isolation of cDNA of an auxin-regulated gene encoding a G protein β subunit-like protein from tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 90 11152–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, T., Fuglsang, A.T., Olsson, A., Bruntrup, I.M., Colling, D.B., Volkmann, D., Sommarin, M., Palmgren, M.G., and Larsson, C. (1997). The 14-3-3 protein interacts directly with the C-terminal region of the plasma membrane H+-ATPase. Plant Cell 9 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, F., Marignani, P.A., Sarbassova, D., Yagi, R., Hall, R.A., Donowitz, M., Hisaminato, A., Fujiwara, T., Ito, Y., Cantley, L.C., and Yaffe M.B. (2000). TAZ: A novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19 6778–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthout, H.A.A.J., and de Boer, A.H. (1994). A fusicoccin binding protein belongs to the family of 14-3-3 brain protein homologs. Plant Cell 6 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat, M.H., Jones, S.N., and Vousden, K.H. (1997). Regulation of p53 stability by Mdm2. Nature 387 299–303. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E.P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242 540–547. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E.P., Wolff, B., Yoshida, M., and Horinouchi, S. (1999). Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96 9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, A., and Dunphy, W.G. (1999). Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 13 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, J., and Omary, M.B. (1996). 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol. 133 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmaier, E.M., and Skoog, F. (1965). Organic growth factor requirements of tobacco tissue culture. Physiol. Plant. 18 100–127. [Google Scholar]
- Liu, D., Bienkowska, J., Petosa, C., Collier, R.J., Fu, H., and Liddington, R. (1995). Crystal structure of the ζ isoform of the 14-3-3 protein. Nature 376 191–194. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., Furnari, B., Mondesert, O., and Russell, P. (1999). Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397 172–175. [DOI] [PubMed] [Google Scholar]
- Lu, G., DeLisle, A.J., de Vetten, N.C., and Ferl, R.J. (1992). Brain proteins in plants: An Arabidopsis homolog to neurotransmitter pathway activators in part of a DNA binding complex. Proc. Natl. Acad. Sci. USA 89 11490–11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z., Zhang, X., Rapp, U., and Avruch, J. (1995). Identification of the 14-3-3 z domains important for self-association and Raf binding. J. Biol. Chem. 270, 23681–23687 [DOI] [PubMed] [Google Scholar]
- Marra, M., Fullone, M.R., Fogliano, V., Pen, J., Mattei, M., Masi, S., and Aducci, P. (1994). The 30-kilodalton protein present in purified fusicoccin receptor preparations is a 14-3-3-like protein. Plant Physiol. 106 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, T., and Soll, J. (2000). 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T.A., Zhang, C.L., and Olson, E.N. (2000). Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead, G., Douglas, P., Morrice, N., Scarabel, M., Aitken, A., and MacKintosh, C. (1996). Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol. 6 1104–1113. [DOI] [PubMed] [Google Scholar]
- Muslin, A.J., Tanner, J.W., Allen, P.M., and Shaw, A.S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84 889–897. [DOI] [PubMed] [Google Scholar]
- Nishi, K., Yoshida, M., Fujiwara, D., Nishikawa, M., Horinouchi, S., and Beppu, T. (1994). Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269 6320–6324. [PubMed] [Google Scholar]
- Oecking, C., Eckerskorn, C., and Weiler, E.W. (1994). The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett. 352 163–166. [DOI] [PubMed] [Google Scholar]
- Oecking, C., Piotrowski, M., Hagemeier, J., and Hageman, K. (1997). Topology and target interaction of the fusicoccin-binding 14-3-3 homologs of Commelina communis. Plant J. 12 441–453. [Google Scholar]
- Ossareh-Nazari, B., Bachelerie, F., and Dargemont, C. (1997). Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278 141–144. [DOI] [PubMed] [Google Scholar]
- Pan, S., Sehnke, P.C., Ferl, R.J., and Gurley, W.B. (1999). Specific interactions with TBP and TFIIB in vitro suggest 14-3-3 proteins may participate in transcriptional regulation when part of a DNA binding complex. Plant Cell 11 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C.-Y., Graves, P.R., Thoma, R.S., Wu, Z., Shaw, A.S., and Piwnica-Worms, H. (1997). Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277 1501–1505. [DOI] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). “Green revolution” genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., Peng, J., and Harberd, N.P. (2000). Plant GRAS and metazoan STATs: One family? BioEssays 22 573–577. [DOI] [PubMed] [Google Scholar]
- Sakai, T., Takahashi, Y., and Nagata, T. (1998). The identification of DNA binding factors specific for as-1-like sequences in auxin-responsive regions of parA, parB and parC. Plant Cell Physiol. 39 731–739. [DOI] [PubMed] [Google Scholar]
- Schultz, T.F., Medina, J., Hill, A., and Quatrano, R.S. (1998). 14-3-3 proteins are part of an abscisic acid–VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell 10 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya, H., Sawada, H., Muramatsu, Y., Shimizu, M., Ohko, K., Yamane, K., and Tsuruo, T. (2000). Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 19 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, V.A. (1992). Gibberellin A1 biosynthesis in Pisum sativum L. II. Biological and biochemical consequences of the le mutation. Plant Physiol. 99 372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S.C., Ganchi, P.A., Ballard, D.W., and Greene, W.C. (1993). NF-κB controls expression of inhibitor IκBα: Evidence for an inducible autoregulatory pathway. Science 259 1912–1915. [DOI] [PubMed] [Google Scholar]
- Sun, T.-p., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., and Nagata, T. (1992). An auxin-regulated gene encoding glutathione S-transferase. Proc. Natl. Acad. Sci. USA 89 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Sakai, T., Ishida, S., and Nagata, T. (1995). Identification of auxin-responsive elements of parB and their expression in apices of shoot and root. Proc. Natl. Acad. Sci. USA 92 6359–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroser, D., Athwal, G.S., and Huber, S.C. (1998). Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 435 110–114. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Luo, Z., and Avruch, J. (1998). A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394 88–92. [DOI] [PubMed] [Google Scholar]
- van Heusden, G.P.H., Wenzel, T.J., Lagendijk, E.L., de Steesma, H.Y., and van den Berg, J.A. (1992). Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett. 302 145–150. [DOI] [PubMed] [Google Scholar]
- Wu, K., Lu, G., Sehnke, P., and Ferl, R.J. (1997). The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch. Biochem. Biophys. 339 2–8. [DOI] [PubMed] [Google Scholar]
- Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., and Gamblin, S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature 376 188–191. [DOI] [PubMed] [Google Scholar]
- Xu, Y.-L., Li, L., Wu, K., Peters, A.J.M., Gage, D.A., and Zeevaart, J.A.D. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.B., Rittinger, K., Volinia, S., Caron, P.R., Aitken, A., Leffers, H., Gamblin, S.J., Smerdon, S.J., and Cantley, L.C. (1997). The structural basis for 14-3-3: Phosphopeptide binding specificity. Cell 91 961–971. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Saito, T., Abe, H., Yamane, H., Murofushi, N., and Kamiya, Y. (1996). Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene synthase B from pumpkin (Cucurbita maxima L.). Plant J. 10 203–213. [DOI] [PubMed] [Google Scholar]
- Yang, J., Winkler, K., Yoshida, M., and Kornbluth, S. (1999). Maintenance of G2 arrest in the Xenopus oocyte: A role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 18 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha, J., Harada, H., Yang, E., Jockel, J., and Korsmeyer, S.J. (1996). Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell 87 619–628. [DOI] [PubMed] [Google Scholar]
- Zhang, C.L., McKinsey, T.A., and Olson, E.N. (2001). The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc. Natl. Acad. Sci. USA 98 7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Wang, H., Liu, D., Liddington, R., and Fu, H. (1997). Raf-1 kinase and exoenzyme S interact with 14-3-3ζ through a common site involving lysine 49. J. Biol. Chem. 272 13717–13724. [DOI] [PubMed] [Google Scholar]