Abstract
An expressed sequence tag–based microarray was used to profile genome expression underlying light control of Arabidopsis development. Qualitatively similar gene expression profiles were observed among seedlings grown in different light qualities, including far-red, red, and blue light, which are mediated primarily by phytochrome A, phytochrome B, and the cryptochromes, respectively. Furthermore, light/dark transitions also triggered similar differential genome expression profiles. Most light treatments also resulted in distinct expression profiles in small fractions of the expressed sequence tags examined. The similarly regulated genes in all light conditions were estimated to account for approximately one-third of the genome, with three-fifths upregulated and two-fifths downregulated by light. Analysis of those light-regulated genes revealed more than 26 cellular pathways that are regulated coordinately by light. Thus, light controls Arabidopsis development through coordinately regulating metabolic and regulatory pathways.
INTRODUCTION
As sessile organisms, higher plants are characterized by a high degree of developmental plasticity in response to environmental cues, thereby optimizing their developmental patterns in a way that maximizes their chances of survival and reproduction. Light is one of the most important environmental factors that govern plant growth and development (Kendrick and Kronenberg, 1994). Besides providing an energy source for the plant via photosynthesis, higher plants have evolved a sophisticated photosensory system that detects the quality, quantity, direction, and duration of light and use this information to control their developmental pattern (Deng and Quail, 1999; Neff et al., 2000). For example, dark-grown (skotomorphogenic) seedlings are characterized by elongated hypocotyls, closed cotyledons on an apical hook, and nonphotosynthetic etioplasts. In contrast, photomorphogenic seedlings have short hypocotyls, expanded cotyledons, and photosynthetically active chloroplasts (Kendrick and Kronenberg, 1994).
A plant's capacity to monitor ambient light and to trigger proper morphological and developmental changes is provided largely by two major types of well-characterized photoreceptors. They are the red/far-red-light–absorbing phytochromes and blue/UV A light–absorbing cryptochromes (Quail et al., 1995; Deng and Quail, 1999; Neff et al., 2000). On the other hand, transient physiological changes such as phototropism and chloroplast high light avoidance responses are modulated by a separate family of blue light receptors, phototropin, and a homologous protein (Briggs and Olney, 2001; Jarillo et al., 2001; Kagawa et al., 2001). For light-controlled development, it is generally assumed that the photoreceptors perceive and interpret incident light and transduce the signals to modulate light-responsive nuclear genes, which direct appropriate growth and developmental responses. Therefore, the contrasting developmental patterns are thought to be mediated primarily by changes in light-regulated gene expression (Terzaghi and Cashmore, 1995; Puente et al., 1996).
Previous studies have revealed probably more than 100 genes whose expression is controlled by light (Terzaghi and Cashmore, 1995; Fankhauser and Chory, 1997; Kuno and Furuya, 2000). However, the dramatic developmental transition during plant photomorphogenesis is likely to involve a much larger number of genes. To date, no systematic study has been done to determine the number and identities of the genes that define plant photomorphogenesis, and the analysis of light-signaling processes and their interactions in plants has focused on a limited set of genes (Somerville and Somerville, 1999). These studies are unable to provide an overall picture of the genome expressions related to photomorphogenesis. Fortunately, semiquantitative methods for genome-wide analysis of gene expression profiles, such as the relatively recently developed cDNA microarray analysis (Schena et al., 1995), can measure the relative expression levels of thousands of genes simultaneously. This technique has been applied to investigate genome-wide gene expression profiles during animal development, such as in Caenorhabditis elegans germline development (Reinke et al., 2000) and Drosophila metamorphosis (White et al., 1999). Plant biologists also have adopted microarray technology to investigate genome expression profiles in several plant processes. They include the circadian regulation of gene expression (Harmer et al., 2000; Schaffer et al., 2001), defense responses (Petersen et al., 2000; Schenk et al., 2000), mechanical wounding response (Reymond et al., 2000), nutrient response (Wang et al., 2000), far-red light regulation of gene expression (Tepperman et al., 2001), blue light regulation of gene expression (Wang et al., 2001), and drought and cold stress responses (Seki et al., 2001).
In the present study, we constructed a microarray with 9216 expressed sequence tags (ESTs) from Arabidopsis. The microarray was used to examine the gene expression profiles of seedlings grown in distinct light conditions and the role of photoreceptors during Arabidopsis photomorphogenesis. Our results showed that approximately one-third of the ESTs in our microarray were regulated twofold or more by light, and at least 26 cellular pathways were coordinately upregulated or downregulated during Arabidopsis photomorphogenesis.
RESULTS
Brief Summary of an Arabidopsis EST Clone–Based Microarray
We constructed a microarray containing 9216 Arabidopsis ESTs that was estimated to represent ∼6120 unique genes (see Methods). Each glass slide contained two copies of the entire array, each of which consisted of 16 subarrays with 24 rows and columns. Negative control genes, including genes of Escherichia coli, yeast, and human origins, were printed on the top of subarrays 1 and 2.
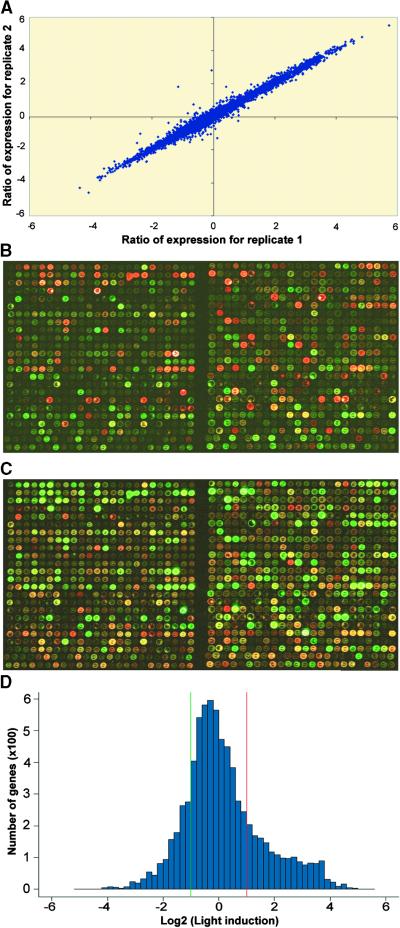
To minimize the inherent variability of the microarray assay (Lee et al., 2000) and to ensure the reliability of our results, at least two microarray slides (four replicates) were used to analyze the mRNA abundance of each sample pair. The first slide was probed with cDNAs labeled with Cy-3 and Cy-5 deoxy UTP. By using independent RNA preparations, the labeling of the sample in each pair was reversed on the second slide to overcome potential artifacts caused by the dye-related differences in labeling efficiency, different laser settings, and nonlinearity of photomultiplier tubes in the scanner. Thus, at least two independent RNA preparations, or four in some cases, were made for each biological sample and were used to prepare labeled probes. The hybridization signal from each of the replicate ESTs were averaged and used for analysis. In our study, the hybridization signals from the replicates were highly reproducible. As illustrated in Figure 1A, two representative replicates of a sample pair from wild type/white light–grown (WT/W) and wild type/dark–grown (WT/D) 6-day-old seedlings were highly reproducible. Figures 1B and 1C show two subarray images with reciprocal labeling schemes in the experiment described above to illustrate the color reverse of hybridization signals. The correlation coefficients from all four replicates in this experiment are summarized in Table 1. The correlation coefficient of the ratios from the four arrays in each experiment was greater than 0.95, suggesting an excellent reproducibility among individual arrays in the same experiment. Therefore, the conclusions derived from this analysis are considered reliable.
Figure 1.
Evaluation of the Microarray Assay.
(A) Scatterplot of signal values from two replicates on microarray. Total RNA from 6-day-old white light– and dark-grown wild-type Arabidopsis seedlings was labeled with Cy5 and Cy3, respectively. The Log2 values of the Cy5-to-Cy3 ratios were plotted for the two replicates.
(B) Overlay image of two subarrays of a microarray hybridized with probes originating from 6-day-old white light–grown wild-type seedlings labeled with Cy5 and 6-day-old dark-grown wild-type seedlings labeled with Cy3.
(C) Overlay image of the same two subarrays as in (B) but hybridized with probes originating from 6-day-old white light–grown wild-type seedlings labeled with Cy3 and 6-day-old dark-grown wild-type seedlings labeled with Cy5.
(D) Distribution of average ratios of expression from white light– and dark-grown wild-type seedlings. Total RNA from 6-day-old white light– and dark-grown wild-type seedlings was labeled reciprocally with Cy3 and Cy5. The ratio for each clone is the average from four replicates.
Table 1.
Correlation Coefficient of Ratios of Data for Four Replicates from Wild-Type Seedlings Grown in White Light Versus Darka
| Replicate | W-Cy5 versus D-Cy3 (1) |
W-Cy5 versus D-Cy3 (2) |
W-Cy3 versus D-Cy5 (1) |
W-Cy3 versus D-Cy5 (2) |
|---|---|---|---|---|
| W-Cy5 versus D-Cy3 (1)b | —c | 0.978 | 0.948 | 0.946 |
| W-Cy5 versus D-Cy3 (2) | 0.978 | — | 0.950 | 0.951 |
| W-Cy3 versus D-Cy5 (1) | 0.948 | 0.950 | — | 0.984 |
| W-Cy3 versus D-Cy5 (2) | 0.946 | 0.951 | 0.984 | — |
a W, white light; D, darkness; Cy3, Cy3-deoxyUTP; Cy5, Cy5-deoxyUTP.
b The numbers 1 and 2 in the parentheses represent replicate 1 and replicate 2 of the same microarray slide.
c N/A, not applicable.
Strategy for Profiling Light-Regulated Gene Expression
To reveal the genome expression profiles specific to photomorphogenesis, photomorphogenic seedlings grown under white light as well as far-red, red, and blue light were compared with dark-grown seedlings. This comparative analysis was aimed at revealing shared as well as unique gene expression patterns triggered by four distinct light quality conditions in the photomorphogenic seedlings.
It is well documented that photomorphogenesis under continuous far-red, red, and blue light is mediated primarily by distinct photoreceptors, phytochrome A, phytochrome B, and cryptochromes (CRY1 and CRY2), respectively. To examine the role of the photoreceptors in the gene expression profile under these various light conditions, the light-regulated gene expression of specific photoreceptor mutants (phyA and phyB) or double mutants (cry1 cry2) was compared with that of wild type grown in the same light condition. This comparison could define the minimal set of genes affected by each photoreceptor under the selected light condition. Furthermore, photoreceptor-overexpressing transgenic lines were used to analyze the effect of the overexpression of photoreceptors on the genome expression profile under specific light quality conditions.
Our microarray assay is validated and confirmed by comparison with previously identified light-regulated genes included in the microarray. As shown in Table 2, both light-upregulated genes, such as RBCS, PC, GAPDH, CHS, and PAL, and light-downregulated genes, including PHYA, PORA, AS, and TUB1, exhibited the expected light-regulated expression patterns. The ratios we obtained for previously analyzed genes in our microarray assay corresponds well with the ratios attained by traditional methods, an indication that our microarray assay is robust.
Table 2.
Microarray Results from Some Well-Known Light- Regulated Genes Reported in the Literature
| Fold Regulated by Light in the Microarraya
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Accession Number | Gene Name | A | B | C | D | E | F | G | Reference |
| N37414 | D1 | 48.0 | 10.1 | 36.0 | 12.8 | 1.9 | 9.1 | −1.4 | Thompson et al. (1983) |
| T76714 | Lhb1B2 | 30.2 | 19.6 | 27.7 | 13.3 | 3.7 | 7.8 | 2.9 | Tobin and Silverthorne (1985) |
| N65655 | Lhb1B1 | 30.2 | 18.2 | 32.1 | 18.3 | 5.0 | 7.9 | 2.4 | Tobin and Silverthorne (1985) |
| R89925 | Lhca5 | 18.5 | 12.1 | 18.9 | 19.2 | 8.2 | 14.4 | 4.7 | Tobin and Silverthorne (1985) |
| N97182 | Lhcb2 | 14.1 | 13.3 | 16.4 | 16.4 | 4.6 | 5.5 | 3.1 | Tobin and Silverthorne (1985) |
| N96851 | Lhca2 | 6.5 | 3.0 | 4.3 | 3.5 | 1.9 | 3.8 | 2.0 | Tobin and Silverthorne (1985) |
| R90003 | PC | 14.6 | 3.6 | 7.7 | 9.0 | 5.9 | 10.2 | 4.1 | Helliwell et al. (1997) |
| AA067471 | Rubisco activase b | 14.5 | 6.5 | 11.5 | 11.5 | 4.2 | 9.0 | 3.8 | Orozco and Ogren (1993) |
| T20492 | RBCS 3b | 14.3 | 3.8 | 10.4 | 9.0 | 6.3 | 11.2 | 5.6 | Tobin and Silverthorne (1985) |
| T45355 | RBCS 2B | 14.2 | 2.9 | 8.4 | 7.1 | 5.4 | 8.6 | 4.8 | Tobin and Silverthorne (1985) |
| R30465 | RBCS 1B | 14.2 | 4.5 | 8.9 | 6.3 | 5.0 | 9.7 | 5.0 | Tobin and Silverthorne (1985) |
| N65785 | RBCS 1A | 10.9 | 5.1 | 4.2 | 8.4 | 4.8 | 7.2 | 4.6 | Tobin and Silverthorne (1985) |
| N37467 | GAPDH | 10.0 | 2.2 | 8.2 | 4.7 | 2.7 | 4.7 | 3.0 | Cerff and Kloppstech (1982) |
| H36324 | CHS | 10.9 | 2.7 | 6.0 | 3.9 | 2.3 | 2.6 | 1.8 | Batschauer et al. (1991) |
| T45207 | PAL1 | 3.0 | −1.3 | 1.5 | 1.2 | 1.3 | 1.2 | 3.1 | Schroder et al. (1979) |
| N65492 | POR A | −2.4 | −2.1 | −1.1 | −2.5 | −2.6 | −1.2 | −0.83 | Apel (1981) |
| T20944 | PHYA | −3.1 | −1.4 | −2.4 | −1.9 | −2.8 | −2.1 | −1.8 | Bruce et al. (1989) |
| N96756 | AS | −5.6 | 1.5 | −2.9 | −3.6 | −3.4 | −20 | −16.7 | Shi et al. (1997) |
| H36556 | AS | −3.1 | −1.3 | −1.7 | −1.6 | −1.4 | −1.6 | −2.2 | Shi et al. (1997) |
| W43629 | TUB1 | −2.7 | −1.9 | −1.3 | −1.8 | −1.8 | −2.2 | −2.0 | Leu et al. (1995) |
a The seven light treatment comparisons are as follows: A, 6-day-old wild-type seedlings grown in white light versus in dark (WT/W versus WT/D); B, 6-day-old wild-type seedlings grown in far-red light versus in dark (WT/FR versus WT/D); C, 6-day-old wild-type seedlings grown in red light versus in dark (WT/R versus WT/D); D, 6-day-old wild-type seedlings grown in blue light versus in dark (WT/B versus WT/D); E, white light–grown 6-day-old wild-type seedlings versus white light–grown 4.5-day-old wild-type seedlings transferred to dark for 36 hr (WT/W versus WT/36D); F, 4.5-day-old dark-grown wild-type seedlings transferred to white light for 36 hr versus 6-day-old dark-grown wild-type seedlings (WT/36W versus WT/D); G, white light–grown 5-week-old wild-type plant leaves versus white light–grown 5-week-old wild-type plant leaves transferred to dark for 36 hr (WTL/W versus WTL/36D).
b Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Light Regulates a Large Number of Genes to Different Extents
Examination of the expression ratios of the EST clones between white light– and dark-grown seedlings revealed the large extent to which light regulates gene expression. As shown in Figure 1D and Table 3, 2976 of the 9216 ESTs (32%) displayed differential expression of at least twofold or more. Among them, 1708 ESTs exhibited light-inducible expression, whereas 1268 ESTs exhibited downregulation by light. General distributions of light-regulated genes regulated by different light qualities and the effect of the respective photoreceptor mutations are summarized in Table 3. For light qualities, there were 1202, 1096, and 853 ESTs induced at least twofold by red, blue, and far-red light, respectively, whereas 950, 616, and 339 ESTs were repressed twofold or more by red, blue, and far-red light, respectively.
Table 3.
Summary of the Number of ESTs Induced or Repressed Twofold or More by Light Qualities, Light/Dark Transitions, and Individual Photoreceptors
| Number of ESTs Induced by Light or Photoreceptor
|
Number of ESTs Repressed by Light or Photoreceptor
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimentsa | Fold >30 | 30 to 20 | 20 to 10 | 10 to 5 | 5 to 3 | 3 to 2 | Total | >30 | 30 to 20 | 20 to 10 | 10 to 5 | 5 to 3 | 3 to 2 | Total |
| WT/W versus WT/D | 3 | 24 | 271 | 428 | 467 | 515 | 1708 | 1 | 3 | 26 | 153 | 373 | 712 | 1268 |
| WT/FR versus WT/D | 0 | 0 | 26 | 121 | 256 | 450 | 853 | 0 | 0 | 0 | 8 | 44 | 287 | 339 |
| WT/R versus WT/D | 3 | 11 | 85 | 277 | 320 | 506 | 1202 | 0 | 1 | 3 | 51 | 249 | 646 | 950 |
| WT/B versus WT/D | 0 | 1 | 71 | 305 | 247 | 472 | 1096 | 0 | 0 | 1 | 40 | 141 | 434 | 616 |
| WT/W versus WT/36D | 0 | 0 | 2 | 148 | 308 | 443 | 901 | 1 | 0 | 5 | 20 | 96 | 399 | 521 |
| WT/36W versus WT/D | 0 | 5 | 68 | 299 | 359 | 578 | 1309 | 2 | 5 | 5 | 80 | 306 | 653 | 1051 |
| WTL/W versus WTL/36D | 1 | 1 | 1 | 74 | 241 | 417 | 735 | 2 | 4 | 10 | 83 | 209 | 472 | 780 |
| WT/FR versus phyA/FR | 0 | 0 | 0 | 12 | 202 | 522 | 736 | 0 | 0 | 1 | 35 | 92 | 359 | 487 |
| WT/R versus phyB/R | 0 | 0 | 0 | 0 | 5 | 97 | 102 | 0 | 0 | 0 | 0 | 8 | 64 | 72 |
| WT/B versus cry1 cry2/B | 0 | 2 | 3 | 185 | 252 | 434 | 876 | 0 | 0 | 2 | 7 | 84 | 290 | 383 |
| PhyAOE/FR versus WT/FR | 9 | 6 | 19 | 24 | 50 | 276 | 384 | 0 | 0 | 1 | 6 | 68 | 189 | 264 |
| PhyBOE/R versus WT/R | 0 | 0 | 0 | 1 | 5 | 66 | 72 | 0 | 0 | 0 | 0 | 7 | 77 | 84 |
| CRY1OE/B versus WT/B | 1 | 0 | 3 | 6 | 49 | 281 | 340 | 0 | 0 | 9 | 30 | 127 | 357 | 523 |
a WT, wild type; W, white light; D, darkness; FR, far-red light; R, red light; B, blue light; 36, 36-hr transition; L, 5-week-old leaves; phyA, phyA mutant; phyB, phyB mutant; cry1 cry2, cry1 and cry2 double mutants; PHYAOE, PHYA overexpression; PHYBOE, PHYB overexpression; CRY1OE, CRY1 overexpression.
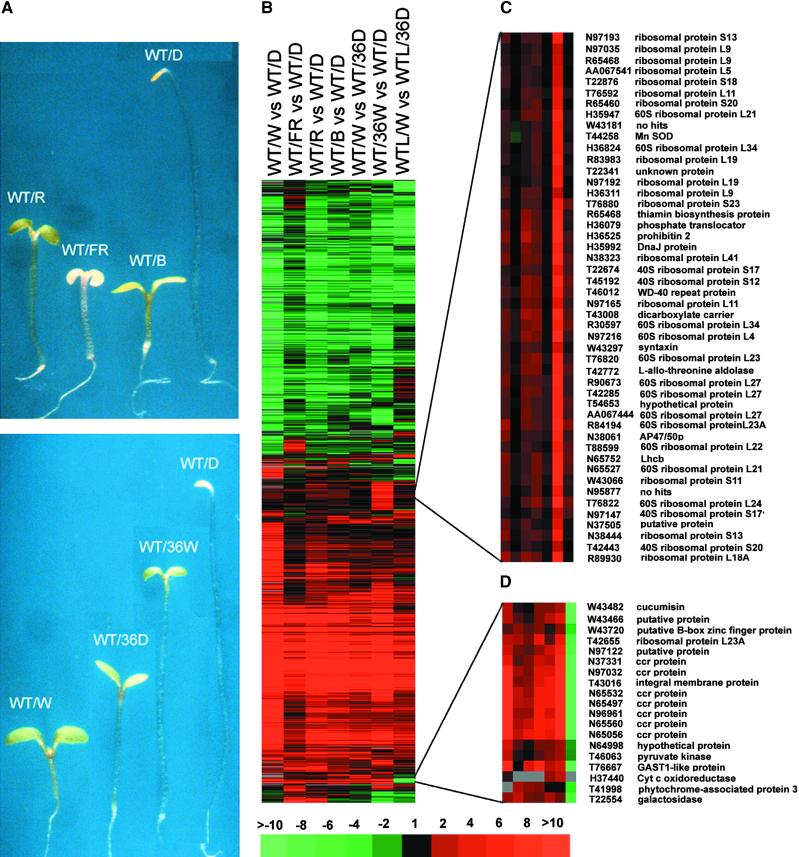
Photomorphogenesis involves a dramatic morphological change. Gene expression profiles from three distinct light/dark transitions were examined to determine how the profiles in light- and dark-grown seedlings were influenced by the light signal per se and by secondary effects resulting from morphological pattern changes (Figure 2A) caused by light (Table 3). The three light/dark transitions were (1) white light–grown seedlings (WT/W) subjected to 36 hr of dark adaptation (WT/36D); (2) dark-grown seedlings (WT/D) transferred to white light for 36 hr (WT/36W); and (3) 5-week-old white light–grown wild-type adult plant leaves (WTL/W) subjected to 36 hr of dark adaptation (WTL/36D). By using twofold change of gene expression as a threshold, the three light/dark transitions described above affected 94, 77, and 88%, respectively, of the total number of genes that exhibited twofold or greater upregulation comparing dark- and white light–grown seedlings (Table 3). Similarly, the dark-to-light transitions affected 88% (WT/W versus WT/36D), 65% (WT/36W versus WT/D), and 79% (WTL/W versus WTL/36D) of clones, respectively, that were downregulated by light comparing dark- and white light–grown seedlings (Table 3). Therefore, most of the differential gene expression patterns observed were caused by light per se, although the morphogenetic pattern change caused by light treatments also contributes to a small but variable extent.
Figure 2.
Representative Seedlings Grown under Defined Light Conditions and Cluster Analysis of Their Light-Regulated Gene Expression.
(A) Wild-type Arabidopsis seedlings grown under different light conditions. WT, wild-type seedlings; W, white light; FR, far-red light; R, red light; B, blue light; 36D, white light–grown seedlings or leaves transferred to darkness for 36 hr; 36W, dark-grown seedlings transferred to white light for 36 hr. All light conditions were continuous illumination. All seedlings were 6 days old at the time of harvesting.
(B) Hierarchical cluster display of expression ratios from wild-type seedlings grown under different light qualities versus dark-grown siblings and seedlings or leaves before and after light/dark transitions. Only those ESTs that exhibited at least twofold differential change in at least one sample pair among the seven pairs tested were included for comparison. There are 4326 EST entries included in the cluster. WTL, leaves of 5-week-old white light–grown wild-type seedlings; vs, versus.
(C) and (D) Two subclusters of ESTs showed distinct patterns of regulation for selected light conditions. SOD, superoxide dismutase; ccr, cytokinin repressed; Cyt, cytochrome.
Gene Expression Pattern Regulated by Light Qualities and Light/Dark Transitions
The gene expression profiles induced by different light qualities and light/dark transitions were compared further by means of cluster analysis (Eisen et al., 1998) in groups of ESTs with similar patterns of expression. To this end, only those ESTs with twofold or greater differential expression in at least one experimental condition were selected and analyzed. As shown in Figure 2B, there was remarkable similarity in gene expression patterns among varying light qualities (WT/W versus WT/D, WT/FR versus WT/D, WT/R versus WT/D, WT/B versus WT/D). The gene expression patterns also were quite similar among the three light/dark transitions (WT/W versus WT/36D, WT/36W versus WT/D, WTL/W versus WTL/36D). Among the ESTs analyzed in Figure 2B, an additional 56 and 123 ESTs exhibited similar upregulation or downregulation in all light conditions, even above the twofold threshold in at least one light condition, but they fell just below the twofold threshold in the white light– and dark-grown seedling comparison (Figure 2 and Table 3). If this group of genes had been considered, the percentage of ESTs regulated similarly by all light conditions would have been 34%, with 1764 upregulated and 1391 downregulated by light. It is evident that the twofold threshold for defining an EST as light regulated may be too stringent. Thus, the number of light-regulated genes in the genome actually may be higher.
Although the overall expression patterns were very similar, a small number of genes were regulated distinctly in different light conditions. The most obvious examples are seen in the far-red light treatment and the adult plant leaves under light/dark transition. Up to 15 and 12% of the ESTs in cluster analysis (Figure 2B) showed qualitatively distinct expression patterns under these conditions, respectively (see supplemental materials at http://plantgenomics.biology.yale.edu/). For example, some genes were upregulated in adult leaves but downregulated in seedlings in all light conditions, whereas other genes were downregulated in adult leaves but upregulated in seedlings by light. On the other hand, some genes were upregulated by far-red light but downregulated by white, red, and blue light. To further illustrate this notion, two subclusters of ESTs with contrasting expression patterns in different light conditions are shown in Figures 2C and 2D. In Figure 2C, a group of genes are particularly highly induced only during the transition of dark-grown seedlings to white light. It is interesting that most of the genes in this group are cytoplasmic ribosomal proteins, indicating a large demand for cytoplasmic protein translation during this particular dark/light transition. Figure 2D shows that a group of genes, including cytokinin repressed proteins, were upregulated in seedlings and downregulated in adult leaves by light.
Role of Phytochrome A in Mediating Far-Red Light Regulation of Gene Expression
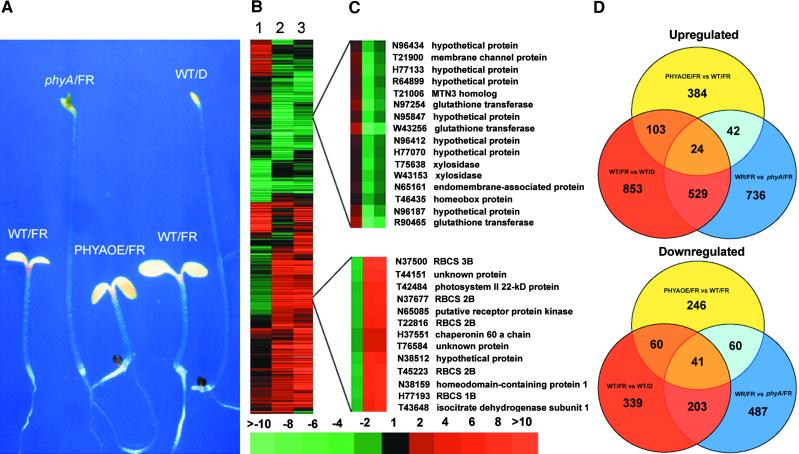
To define the role of phytochrome A in mediating the far-red light regulation of genome expression, we generated gene expression profiles of wild-type, phyA null mutant (Whitelam et al., 1993), and phyA overexpressor (Boylan and Quail, 1991) seedlings that were grown in far-red light (Figure 3A). The expression profiles were compared with those of the far-red-light– and dark-grown wild-type seedlings and subjected to cluster analysis (Figure 3B). The gene expression profiles in far-red-light–grown wild-type seedlings (Figure 3B, lane 3, WT/FR versus WT/D) are remarkably similar to those of phytochrome A–mediated gene expression under far-red light (Figure 3B, lane 2, WT/FR versus phyA/FR). It is clear that phytochrome A plays a major role in the far-red light regulation of gene expression, because the absence of phytochrome A abolishes most of the far-red-light–triggered gene expression pattern changes (Figure 3A). Indeed, when comparing the number of ESTs that displayed more than a twofold change in expression (Figure 3D), ∼62 and 60% of clones that were far-red light upregulated and downregulated, respectively, also were included in the set of phytochrome A null mutant affected ESTs (Figure 3D). This imperfect overlapping of the far-red-light– and phytochrome A–regulated genes in Figure 3D largely reflects quantitative differences in differential gene expression (Figure 3B). In general, our conclusion is consistent with a recently reported study of phyA's role in far-red light regulation of gene expression using distinct gene chip technology (Tepperman et al., 2001).
Figure 3.
Developmental Characteristics of Far-Red-Light–Grown Seedlings and Cluster Analysis of Phytochrome A–Mediated Far-Red Light Regulation of Gene Expression in Arabidopsis Seedlings.
(A) Phenotypic characteristics of 6-day-old wild type (WT), phyA mutant, and phytochrome A overexpression line (PHYAOE) grown under far-red light (FR). A 6-day-old dark-grown seedling (WT/D) was included for comparison. Wild types of both Landsberg erecta and No-O ecotypes were used for direct comparison with the phyA mutant and PHYAOE seedlings in their respective ecotypes.
(B) Overview of the hierarchical cluster display. Lane 1, expression ratios of far-red-light–grown phytochrome A overexpression and wild-type seedlings (PHYAOE/FR versus WT/FR). Lane 2, expression ratios of far-red-light–grown wild-type and phyA mutant seedlings (WT/FR versus phyA/FR). Lane 3, expression ratios of far-red-light– and dark-grown wild-type seedlings (WT/FR versus WT/D). A total of 2062 ESTs that had at least twofold differential expression in one of the three sample pairs were included in the cluster.
(C) Two sample subclusters of ESTs displaying antagonistic regulation by phytochrome A overexpression and phytochrome A–mediated far-red light regulation in wild type.
(D) Interloping diagrams of the number of differentially expressed EST clones that exhibited twofold or greater upregulation (induction) or downregulation (repression) for each of the three sample pairs. The numbers in the overlapping areas indicate the shared number of EST clones that exhibited twofold or greater differential expression in either two or three sample pairs. vs, versus.
Surprisingly, overexpression of phytochrome A in seedlings does not necessarily result in the enhancement of phytochrome A–mediated far-red light upregulation or downregulation of gene expression (Figure 3B). Examination of the expression of ESTs downregulated by far-red light shows that overexpression of phytochrome A enhanced their downregulation by far-red light in ∼40% of cases, caused upregulation by far-red light in another 40% of cases, and exhibited minimal effect in the remaining 20% of cases. A parallel effect was observed when the expression of ESTs upregulated by far-red light was examined. Two subclusters of genes in which phytochrome A overexpression antagonistically affected far-red light regulation are shown in Figure 3C. The antagonistic effects of phytochrome A overexpression on the far-red-light–regulated genes are largely responsible for the low degree of overlap with the far-red-light–regulated ESTs in wild type (Figure 3D), because those ESTs exhibited twofold or greater differential expression by phytochrome A overexpression.
It is worth noting that phytochrome A overexpression resulted in a dramatic enhancement of far-red light inducibility for some genes. For example, there were nine EST clones whose transcripts were induced more than 30-fold by phytochrome A overexpression relative to the wild-type seedlings in far-red light (Table 3 and supplemental materials at http://plantgenomics.biology.yale.edu/).
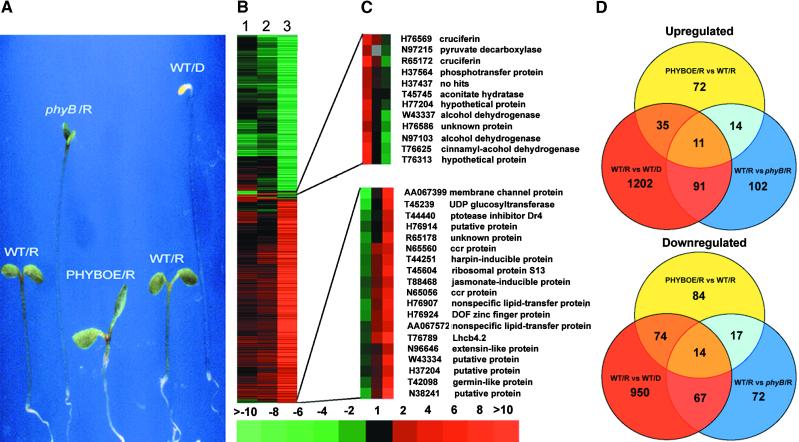
Role of Phytochrome B in Mediating Red Light Regulation of Gene Expression
To define the role of phytochrome B in red light–dependent gene expression, we performed a similar analysis of the changes in gene expression patterns in a phyB null mutant and an overexpression strain. As expected (Figure 4A), compared with wild-type seedlings, the phyB null mutant seedlings displayed a long hypocotyl and small cotyledons in red light (Koornneef et al., 1980; Reed et al., 1993), whereas the seedlings that overexpressed phytochrome B (PHYBOE) exhibited exaggerated photomorphogenic development (Wagner et al., 1991). Interestingly, cluster analysis indicated that although phytochrome B plays a role in mediating the vast majority (>90%) of red light–regulated gene expression (Figure 4B), the extent to which the null phyB mutation affects the regulation is quite limited. The results show that the phyB mutation affects only 102 and 72 ESTs upregulated or downregulated, respectively, by at least twofold in red light (Figure 4D). Essentially all of these genes are subject to red light regulation in wild-type seedlings. For the majority of red light–regulated genes, the phyB mutation only reduced the extent of the red light regulation (Figure 4B). This seemingly limited effect of phytochrome B on the red light regulation of gene expression is consistent with the fact that other phytochromes, particularly phytochrome D, play a redundant role in red light (Deng and Quail, 1999).
Figure 4.
Developmental Characteristics of Red Light–Grown Seedlings and Cluster Analysis of Phytochrome B–Mediated Red Light Regulation of Gene Expression in Arabidopsis Seedlings.
(A) Phenotypic characteristics of 6-day-old wild type (WT), phyB mutant, and phytochrome B overexpression line (PHYBOE) grown under red light (R). A 6-day-old dark-grown seedling (WT/D) was included for comparison. Wild types of both Landsberg erecta and No-O ecotypes were used for direct comparison with the phyB mutant and PHYBOE seedlings in their respective ecotypes.
(B) Overview of the hierarchical cluster display. Lane 1, expression ratios of red light–grown phytochrome B overexpression and wild-type seedlings (PHYBOE/R versus WT/R). Lane 2, expression ratios of red light–grown wild-type and phyB mutant seedlings (WT/R versus phyB/R). Lane 3, expression ratios of red-light– and dark-grown wild-type seedlings (WT/R versus WT/D). A total of 2204 EST entries that had at least twofold differential expression in one of the three sample pairs were included in the cluster.
(C) Two sample subclusters of ESTs displaying antagonistic regulation by phytochrome B overexpression and phytochrome B–mediated red light regulation in wild type. ccr, cytokinin repressed.
(D) Interloping diagrams of the number of differentially expressed EST clones that exhibited twofold or greater upregulation (induction) or downregulation (repression) for each of the three sample pairs. The numbers in the overlapping areas indicate the shared number of EST clones that exhibited twofold or greater differential expression in either two or three sample pairs. vs, versus.
For the vast majority of the ESTs whose expression was regulated by red light, phytochrome B overexpression either enhanced the red light regulation or had minimal effect. However, the qualitative degree of this phytochrome B overexpression effect on each EST was relatively small, with only 72 and 84 ESTs exceeding the twofold threshold of change (Figure 4D). Among the 156 ESTs whose expression was strongly affected by phytochrome B overexpression, 109 and 14 ESTs were regulated in the same or the opposite manner, respectively, by red light in wild type. On the other hand, 33 ESTs displayed little or no red light regulation in wild type. Two representative groups of these genes whose red light regulation were strongly affected by phytochrome B overexpression are illustrated in Figure 4C as examples. The top subcluster of 12 ESTs displayed an antagonistic effect as a result of phytochrome B overexpression relative to normal red light downregulation in wild type, whereas the bottom subcluster of 22 ESTs exhibited an antagonistic effect as a result of phytochrome B overexpression relative to normal red light upregulation in wild type.
Role of Cryptochromes in Mediating Blue Light Regulation of Gene Expression
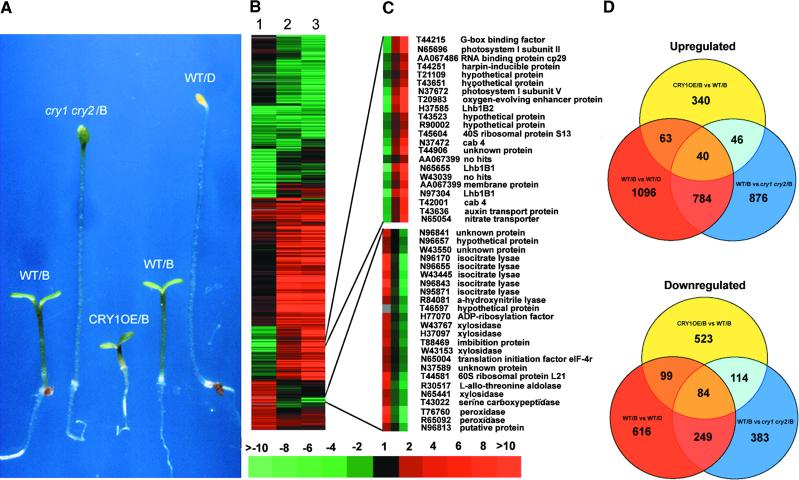
To define the role of the cryptochromes (CRY1 and CRY2) in mediating blue light–regulated gene expression, we examined the effect of the cry1 cry2 double mutation and CRY1 overexpression on gene expression profiles in blue light. As shown in Figure 5A and consistent with previous reports, the double mutant seedlings displayed diminished sensitivity to blue light (Mockler et al., 1999), whereas the overexpression of CRY1 enhanced photomorphogenic development under blue light conditions (Lin et al., 1996). As shown in Figure 5B, a comparison of the blue light–regulated gene expression profiles in wild type (lane 3, WT/B versus WT/D) and the blue light effect attributed to the two cryptochromes (lane 2, WT/B versus cry1 cry2/B) revealed largely similar patterns. Examination of those ESTs that exhibited twofold or greater differential expression (Figure 5D) showed a large overlap between the blue light effect in wild type and the cryptochrome-mediated effect. These data indicate that the two partially redundant cryptochromes are the photoreceptors responsible for mediating the blue light effect on gene expression.
Figure 5.
Developmental Characteristics of Blue Light–Grown Seedlings and Cluster Analysis of Cryptochrome-Mediated Blue Light Regulation of Gene Expression in Arabidopsis Seedlings.
(A) Phenotypic characteristics of 6-day-old wild type (WT), cry1 cry2 double mutant, and cryptochrome 1 overexpression line (CRY1OE) grown under blue light (B). A 6-day-old dark-grown seedling (WT/D) was included for comparison. Wild types of both Columbia and Wassilewskija ecotypes were used for direct comparison with the cry1 cry2 double mutant and CRY1OE seedlings in their respective ecotypes.
(B) Overview of the hierarchical cluster display. Lane 1, expression ratios of blue light–grown cryptochrome 1 overexpression and wild-type seedlings (CRY1OE/B versus WT/B). Lane 2, expression ratios of blue light–grown wild-type and cry1 cry2 double mutant seedlings (WT/B versus cry1 cry2/B). Lane 3, expression ratios of blue light– and dark-grown wild-type seedlings (WT/B versus WT/D). A total of 2444 EST entries that had at least twofold differential expression in one of the three sample pairs were included in the cluster.
(C) Two sample subclusters of ESTs displaying antagonistic regulation by cryptochrome 1 overexpression and cryptochrome-mediated blue light regulation in wild type.
(D) Interloping diagrams of the number of differentially expressed EST clones that exhibited twofold or greater upregulation (induction) or downregulation (repression) for each of the three sample pairs. The numbers in the overlapping areas indicate the shared number of EST clones that exhibited twofold or greater differential expression in either two or three sample pairs. vs, versus.
As shown in Figure 5B, CRY1 overexpression (CRY1OE) affected the blue light regulation of EST expression in three different manners. For ∼75% of the ESTs, CRY1 overexpression had either a negligible effect or resulted in enhanced blue light regulation. In contrast, CRY1 overexpression antagonistically affected the blue light–induced expression of 18% of ESTs. Finally, CRY1 overexpression resulted in the upregulation of expression of 7% of the ESTs, whereas EST expression levels were affected marginally or not affected by blue light in wild type. Two sample subclusters of those ESTs with expression levels responding antagonistically to CRY1 overexpression are illustrated in Figure 5C. Like phyA overexpression, the overlap is relatively low between the ESTs whose expression was regulated by more than twofold in the three sets of experiments (Figure 5D).
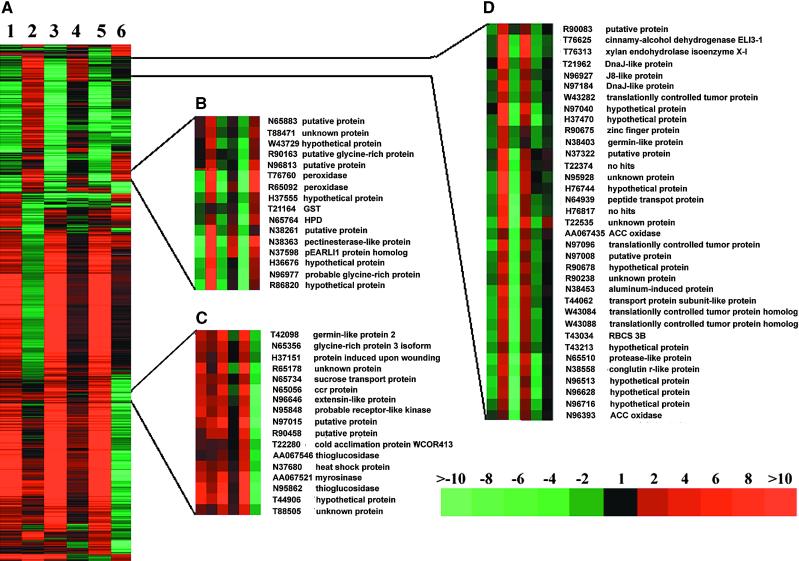
Distinct Target Gene Groups Are Affected Antagonistically by Overexpression of Individual Photoreceptors
Although overexpression of phytochrome A, phytochrome B, and cryptochrome 1 resulted in antagonistic effects on some light-regulated gene expression under specific light qualities (Figures 3 to 5), there are great differences in the number of ESTs they affected. To further examine the nature of the antagonistic regulation, we set a differential value of 2 for the ratios of the light-regulated expression changes and photoreceptor overexpression–dependent expression changes and used this as a cutoff definition for antagonistically affected ESTs. All of the ESTs that exhibited an-tagonistic effects on light-regulated expression by the overexpression of at least one photoreceptor according to this standard were subjected to cluster analysis (Figure 6). It is important to note that the ESTs that exhibited an an-tagonistic effect from the overexpression of the three photoreceptors were largely nonoverlapping (Figure 6). For example, ESTs for only eight genes were downregulated by distinct light quality in wild type and were regulated antagonistically by overexpression of phytochromes A and B and cryptochrome 1 under the corresponding light quality. There were 42, 22, and 14 ESTs that showed antagonistic regulation and that were shared by overexpressing phytochrome A and phytochrome B, phytochrome A and cryptochrome 1, and phytochrome B and cryptochrome 1, respectively. Overexpression of phytochrome A, phytochrome B, and cryptochrome 1 in their respective light qualities resulted in antagonistic effects on 477, 38, and 400 distinct ESTs, respectively. For example, overexpression of CRY1 resulted in an antagonistic effect on blue light upregulation of all EST clones for CAB genes, whereas overexpression of phytochrome A resulted in an antagonistic effect on the far-red upregulation of all ESTs for rbcS genes.
Figure 6.
Cluster Analysis of ESTs Displaying Antagonistic Effects by Overexpression of Individual Photoreceptors.
(A) Overview of the hierarchical cluster display. Lane 1, expression ratios of far-red-light– and dark-grown wild-type seedlings (WT/FR versus WT/D). Lane 2, expression ratios of far-red-light–grown phytochrome A overexpression and wild-type seedlings (PHYAOE/FR versus WT/FR). Lane 3, expression ratios of red light– and dark-grown wild-type seedlings (WT/R versus WT/D). Lane 4, expression ratios of red light–grown phytochrome B overexpression and wild-type seedlings (PHYBOE/R versus WT/R). Lane 5, expression ratios of blue light– and dark-grown wild-type seedlings (WT/B versus WT/D). Lane 6, expression ratios of blue light-grown cryptochrome 1 overexpression and wild-type seedlings (CRY1OE/B versus WT/B). A total of 977 ESTs that had at least twofold expression changes in both light-regulated expression and antagonistic effect in a photoreceptor overexpression line were included in the cluster.
(B), (C), and (D) show three representative subclusters that contain ESTs displaying distinct patterns of antagonistic effects by different photoreceptor overexpression. The accession numbers and gene identities are listed. Subcluster B contains ESTs that displayed downregulation in all three light quality conditions and antagonist effects by overexpression of each of the three photoreceptors. Subcluster C shows ESTs that were upregulated in all three light quality conditions but were positively or antagonistically regulated by overexpression of phytochrome A and cryptochrome 1. The effects of phytochrome B on this group of ESTs were either nonantagonistic or antagonistic. Subcluster D shows ESTs that were downregulated by three light quality conditions and that were antagonistically affected by overexpression of both phytochrome A and phytochrome B but that were minimally affected by cryptochrome 1 overexpression. GST, glutathione S-transferase; ccr, cytokinin repressed; ACC, 1-aminocyclopropane-1-carboxylic acid; HPD, 4-hydroxyphenypyruvatev dioxygenase.
The variable effects of photoreceptor overexpression may reflect the difference in sensitivity of individual genes in response to distinct light quality regulation. For instance, some genes already were maximally induced by the responsible photoreceptors in wild type, whereas others still were able to respond to stronger signals in the overexpression seedlings. For the antagonistically regulated genes, it can be assumed that the genes already were maximally induced under our experimental light conditions in wild type. Overdose of light signaling as a result of photoreceptor overexpression may cause a reduction from the maximal degree of light regulation, thus resulting in the antagonistic effect observed. In addition to this hypothesis of “differential gene sensitivity to light signaling dosage,” there are two other mutually nonexclusive possibilities. One is that the ectopic photoreceptor overexpression by the 35S promoter used in these experiments resulted in novel light regulation of gene expression. The other is that for some genes, excess light signals caused by the overabundance of photoreceptors may mimic light stress responses under high intensity light environments. If the latter were the case, it would suggest that stress responses under different light qualities are quite distinct with regard to their effects on target gene expression.
Functional Classification of Light-Regulated Genes
The gene expression profiling described above suggested that the expression patterns exhibited under different light qualities that are mediated by distinct photoreceptors and during multiple light/dark transitions are largely similar except for a small fraction of the ESTs (Figures 2B, 3B, 4B, and 5B) that display characteristic gene expression patterns. Therefore, it is possible to examine the cellular pathways and processes that these ESTs, which share common light-regulated patterns, represent. Because the white light– and dark-grown seedlings exhibited the strongest light effects in gene expression changes, we focus our attention on the one-third of the 9216 ESTs that displayed twofold or greater differential expression in this light treatment pair. This group of ESTs represents ∼2000 genes, and 400 (20%) of them can be assigned a cellular or biochemical function based on their identity or strong sequence homology with known genes. The ∼400 genes represent a wide spectrum of cellular and biochemical functions, ranging from DNA replication to transcription, metabolism, protein degradation, plant defense, and developmental regulation (Table 4 and supplemental materials at http://plantgenomics.biology.yale.edu/).
Table 4.
Number of Clones Involved in Different Functional Groups Upregulated or Downregulated at Least Two-Fold by White Light
| Pathways | Gene Number |
Representative Gene Namesa |
|---|---|---|
| Upregulated | ||
| Photosynthetic light reactions |
56 | 17 CAB genes, 15 PSI proteins, 14 PSII proteins, 10 proteins involved in electron transport and ATP synthesis |
| Photosynthetic carbon metabolism |
25 | 4 RBCS genes, 2 phosphoglyceate kinases, 2 GAPDHs, triose-phosphate isomerase, aldolase, fructose-1,6-phosphate phosphatase, 3 fructose-1,6-bisphosphate aldolases, transketolase, sedoheptulose 1,7-bisphosphate phosphatase, ribulose-5-phosphate epimerase, 2 ribose-5-phosphate isomerases, ribulose-5-phosphate kinase, ferredoxin, 2 thioredoxins, 2 ferredoxin-thioredoxin reductases, Rubisco activase |
| Starch synthesis | 3 | Fructose-1,6-bisphosphate aldolase, fructose-1,6-bisphosphate phosphatase, pyrophosphorylase |
| Sucrose synthesis | 5 | Phosphate/triose phosphate translocator, cytosolic triose-phosphate isomerase, cytosolic fructose-1,6-bisphosphate aldolase, phosphofructokinase, sucrose synthase |
| Photorespiration | 8 | 4 RBCS genes, glycolate oxidase, catalase, hydroxypyruvate reductase, glycine decarboxylase |
| Glycolysis | 5 | Fructokinase, triose phosphate isomerase, pyruvate kinase, pyruvate decarboxylase, alcohol dehydrogenase |
| Trichloroacetic acid cycle | 5 | Pyruvate dehydrogenase, citrate synthase, isocitrate dehydrogenase, phosphopyruvate hydratase, malate dehydrogenase |
| Cell wall synthesis and cell wall protein |
8 | Cellulose synthase, glucosyltransferase, β-1,2-xylosyltransferase, UDP-glucose:glycoprotein glucosyltransferase, cell wall–plasma membrane linker protein, proline-rich protein, extensin, AGP |
| Protein synthesis in chloroplast |
56 | 11 50S ribosomal proteins, 4 30S ribosomal proteins, 7 other ribosomal proteins, 9 initiation and elongation factors, 9 aminoacyl-tRNA synthetases, 6 RNA helicases, 10 chaperonines |
| Protein synthesis in cytoplasm |
60 | 15 60S ribosomal proteins, 3 40S ribosomal proteins, 8 other ribosomal proteins, 9 initiation and elongation factors, 9 aminoacyl-tRNA synthetases, 6 RNA helicases, 10 chaperonins |
| Phenylpropanoid biosynthesis |
8 | PAL1, CHS, caffeoyl-CoA O-methyltransferase, cinnamyl-alcohol dehydrogenase, O-methyltransferase, GST, glutathione-conjugate transporters, cytochrome P450 |
| Amino acid synthesis pathways |
15 | Dehydrogenase, glutathione peroxidase, Δ-1-pyrroline-5-carboxylate synthetase, phosphoribosylglycinamide synthetase, histidinol dehydrogenase, acetolactate synthase, aminotransferase, adenine phosphoribosylanthranilate transferase, phosphoribosylanthranilate transferase, uridylyl transferase, glutamine synthetase, glutamate/ornithine acetyltransferase, 3-isopropylmalate dehydratase, glutathione reductase, tryptophan synthase |
| Chlorophyll and heme synthesis |
4 | Ferrochelatase, glutamyl-tRNA reductase, protoporphyrinogen IX oxidase, glutamate-1-semialdehyde 2,1-aminomutase 1 |
| Transcription factors | 15 | G-box binding factor, B-box zinc finger, 2 bHLH transcription factors, 3 MYB-related transcription factors, SCARECROW-like factor, zinc finger transcription factor–like protein, AP2 domain transcription factor-like protein, bZIP transcription factor, transcription factor HUA2, transcription factor II homolog, homeobox leucine zipper protein, putative transcription factor |
| Uniquitin-proteasome pathway |
23 | 4 polyuniquitin and ubiquitin genes, 9 26S proteasome subunits, 2 proteasome subunit–related proteins, E1, E2, and 6 E3 genes |
| Downregulated | ||
| Ethylene biosynthesis | 3 | ACC synthase, ACC oxidase, SAM synthase |
| BR biosynthesis | 4 | 24-sterol C-methyltransferase, C-8,7-sterol isomerase, 3-β-hydroxysteroid dehydrogenases, steroid sulfotransferase |
| Cell wall degradation and cell wall proteins |
26 | Cellulase, XET, 2 expansins, xylosidase, xyloglucan endo-1,4-β-d-glucanase, endo-1,4-β-d-glucanase, 1,3-β-glucanase, xylose isomerase, pectinesterase, pectate lyase, 7 peroxidases, 2 extensin homologs, glycine-rich protein, AGP4, 2 proline-rich cell wall proteins, hydroxyproline-rich protein, pollen surface protein homolog |
| Water transport across tonoplast |
5 | 5 aquaporins in tonoplast |
| Water transport across plasma membrane |
5 | 5 water channel and membrane intrinsic proteins in plasma membrane |
| Sulfur assimilation | 9 | 2 GSTs, sulfate adenylyltransferase, sulfate transporter, phytochelatin synthase, cysteine synthase, cytochrome P450 monooxygenase, 2 ABC transporters |
| Nitrogen assimilation | 9 | 3 nitrate transporters, 2 AS proteins, glutamine synthetase, glutamate dehydrogenase, serine carboxypeptidase, cysteine synthase |
| Fatty acid oxidation | 5 | Glycerol-3-phosphate dehydrogenase, inorganic pyrophosphatase, ketoacyl-CoA thiolase, malic enzyme, acyl-CoA oxidase |
| Glyoxylate cycle | 4 | isocitrate lyase, citrate synthase, malate synthase, malate dehydrogenase |
| Transcription factors | 18 | 3 AP2 domain transcription factors, 2 MYB-related transcription factors, transcription factor L2, transcription factor tga1, transcription factor EREBP, MADS box transcription factor–like protein, TATA box binding protein, transcription factor–like protein, putative HLH DNA binding protein, homeobox leucine zipper proteins HAT4 and HAT5, homeodomain transcription factor–like protein, CONSTANS-like B box zinc finger protein–like protein, abscisic acid–responsive element binding factor, homeobox protein |
| Uniquitin-proteasome pathway |
16 | 6 polyuniquitin and ubiquitin genes, 2 26S proteasome subunits, 5 E2 and 3 E3 genes |
a PSI and PSII, photosystems I and II; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; GST, glutathione S-transferase; ACC, 1-aminocyclopropane-1-carboxylic acid; SAM, S-adenosyl methionine.
In assessing whether a cellular pathway is regulated coordinately by light, we set the criterion that all genes involved in a specific pathway must be included in the microarray and be regulated similarly. Furthermore, we defined a pathway as “light regulated” only if we found ESTs corresponding to at least three similarly regulated genes in the pathway. These strict criteria are essential because the EST collection used to fabricate the microarray has only slightly less than 70% with correct identity (see Methods); however, many light-regulated pathways will be missed. With this strict requirement, the data indicated that at least 15 and 11 pathways were coordinately upregulated or downregulated by light, respectively, in dark- and white light–grown seedlings (Table 4). Cluster analysis (Figures 2B, 3B, 4B, and 5B) of the genes in all of the 26 pathways demonstrated that they exhibited similar differential expression in other specific light treatments and were mediated by specific photoreceptors in distinct light quality conditions (see supplemental materials at http://plantgenomics.biology.yale.edu/). However, different pathways exhibited variable sensitivity to light signals of distinct qualities. For example, genes in most of the metabolic pathways shown in Table 4 were more sensitive to red and blue light than to far-red light, whereas genes involved in phenylpropanoid biosynthesis, chlorophyll/heme biosynthesis, amino acid biosynthesis, ethylene biosynthesis, brassinosteroids (BR) biosynthesis, and the glyoxylate cycle were more sensitive to far-red light than to red or blue light.
It is likely that there are many other pathways regulated by light that were not elucidated in this study. First, the microarray contains ∼6120 unique gene probes, slightly less than one-quarter of the Arabidopsis genome coding capacity. Second, the functions of the majority of the ESTs that define light-regulated genes are not known at this time. Third, many pathways that are represented by only one or two genes on the microarray were not included. Therefore, we are drastically underestimating the number of cellular processes that are regulated coordinately by light. This is reflected well by the fact that more than one-third of the ESTs demonstrated at least a twofold or greater differential expression between light- and dark-grown seedlings.
DISCUSSION
In this study, we systematically investigated the genome-wide expression changes during light-regulated Arabidopsis development by a newly constructed cDNA microarray. This approach allowed us to examine the light-regulated expression of ∼6120 genes simultaneously. Our conservative estimation indicates that one-third of the genes examined showed significant (twofold or greater) and reproducible differential expression between white light– and dark-grown seedlings (Figures 1D and 2B, Table 3). Because our study identified essentially all previously known light-regulated genes included in our microarray (Table 2), it suggests that our microarray data are highly reliable. To date, around 100 individual genes have been reported to be regulated by light using traditional approaches (Terzaghi and Cashmore, 1995; Fankhauser and Chory, 1997; Kuno and Furuya, 2000). Thus, our work has documented the vast extent of light regulation of gene expression in higher plants. It is interesting that apart from more than 1700 ESTs (∼1150 genes) that were upregulated by light, there are more than 1200 ESTs (∼800 genes) that were downregulated by light (Table 3). Because only a handful of light-downregulated genes have been reported in the literature to date, this greatly extends our current knowledge about genes downregulated by light and indicates that light-downregulated genes are almost as common as light-upregulated genes.
The degree of gene expression changes between light- and dark-grown seedlings provides direct support for the notion that distinct gene expression patterns define skotomorphogenesis and photomorphogenesis. Because the microarray contains only ∼6120 genes, approximately one-quarter of the total Arabidopsis genes (The Arabidopsis Genome Initiative, 2000), we extrapolated that there may be more than 8000 Arabidopsis genes whose expression is regulated by light. In addition, our results provide several novel insights regarding the role of light in the control of gene expression and plant developmental patterns.
Light Induces Characteristic Patterns of Genome Expression
Comparison of the gene expression profiles of seedlings grown under white, far-red, red, and blue light revealed a very similar pattern of gene expression for the vast majority of ESTs (Figure 2B). Thus, different qualities of light appear to trigger largely similar genome expression patterns, which is consistent with the fact that all light quality can induce photomorphogenic development. It has been well established that distinct photoreceptors are used by plants to sense distinct light signals. Thus, it is plausible that different wavelengths of light, perceived by distinct photoreceptors, are transduced to regulate the same developmental switch (i.e., to switch on the photomorphogenic gene expression pattern). Although the light treatments, both light qualities and light/dark transitions, used in this study are associated with photomorphogenic development to variable degrees, light-regulated gene expression patterns for the vast majority of genes are very similar qualitatively and independent of light-induced morphological changes. Therefore, it is logical to suggest that light-regulated gene expression patterns are not the result of light-induced morphological changes but a prerequisite for the light-induced developmental patterns.
Despite this finding, for most of the light treatments, there is a small fraction of genes whose expression seems to be distinct. It is possible that it is this small fraction of genes whose expression may be important to define the characteristic physiological and developmental patterns associated with each light condition. Our results also indicated that the effect of photoreceptor overexpression on light-regulated gene expression is rather complicated. Large numbers of the far-red-light– and blue light–regulated genes were affected antagonistically by phytochrome A or CRY1 overexpression. To fully understand these overexpression effects, it will be necessary to determine what portion of those effects are caused by ectopic expression of the photoreceptors and what portion are attributable to high dose photoreceptor signaling.
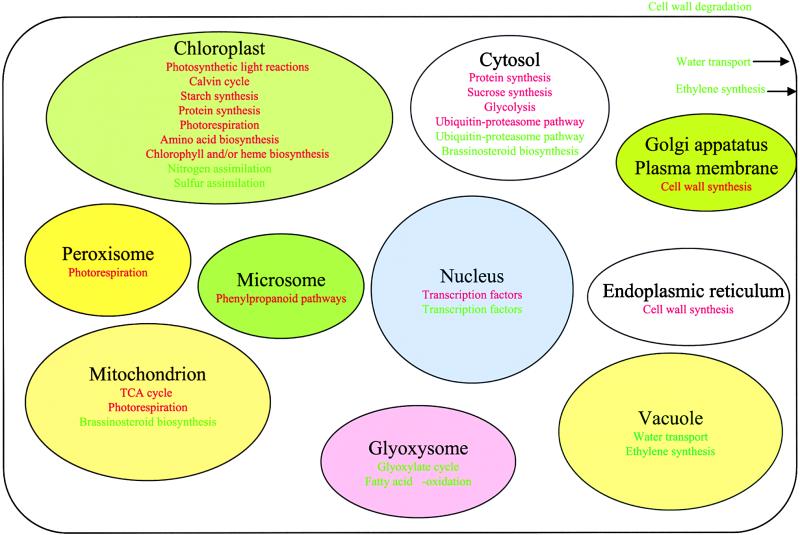
Many Cellular Pathways Are Regulated Cocoordinately by Light
Examination of the light-regulated genes revealed that at least 26 cellular pathways were regulated cocoordinately by light (Figure 7). Some of them were activated by light, whereas others were repressed by light. As expected, all photosynthetic genes, including light and dark reaction components and starch and sucrose biosynthesis pathways, were among the light-activated genes. In particular, the genes encoding light reaction components were among the strongest light-activated genes. Light-promoted photosynthesis results in the production and export of carbohydrates from the chloroplasts to the cytosol. The carbohydrates are further oxidized through glycolysis and the trichloroacetic acid cycle to provide energy and a carbon source for many biosynthetic pathways. Therefore, it is no surprise that both the glycolysis and the trichloroacetic acid cycle were upregulated by light. This is consistent with the light upregulation of the protein synthesis machinery both in the chloroplast and cytoplasm, the cell wall biosynthetic pathways, and the photorespiration pathway to protect plant organelles from light injury. Also, upregulation of phenylpropanoid biosynthetic pathways would be consistent with the production of many secondary metabolites, including photoprotective pigments to protect plant cells from light injury and lignins and lignans to strengthen the hypocotyls.
Figure 7.
Diagrammatic Summary of Cellular Metabolic and Regulatory Pathways Controlled by Light in Arabidopsis.
The cellular metabolic and regulatory pathways that are upregulated by light are listed in red type, whereas the cellular metabolic and regulatory pathways that are downregulated by light are listed in green type. Note that some pathways involve multiple cellular compartments. TCA, trichloroacetic acid.
Most of the cellular pathways that were downregulated also are consistent with the well-characterized developmental pattern. For example, when seedlings germinated in soil (darkness) reached the light, cell elongation was inhibited. This is likely achieved by light inhibition of the pathways responsible for cell wall loosening and degradation and water transport into vacuoles (see below for details). Reduction of cell wall loosening would help to build a strong hypocotyl to survive wind and pathogen attack in the open environment. Because all three key enzymes in ethylene biosynthesis were downregulated by light, the apical hook, which is controlled by ethylene (Kieber, 1997), was opened in the light. Also, because enough carbohydrates and energy were produced by photosynthesis and carbohydrate oxidation, glyoxylate cycle and fatty acid oxidation pathways that generate carbohydrate and energy from lipid storage in darkness (Eastmond and Graham, 2001) were repressed by light.
It has been shown that the energy for assimilation of nitrogen and sulfur comes from photosynthesis (Lam et al., 1996; Leustek et al., 2000). In this study, we verified that the genes involved in nitrogen and sulfur assimilation were downregulated by light. Interestingly, some of these genes also are under clock control and are expressed at the peak toward the end of the day (Harmer et al., 2000). The downregulation of nitrogen and sulfur assimilation by light suggested that the assimilation activities for carbon, nitrogen, and sulfur might be separated by the light/dark transition. Carbon assimilation takes place in the light (during daytime), whereas plants use the stored energy to assimilate nitrogen and sulfur in the dark (during nighttime).
In addition, we observed that the ubiquitin-proteasome protein degradation pathway also was regulated by light. Ubiquitin 5, some family members of the 26S proteasome subunits, and some E1, E2, and E3 (cullin, F-box protein, and Ring finger protein) proteins were upregulated by light, whereas ubiquitin 3 and 10, some other family members of the 26S proteasome subunits, and some E2 proteins were downregulated by light. Among the transcription factors, some were upregulated and others were downregulated by light.
It is worth mentioning that we have focused our discussion on genes involved in metabolism, primarily because these processes are the best understood in plants (Bevan et al., 1998, 2001). However, numerous genes with probable regulatory roles, such as protein kinases and phosphatases and cell skeleton proteins, also have been found to be regulated by light. In addition, large numbers of the light-regulated genes found in this experiment are completely uncharacterized. These light-regulated genes undoubtedly play important roles in pathways not mentioned here.
Brassinosteroid Biosynthesis Pathway Was Downregulated by Light
Brassinosteroids (BRs) are plant growth-promoting hormones found in pollen, seed, and young vegetative tissue throughout the plant kingdom (Schumacher and Chory, 2000). BRs have many effects on plants (e.g., cell elongation, bending, cell division, and reproductive and vascular development) (Clouse and Sasse, 1998). In Arabidopsis, BR mutants show a deetiolated (COP) phenotype in which dark-grown seedlings exhibit the short hypocotyls and open cotyledons characteristic of light-grown plants (Li et al., 1996; Klahre et al., 1998). It is also reported that treatment of dark-grown seedlings with a BR biosynthesis inhibitor induced characteristics of light-grown plants (Nagata et al., 2000). In our experiments, we verified that all four genes for the BR biosynthesis pathway included in our microarray (Table 4) were downregulated by light. Those results suggested that BR plays an important role in plant photomorphogenesis. High levels of BR are needed for skotomorphogenesis in the dark, whereas BR biosynthesis was inhibited by light, which contributes to overall photomorphogenesis in the light.
Key Enzymes Involved in Cell Wall Loosening Were Downregulated by Light
Light control of plant development is illustrated most dramatically by the inhibition of hypocotyl elongation and cotyledon expansion (von Arnim and Deng, 1996). Plant cells are enclosed by the cell wall, which defines the shape and size of cells. The elongation of hypocotyl cells is dependent on cell wall relaxation and expansion and water influx into plant vacuolar compartments (McCann and Roberts, 1994). The former is controlled by a series of cell wall–loosening/hydrolytic enzymes (Cosgrove, 2001), and the latter is controlled by the water channel protein aquaporin (Maurel and Chrispeels, 2001). In the present study, we found that the genes for more than nine types of cell wall hydrolytic enzymes were downregulated by light. Among them, three enzymes have been suggested to have possible wall-loosing activities (Nicol et al., 1998; Campbell and Braam, 1999; Cosgrove, 1999). The first is xyloglucan endotransglycosylase (XET; accession number N96608), which showed endolytic cleavage of the xyloglucan tethers in the cell wall (Campbell and Braam, 1999). The expression of this gene was downregulated more than 32-fold by white light compared with dark- and light-grown seedlings. The second is expansin (accession number R29778), which provides hydrogen bonds between cellulose and the load-bearing cross- link glycans (Cosgrove, 1999). The third is endo-1,4-β-d-glu-canase (accession numbers W43495 and W43496), which hydrolyze 1,4-β linkages adjacent to unsubstituted glucose residues (Nicol et al., 1998). Interestingly, the expression of both XET and endo-1,4-β-d-glucanase was induced by BR (Zurek and Clouse, 1994; Nicol et al., 1998). In addition, we also found that more than 15 different EST clones encoding tonoplastic aquaporin were downregulated by light. Almost all of these genes encoding hydrolytic enzymes and water channel proteins were inhibited by every quality of light and during dark/light transitions. For example, when dark-grown seedlings were transferred to light for 36 hr, XET gene expression was inhibited by 22.7-fold, whereas when dark-grown seedlings or adult plants were transferred to dark for 36 hr, XET gene expression activity was induced by 38.5- and 4.6-fold, respectively. These results substantiate a coordinated regulation of all pathways for the proper regulation of cell expansion in changing light environments.
METHODS
Plant Materials and Growth Conditions
The wild type used in light/dark transition was in the Arabidopsis thaliana Columbia ecotype. The photoreceptor single and double mutants used were phyA-1 in the Landsberg erecta ecotype (Whitelam et al., 1993), phyB-B064 in the Landsberg erecta ecotype (Koornneef et al., 1980; Reed et al., 1993), and cry1-304 cry2-1 in the Columbia ecotype (Mockler et al., 1999). Transgenic lines overexpressing the photoreceptors have been described for PHYA in the No-O ecotype (Boylan and Quail, 1991), PHYB in the NO-O ecotype (Wagner et al., 1991), and CRY1 in ecotype Wassilewskija (Lin et al., 1996). Surface sterilization and cold treatment of the seed were as described previously (Ang and Deng, 1994). Arabidopsis seedlings were grown on growth medium (GM) agar plates containing 1% sucrose. Unless stated otherwise, the seedlings were grown in continuous white, red, far-red, or blue light or darkness for 6 days. The white light intensity used was 152 μmol·m−2·sec−1. The colored light growth chambers (E-30LED2/3; Percival Scientific, Perry, IA) have intensities of 16.2 μmol·m−2·sec−1 for blue light, 108.5 μmol·m−2·sec−1 for red light, and 160.8 μmol·m−2· sec−1 for far-red light.
For all light shift experiments, seedlings were grown in continuous white light or darkness for 4.5 days. Seedlings then were transferred to the opposite light condition for 36 hr. After growth in 16 hr of light/8 hr of dark for 4 weeks, adult plants were transferred to continuous white light for 5 days before transfer to darkness for 36 hr.
Expressed Sequence Tag (EST) cDNA Clones and Microarray Preparation
The Arabidopsis 9.2K array (http://info.med.yale.edu/wmkeck/dna_arrays.htm) was generated from the first 9216 ESTs of the Arabidopsis Biological Resources Center's 11,521 Arabidopsis cDNA bacterial clone set, which was provided originally by Michigan State University. A comparison of those EST accession numbers with The Institute for Genomic Research (http://www.tigr.org) unique EST database revealed that 9079 of our 9216 EST clones define 6036 unique gene accessions. The rest, 137 EST sequences, have no Institute for Genomic Research match and were estimated to represent 90 unique gene accessions (2:3 ratio). Thus, we estimate that we have ∼6120 unique genes represented in our microarray. A sample of more than 200 EST polymerase chain reaction (PCR) products generated from the clones were chosen randomly and sequenced, and ∼70% of the EST sequences match the expected identities. In addition, the plasmid DNA from which 90 of the PCR products were generated was sequenced and exhibited 80% match to the predicted EST sequence. These data are consistent with the notion that up to 10% of the 30% mismatch in PCR products may be the result of heterogeneity introduced at the level of the PCR, and the remaining 20% may be caused by plasmid heterogeneity. Approximately 70% of the ESTs sequenced as both PCR product and plasmid DNA showed perfect sequence alignment (see supplemental materials at http://info.med.yale.edu/wmkeck/dna_arrays.htm).
The PCR products were generated from 96-well plasmid DNA using the following primer pairs: forward primer M13-20, 5′-GTAAAA-CGACGGCCAGT-3′; reverse primer M13, 5′-GGAAACAGCTATGACCATG-3′. A simple ethanol/isopropanol precipitation was used for PCR product cleanup. A sample of each cleaned up product was run on an agarose gel for quality control before arraying. Seventy-five percent of the PCR products were single bands, 22% had multiple bands, and 13% of the products showed lower than average PCR product concentration. The PCR failure rate was 2%. The products were resuspended in 100 μL of 3 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), and 5 μL was transferred using a Robbins Hydra microdispenser (Sunnyvale, CA) with Twister-H from 96-well plates to printing source plates (384-well plates; Genetix V-bottom, flat). The 384-well print plate sets were dried with a Speed-Vac (Savant Instruments, Holbrook, NY), closed with aluminum foil adhesive covers, and stored until ready to print, at which time the PCR products were resuspended in 5 μL of distilled H2O. cDNA arrays were printed on poly-l-lysine–coated glass microscope slides made at the Keck DNA Microarray Resource exactly according to P. Brown's protocol (see MGuide Protocols at http://cmgm.stanford.edu/pbrown/protocols/index.html). The Arabidopsis 9.2K array was printed with a GeneMachines Omnigrid arrayer using 16 Major Precision split pins. The array spots are 175 μm center-to-center distance, and spots average 125 μm in diameter. After printing, the arrays were rehydrated over water vapor until they just glistened and were then snap dried on an inverted heat block. They were then cross-linked under UV light in a Stratalinker (Stratagene). Free amine groups on the slide were blocked by treatment with succinic anhydride. The slides then were dipped in 95% ethanol for 2 min and spun dry. The control genes, including green fluorescent protein, globin, luciferase, kanamycin/neomycin phosphotransferase (nptII), B-cell receptor, insulin-like growth factor, tyrosine phosphatase, and PHI-X individual fragment, were printed on the top of subarrays 1 and 2. For details, see http://info.med.yale.edu/wmkeck/dna_arrays.htm.
RNA Preparation and Fluorescent Labeling of Probe
Total RNA was extracted from the whole seedlings or leaves of adult plants using the Qiagen RNeasy Plant Mini Prep kit. At least two independent RNA preparations for each biological sample were made and used for probe synthesis. The RNA was labeled by direct incorporation of Cy-3– or Cy-5–conjugated deoxy UTP (Amersham Pharmacia Biotech, Piscataway, NJ) during reverse transcription as follows: 100 μg of total RNA and 1 ng of each control RNA was combined with 4 μg of anchored oligo(dT) 19-mer and 5 μg of random hexamers, heated to 65°C for 5 min, and then cooled on ice. To this mixture, the remaining components were added to obtain the following reaction mixture in a total volume of 42 μL: 1 × Superscript II reverse transcriptase buffer (Life Technologies, Grand Island, NY), 2 μL of Superscript II reverse transcriptase, 1 μL of RNAsin (Promega, Madison, WI), 100 μM Cy-3 or Cy-5 deoxy UTP, 500 μM each deoxy ATP, deoxy CTP, and deoxy GTP, and 200 μM deoxy thymine triphosphate (TTP). After a 60-min incubation at 42°C, 2 μL of Superscript II reverse transcriptase was added again to the mixture and incubated at 42°C for another 120 min. The reaction was stopped by adding 5 μL of 0.5 M EDTA and incubating at 94°C for 3 min, and RNA was hydrolyzed by adding 10 μL of 1 M NaOH and incubating at 65°C for 20 min. This reaction was neutralized by adding 6 μL of 1 M HCl and 2 μL of 1 M HCl-Tris, pH 7.5. The labeled cDNA was purified from the unincorporated dye molecules by adding 400 μL of water and spinning through a Microcon YM-30 filter (Millipore, Bedford, MA) for 7 min at 11,000g, after which it was washed again two times. The purified, labeled probe was concentrated to a final volume of 7 μL.
Hybridization, Washing, and Scanning
For hybridization to the Arabidopsis array, we combined the labeled probe with 1 μL of 20 × SSPE (1 × SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 1 μL of blocking solution, and 16 μL of hybridization solution to a final volume of 25 μL. This mixture was incubated at 90°C for 2 min, cooled on ice, applied to the prehybridized microarray, covered with a cover slip, and incubated in a GeneMachines hybridization chamber at 42°C air for 16 to 20 hr. After 16 to 20 hr of incubation, the array was washed with 2 × SSC and 0.1% SDS, 0.2 × SSC and 0.1% SDS, 0.2 × SSC, and 0.02 × SSC for 10 min at room temperature. After washing, the slide was dried by spinning at 1100g for 5 min.
Hybridized microarray slides were scanned at 532 nm (Cy3) and 635 nm (Cy5) wavelengths with an Axon GenePix 4000A scanner (Foster City, CA) at 10-nm resolution, generating two separate TIFF images. Photomultiplier tube (PMT) voltages were adjusted manually to minimize background and reduce the percentage of spots on the array with saturated signal values. The normalization of the two channels with respect to signal intensity also was achieved by adjusting the PMT voltage settings. We chose the PMT voltages that resulted in a signal ratio of Cy3:Cy5 for the majority of control genes as close to 1.0 as possible.
Data Analysis
Spot intensities were quantified using Axon GenePix Pro 3 image analysis software. The channel ratios were measured using the GenePix Pro 3 median of ratio method, and they were normalized using the corresponding GenePix default normalization factor. To merge the replicated GenePix Pro 3 output data files (.gpr files) in a reasonable way, we developed a computer program called GPMERGE (http://bioinformatics.med.yale.edu/software.html). With this program, we pooled four replicated data sets of each experiment. A number of quality control procedures were conducted before data points from the four replicates of two independent biological sample sets were averaged. First, all spots flagged Bad or Not Found by GenePix software were removed from the final data analysis. Second, a very simple outlier searching algorithm was incorporated into GPMERGE that defined as outliers and eliminated from the analysis those spots that exhibited a large difference between the ratio mean and the ratio median. Third, only those spots that met both of the following conditions were considered for further data analysis: (1) spot signals were higher than the array backgrounds for both channels, and (2) the signal was twofold higher than the background at least for one channel.
Different types of analyses were used to identify and match expression patterns within or across the experimental groups. Within each group, a hierarchical clustering analysis was performed as described by Eisen et al. (1998). Only those genes that had more than twofold changes in expression in at least one of the experiment sets were used in the cluster analysis.
Acknowledgments
We thank Magnus Holm and Haiyang Wang for reading and commenting on this manuscript and Dr. Garry Whitelam, Dr. Peter Quail, Dr. Anthony Cashmore, and Dr. Chentao Lin for photoreceptor mutant and overexpression strains. We also thank Dr. Kenneth Williams and the staff of the DNA Microarray Resource of the HHMI Biopolymer/ W.M. Keck Biotechnology Resource Laboratory for the production of the microarrays used in this study. The microarrays described in this work are available to all researchers through the DNA Microarray Resource at http://info.med.yale.edu/wmkeck/dna_arrays.htm. This work was supported by National Institutes of Health Grant No. GM-47850 to X.W.D., a grant from the National Program for Transgenic Plants from China (J99-A-001), and National Institutes of Health Grant No. GM59507 to H.Z. X.W.D. is a National Science Foundation Presidential Faculty Fellow.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010229.
Footnotes
Online version contains Web-only data.
References
- Ang, L.H., and Deng, X.W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K. (1981). The protochlorophyllide homochrome of barley (Hordeum vulgare L.) phytochrome-induced decrease of translatable mRNA coding for the NADPH:protochlorophyllide oxidoreductase. Eur. J. Biochem. 120, 89–93. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Batschauer, A., Ehmann, B., and Schafer, E. (1991). Cloning and characterization of a chalcone synthase gene from mustard and its light-dependent expression. Plant Mol. Biol. 16, 175–185. [DOI] [PubMed] [Google Scholar]
- Bevan, M., et al., (1998). Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 484–488. [DOI] [PubMed] [Google Scholar]
- Bevan, M., Mayer, K., White, O., Eisen, J.A., Preuss, D., Bureau, T., Salzberg, S., and Mewes, H.W. (2001). Sequence and analysis of the Arabidopsis genome. Curr. Opin. Plant Biol. 4, 105–110. [DOI] [PubMed] [Google Scholar]
- Boylan, M.Y., and Quail, P.H. (1991). Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88, 10806–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., and Olney, M.A. (2001). Photoreceptors in plant photomorphogenesis to date: Five photochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, W.B., Christensen, A.H., Klein, T., Fromm, M., and Quail, P.H. (1989). Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc. Natl. Acad. Sci. USA 86, 9692–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, P., and Braam, J. (1999). Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 9, 361–366. [DOI] [PubMed] [Google Scholar]
- Cerff, R., and Kloppstech, K. (1982). Structural diversity and differential light control mRNAs coding for angiosperm glyceraldehydes-3-phosphate dehydrogenases. Proc. Natl. Acad. Sci. USA 79, 7624–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). Brassinosteroids: Essential regulation of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2001). Wall structure and wall loosening: A look backwards and forwards. Plant Physiol. 125, 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.W., and Quail, P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., and Graham, I.A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6, 72–77. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of a key pathway in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Webster, C.I., and Gray, J.C. (1997). Light-regulated expression of the pea plastocyanin gene is mediated by elements within the transcribed region of the gene. Plant J. 12, 499–506. [DOI] [PubMed] [Google Scholar]
- Jarillo, J.A., Gabrys, H., Capel, J., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410, 952–954. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K., and Wada, M. (2001). Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kieber, J.J. (1997). The ethylene response pathway in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 277–296. [DOI] [PubMed] [Google Scholar]
- Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., Yoshida, S., and Chua, N.H. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Kuno, N., and Furuya, M. (2000). Phytochrome regulation of nuclear gene expression in plants. Semin. Cell Dev. Biol. 11, 485–493. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., Oliverira, C.I.C., Melo-Oliverira, O.R., and Coruzzi, G.M. (1996). The molecular-genetics of nitrogen assimilation into amino acid in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. [DOI] [PubMed] [Google Scholar]
- Lee, M.L., Kuo, F.C., Whitmore, G.A., and Sklar, J. (2000). Importance of replication in microarray gene expression studies: Statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl. Acad. Sci. USA 97, 9834–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, W.M., Cao, X.L., Wilson, T.J., Sunstad, D.P., and Chua, N.H. (1995). Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in Arabidopsis. Plant Cell 7, 2187–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek, T., Martin, M.N., Bick, J.A., and Davies, J.P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 141–165. [DOI] [PubMed] [Google Scholar]
- Li, J., Nagpal, P., Vitart, V., McMorris, T.C., and Chory, J. (1996). A role for brassinosteroids in light-dependent of Arabidopsis. Science 272, 398–401. [DOI] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., and Cashmore, A.R. (1996). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10, 893–902. [DOI] [PubMed] [Google Scholar]
- Maurel, C., and Chrispeels, M.J. (2001). Aquaporins: A molecular entry into plant water relations. Plant Physiol. 125, 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, M.C., and Roberts, K. (1994). Changes in cell wall architecture during cell elongation. J. Exp. Bot. 45, 1683–1691. [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Nagata, N., Min, Y.K., Nakano, T., Asami, T., and Yoshida, S. (2000). Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinazole, induces some characteristics of light-grown plants. Planta 211, 781–790. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, B.M., and Ogren, W.L. (1993). Localization of light-inducible and tissue-specific regions of the spinach ribulose bisphosphate carboxylase/oxygenase (rubisco) activase promoter in transgenic tobacco plants. Plant Mol. Biol. 23, 1129–1138. [DOI] [PubMed] [Google Scholar]
- Petersen, M., et al., (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Puente, P., Wei, N., and Deng, X.W. (1996). Combinatorial interplay of promoter elements constitutes minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15, 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.M., Punita, N., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, V., Smith, H.E., Nance, J., Wang, J., Doren, A., Begley, R., Jones, S.J.M., Davis, E.B., Scherer, S., Ward, S., and Kim, S.K. (2000). A global profile of germline gene expression in C. elegans. Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, J., Kreuzaler, F., Schafer, E., and Hahlbrock, K. (1979). Concomitant induction of phenylalanine ammonia-lyase and flavanone synthase mRNAs in irradiated plant cells. J. Biol. Chem. 254, 57–65. [PubMed] [Google Scholar]
- Schumacher, K., and Chory, J. (2000). Brassinosteroid signal transduction: Still casting the actors. Curr. Opin. Plant Biol. 3, 79–84. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Caminic, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L.F., Twary, S.N., Yoshiako, H., Gregerson, R.G., Miller, S.S., Samac, D.A., Gantt, J.S., Unkefer, P.J., and Vance, C.P. (1997). Nitrogen assimilation in alfalfa: Isolation and characterization of an asparagine synthetase gene showing enhanced expression in root nodules and dark-adapted leaves. Plant Cell 9, 1339–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C., and Somerville, S. (1999). Plant functional geno-mics. Science 285, 380–383. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi, W.B., and Cashmore, A.R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474. [Google Scholar]
- Thompson, W.F., Everett, M., Polans, N.O., Jorgensen, R.A., and Palmer, J.D. (1983). Phytochrome control of RNA levels in developing pea and mung-bean leaves. Planta 158, 487–500.Q13 [DOI] [PubMed] [Google Scholar]
- Tobin, E.M., and Silverthorne, J. (1985). Light regulation of gene expression in higher plants. Annu. Rev. Plant Physiol. 36, 569–593. [Google Scholar]
- von Arnim, A., and Deng, X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 215–243. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Tepperman, J.M., and Quail, P.H. (1991). Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell 3, 1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Guegler, K., LaBrie, S.T., and Crawford, N.M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochomes with COP1 in light control development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- White, K.P., Rifkin, S.A., Hurban, P., and Hogness, D.S. (1999). Microarray analysis of Drosophila development during metamorphosis. Science 286, 2179–2184. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, NP. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 767–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek, D.M., and Clouse, S.D. (1994). Molecular cloning and characterization of a brassinosteroid-regulated gene from the elongating soybean (Glycine max L.) epicotyl. Plant Physiol. 104, 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]