Abstract
To understand the biogenesis of the plastid cytochrome b6f complex and to identify the underlying auxiliary factors, we have characterized the nuclear mutant hcf164 of Arabidopsis and isolated the affected gene. The mutant shows a high chlorophyll fluorescence phenotype and is severely deficient in the accumulation of the cytochrome b6f complex subunits. In vivo protein labeling experiments indicated that the mutation acts post-translationally by interfering with the assembly of the complex. Because of its T-DNA tag, the corresponding gene was cloned and its identity confirmed by complementation of homozygous mutant plants. HCF164 encodes a thioredoxin-like protein that possesses disulfide reductase activity. The protein was found in the chloroplast, where it is anchored to the thylakoid membrane at its lumenal side. HCF164 is closely related to the thioredoxin-like protein TxlA of Synechocystis sp PCC6803, most probably reflecting its evolutionary origin. The protein also shows a limited similarity to the eubacterial CcsX and CcmG proteins, which are required for the maturation of periplasmic c-type cytochromes. The putative roles of HCF164 for the assembly of the cytochrome b6f complex are discussed.
INTRODUCTION
The cytochrome b6f complex of the thylakoid membranes of higher plants takes a central position in photosynthetic electron transport. It functions as a plastoquinol-plastocyanin-oxidoreductase linking photosystems I and II (PSI and PSII). It also is involved in cyclic electron transport around PSI and acts as a proton translocase. The complex is composed of the four major subunits cytochrome f (petA), cytochrome b6 (petB), the Rieske-FeS protein (petC), and PetD (petD) and the four small subunits PetG, PetL, PetM, and PetN. A projection map at 8 Å resolution showed the presence of cytochrome b6f complex dimers that are believed to be the functional form (Mosser et al., 1997).
The cytochrome b6f complex consists of nucleus- and plastid-encoded subunits. Although the Rieske-FeS protein and PetM are encoded by the nucleus, all other subunits are encoded by the plastome. The biogenesis of the complex, therefore, requires coordinated gene expression in the nucleus and chloroplast. Several auxiliary and regulatory factors are necessary to ensure that the biogenesis of the cytochrome b6f complex proceeds in an ordered manner (reviewed by Rochaix, 1996; Wollman et al., 1999): factors that regulate processing and splicing of the plastome-encoded RNAs that are mostly transcribed in polycistronic transcription units (Barkan et al., 1994; Jenkins et al., 1997; Fisk et al., 1999); factors that control translation and the stoichiometric accumulation of the various subunits (Barkan, 1993; Kuras and Wollman, 1994; Choquet et al., 1998); and factors that target the proteins to their destinations (Voelker and Barkan, 1995a; Robinson and Mant, 1997). Finally, cofactors have to be attached to the cytochromes and the Rieske-FeS protein (Kuras et al., 1997; Xie et al., 1998), and the holosubunits have to be assembled into a functional complex.
There is convincing evidence that most of these factors are encoded in the nucleus. Nuclear mutants with defects in the cytochrome b6f complex, therefore, can serve to identify such factors. These mutants have reduced or completely blocked electron transport and can be identified on the basis of their high chlorophyll fluorescence phenotype (hcf). This phenotype arises when the absorbed light energy cannot be used for photosynthesis and is emitted as red fluorescent light (Miles, 1994).
Here we report on the mutant hcf164 of Arabidopsis, which was generated by T-DNA insertional mutagenesis (Bechtold et al., 1993) and which is deficient specifically in the biogenesis of the cytochrome b6f complex. None of the major subunits of the complex accumulate, but they are synthesized. Therefore, the gene disrupted in this mutant encodes a factor that is essential for the assembly of the complex or its stability. The corresponding gene, HCF164, could be isolated because of its T-DNA tag. It encodes a thioredoxin-like protein that functions biochemically as a disulfide oxidoreductase. HCF164 is located inside the chloroplast and is anchored to the thylakoid membrane at its lumenal side. The protein is closely related to the thioredoxin-like protein TxlA of cyanobacteria. Additionally, there is limited homology with the eubacterial proteins CcsX of Bordetella pertussis and CcmG of Escherichia coli and other Gram-negative bacteria, both of which are involved in the maturation of periplasmic c-type cytochromes.
RESULTS
hcf164 Is Affected in the Intersystem Electron Transport Chain
The hcf164 mutant of Arabidopsis ecotype Wassilewskija was generated by T-DNA insertional mutagenesis (Bechtold et al., 1993) and selected from a collection of the Institute National de la Recherche Agronomique (Versailles, France). hcf164 segregated as a single recessive mutation. Cosegregation with the phosphinotricin resistance marker of the T-DNA indicated that the mutation was tagged by the T-DNA insertion. The mutant plants showed an hcf phenotype and were not able to grow photoautotrophically. They could be maintained, however, on sucrose-supplemented medium, but they did not develop any flowers.
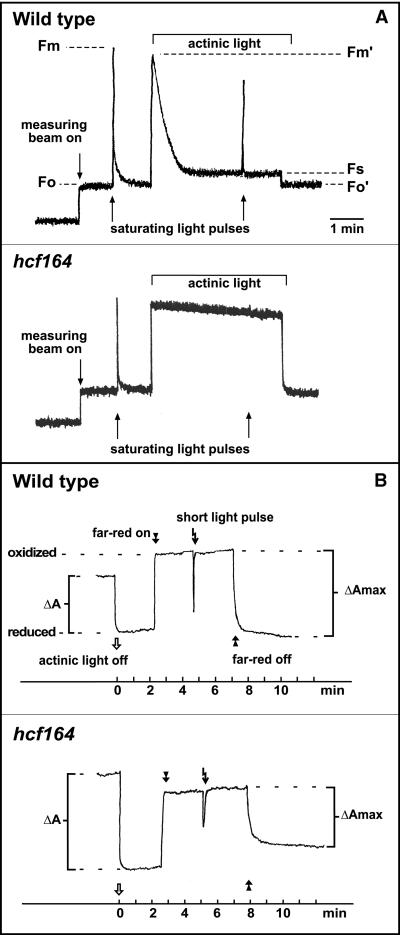
Because the hcf phenotype indicated decreased photosynthetic electron transport rates, we measured chlorophyll fluorescence induction (Figure 1A). The ratio (Fv/Fm) of variable fluorescence (Fv = Fm − Fo; where Fv is variable fluorescence, Fm is maximum fluorescence, and Fo is minimal fluorescence) to maximum fluorescence in hcf164 (Fv/Fm = 0.76 ± 0.02) was comparable to that in wild type (Fv/Fm = 0.79 ± 0.01). This revealed the presence of an active PSII in hcf164. In contrast, photochemical quenching (qP = [Fm′ − Fs]/[Fm′ − Fo′]; where Fs is steady-state fluorescence) was drastically reduced in hcf164 (qP = 0.16 ± 0.02) compared with wild-type plants (qP = 0.88 ± 0.03), suggesting that the electrons released by PSII accumulated in the plastoquinone pool and were not transported downstream of the photosynthetic electron transport chain. This could be attributable to a defect in intersystem electron transport or in PSI. To distinguish between these two possibilities, we studied PSI function by measuring the absorbance kinetics of P700 at 830 nm (Figure 1B). A clear absorbance change induced by far-red light similar to that in wild-type indicated that PSI was functional in hcf164. Application of a white light pulse in far-red background led to a short re-reduction of PSI by electrons released from PSII. This re-reduction was diminished in hcf164. Together, the spectroscopic data indicate that the defect is located in the intersystem electron transport chain and not in one of the two photosystems.
Figure 1.
Spectroscopic Analyses of hcf164 Mutants and Wild-Type Plants.
Measurements of chlorophyll fluorescence induction (A) and P700 absorbance kinetics (B) were performed with 2-week-old plants as described in Methods.
Subunits of the Cytochrome b6f Complex Do Not Accumulate in hcf164
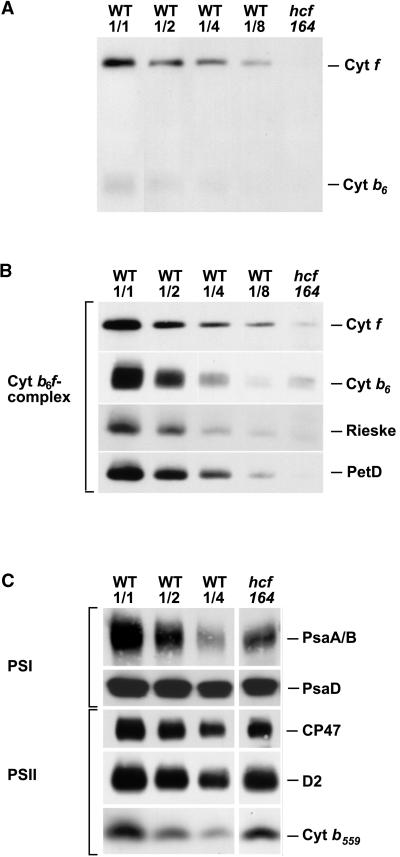
A block in the intersystem electron flow could be the result of a defect in the cytochrome b6f complex. To address this possibility, we assayed thylakoid membranes of wild-type and mutants for the presence of cytochrome f and cytochrome b6 holoproteins by virtue of their heme-associated peroxidase activities. Figure 2A shows that cytochrome f, with its covalently bound heme, can be detected clearly even when only 2.5 μg of total chloroplast proteins was analyzed. The peroxidase activity of cytochrome b6 is drastically lower; therefore, at least 5 μg of chloroplast proteins have to be assayed. Figure 2A reveals that in mutant thylakoids, both proteins are below the level of detection—that is, their amounts are significantly less than 12.5% (cytochrome f) and 25% (cytochrome b6) of the wild-type levels analyzed.
Figure 2.
Analysis of Photosynthetic Membrane Proteins from the Mutant hcf164 and the Wild-Type (WT).
Twenty micrograms (hcf164, WT1/1), 10 μg (WT1/2), 5 μg (WT1/4), and 2.5 μg (WT1/8) of chloroplast proteins were size separated by SDS-PAGE and electroblotted onto nitrocellulose membranes.
(A) Detection of the heme binding proteins cytochrome f and cytochrome b6 by enhanced chemiluminescence.
(B) Immunoblot analysis of the cytochrome b6f subunits cytochrome f, cytochrome b6, the Rieske-FeS protein (Rieske), and PetD.
(C) Immunodetection of the subunits PsaA/B and PsaD of PSI, the inner antenna protein CP47, and the reaction center proteins D2 and cytochrome b559 of PSII.
Cyt, cytochrome.
To measure the steady state levels of the subunits of the cytochrome b6f complex directly, immunoblot analyses were performed. Under the experimental conditions used, the antisera to the subunits cytochrome f, the Rieske-FeS protein, and PetD detected less than 12.5% of the wild-type level in hcf164 (Figure 2B). Cytochrome b6 accumulated to amounts between 12.5 and 25%. These data indicate that the accumulation of the cytochrome b6f complex is impaired considerably in the mutant.
In parallel, we analyzed the protein levels of the two photosystems. Representative PSI polypeptides (PsaA/B and PsaD) were reduced to 30 to 50% of wild-type amounts. For PSII representatives, we tested CP47, D2, and cytochrome b559. These polypeptides were reduced to 50 to 70% in hcf164 (Figure 2C). In contrast, when plants were grown under very low light intensities (PPFD = 5 μmol·m−2·sec−1), neither PSI nor PSII subunits were reduced in hcf164 mutants, whereas the levels of the subunits of the cytochrome b6f complex still were diminished (data not shown). This suggests that the reduction in PSI and PSII subunits most likely are a secondary effect of the mutation.
We conclude from the spectroscopic analyses, the heme staining, and the immunoblot experiments that the hcf164 mutation affects primarily the cytochrome b6f complex and that this complex as a whole is reduced significantly in mutant thylakoids.
Plastome-Encoded Subunits of the Cytochrome b6f Complex Are Synthesized in hcf164
To determine whether the reduced amounts of polypeptides of the cytochrome b6f complex are caused by impaired RNA accumulation, RNA gel blot hybridizations with probes for the four major subunits of the cytochrome b6f complex were performed. The plastid-encoded genes petA (cytochrome f), petB (cytochrome b6), and petD (PetD) are transcribed in polycistronic transcription units resulting in complex RNA patterns (Barkan, 1988; Westhoff and Herrmann, 1988; Gray, 1992). In contrast, the nucleus-encoded petC (Rieske-FeS protein) is transcribed into a monocistronic transcript. No differences in transcript patterns and accumulation were observed between mutant and wild-type, indicating that the processes of transcription, RNA processing, and RNA stability are not affected by the mutational lesion (data not shown).
To determine whether the impaired accumulation of the subunits of the cytochrome b6f complex is caused by decreased translation or protein stability, the rates of synthesis of chloroplast-encoded thylakoid membrane proteins were studied by pulse labeling of mutant leaves with 35S-methionine in the presence of cycloheximide as an inhibitor of cytoplasmic translation.
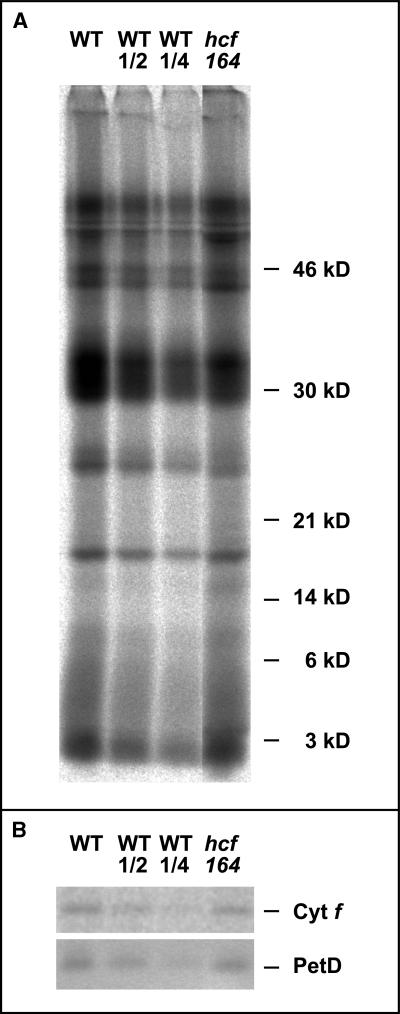
Figure 3A shows that the protein labeling patterns in mutant and wild-type plants were similar. Because the subunits of the cytochrome b6f complex could not be identified easily in the protein patterns, immunoprecipitation experiments were performed. After 30 min of labeling, membrane proteins were extracted from the leaves and individual subunits of the cytochrome b6f complex were immunoprecipitated using specific antisera raised against cytochrome f, cytochrome b6, and PetD. To ensure a quantitative immunoprecipitation, a surplus of the corresponding antibodies was used in the assay and, as a control, the supernatants from the first immunoprecipitation were subjected to a second round of immunoprecipitation. These experiments revealed that all proteins could be precipitated quantitatively from wild-type and mutant extracts in the first round of immunoprecipitation (data not shown).
Figure 3.
In Vivo Protein Synthesis in hcf164 Mutants and Wild-Type (WT) Plants.
Primary leaves of 15-day-old plants were radiolabeled with 35S-methionine for 30 min.
(A) Membrane proteins were isolated, separated by SDS-PAGE, and analyzed by autoradiography as described in Methods. Proteins equivalent to 100,000 incorporated cpm or the indicated dilutions of wild-type were loaded in each lane.
(B) Membrane proteins were solubilized, and the protein equivalent of 200,000 incorporated cpm was used for immunoprecipitation with antisera against cytochrome f (Cyt f) and the PetD subunit. Lanes WT and hcf164 were loaded with 50% of the appropriate immunoprecipitation reaction, and lanes WT1/2 and WT1/4 were loaded with 25 and 12.5% of the WT precipitation, respectively. For experimental details, see Methods.
Gel electrophoretic analysis of the immunoprecipitates showed that both cytochrome f and PetD are synthesized in the mutant plants (Figure 3B). Only one band of cytochrome f could be precipitated from the mutant protein extracts. Its apparent molecular weight was indistinguishable from that obtained with wild-type extracts, suggesting that in the mutant the precursor of cytochrome f is processed correctly at the N-terminus (Gray, 1992). The antiserum against cytochrome b6 was not able to immunoprecipitate radiolabeled protein; thus, the synthesis of this subunit could not be analyzed. The quantitative analysis of the immunoprecipitates by phosphorimaging revealed that the accumulation of radioactive label in cytochrome f and PetD is comparable in wild-type and hcf164 mutant plants (Figure 3B). We conclude, therefore, that the hcf164 mutation does not affect primarily the synthesis of the subunits of the cytochrome b6f complex but rather leads to an increased rate of their degradation.
Molecular Cloning of HCF164
Cosegregation of the hcf164 mutation with the phosphinotricin resistance marker of the T-DNA indicated the presence of a single T-DNA tagging the mutated gene. This was confirmed by DNA gel blot analysis (data not shown). To isolate the genomic sequences flanking the left border of the T-DNA, inverse polymerase chain reaction (PCR) was used (Ochman et al., 1993). The amplified sequences were cloned, sequenced, and subjected to database searches. One hundred percent identity was found between the isolated PCR fragment and a putative gene (GenBank accession number CAB16778) on chromosome IV of Arabidopsis. This sequence information was used to design primers, and an HCF164 cDNA was amplified by reverse transcriptase–mediated PCR. The cDNA clone contained an open reading frame of 783 nucleotides. The ATG start codon is preceded by a stop codon, indicating that the isolated cDNA harbors the complete HCF164 coding sequence. In the mutant line, the T-DNA insertion maps 34 bp upstream of the ATG start codon.
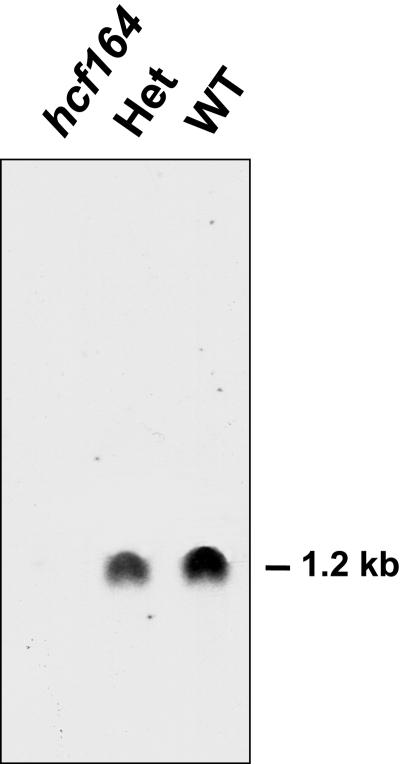
To study the effect of the T-DNA insertion on HCF164 expression, the HCF164 cDNA was used as a probe in RNA gel blot hybridization. Figure 4 shows that in wild-type RNA, a single transcript of 1.2 kb was present. This HCF164 transcript still could be detected in heterozygous plants, although at lower levels, but it was completely absent in RNA from homozygous mutant plants.
Figure 4.
HCF164 Transcript Analysis of Wild-Type, Heterozygous, and Homozygous hcf164 Mutant Plants.
Ten micrograms of poly(A+) RNA of 3-week-old wild-type (WT), heterozygous (Het), and homozygous hcf164 mutant plants was separated on a denaturing agarose gel, transferred to a nylon membrane, and hybridized with a radiolabeled HCF164 cDNA probe.
To prove that the HCF164 gene disruption was responsible for the mutant phenotype, the isolated cDNA was used for complementation analysis. The HCF164 cDNA was fused to the 35S promoter of the Cauliflower mosaic virus in the plant transformation vector pPCV91 (Strizhov et al., 1996), and roots of homozygous hcf164 mutants were transformed by means of Agrobacterium tumefaciens (Koncz et al., 1994). Forty-five independent transgenic plants were regenerated. They were able to grow photoautotrophically, and their chlorophyll fluorescence induction kinetics were indistinguishable from those of wild-type plants (data not shown). We conclude from this complementation experiment that the T-DNA insertion in the HCF164 gene is responsible for the mutant phenotype and that the isolated cDNA encodes a functional HCF164 protein.
HCF164 Encodes a Thioredoxin-Like Protein
The HCF164 cDNA encodes a protein of 261 amino acids with a predicted molecular mass of 29 kD. Hydropathy analysis revealed that the protein is composed of three domains: an N-terminal hydrophilic domain up to amino acid 97; a hydrophobic part from amino acids 98 to 113; and a large C-terminal hydrophilic part from amino acids 114 to 261 (data not shown). The first two domains exhibit typical features of plastid transit peptides of lumenal proteins, suggesting that the HCF164 protein is localized in the chloroplast. The C-terminal hydrophilic part contains a WCxxC motif (boxed in Figure 5), which is a typical signature of the active site of thioredoxins. Database searches and protein sequence alignments revealed that this segment of the HCF164 protein exhibits significant similarities to thioredoxins, thioredoxin-like proteins, and protein disulfide isomerases from various organisms. The strongest similarity was observed with the thioredoxin-like protein TxlA of Synechocystis sp PCC6803 (44% identity, 59% similarity) and its homologs of Synechococcus sp PCC7249 (38% identity, 57% similarity) and Anabaena sp PCC7120 (33% identity, 53% similarity) (Figure 5). In addition, we noted a limited sequence similarity between HCF164 and the thioredoxin-like proteins CcsX of B. pertussis (Beckett et al., 2000) (15% identity, 28% similarity) and CcmG of E. coli (15% identity, 32% similarity) and its homologs in other Gram-negative bacteria (Beckman and Kranz, 1993; Vargas et al., 1994; Fabianek et al., 1997, 1998). CcsX and CcmG are periplasmic proteins that are involved in the maturation of c-type cytochromes (Fabianek et al., 1998; Beckett et al., 2000).
Figure 5.
Amino Acid Sequence Alignment of HCF164 and Thioredoxin-Like Proteins.
Multiple alignment of the amino acid sequences of HCF164 and the TxlA homologs of Synechocystis sp PCC6803 (accession number P73920), Synechococcus sp PCC7942 (accession number P35088), and Anabaena sp (Cyanobase, Kazusa Research Institute, Kisarazu, Japan). Identical amino acids are marked with asterisks, and conserved exchanges are marked with dots. The thioredoxin motif is boxed, and the hydrophobic region is shaded.
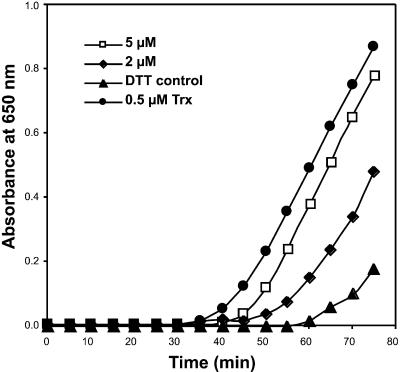
Recombinant HCF164 Protein Exhibits Disulfide Reductase Activity in Vitro
To determine whether the HCF164 protein shows disulfide reductase activity, insulin reduction assays were performed. Such assays are used commonly to test proteins for thioredoxin activities (Holmgren, 1979). In the presence of DTT, thioredoxin reduces the intermolecular disulfide bonds between the insulin A and B chains, and as a consequence, the insoluble B chain precipitates. The assay was performed with recombinant HCF164, which lacked the N-terminal segment up to amino acid 115. Figure 6 shows that recombinant HCF164 was able to accelerate the reduction of insulin in the presence of DTT. Under the same assay conditions, purified Spirulina sp thioredoxin also catalyzed the insulin reduction. This demonstrates that HCF164 possesses disulfide reductase activity in vitro. However, compared with the purified thioredoxin, the disulfide reductase activity of HCF164 is 1 to 2 orders of magnitude lower.
Figure 6.
Measurement of Disulfide Reductase Activity of Recombinant HCF164 Using the Turbidimetric Assay of Insulin Reduction.
The incubation mixture contained 100 mM sodium phosphate, pH 7.0, 1 mM EDTA, 1 mg/mL insulin, 0.35 mM DTT, and 2 or 5 μM HCF164 protein. As a positive control, insulin reduction by 0.5 μM thioredoxin from Spirulina sp was assayed. The nonenzymatic insulin reduction by DTT served as a negative control.
HCF164 Is a Thylakoid Membrane Protein with Its Hydrophilic C-Terminal Part Exposed to the Lumen
The sequence analysis suggested that the HCF164 protein is a plastid protein to be targeted into the thylakoid lumen (Robinson and Mant, 1997): the protein starts with the motif MA and possesses a serine-rich region from amino acids 62 to 84, which is typical for plastid transit peptides. The lu-menal targeting is suggested by the presence of a twin arginine motif at amino acids 96/97, which is diagnostic for the ΔpH-dependent translocation pathway (Hynds et al., 2000), and furthermore by the hydrophobic domain of 16 amino acids directly after the twin arginines.
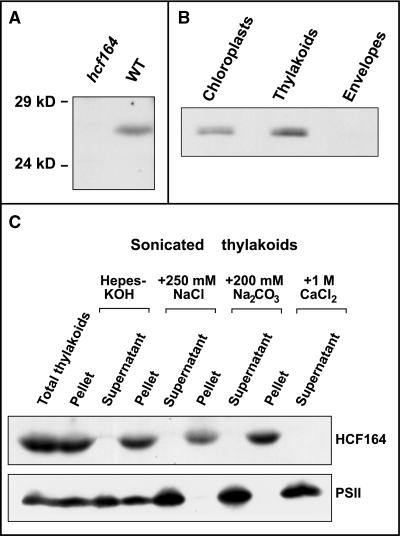
To determine the intracellular localization of HCF164, a polyclonal antiserum was raised against recombinant HCF164 protein (amino acids 85 to 261). The affinity-purified antiserum detected a 26-kD protein in wild-type leaf protein extracts (Figure 7A). No protein of that size could be detected in leaf protein extracts of the hcf164 mutant, which was to be expected from the RNA gel blot analysis described above.
Figure 7.
Immunolocalization of the HCF164 Protein.
(A) Immunoblot analysis of the HCF164 protein from wild-type (WT) and mutant. Total membrane proteins were isolated from Arabidopsis leaves, separated by SDS-PAGE, and analyzed by immunoblotting. Each lane was loaded with a protein equivalent of 10 μg of chlorophyll.
(B) Chloroplast localization of HCF164. Chloroplast proteins isolated from spinach were fractionated by a sucrose step gradient as described in Methods. In each lane, 30 μg of protein was separated by SDS-PAGE in the presence of 6 M urea and analyzed by immunoblotting.
(C) Salt washing of thylakoid membranes. Spinach thylakoids were sonicated in the salt conditions indicated and separated into membrane and soluble proteins by centrifugation. The protein equivalent of 10 μg of chlorophyll was analyzed. The 23-kD lumenal protein of PSII served as a marker for a membrane-associated but releasable hydrophilic thylakoid protein.
To prove the assumption that HCF164 is a chloroplast protein, intact chloroplasts were prepared from spinach and fractionated into envelopes and thylakoid membranes, and the protein fractions obtained were subjected to immunoblot analyses. Figure 7B demonstrates that HCF164 is detectable only in the thylakoid membrane and not in the envelope fraction. Washing of sonicated thylakoids with 0.25 M NaCl, 0.2 M Na2CO3, or 1 M CaCl2 did not release HCF164 from the membrane, in contrast to the 23-kD extrinsic PSII subunit that was used as a control (Figure 7C). This finding supports the conclusion that the HCF164 protein is firmly associated with the thylakoid membrane. The only hydrophobic part of HCF164 that could bind to the membrane is part of the transit peptide. We conclude, therefore, that this segment is still present in the mature HCF164 protein and serves as a membrane anchor of the otherwise hydrophilic protein.
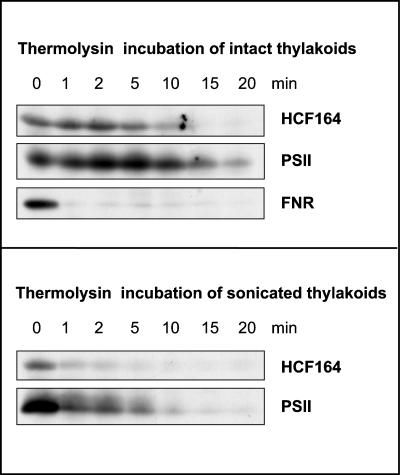
To investigate whether the large hydrophilic part of HCF164 is located toward the stroma or the thylakoid lumen, thermolysin protection assays were performed (Figure 8). When intact thylakoids were incubated with thermolysin, the HCF164 protein remained unaffected for at least 5 min of incubation. The lumenal 23-kD subunit of PSII showed the same protection, whereas the stromal ferredoxin NADP oxidoreductase was degraded within 1 min of incubation. In contrast, when sonicated thylakoids that contained ∼50% right-side out and 50% inside-out vesicles were treated with thermolysin, the HCF164 protein and the 23-kD PSII subunit were no longer protected and thus were degraded rapidly within 1 min of incubation.
Figure 8.
Lumenal Localization of the HCF164 Protein.
Intact and sonicated spinach thylakoids were protease treated with thermolysin (final concentration, 0.1 mg/mL). The reaction was stopped at the times indicated by the addition of EDTA to a final concentration of 50 mM. Membranes were recovered by centrifugation, and the equivalent of 10 μg of chlorophyll was loaded in each lane for immunoblot analysis. Ferredoxin NADP oxidoreductase (FNR) was used as a control for a peripheral thylakoid protein that is exposed to the stromal side, and the 23-kD protein of PSII was used as control for a lumenal protein.
We conclude from these data that the HCF164 protein is inserted into the thylakoid membrane via its N-terminal hydrophobic segment and that the C-terminal hydrophilic part that harbors the thioredoxin motif extends into the thylakoid lumen.
DISCUSSION
In recent years, a number of nuclear mutations affecting the biogenesis of the cytochrome b6f complex have been characterized in the higher plants Arabidopsis (Dinkins et al., 1994; Meurer et al., 1996), maize (Barkan et al., 1994; Voelker and Barkan, 1995b; Fisk et al., 1999), and Lemna perpusilla (Bruce and Malkin, 1991) as well as in the green alga Chlamydomonas reinhardtii (Howe and Merchant, 1992; Kuras and Wollman, 1994; Gumpel et al., 1995; Howe et al., 1995) (reviewed by Goldschmidt-Clermont, 1998; Wollman et al., 1999). In most cases, the corresponding gene has not been identified; as a consequence, the corresponding factor could not be characterized. We report here on the nuclear mutant hcf164 of Arabidopsis, which allowed us to isolate a gene that is involved in the biogenesis of the cytochrome b6f complex in chloroplasts of higher plants.
The mutant hcf164 displayed a severe deficiency in the cytochrome b6f complex. The accumulation of its component subunits was reduced to less than 10 to 20% of wild-type amounts. This is in line with spectroscopic data revealing a defect in intersystem electron transport rates. There also was a reduction in the amounts of PSI and PSII complexes; the remaining complexes, however, were fully functional, as inferred from spectroscopic analyses. Wild-type levels of PSI and PSII subunits were observed when hcf164 mutants were grown under very low light intensities. We conclude, therefore, that the reduction of these complexes most likely is a secondary effect of the mutation and that hcf164 has to be classified as a mutant affected specifically in the cytochrome b6f complex.
Cytochrome b559 of PSII accumulated to normal amounts in hcf164 mutants grown under very-low-light conditions. This indicates that the loss of the cytochrome b6f complex is not a result of a defect in heme biosynthesis. In vivo labeling of chloroplast proteins showed no principal difference between wild-type and mutant, and the overall rate of plastid protein synthesis in hcf164 was comparable to that in the wild-type. Immunoprecipitation experiments demonstrated that at least the synthesis of cytochrome f and PetD was similar in the wild-type and mutant, suggesting that the translation of these subunits is not affected primarily by the mutation. Therefore, we conclude that HCF164 operates at the post-translational level and is involved in the maturation or the assembly of the cytochrome b6f complex.
HCF164 encodes a thioredoxin-like protein. In accordance with this fact, the protein behaves biochemically as a thiol disulfide oxidoreductase. The cyanobacterial thioredoxin-like TxlA proteins were found to be the closest relatives of HCF164. This finding shows that HCF164 is a thioredoxin-like protein with an evolutionary origin in the cyanobacterial ancestor of chloroplasts. The function of the TxlA proteins is not yet clear. Partial gene knockouts yielded a severe defect in photoautotrophic growth (Collier and Grossman, 1995). However, the results remained inconclusive because the cells grew poorly, and therefore detailed biochemical analyses could not be performed.
Interestingly, HCF164 also showed a limited similarity to the thioredoxin-like proteins CcsX of the β proteobacterium B. pertussis (15% identity, 28% similarity) and CcmG of E. coli (15% identity, 32% similarity) and its homologs in Gram-negative bacteria. Both proteins are necessary for periplasmic thiol reduction and are believed to reduce the cysteine thiols of the heme binding motif (CxxCH) in apocytochrome c before the covalent binding of the heme group (Beckman and Kranz, 1993; Vargas et al., 1994; Fabianek et al., 1997, 1998; Page and Ferguson, 1997; Page et al., 1998; Beckett et al., 2000). Three distinct pathways, designated systems I, II, and III, with different assembly proteins have evolved for the covalent heme attachment to apocytochrome c (Kranz et al., 1998). CcmG participates in system I cytochrome c biogenesis, which is active in α and γ proteobacteria, plant and protozoal mitochondria, and some archaea. CcsX is associated with the system II pathway, which operates in Gram-positive bacteria, some β, ɛ, and δ proteobacteria, and cyanobacteria and chloroplasts.
Is HCF164 a component of the system II–type pathway of cytochrome c biogenesis in plastids? The protein localization experiments support this notion. HCF164 is a thylakoid protein that is anchored to the membrane, presumably via its uncleaved signal peptide, and whose major part extends into the lumen. Thus, it shows the same topology as CcsX/CcmG proteins that extend into the periplasm, the equivalent of the thylakoid lumen, and that are anchored to their membranes via hydrophobic patches of their uncleaved signal sequences (Monika et al., 1997; Fabianek et al., 1997, 1998; Beckett et al., 2000). If this functional assignment is correct, HCF164 is the functional equivalent of CcsX in a system II–type pathway of cytochrome c biogenesis in higher plants and is involved in the maturation of a c-type cytochrome (most likely cytochrome f).
Although these suggestions are persuasive, the results of the in vivo protein synthesis experiments do not corroborate the hypothesis that HCF164 represents a cytochrome f maturation factor. These experiments revealed that cytochrome f is synthesized and accumulates normally in the mutant during the labeling experiment (30 min). Provided that covalent binding of the heme group does not occur, the detected protein should be apocytochrome f. In Chlamydomonas, however, it has been shown that the inability to convert apocytochrome f to holocytochrome f results in increased turnover of the apoprotein, with a half-life of ∼10 to 25 min (Howe et al., 1995; Kuras et al., 1997). Because of this discrepancy, we cannot exclude that HCF164 is necessary for processes other than cytochrome f maturation. For instance, it could be involved in heme attachment to cytochrome b6. For this cytochrome, it has been suggested that one of the two heme groups is bound covalently to the protein (Kuras et al., 1997). Mutational analysis with Chlamydomonas showed that nuclear factors are necessary to catalyze this cytochrome b6 maturation process (Nakamoto et al., 2000); unfortunately, none of these factors has been cloned yet. HCF164 might be one of these unidentified proteins. Alternately, HCF164 could be a chaperone-like factor that is specific for the assembly of the cytochrome b6f complex. An increasing number of such protein complex–specific assembly factors are being discovered in eubacteria and their evolutionary descendants, the mitochondria and chloroplasts (Glerum et al., 1997; Suzuki et al., 1997; Meurer et al., 1998). HCF164 is unlikely to be a stoichiometric component of the cytochrome b6f complex. Centrifugation of detergent-treated thylakoid membranes on sucrose density gradients revealed that HCF164 and the cytochrome b6f complex do not comigrate (data not shown). Moreover, a stoichiometric component of the cytochrome b6f complex of ∼26 kD would not have escaped detection in earlier investigations aimed at the protein chemical characterization of that complex (Hauska et al., 1996).
Together, the evidence discussed here and the reports of other groups lead us to favor the model that HCF164 is the functional equivalent of CcsX that is involved in the maturation of cytochrome f. Whether the protein has other functions during the biogenesis of the cytochrome b6f complex, for instance, the attachment of the heme group to cytochrome b6, must be left for future experimentation.
METHODS
Mutant Isolation and Growth Conditions
The hcf164 mutant was recovered from a collection of T-DNA insertion lines of Arabidopsis thaliana ecotype Wassilewskija produced by Nicole Bechtold and co-workers (Institut National de la Récherche Agronomique, Versailles, France). Selection of the mutant and its propagation was essentially as described by Meurer et al. (1998) except that phosphinotricin (10 mg/L final concentration) was used for mutant selection.
Spectroscopic Measurements
Chlorophyll fluorescence and P700 absorbance measurements were performed as described by Meurer et al. (1996) except that for the P700 absorbance kinetics, the pulse amplitude modulated fluorometer was modified with a P700 dual-wavelength emitter/detector unit (Waltz, Effeltrich, Germany).
PAGE, Heme Staining, and Immunoblot Analysis of Proteins
Isolation of protein extracts from wild-type and mutant leaves, electrophoresis of proteins on SDS-polyacrylamide gels (Laemmli, 1970; Schägger and von Jagow, 1987), and immunodecoration of electroblotted proteins were performed according to the methods described by Meurer et al. (1996)(1998) with the following variations. Plant material was ground for 20 sec in a Waring blender (capacity, 70 mL) with 25 mL of homogenization buffer (350 mM sucrose, 50 mM Tris-HCl, pH 8.0, 7 mM EDTA, and 10 mM DTT). After filtration through two layers of Miracloth, the homogenate was centrifuged for 10 min at 4000g (HB4 rotor; Sorvall, Norwalk, CT). The pellet containing the chloroplasts was resuspended in homogenization buffer without saccharose and adjusted to a final concentration of 2 to 5 mg protein/mL. For the detection of heme-containing proteins, samples were treated as described for the immunoblotting procedures. Proteins were electroblotted onto nitrocellulose filters, and these were incubated immediately for 30 min with the reagent of the chemiluminescence assay (ECL Plus; Amersham Pharmacia Biotech, Uppsala, Sweden). Exposure times of 1 hr produced a visible signal for the lowest concentration of the dilution series.
In Vivo Labeling of Proteins and Immunoprecipitation
Primary leaves of 20 15-day-old plants were preincubated in 200 μg/mL cycloheximide for 30 min and radiolabeled with 5 μCi/μL 35S-methionine (specific activity >1000 Ci/mmol; Amersham Pharmacia Biotech) in the presence of 100 μg/mL cycloheximide for 30 min at 25°C. After washing five times with homogenization buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM EDTA), leaves were ground with a conical stainless steel rod in an Eppendorf tube with 300 μL of the same buffer. Membranes were pelleted by centrifugation at 15,000g for 10 min and resuspended in 100 μL of homogenization buffer. SDS was added to a final concentration of 2% (w/v), and proteins were solubilized for 10 min at 25°C followed by a 30-sec incubation at 70°C. For immunoprecipitation, aliquots equivalent to 200,000 cpm of incorporated label were diluted 10-fold with immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% [v/v] Nonidet P-40) and incubated with 5 μL of specific antiserum at 4°C overnight. Antibody-antigen complexes were adsorbed by adding 30 μL (bed volume) of protein A–Sepharose (Amersham Pharmacia Biotech) and by incubation of the suspension for 1 hr at 20°C under slow rotation. The protein A–Sepharose then was collected by centrifugation at 900g for 5 min and washed four times with immunoprecipitation buffer. Antibody-antigen-protein A complexes were dissociated by incubation in gel loading buffer (Laemmli, 1970) for 30 sec at 70°C. To ensure a quantitative precipitation of the proteins, the supernatants from the first immunoprecipitation were treated a second time with the same amount of antiserum. Proteins were electrophoresed by PAGE according to Laemmli (1970). Radiolabeled proteins were detected on the dried gels by autoradiography and quantified using a BioImager (BioImaging Analyser BAS-1800; Fuji Photo Film Co., Düsseldorf, Germany) and the software packages Image Reader BAS 1.4 and Image Gauge 3.0 (Fuji Photo Film Co., Düsseldorf, Germany).
RNA Gel Blot Analysis
Isolation of RNA, electrophoresis of glyoxylated RNA on agarose gels, and RNA gel blot hybridization analysis were performed as described elsewhere (Westhoff et al., 1991; Meurer et al., 1996). For isolation of poly(A+) RNA, the total RNA was treated with Oligotex (Amersham Pharmacia Biotech) according to the protocol supplied by the manufacturer.
Inverse Polymerase Chain Reaction Amplification of T-DNA Flanking Genomic Sequences, Cloning of the HCF164 cDNA, and Mutant Complementation
Genomic DNA was isolated from 3-week-old mutants according to Dellaporta et al. (1983), restricted with EcoRI, and self-ligated. Standard polymerase chain reaction (PCR) reactions with the primers LB-TAG5 (5′-CTACAAATTGCCTTTTCTTATCGA-3′) and LB-TAG17 (5′-GTCCGCTCTACCGAAAGTTACG-3′) resulted in a 2040-bp product. This was reamplified with the nested primers LB-TAG14 (5′-GGTAATAGGACACTGGGATTCGTC-3′) and LB-TAG18 (5′-CGACAACATGTCGAGGCTCAGC-3′), yielding a 1820-bp DNA fragment. The PCR product was cloned into the EcoRV site of pBluescript II KS+ (Stratagene, La Jolla, CA) as described (Marchuk et al., 1991), and the resulting clone, pnAt-HCF164, was sequenced. Based on genomic sequences that matched pnAt-HCF164 (EMBL accession numbers Z99707 and CAB16778.1), the primers 164FOR (5′-GCGGATCCCGACGATGGCTCGCTTAG-3′) and 164REV (5′-CGGATCCGCATGATAGTGATGCCACTC-3′) were designed and used for reverse transcriptase–mediated PCR amplification of the complete HCF164 coding sequence from ecotype Wassilewskija. Because the primers 164FOR and 164REV contained a BamHI restriction site, the PCR product of 814 bp was cloned into the BamHI site of pBluescript II KS+, and several of the resulting cDNA clones were sequenced.
One of the clones with a correct HCF164 sequence, pcAt-HCF164, was used for the complementation of homozygous hcf164 plants by Agrobacterium tumefaciens–mediated root transformation essentially as described by Meurer et al. (1998). Forty-five independent transgenic plants were recovered and analyzed by chlorophyll fluorescence induction for a complemented mutant phenotype.
Enzymatic Analysis of Recombinant HCF164
The nucleotide sequences encoding the soluble part of HCF164 (amino acids 116 to 261, corresponding to nucleotide positions 380 to 788 of pcAt-HCF164) were amplified by PCR using the primers 5′-AGGATTGCTAGCGATTTTGGGATTTCTTTGAAGG-3′ and 5′-TACTCAAGCTTATCCATGGCTTAAGGGATC-3′. The resulting DNA fragment was cleaved with NheI and HindIII and fused in frame with the signal peptide sequence present in the expression plasmid pAH33 (Seidler, 1994). The resulting plasmid, pEX-HCF164, when introduced into Escherichia coli, directed the soluble recombinant HCF164 protein into the periplasmic space. An N-terminal His tag allowed purification by metal chelate affinity chromatography.
Expression of the protein was performed in E. coli strain SF110 (Meerman and Georgiou, 1994) in tryptone phosphate (TP) medium (2% [w/v] tryptone, 1.5% [w/v] yeast extract, 0.8% [w/v] NaCl, 0.2% [w/v] Na2HPO4, and 0.1% [w/v] KH2PO4) (Moore et al., 1993). Cells were grown at 25°C to OD600 = 0.6, and the expression of recombinant HCF164 was induced by adding isopropylthio-β-d-galactoside to a final concentration of 0.5 mM. Growth was continued for another 12 to 14 hr, and periplasmic proteins were isolated as described by Seidler (1994), except that lysozyme was omitted. The periplasmic protein fraction was dialyzed against buffer A (20 mM Hepes, pH 7.5, and 500 mM NaCl) and loaded onto a metal-affinity chromatography column (Chelating Sepharose FF; 1.5 × 12 cm; Amersham Pharmacia Biotech) precharged with Zn2+. The column was washed with 30 mL of buffer A containing 20 mM imidazole. Elution of the His-tagged HCF164 was performed with a linear gradient of 20 to 350 mM imidazole in 150 mL of buffer A.
The recombinant HCF164 protein was assayed for disulfide reductase activity according to Holmgren (1979). The reaction mixture with a final volume of 1 mL contained 100 mM sodium phosphate, pH 7.0, 1 mM EDTA, 1 mg/mL insulin (bovine insulin; Sigma, München, Germany), and 2 or 5 μM recombinant HCF164 protein. The reaction was started by the addition of 0.35 mM DTT and performed at room temperature. The insulin disulfide reduction was measured by the absorbance change at 650 nm. The nonenzymatic reduction of insulin by DTT served as a negative control, and as a positive control, 0.5 μM thioredoxin from Spirulina sp (Sigma, Deisenhofen, Germany) was used.
Antiserum Production
The nucleotide sequence encoding amino acids 85 to 261 of the HCF164 protein (nucleotide positions 258 to 788 of pcAt-HCF164) was fused in frame to the glutathione S-transferase sequence of the expression vector pGEX-4T3 (Amersham Pharmacia Biotech), and the construct was transformed into E. coli strain BL21(DE3) pLysE. Expression of the 46-kD glutathione S-transferase HCF164 fusion protein was induced with 1 mM isopropylthio-β-d-galactoside. The E. coli cells were recovered by centrifugation and resuspended in 50 mM Tris-HCl, pH 7.9, 100 mM NaCl, 0.1 mM EDTA, and 0.01% (w/v) Triton X-100. After incubation for 30 min at room temperature in the presence of lysozyme at a final concentration of 1 mg/mL, the cell suspension was sonicated four times for 30 sec at an output intensity of 7 (Branson Sonifier B12; Branson, Danbury, CT) while kept on ice. Because the fusion protein was found exclusively in inclusion bodies, the sonicated cell suspension was centrifuged and the pellet was solubilized in 2% (w/v) SDS, 10% (v/v) 2-mercaptoethanol, 62.5 mM Tris-HCl, pH 6.8, and 10% (w/v) glycerol. This crude fraction was subjected to preparative PAGE (Hager and Burgess, 1980). The fusion protein was eluted in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 0.75% (w/v) N-lauryl-sarcosine, and 0.5% (v/v) 2-mercaptoethanol. After adjusting the eluate to a final concentration of 1.5% (v/v) Triton X-100, the fusion protein was cleaved with thrombin (Amersham Pharmacia Biotech). The released HCF164 protein was purified again by preparative PAGE and finally dissolved in 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3, 2.7 mM KCl, 140 mM NaCl, and 0.2% (w/v) SDS. A polyclonal antiserum was raised in rabbits and affinity purified on an HCF164 protein column by BioGenes (Berlin, Germany).
Immunolocalization Studies with Protein Gel Blots
The intracellular localization of the HCF164 protein was determined by immunoblot analysis. Thylakoid membranes and envelope fractions were prepared by fractionation of lysed spinach chloroplasts on a discontinuous sucrose density gradient according to Cline et al. (1981).
The membrane association of HCF164 was analyzed by salt washing experiments. Spinach thylakoids were resuspended to a final concentration of 50 μg/mL chlorophyll in 10 mM Hepes/KOH, pH 8.0, 10 mM MgCl2, 330 mM sorbitol, 2 mM phenylmethylsulfonyl fluoride, and 2 mM benzamidine supplemented with 250 mM NaCl, 200 mM Na2CO3, or 1 M CaCl2, respectively, without further supplement as a control. These suspensions were sonicated three times for 15 sec at an output intensity of 5 (Branson Sonifier B12) while kept on ice. The membrane vesicles were separated from the soluble components by ultracentrifugation at 100,000g for 2 hr at 4°C. Proteins of the supernatant were recovered by trichloroacetic acid precipitation (15% final concentration). Protein equivalents of 10 μg of chlorophyll of the soluble (supernatant) and membranous proteins were subjected to immunoblot analyses using the HCF164 antiserum.
The intraorganellar location of HCF164 was studied further by protease protection studies using thermolysin as a probe as described by Meurer et al. (1998).
Accession Number
The nucleotide sequence of the HCF164 cDNA has been submitted to the DDBJ/EMBL/GenBank databases under accession number AJ293262.
Acknowledgments
The authors are grateful to Richard J. Berzborn (Ruhr-Universität, Bochum), Günter Hauska (Universität Regensburg, Regensburg), and Ralf Bernd Klösgen (Martin-Luther-Universität, Halle-Wittenberg) for the gift of antisera. We thank George Georgiou (University of Texas, Austin) for providing the E. coli strain SF110. This research was supported by a grant from the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 189) to K.M.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010245.
References
- Barkan, A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium–mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316 1194–1199. [DOI] [PubMed] [Google Scholar]
- Beckett, C.S., Loughman, J.A., Karberg, K.A., Donato, G.M., Goldman W.E., and Kranz, R.G. (2000). Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38 465–481. [DOI] [PubMed] [Google Scholar]
- Beckman, D.L., and Kranz, R.G. (1993). Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc. Natl. Acad. Sci. USA 90 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, B.D., and Malkin, R. (1991). Biosynthesis of the chloroplast cytochrome b6f complex: Studies in a photosynthetic mutant of Lemna. Plant Cell 3 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., Stern, D.B., Wostrikoff, K., Kuras, R., Girard-Bascou, J., and Wollman, F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., Andrews, J., Mersey, B., Newcomb, E.H., and Keegstra, K. (1981). Separation and characterization of inner and outer envelope membranes of pea chloroplasts. Proc. Natl. Acad. Sci. USA 78 3595–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, J.L., and Grossman, A.R. (1995). Disruption of a gene encoding a novel thioredoxin-like protein alters the cyanobacterial photosynthetic apparatus. J. Bacteriol. 177 3269–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Dinkins, R.D., Bandaranayake, H., Green, B.R., and Griffith, A.J. (1994). A nuclear photosynthetic electron transport mutant of Arabidopsis thaliana with altered expression of the chloroplast petA gene. Curr. Genet. 25 282–288. [DOI] [PubMed] [Google Scholar]
- Fabianek, R.A., Huber-Wunderlich, M., Glockshuber, R., Kunzler, P., Hennecke, H., and Thöny-Meyer, L. (1997). Characterization of the Bradyrhizobium japonicum CycY protein, a membrane-anchored periplasmic thioredoxin that may play a role as a reductant in the biogenesis of c-type cytochromes. J. Biol. Chem. 272 4467–4473. [DOI] [PubMed] [Google Scholar]
- Fabianek, R.A., Hennecke, H., and Thöny-Meyer, L. (1998). The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J. Bacteriol. 180 1947–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between mitochondrial and chloroplast gene expression. EMBO J. 18 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum, D.M., Muroff, I., Jin, C., and Tzagaloff, A. (1997). COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 272 19088–19094. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177 115–180. [DOI] [PubMed] [Google Scholar]
- Gray, J.C. (1992). Cytochrome f: Structure, function and biosynthesis. Photosynth. Res. 34 359–374. [DOI] [PubMed] [Google Scholar]
- Gumpel, N.J., Ralley, L., Girard-Bascou, J., Wollman, F.A., Nugent, J.H.A., and Purton, S. (1995). Nuclear mutants of Chlamydomonas reinhardtii defective in the biogenesis of the cytochrome b6f complex. Plant Mol. Biol. 29 921–932. [DOI] [PubMed] [Google Scholar]
- Hager, D.A., and Burgess, R.R. (1980). Elution of proteins from sodium dodecyl sulfate polyacrylamide gels, removal of sodium dodecyl sulfate and renaturation of enzymatic activity: Results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase and other enzymes. Anal. Biochem. 109 1676–1680. [DOI] [PubMed] [Google Scholar]
- Hauska, G., Schütz, M., and Büttner, M. (1996). The cytochrome b6f complex: Composition structure and function. In Oxygenic Photosynthesis: The Light Reactions, D.R. Ort and C.F. Yocum, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 377–398.
- Holmgren, A. (1979). Thioredoxin catalyses the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254 9627–9632. [PubMed] [Google Scholar]
- Howe, G., and Merchant, S. (1992). The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J. 11 2789–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G., Mets, L., and Merchant, S. (1995). Biosynthesis of cytochrome f in Chlamydomonas reinhardtii: Analysis of the pathway in gabaculine-treated cells and in the heme attachment mutant B6. Mol. Gen. Genet. 246 156–165. [DOI] [PubMed] [Google Scholar]
- Hynds, P.J., Plücken, H., Westhoff, P., and Robinson, C. (2000). Different lumen-targeting pathways for nuclear-encoded versus cyanobacterial/plastid-encoded HCF136 proteins. FEBS Lett. 467 97–100. [DOI] [PubMed] [Google Scholar]
- Jenkins, B.D., Kulhanek, D.J., and Barkan, A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, Vol B2, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–22.
- Kranz, R., Lill, R., Goldman, B., Bonnard, G., and Merchant, S. (1998). Molecular mechanisms of cytochrome c biogenesis: Three distinct systems. Mol. Microbiol. 29 383–396. [DOI] [PubMed] [Google Scholar]
- Kuras, R., and Wollman, F.A. (1994). The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, R., de Vitry, C., Choquet, Y., Girard-Bascou, J., Culler, D., Buschlen, S., Merchant, S., and Wollman, F.A. (1997). Molecular and genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272 32427–32435. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Marchuk, D., Drumm, M., Saulino, A., and Collins, F.S. (1991). Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerman, H.J., and Georgiou, G. (1994). Construction and characterization of a set of E. coli strains deficient in all known loci affecting the proteolytic stability of secreted recombinant proteins. Biotechnology 12 1107–1110. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Meierhoff, K., and Westhoff, P. (1996). Isolation of high chlorophyll fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and Northern hybridisation. Planta 198 385–396. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, D. (1994). The use of high chlorophyll fluorescence photosynthetic mutants in the analysis of thylakoid membrane assembly and function. Maydica 39 35–45. [Google Scholar]
- Monika, E.M., Goldman, B.S., Beckman, D.L., and Kranz, R.G. (1997). A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis: In vitro and in vivo studies. J. Mol. Biol. 271 679–692. [DOI] [PubMed] [Google Scholar]
- Moore, J.T., Uppal, A., Maley, F., and Maley, G.F. (1993). Overcoming inclusion body formation in a high-level expression system. Protein Exp. Purif. 4 160–163. [DOI] [PubMed] [Google Scholar]
- Mosser, G., Breyton, C., Olofsson, A., Popot, J.L., and Rigaud, J.L. (1997). Projection map of cytochrome b6f complex at 8 Å resolution. J. Biol. Chem. 272 20263–20268. [DOI] [PubMed] [Google Scholar]
- Nakamoto, S.S., Hamel, P., and Merchant, S. (2000). Assembly of chloroplast cytochromes b and c. Biochimie 82 603–614. [DOI] [PubMed] [Google Scholar]
- Ochman, H., Ayaly, F.H., and Hartl, D.L. (1993). Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 218 309–321. [DOI] [PubMed] [Google Scholar]
- Page, M.D., and Ferguson, S.J. (1997). Paracoccus denitrificans CcmG is a periplasmic protein-disulfide oxidoreductase required for c- and aa3-type cytochrome biogenesis: Evidence for reductase role in vivo. Mol. Microbiol. 24 977–990. [DOI] [PubMed] [Google Scholar]
- Page, M.D., Sambongi, Y., and Ferguson, S.J. (1998). Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem. Sci. 23 103–108. [DOI] [PubMed] [Google Scholar]
- Robinson, C., and Mant, A. (1997). Targeting of proteins into and across the thylakoid membrane. Trends Plant Sci. 2 431–437. [Google Scholar]
- Rochaix, J.D. (1996). Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol. 32 327–341. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1–100 kDa. Anal. Biochem. 166 368–379. [DOI] [PubMed] [Google Scholar]
- Seidler, A. (1994). Introduction of a histidine tail at the N-terminus of a secretory protein expressed in Escherichia coli. Protein Eng. 7 1277–1280. [DOI] [PubMed] [Google Scholar]
- Strizhov, N., Keller, M., Mathur, J., Koncz-Kálmán, Z., Bosch, D., Prudovsky, E., Schell, J., Sneh, B., Koncz, C., and Zilberstein, A. (1996). A synthetic cryIC gene, encoding a Bacillus thuringiensis δ-endotoxin, confers Spodoptera resistance in alfalfa and tobacco. Proc. Natl. Acad. Sci. USA 93 15012–15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, C.K., Rep, M., Van Dijl, J.M., Suda, K., Grivell, L.A., and Schatz, G. (1997). ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 22 118–123. [DOI] [PubMed] [Google Scholar]
- Vargas, C., Wu, G., Davies, A.E., and Downie, J.A. (1994). Identification of a gene encoding a thioredoxin-like product necessary for cytochrome c biosynthesis and symbiotic nitrogen fixation in Rhizobium leguminosarum. J. Bacteriol. 176 4117–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, R., and Barkan, A. (1995. a). Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 14 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, R., and Barkan, A. (1995. b). Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol. Gen. Genet. 249 507–514. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., and Herrmann, R.G. (1988). Complex RNA maturation in chloroplasts: The psbB operon from spinach. Eur. J. Biochem. 171 551–564. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., Offermann-Steinhard, K., Höfer, M., Eskins, K., Oswald, A., and Streubel, M. (1991). Differential accumulation of plastid transcripts encoding photosystem II components in the mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants. Planta 184 377–388. [DOI] [PubMed] [Google Scholar]
- Wollman, F.A., Limor, M., and Nechushtai, R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411 21–85. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Culler, D., Dreyfuss, B.W., Kuras, R., Wollman, F.A., Girard-Bascou, J., and Merchant, S. (1998). Genetic analysis of c-type cytochrome assembly in Chlamydomonas reinhardtii: One chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]