Abstract
The bioactive isoflavonoids of the Leguminosae often are methylated on the 4′-position of their B-rings. Paradoxically, reverse genetic evidence implicates alfalfa isoflavone O-methyltransferase (IOMT) in the biosynthesis of 4′-O-methylated isoflavonoids such as the phytoalexin medicarpin in vivo, whereas biochemical studies indicate that IOMT has strict specificity for methylation of the A-ring 7-hydroxyl of daidzein, the presumed substrate for O-methylation, in vitro. Radiolabeling and isotope dilution studies now confirm that daidzein is not an intermediate in isoflavonoid phytoalexin biosynthesis in alfalfa. Furthermore, protein gel blot analysis and confocal microscopy of a transiently expressed IOMT–green fluorescent protein fusion in alfalfa leaves show that the operationally soluble IOMT localizes to endomembranes after elicitation of the isoflavonoid pathway. We propose that IOMT colocalizes with the endoplasmic reticulum–associated isoflavone synthase cytochrome P450 to ensure rapid B-ring methylation of the unstable 2,4′,7-trihydroxyisoflavanone product of isoflavone synthase, thereby preventing its dehydration to daidzein and subsequent A-ring methylation by free IOMT. In this way, metabolic channeling at the entry point into isoflavonoid phytoalexin biosynthesis protects an unstable intermediate from an unproductive metabolic conversion.
INTRODUCTION
The organization of plant natural product biosynthetic enzymes into metabolic complexes, known as metabolic channels or metabolons, is becoming increasingly apparent (Winkel-Shirley, 1999). Metabolic channeling facilitates rapid conversion of the product of one enzyme by the subsequent enzyme of the channel. This can overcome kinetic constraints imposed by the diffusion of metabolites within the cytosol and can protect unstable intermediates from spontaneous breakdown or potentially competing enzymatic reactions (Srere, 1987). It also can confound attempts to engineer pathways by the introduction of transgenes that encode enzymes acting on intermediates that are channeled (Dixon, 2000).
Metabolic channeling in plants can be demonstrated directly by precursor–product labeling studies or indirectly by the demonstration of physical associations between consecutive pathway enzymes. Using these approaches, either singly or in combination, channeling has been implicated in a variety of plant natural product pathways. In phenylpropanoid biosynthesis, channeling has been demonstrated at the phenylalanine ammonia-lyase/cinnamate 4-hydroxylase reactions at the entry point into the pathway (Czichi and Kindl, 1975, 1977; Rasmussen and Dixon, 1999), at the early stages of the flavonoid branch involving chalcone synthase, chalcone isomerase, flavonol 3-hydroxylase, and dihydroflavonol 4-reductase in Arabidopsis (Burbulis and Winkel-Shirley, 1999; Winkel-Shirley, 1999), and in the late stages of condensed tannin biosynthesis in Onobrychis viciifolia (Singh et al., 1997). Other notable examples include cyanogenic glycoside biosynthesis in sorghum (Kahn et al., 1997) and the proposal that channeling is necessary to account for the control of the biosynthesis of different classes of isoprenoid compounds via the mevalonate pathway (Chappell, 1995).
Isoflavonoids constitute a class of plant natural products with important activities for biotechnological applications, including health promotion in humans and antimicrobial activity against plant pathogens (Dixon, 1999). The entry point enzyme into the isoflavonoid pathway, 2-hydroxyisoflavanone synthase (2-HIS; also known as isoflavone synthase [IFS]), is a membrane-bound cytochrome P450 (Kochs and Grisebach, 1986). 2-HIS is attached to the outer surface of the endoplasmic reticulum (ER) by an N-terminal membrane anchor. It catalyzes NADPH-dependent hydroxylation of the 2-position of the flavanones liquiritigenin (7,4′-dihydroxyflavanone; Figure 1) and naringenin (5,7,4′-trihydroxyflavanone) with accompanying B-ring migration to yield unstable 2-hydroxyisoflavanone intermediates (Figure 1) that can undergo spontaneous or enzyme-catalyzed dehydration to the corresponding isoflavones (Kochs and Grisebach, 1986; Hakamatsuka et al., 1998). 2-HIS has been cloned recently from soybean, licorice, and several other legumes (Akashi et al., 1999; Steele et al., 1999; Jung et al., 2000). Soybean 2-HIS, when assayed in insect cell microsomes, produces the isoflavones daidzein and genistein (5,7,4′-trihydroxyisoflavone) from their corresponding flavanones liquiritigenin or naringenin without measurable accumulation of 2-hydroxyisoflavanone (Steele et al., 1999).
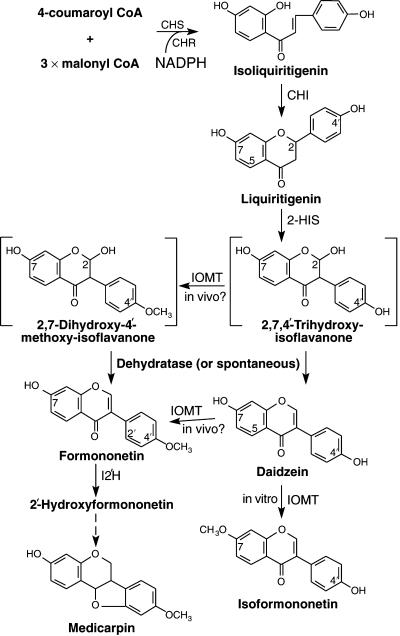
Figure 1.
Possible Pathways for the Biosynthesis of Isoflavonoids in Alfalfa.
The enzymes are chalcone synthase (CHS), chalcone reductase (CHR), chalcone isomerase (CHI), 2-hydroxyisoflavanone synthase (2-HIS), isoflavone O-methyltransferase (IOMT), and isoflavone 2′-hydroxylase (I2′H). The compounds shown in square brackets are unstable intermediates.
The next enzymatic step specific for isoflavonoid biosynthesis in alfalfa is methylation of the B-ring 4′-hydroxyl group to yield formononetin (Figure 1). However, previous studies in alfalfa and other species that produce 4′-O-methylated isoflavonoids have failed to identify enzymes capable of methylating the B-ring 4′-hydroxyl group of daidzein in vitro. All isoflavone O-methyltransferases characterized to date catalyze methylation of the A-ring 7-hydroxyl group of isoflavones (Hagmann and Grisebach, 1984; Edwards and Dixon, 1991). Thus, alfalfa isoflavone O-methyltransferase (IOMT), either purified from plants or expressed as a recombinant enzyme in Escherichia coli, converts daidzein to isoformononetin (Figure 1) in vitro (Edwards and Dixon, 1991; He and Dixon, 1996; He et al., 1998). Isoformononetin is undetectable in alfalfa. However, radiolabeled daidzein is a poor precursor of the phytoalexin medicarpin in abiotic elicitor-treated alfalfa seedlings, whereas formononetin is incorporated readily into medicarpin (Dewick and Martin, 1979). This led to the suggestion that B-ring migration in isoflavone biosynthesis in alfalfa is associated tightly with, and might even require, the 4′-O-methylation reaction (Dewick and Martin, 1979), although this has been challenged in view of the demonstrable IFS activity in vitro with no requirement for methylation (Dixon, 1999).
Alfalfa contains a small family of very closely related IOMT genes with ∼99% amino acid sequence identity (He et al., 1998). Transgenic alfalfa plants with strong constitutive expression of IOMT8 exhibit a greater and more rapid accumulation of 4′-O-methylated isoflavonoids (including medicarpin) than do control plants (He and Dixon, 2000). Thus, IOMT appears to methylate the 4′-position of isoflavonoids in vivo and acts as a flux-determining enzyme for isoflavonoid phytoalexin biosynthesis when overexpressed in alfalfa.
The cocrystal structure of IOMT8 with its substrates indicates that the 7-hydroxyl of daidzein is positioned close to the methyl group of the methyl donor S-adenosyl-l-methionine (SAM), supporting the 7-O-methylation of daidzein as observed in vitro (Zubieta et al., 2001). However, modeling studies indicate that one of the four potential stereoisomers of 2,7,4′-trihydroxyisoflavanone (Figure 1), the unstable product of 2-HIS, can fit into the active site in such a way that the 4′-hydroxyl group faces the methyl group of SAM. Binding of this intermediate may be more stable thermodynamically than binding of daidzein (Zubieta et al., 2001). Furthermore, it has been demonstrated recently that 2,4′,7-trihydroxyisoflavanone can be converted to formononetin by a cell-free extract from elicited licorice cells (Akashi et al., 2000). 2,4′,7-Trihydroxyisoflavanone therefore is a potential substrate of IOMT in vivo, and this presents an alternative route to formononetin that would bypass daidzein (Figure 1). However, if this model is correct, how the cell avoids making isoformononetin by the concerted actions of 2-HIS and IOMT is not clear.
Here, we provide new evidence that both reveals and provides a rationale for metabolic channeling in the biosyn-thesis of formononetin from liquiritigenin in alfalfa. This channeling features the elicitor-induced association of IOMT with microsomal membranes that allows IOMT to “capture” the 2-hydroxyisoflavanone intermediate of 2-HIS before its dehydration to daidzein. This model explains previously described paradoxical data regarding the involvement of daidzein in isoflavonoid biosynthesis (reviewed by Dixon, 1999) and the apparently different regiospecificities of alfalfa IOMT in vivo and in vitro (He et al., 1998; He and Dixon, 2000).
RESULTS
Incorporation of 3H-Liquiritigenin into Methylated Isoflavonoids in Alfalfa Cell Suspension Cultures
The involvement of daidzein in the biosynthesis of medicarpin was assessed using an isotope dilution approach. Alfalfa cell suspension cultures were treated with a crude polysaccharide elicitor preparation from yeast cell walls to induce the accumulation of glucosides of formononetin and medicarpin, as shown in Figures 2A and 2B. Elicited cells were fed 3H-liquiritigenin for 48 hr, and isoflavonoids then were extracted and treated with β-glucosidase to release the corresponding aglycones before fractionation by HPLC. Peak fractions were collected, and incorporation of 3H into the downstream metabolites formononetin, 2′-hydroxyformononetin, and medicarpin was measured by liquid scintillation counting, as shown in Figure 2C. The percentage incorporation of label into formononetin, 2′-hydroxyformononetin, and medicarpin in the experiment shown in Figure 2C was 0.8, 0.2, and 0.3%, respectively (Table 1, experiment I). No label appeared in daidzein, although this compound was resolved poorly from the large peak of labeled liquiritigenin. Very similar results were obtained in an independent experiment using a different cell culture batch (Table 1, experiment II). Simultaneous feeding of unlabeled daidzein with 3H-liquiritigenin had no effect on the percentage incorporation or isotopic dilution of 3H label into formononetin, 2′-hydroxyformononetin, or medicarpin (Table 1) or on the levels of these compounds (Figure 2D). In contrast, simultaneous feeding of unlabeled formononetin with 3H-liquiritigenin had two effects: to increase the production of both 2′-hydroxyformononetin and medicarpin, as shown by comparison of Figures 2D and 2E, and to greatly decrease the specific activities of, and increase the isotope dilution into, these compounds (Table 1). These results suggest that daidzein is not an intermediate in the biosynthesis of formononetin and medicarpin, whereas formononetin is an intermediate in medicarpin biosynthesis.
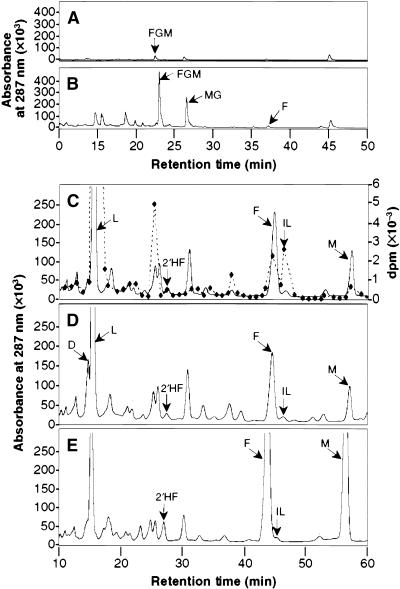
Figure 2.
HPLC Analysis of Isoflavonoids from Alfalfa Cell Cultures.
Compounds were detected by UV light absorbance at 287 nm.
(A) Nonhydrolyzed extract from an unelicited cell culture.
(B) Nonhydrolyzed extract from a culture 48 hr after exposure to yeast elicitor.
(C) Portion of the same extract as in (B) after hydrolysis with β-glucosidase. The dashed line represents radioactivity in compounds after feeding 3H-liquiritigenin.
(D) Hydrolyzed extract from elicited cells that had been fed unlabeled daidzein. The daidzein is not resolved fully from the large peak of liquiritigenin with slightly higher retention time.
(E) Hydrolyzed extract from elicited cells that had been fed unlabeled formononetin.
The compounds are as follows: formononetin glucosyl malonate (FGM), medicarpin glucoside (MG), formononetin (F), liquiritigenin (L), 2′-hydroxyformononetin (2′HF), isoliquiritigenin (IL), medicarpin (M), daidzein (D). Chromatography was performed using gradient I (see Methods) in (A) and (B) and gradient II in (C) to (E).
Table 1.
Incorporation of 3H-Liquiritigenin into Formononetin, 2′-Hydroxyformononetin, and Medicarpin in Elicited Alfalfa Cell Suspension Culturesa
| Formononetin
|
2′-Hydroxyformononetin
|
Medicarpin
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unlabeled Compound Fed |
Content (nmol/g FW)b |
Specific Activity (dpm/nmol) |
Dilution | Incorporation (%) |
Content (nmol/g FW)b |
Specific Activity (dpm/nmol) |
Dilution | Incorporation (%) |
Content (nmol/g FW)b |
Specific Activity (dpm/nmol) |
Dilution | Incorporation (%) |
| Experiment I | ||||||||||||
| None | 42.9 | 87.7 ± 2.5 | 51.7 | 0.8 | 1.0 | 913.5 ± 37.7 | 4.9 | 0.2 | 15.9 | 75.5 ± 0.3 | 61.4 | 0.3 |
| Daidzein | 34.5 | 81.8 ± 1.4 | 57.2 | 0.6 | 0.9 | 954.5 ± 4.5 | 4.9 | 0.2 | 12.6 | 79.1 ± 5.4 | 55.8 | 0.2 |
| Formononetin | 93.3 | 24.7 ± 0.0 | 187.0 | 0.8 | 4.5 | 135.4 ± 1.3 | 34.6 | 0.2 | 92.1 | 10.0 ± 0.1 | 466.8 | 0.3 |
| Experiment II | ||||||||||||
| None | 39.3 | 68.6 ± 2.3 | 48.7 | 0.8 | 0.9 | 660.8 ± 43.5 | 5.1 | 0.2 | 17.9 | 126.9 ± 22.6 | 26.3 | 0.8 |
| Daidzein | 39.9 | 68.6 ± 18.8 | 48.6 | 0.8 | 1.0 | 742.7 ± 95.4 | 4.5 | 0.2 | 17.9 | 152.4 ± 11.6 | 21.9 | 0.7 |
| Formononetin | 88.3 | 27.1 ± 0.9 | 123.3 | 0.8 | 3.2 | 147.8 ± 7.0 | 22.5 | 0.2 | 138.9 | 16.1 ± 0.7 | 208.3 | 0.7 |
a Cells were fed liquiritigenin alone or liquiritigenin plus unlabeled daidzein or formononetin. After 48 hr, metabolites were resolved by HPLC, quantified by UV light absorption, and incorporation of 3H was determined by liquid scintillation counting. The experiment was repeated with a different batch of alfalfa cell suspension culture, and the results of both experiments are given. Data represent mean and standard deviations from two to four independent determinations for each experiment. Incorporation represents the percentage of the total radioactivity fed that was recovered in a particular compound. Specific activity indicates the amount of radioactivity per nanomole of recovered product, and isotope dilution is the specific activity of the 3H-liquiritigenin fed to the cultures divided by the specific activity of the recovered product. Isoflavone conjugates present in the extracts were hydrolyzed to the aglycones before HPLC analysis.
b FW, fresh weight.
Three important control experiments were performed to validate the data in Figure 2 and Table 1. First, the HPLC gradient (II) used to profile the full spectrum of isoflavonoid compounds in glucosidase-digested alfalfa cell extracts does not give baseline resolution of liquiritigenin and daidzein, as seen in Figure 2D. To confirm that no label was incorporated into daidzein in the feeding experiment described above, labeled glucosidase-digested samples were rechromatographed using a solvent gradient (III) incorporating an isocratic stage that resolves liquiritigenin from daidzein, as shown in Figure 3C. No daidzein was observed in cultures to which the isoflavone had not been fed (Figure 3A). A daidzein peak was recovered in cultures fed unlabeled daidzein (Figure 3B), but no 3H label from liquiritigenin was associated with this peak. This result was confirmed using thin-layer chromatography to resolve liquiritigenin from daidzein (data not shown). Thus, feeding 3H-liquiritigenin results in the labeling of formononetin and downstream metabolites but no labeling of daidzein, consistent with daidzein not being an intermediate in formononetin and medicarpin biosynthesis in alfalfa.
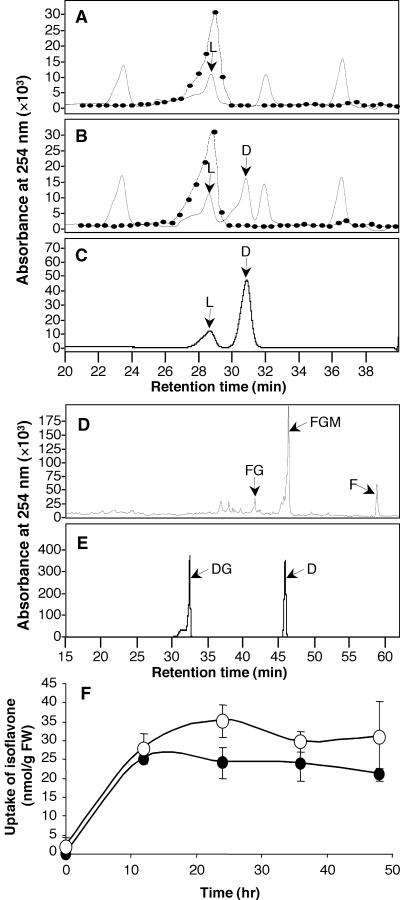
Figure 3.
Uptake of Daidzein and Formononetin in Elicited Alfalfa Cell Suspension Cultures.
Compounds were detected by UV light absorbance at 254 nm.
(A) to (C) Demonstration that alfalfa cells lack an endogenous pool of daidzein and do not convert 3H-liquiritigenin to daidzein. (A) Portion of an HPLC trace showing the elution of 3H-liquiritigenin (L) in an extract from an elicited culture. (B) Like (A) but from a parallel culture that had been fed unlabeled daidzein. Note the lack of incorporation of radiolabel (dotted lines in [A] and [B]) into daidzein (D). (C) Liquiritigenin and daidzein standards.
(D) and (E) Demonstration that exogenously supplied daidzein is not glucosylated to daidzin in alfalfa cell cultures. (D) A nonhydrolyzed extract from elicited cells that had been fed unlabeled daidzein. FG, formononetin glucoside; FGM, formononetin glucosyl malonate; F, formononetin. (E) Daidzin (DG) and daidzein (D) standards.
(F) Uptake of daidzein (closed circles) and formononetin (open circles) in elicited alfalfa cell suspension cultures. FW, fresh weight.
Chromatography was performed using gradient III in (A) to (C) and gradient IV in (D) and (E).
Error bars in (F) indicate ±sd.
The second control experiment was performed to ensure that added unlabeled daidzein was not removed rapidly by glucosylation to daidzin (daidzein 7-O-glucoside), thereby compromising the isotope dilution experiments. HPLC analysis of nonhydrolyzed extracts from daidzein-fed cells revealed formononetin, formononetin glucoside, and formononetin glucoside malonate, but no daidzin, as shown in Figure 3D (standards are shown in Figure 3F). With the HPLC gradient used in this analysis, the added daidzein cochromatographed with the peak of endogenous formononetin glucoside.
Finally, a comparison was made of the uptake kinetics of the unlabeled daidzein and formononetin fed to the cultures. In the latter case, the size of the endogenous pool at each time point was subtracted. Figure 3F shows that maximum levels of both compounds in the cells were attained after 12 to 24 hr of feeding and that, importantly, the cellular concentrations of added daidzein and formononetin were very similar throughout the time course. Thus, the inability of daidzein compared with formononetin to decrease the incorporation of label into downstream products is not the result of the differential uptake of the two compounds.
Together, the data in Table 1 and Figures 2 and 3 indicate that daidzein is not formed from liquiritigenin in elicited alfalfa cells and that formononetin, but not daidzein, is an intermediate in the biosynthesis of medicarpin via 2′-hydroxyformononetin (Figure 1). The apparently paradoxical increase in the specific activity of 2′-hydroxyformononetin compared with that of formononetin (Table 1) is attributable to the fact that the formononetin pool being measured in this experiment includes contributions from formononetin released from preexisting (and therefore unlabeled) constitutive pools of formononetin conjugates. The lack of the expected reduction in the percentage incorporation of 3H into 2′-hydroxyformononetin and medicarpin in cells fed unlabeled formononetin might be explained by a compensatory stimulation of total flux into the pathway by this compound.
Formation of Daidzein and Methylated Isoflavones in Alfalfa Leaf and Cell Culture Microsomes
Daidzein is formed from liquiritigenin by IFS in microsomal preparations from elicited alfalfa cell cultures (Kessmann et al., 1990). Therefore, we examined whether alfalfa microsomes also contain a SAM-dependent IOMT activity for the formation of formononetin. Microsomes were analyzed from two different tissue sources chosen to allow analysis of the effects of increasing the expression level of IOMT. Leaves were extracted from empty vector–transformed control alfalfa plants and plants transformed with the full-length alfalfa IOMT8 cDNA, as described previously by He and Dixon (2000), and from untreated and elicited alfalfa cell suspensions. Two substrates were used: daidzein, to determine whether an enzyme with the same substrate specificity but different regiospecificity from the previously characterized IOMT is present in microsomes; and liquiritigenin, to determine whether the substrate for isoflavonoid methylation in microsomes might be either 2,4′,7-trihydroxyisoflavanone or daidzein generated by IFS and passed on to the methyltransferase in a channeled reaction.
Total, soluble, and microsomal fractions were prepared from leaves of control (empty vector–transformed) and IOMT8-overexpressing alfalfa that had been exposed previously to water or to 0.3 mM copper chloride to induce IFS and other enzymes of the isoflavonoid pathway. First, daidzein and 3H-SAM were supplied as substrates to confirm the regiospecificity of soluble IOMT against daidzein and to determine whether any enzyme in the microsomes could methylate daidzein. Daidzein was methylated to yield isoformononetin by total and soluble fractions, but no conversion was detected in the microsomal fractions, as shown in Figure 4A. Production of isoformononetin was higher using leaf extracts from plants transformed with alfalfa IOMT8 than with leaf extracts from control plants, and it was increased further if the plants had been elicited first (Figure 4A). The same subcellular distribution and elicitor inducibility was shown for isoflavone 7-O-methyltransferase activity in alfalfa cell suspension cultures, as shown in Figure 4B. These results confirm the activity of alfalfa IOMT as an elicitor-inducible, operationally soluble enzyme with 7-position specificity for daidzein in vitro.
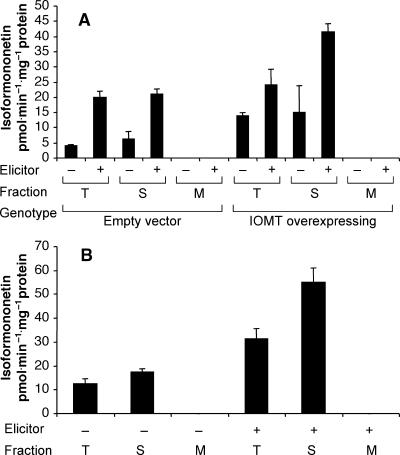
Figure 4.
O-Methylation of Daidzein in Soluble and Microsomal Extracts from Alfalfa Leaves and Cell Suspension Cultures.
(A) Total (T), soluble (S), and microsomal (M) extracts from leaves of unelicited (−) and copper chloride–elicited (+) control (empty vector) and IOMT-overexpressing alfalfa plants were incubated with daidzein and 3H-SAM, and incorporation of label into isoformononetin was determined after HPLC separation. Bars represent means and standard deviations for two to four independent determinations.
(B) Total, soluble, and microsomal extracts were prepared from unelicited (−) and yeast-elicited (+) alfalfa cell suspension cultures and assayed for methylation of daidzein as in (A).
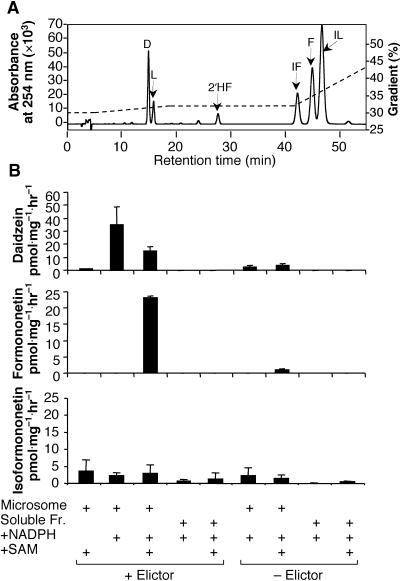
To determine whether formononetin could be produced by the coupled activities of 2-HIS and microsomal IOMT, microsomal and soluble fractions from yeast elicitor–treated or untreated cell cultures were incubated with 3H-liquiritigenin in the presence or absence of NADPH (an essential cofactor for 2-HIS) and SAM. Labeled isoliquiritigenin (2′,4,4′-trihydroxychalcone; Figure 1) was formed by the isomerization of 3H-liquiritigenin in all in vitro experiments. Therefore, the products were analyzed by HPLC using a different elution gradient (gradient II; see Methods) that resolves formononetin, isoformononetin, and isoliquiritigenin, as shown in Figure 5A. Feeding microsomes from elicited cells with 3H-liquiritigenin and SAM in the absence of NADPH led to very little label in either daidzein or formononetin. The addition of NADPH in the absence of SAM led to the production of daidzein but not formononetin, whereas labeled daidzein decreased and labeled formononetin was produced after the addition of both NADPH and SAM, as shown in Figure 5B. A labeled product coeluting with isoformononetin was produced at low levels when 3H-liquiritigenin was incubated with microsomal extracts (Figure 5B). Very little incorporation of liquiritigenin into daidzein or formononetin was observed in microsomal extracts from unelicited cells, in which 2-HIS was not induced. Likewise, liquiritigenin underwent little conversion when incubated with soluble fractions that lack the membrane-associated 2-HIS. Thus, in the absence of the methyl group donor SAM, elicitor-induced 2-HIS catalyzed the aryl migration of liquiritigenin to yield daidzein via the dehydration of 2-hydroxyisoflavanone. However, the production of daidzein decreased in the presence of SAM, concomitant with the production of formononetin. Very little 7-O-methylation of daidzein by IOMT to yield isoformononetin occurred, even though IOMT is induced strongly by elicitation (He et al., 1998). These results indicate that the 2-hydroxyisoflavanone intermediate of 2-HIS is methylated rapidly in the presence of SAM, followed by dehydration to yield formononetin, and that daidzein and its in vitro methylation product isoformononetin are not produced. This finding suggests that 2-HIS and IOMT are in close physical proximity.
Figure 5.
Incorporation of 3H-Liquiritigenin into Daidzein, Formononetin, and Isoformononetin in Elicited Alfalfa Cell Suspension Microsomes in Vitro.
(A) HPLC trace showing separation of standards of daidzein (D), liquiritigenin (L), 2′-hydroxyformononetin (2′HF), isoformononetin (IF), formononetin (F), and isoliquiritigenin (IL). Chromatography was performed using gradient II.
(B) Formation of daidzein, formononetin, and isoformononetin from 3H-liquiritigenin by alfalfa cell culture microsomes or soluble supernatant incubated in the presence or absence of NADPH and/or SAM. Bars represent means and standard deviations for two to four independent determinations. The amount of label in isoformononetin may be an overestimate caused by the variable presence of a second, unidentified compound coeluting with isoformononetin in the cell extracts. Chromatography was performed using gradient II. Fr., fraction.
Isoflavone 7-O-Methyltransferase Associates with Endomembranes after the Elicitation of Isoflavonoid Biosynthesis
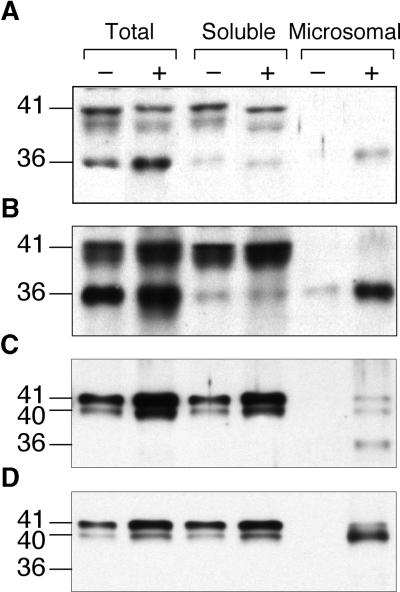
To determine whether we could detect IOMT protein in microsomal membranes from alfalfa, protein gel blot analysis was first performed on total, soluble, and microsomal extracts from untreated and copper chloride–treated leaves from control (empty vector–transformed) and IOMT8-overexpressing transgenic alfalfa. Because of the near identity of the open reading frames of the different IOMT genes in alfalfa (He et al., 1998), the polyclonal antiserum raised against recombinant IOMT8 would be expected to cross-react with all IOMT isoforms. The results shown in Figures 6A and 6B indicate the presence of two protein bands, at 41 and 36 kD (as assessed by comparison with the mobilities of prestained molecular mass markers), that react strongly with anti-alfalfa IOMT8 serum. The 41-kD band corresponds to the apparent molecular mass of the native IOMT8 subunit (He et al., 1998). The band of ∼40 kD that runs slightly below that corresponding to the native IOMT8 subunit may represent the product of a second alfalfa IOMT gene, IOMT2, that has a nine–amino acid deletion at the C terminus compared with IOMT8 (He and Dixon, 1997), or it may be a degradation product of IOMT. Levels of both the 41- and 36-kD bands were much higher in extracts from the IOMT8-overexpressing line than in the corresponding extracts from the control line, consistent with both bands representing the IOMT protein. The 41-kD protein was not found in the microsomal fraction, whereas the 36-kD protein was found at low levels in the soluble and microsomal fractions from the unelicited IOMT8-overexpressing line and was increased strongly in the microsomal fraction after the elicitation of this line. A similar pattern was seen in fractions from the control line, except that there was no IOMT signal in the microsomal fraction in the absence of elicitation.
Figure 6.
Protein Gel Blot Analysis of IOMT in Total, Soluble, and Microsomal Fractions from Control (−) and Elicited (+) Alfalfa Leaves and Cell Suspension Cultures.
(A) and (B) Leaves were either from control plants (harboring an empty vector) (A) or from a transgenic line overexpressing alfalfa IOMT8 (B). Leaves were elicited with copper chloride. IOMT activities (production of isoformononetin from daidzein) in the total extract from leaves of the two unelicited plants were 3.75 pmol·min−1·mg−1 protein (A) and 27.72 pmol·min−1·mg−1 protein (B).
(C) and (D) Protein gel blots of extracts from control (C) and yeast elicitor–treated (D) alfalfa cell suspension cultures.
Microsomal extracts in (D) were prepared in a buffer containing 28 mM 2-mercaptoethanol and 10 mM EDTA. Protein loadings in (A) and (B) were designed to give protein amounts equivalent to equal initial amounts of leaf fresh weight per lane: 46 μg of total protein, 38 μg of soluble protein, and 9 μg of microsomal protein. In (C) and (D), 5 μg of protein was loaded in each lane.
Similar analysis of untreated and yeast elicitor–treated cell suspension cultures (Figure 6C) revealed a 41/40-kD IOMT doublet in the total and soluble fractions, the levels of which were induced by elicitor. No signal was detected in the microsomal fraction from unelicited cells, whereas microsomes from elicited cells contained immunoreactive proteins corresponding to the 41/40-kD doublet and the 36-kD band, as found in the microsomes from elicited leaves. If the cell culture microsomes were prepared in a buffer with 28 mM 2-mercaptoethanol and 10 mM EDTA (a combination of conditions that had been used previously for the optimal extraction of IOMT activity [Hagmann and Grisebach, 1984; He and Dixon, 1996]), the 36-kD band was no longer visible and the 41/40-kD doublet was of increased intensity, as shown in Figure 6D. This finding suggests that the 36-kD protein is in fact a degradation product of IOMT. However, careful preparation of leaf microsomes in the presence of high 2-mer-captoethanol, EDTA, and a protease inhibitor cocktail failed to prevent the appearance of the 36-kD protein (data not shown), suggesting that the degradation of IOMT in elicited leaves may occur before extraction and analysis.
These data suggest that elicitation leads to the localization of IOMT to membranes, where it might be involved in the channeled formation of formononetin as part of a complex with 2-HIS. To independently confirm that IOMT can assume membrane localization after elicitation, we fused the coding regions of alfalfa IOMT8 and Medicago truncatula 2-HIS to enhanced green fluorescent protein (EGFP) for bombardment into elicited and untreated alfalfa leaves under the control of a double-enhanced 35S promoter of cauliflower mosaic virus. Control constructs consisted of double-enhanced 35S promoter–driven EGFP alone or 35S promoter–driven modified green fluorescent protein (mGFP) with a C-terminal HDEL ER retention signal (Haseloff et al., 1997). The localization patterns of EGFP and the EGFP fusion proteins then were determined by laser scanning confocal microscopy.
Figures 7B and 7F show that IOMT8–EGFP expressed in unelicited alfalfa leaf vein or epidermal cells exhibited a diffuse distribution throughout the cytoplasm, in a manner similar to free EGFP, as shown in Figures 7D and 7H for vein and epidermal cells, respectively. Bombardment of the free EGFP construct into preelicited alfalfa leaves resulted in the same diffuse cellular distribution of EGFP fluorescence that was observed in unelicited leaves, as shown in Figures 7C (vein cell) and 7G (epidermal cell). In contrast, IOMT8–EGFP localized to narrow strands within the cell and lost most of the diffuse cytoplasmic staining after bombardment into preelicited leaves, as shown in Figures 7A (vein cell) and 7E (epidermal cell). The arrows in Figure 7A point to the narrow endomembrane strands, which are very distinct from the broader cytoplasmic strands seen in many of the images. This distribution was not observed, however, when IOMT8–EGFP was expressed in unelicited leaves (Figures 7B and 7F). The distribution of IOMT8–EGFP in elicited leaf cells was very similar to that of mGFP fused to a C-terminal HDEL ER retention signal expressed in either unelicited (Figure 7I) or elicited (Figure 7K) leaf cells. This included both a reticulate pattern and localization around, but not within, the nucleus, as shown by comparing Figures 7I and 7K (perinuclear and reticulate distribution of mGFP–HDEL) with Figures 7J and 7L (perinuclear and reticulate distribution of IOMT8–EGFP in elicited leaves). A very similar distribution of GFP fluorescence has been shown to be associated with ER localization of GFP in a genetic screen for ER-targeted proteins in Arabidopsis (Cutler et al., 2000).
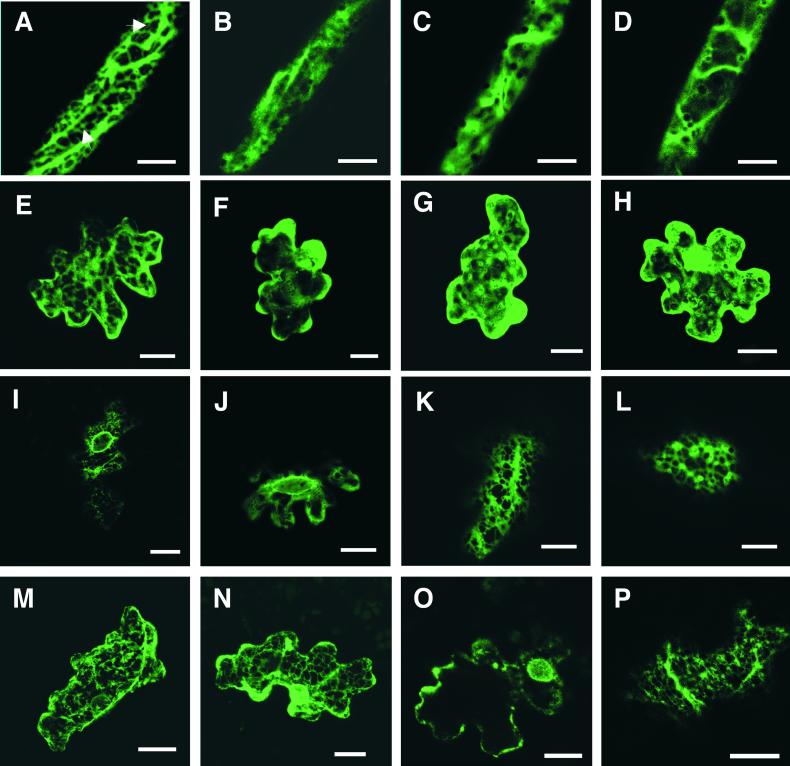
Figure 7.
Confocal Laser Scanning Microscopy Showing Subcellular Fluorescence Distribution from IOMT8–EGFP, 2-HIS–EGFP, mGFP–HDEL, and Control EGFP Expressed Transiently in Alfalfa Leaf Cells.
(A) to (D) Projections of 25 0.36-μm-thick optical sections from alfalfa leaf vein cells. (A) Distribution of IOMT8–EGFP fluorescence in an elicited vein cell showing cortical ER localization (arrows). (B) Diffuse distribution of IOMT8–EGFP fluorescence in the cytoplasm of an unelicited vein cell. (C) and (D) Diffuse distribution of control EGFP fluorescence in the cytoplasm of an elicited (C) or unelicited (D) leaf vein cell.
(E) to (H) Projections of 30 0.36-μm-thick optical sections from alfalfa leaf epidermal cells. (E) Cortical ER localization of IOMT8–EGFP fluorescence in an elicited epidermal cell. (F) Diffuse cytoplasmic distribution of IOMT8–EGFP fluorescence in an unelicited epidermal cell. (G) and (H) Distribution of control EGFP fluorescence in the cytoplasm of either elicited (G) or unelicited (H) epidermal cells.
(I) and (J) Single optical sections at the plane of the nucleus from an unelicited leaf epidermal cell expressing mGFP–HDEL (I) or an elicited epidermal cell expressing IOMT8–EGFP (J); note the perinuclear fluorescence patterns.
(K) and (L) Distribution of fluorescence on the cortical ER in single optical sections of elicited epidermal cells expressing mGFP–HDEL (K) or IOMT8–EGFP (L).
(M) and (N) Projections of 25 0.36-μm-thick sections from unelicited alfalfa vein (M) and epidermal (N) cells expressing 2-HIS–EGFP showing cortical ER and perinuclear fluorescence distribution.
(O) and (P) Single optical sections from elicited alfalfa epidermal cells expressing 2-HIS–EGFP illustrating perinuclear (O) and reticulate (P) fluorescence distribution.
Bars = 20 μm.
The M. truncatula 2-HIS cDNA encodes a type IIb membrane protein with an N-terminal membrane anchor domain typical of ER-localized cytochrome P450 enzymes. Alfalfa leaf cells transiently expressing 2-HIS–EGFP displayed both the typical perinuclear and reticulate cortical fluorescence patterns indicative of ER localization. This was observed in both vein (Figure 7M) and epidermal (Figure 7N) cells from unelicited leaves. The same pattern was observed in cells from elicited leaves (Figures 7O and 7P). Thus, IOMT appears to share similar subcellular localization to 2-HIS, the enzyme with which it is functionally associated.
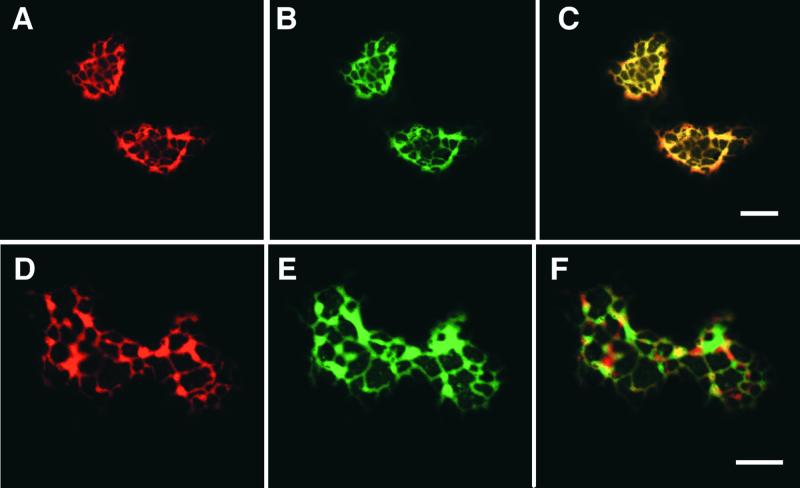
We attempted to demonstrate directly the colocalization of IOMT with 2-HIS by cobombardment of alfalfa leaf cells with IOMT8–EGFP and 2-HIS fused to red fluorescent protein (RFP) or with IOMT8–RFP and 2-HIS–EGFP. However, fusion of large proteins with DsRed RFP, an obligate tetramer, can lead to problems with aggregation and abnormal fluorescence (Baird et al., 2000), and expression of IOMT8–RFP led to the production of large fluorescent aggregates, whereas the 2-HIS–RFP fusion protein was not expressed in the alfalfa cells. Therefore, we used confocal microscopy to compare the localization, within the same cell, of IOMT8–EGFP with that of RFP targeted to the ER by way of a C-terminal HDEL sequence. Figure 8 shows the results of this colocalization experiment in elicited alfalfa epidermal cells using either simultaneous excitation of EGFP and RFP ([A] to [C]) or independent excitation with data capture and computer merging of the images ([D] to [F]). RFP–HDEL showed the same reticulate ER localization (Figures 8A and 8D) that was observed for mGFP–HDEL in Figure 7K. The same pattern was observed for IOMT8–EGFP (Figures 8B and 8E), and superimposition of the images yielded a yellow image indicative of close colocalization, as seen in Figures 8C and 8F. The data in Figure 8C are suggestive of almost complete colocalization, whereas the areas of green fluorescence in Figure 8F indicate areas where IOMT is expressed that do not correspond to RFP–HDEL; therefore, these may represent residual cytosolic IOMT8–EGFP protein.
Figure 8.
Colocalization of IOMT8–EGFP with ER-Targeted RFP–HDEL in Alfalfa Leaf Epidermal Cells.
All images are from single 0.36-μm optical sections. The sections shown in (A) to (C) were excited simultaneously for RFP and GFP emission, and emitted light was recorded at the emission band for RFP (A), EGFP (B), or at the emission wavelengths for both RFP and EGFP (C). The sections shown in (D) to (F) were excited singly for RFP (D) and EGFP (E), and the captured images are superimposed in (F). Bars = 20 μm.
DISCUSSION
Daidzein Is Not an Intermediate in Formononetin Biosynthesis
The paradox surrounding the mechanism of 4′-O-methylation of isoflavones can be explained once it is realized that daidzein is not itself an intermediate in the biosynthesis of 4′-O-methylated isoflavonoids such as the phytoalexin medicarpin. This was suggested many years ago based on the poor incorporation of 3H-daidzein into medicarpin in copper chloride–elicited alfalfa seedlings (Dewick and Martin, 1979), but it was apparently contradicted by the demonstration of the enzymatic production of daidzein by IFS preparations (Hagmann and Grisebach, 1984; Kessmann et al., 1990) and by the fact that mutants of subterranean clover lacking 4′-O-methylation accumulate daidzein and genistein (Wong and Francis, 1968). The present results provide new evidence, based on radiolabeling and isotope dilution experiments, to confirm that daidzein is not involved in the synthesis of 4′-O-methylated isoflavonoids using a cell culture system from which the enzymatic synthesis of daidzein nevertheless could be demonstrated in microsomal extracts. Daidzein is not synthesized from labeled liquiritigenin, nor does its exogenous addition decrease the incorporation of label from liquiritigenin into downstream isoflavonoids.
It now appears that the synthesis of formononetin occurs by methylation of the 4′-hydroxyl group of the unstable trihydroxyisoflavanone product of 2-HIS catalyzed by IOMT. This model is supported by several independent observations. First, the conversion of liquiritigenin to formononetin in microsomes from elicited alfalfa cells demonstrates that isoflavone O-methylation, although not involving daidzein, nevertheless is associated with the microsomally catalyzed aryl migration reaction that, in the absence of IOMT and SAM, produces daidzein from liquiritigenin. Second, the conversion of liquiritigenin to formononetin in yeast microsomes expressing licorice 2-HIS in the presence of licorice cell-free extracts and SAM, and the demonstration of the ability of these extracts to form formononetin from 2,7,4′-trihydroxyisoflavanone and SAM (Akashi et al., 2000), confirm the existence of an O-methyltransferase active against 2,7,4′-trihydroxyisoflavanone. Third, the demonstration that transgenic plants overexpressing the enzyme shown previously to catalyze the 7-O-methylation of daidzein accumulate increased levels of formononetin and medicarpin (He and Dixon, 2000), and the increased production of formononetin by microsomal membranes isolated from leaves of transgenic plants overexpressing IOMT, together indicate that an enzyme with 7-position specificity in vitro is the O-methyltransferase involved in formononetin biosynthesis (via 2,4′,7-trihydroxyisoflavanone) in vivo, even though we have been unable to show direct activity of IOMT with 2,4′,7-trihydroxyisoflavanone in vitro because of our inability to detect and thereby isolate the intermediate. In reactions using recombinant 2-HIS in insect cell microsomes, liquiritigenin is converted directly to daidzein (Steele et al., 1999). Finally, the solution of the crystal structure of IOMT, with its implication for binding of 2,4′,7-trihydroxyisoflavanone to the active site in an orientation favorable for 4′-O-methylation (Zubieta et al., 2001), provides an explanation for the different substrate specificities of IOMT in vivo and in vitro. The new pathway to formononetin in vivo is consistent with the accumulation of nonmethylated isoflavones in putatively O-methyltransferase–deficient clover (Wong and Francis, 1968), because in this case the dehydration of 2-hydroxyisoflavanone to isoflavone would occur in the absence of methylation.
IOMT Localizes to Microsomal Membranes after Elicitation
Both biochemical fractionation and confocal microscopy studies clearly indicate the localization of IOMT to membranes, but only after elicitation. The fluorescence colocalization experiments using IOMT8–EGFP and RFP–HDEL indicate that the membrane localization is to the ER. We believe that the simultaneous excitation experiment correctly reports colocalization because, in previous work, simultaneous excitation at 488 and 568 nm led to almost no crossover from DsRed emission into the EGFP channel, and the crossover from EGFP emission into the DsRed channel was consistently 5% or less (Jackobs et al., 2000). To avoid any chromatic aberration caused by the fluorescence bleach between red and green under simultaneous excitation, the fluorescence of IOMT8–EGFP and RFP–HDEL at the same cell section were captured separately using individual excitation channels and the projections were merged. This confirmed the colocalization of IOMT8–EGFP and RFP–HDEL to reticulate endomembranes.
Our confocal microscopy and transgenic plant studies used the alfalfa IOMT8 gene. IOMT is encoded by a small family of up to four genes in alfalfa (He et al., 1998), and four different IOMT cDNA clones have been characterized from this species. IOMT8, IOMT6, and IOMT9 are 98.9% identical to one another at the amino acid level, and they differ only in regions that are not involved in catalysis, by reference to the three-dimensional structure and reaction mechanism of IOMT8 (Zubieta et al., 2001). They may represent allelic variants in autotetraploid alfalfa. A fourth cDNA, IOMT2, encodes a slightly truncated protein that is 99.7% identical to IOMT8; like IOMT8, it has been shown to possess strict 7-O-methylation specificity when expressed in E. coli (He and Dixon, 1997). Biochemical fractionation studies have shown two forms of IOMT in alfalfa separable by ion-exchange chromatography; both show 7-position specificity, and it has not proven possible to characterize an activity, either soluble or microsomal, that methylates the 4′-hydroxyl of daidzein in vitro (Edwards and Dixon, 1991; He and Dixon, 1996). Thus, it is unlikely that the 36-kD protein observed in alfalfa leaf microsomes, the level of which increases in parallel with the 42-kD intact IOMT8 subunit in transgenic plants overexpressing IOMT8, is a novel form of IOMT with 4′ specificity, unless its structure is such that it retains strong cross-reactivity to anti-IOMT8 serum while at the same time possessing relatively low nucleotide similarity. Rather, the 4′ specificity most likely reflects the true in vivo activity of the IOMT gene products characterized previously.
We propose that colocalization of IOMT with the 2-HIS cytochrome P450 on membranes of the ER enables the OMT to capture the unstable trihydroxyisoflavanone product of 2-HIS and thereby prevent its dehydration to daidzein, as shown in the model in Figure 9. The dehydration of isoflavanone to isoflavone may be enzymatic (Hakamatsuka et al., 1998). However, it also can occur spontaneously in vivo, as demonstrated by the accumulation of isoflavone in Arabidopsis plants transformed solely with soybean 2-HIS (Jung et al., 2000; C.-J. Liu and R.A. Dixon, unpublished results), and this may be why the close association between IOMT and 2-HIS is necessary. The membrane localization of IOMT provides an explanation for why daidzein is not produced in vivo during phytoalexin biosynthesis in alfalfa and why there is no 7-O-methylation of daidzein in vivo by the enzyme characterized previously as a 7-O-methyltransferase in vitro.
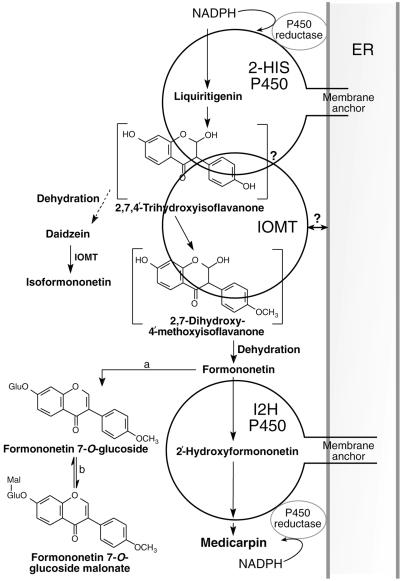
Figure 9.
Model for Metabolic Channeling in the Biosynthesis of Formononetin.
IOMT is shown associating with the membrane-bound 2-HIS P450 to capture the unstable 2,7,4′-hydroxyisoflavanone, thereby preventing its dehydration to daidzein and subsequent conversion to isoformononetin by the 7-position specificity of free IOMT. Subsequently, formononetin is either metabolized by 2′-hydroxylation involving a second membrane-associated cytochrome P450 (I2′H) en route to the phytoalexin medicarpin or converted to formononetin glucosyl malonate via formononetin glucoside. a, formononetin 7-O-glucosyl transferase; b, formononetin glucoside malonyl transferase. The question marks refer to the two models for interaction of IOMT with 2-HIS: direct interaction with the 2-HIS protein or close association by targeting of IOMT to the outside of the ER membrane.
The product of IOMT methylation, 2,7-dihydroxy-4′-methoxyisoflavanone, dehydrates to yield formononetin, a bona fide precursor of medicarpin. Dewick and Martin (1979) therefore were correct in proposing a tight linkage between the aryl migration and B-ring methylation reactions of isoflavone biosynthesis, although their model required methylation as an integral part of ring migration. If the association between 2-HIS and IOMT did not occur, the dehydration of trihydroxyisoflavanone would result in the formation of daidzein, which would be methylated by cytosolic IOMT to isoformononetin, a compound that cannot contribute further to isoflavonoid biosynthesis in alfalfa. The fact that isoformononetin is not observed in alfalfa tissue extracts, even though a portion of the IOMT appears to remain cytosolic, as seen from both our immunoblot and confocal microscopy analyses, can be explained readily by our model, in which metabolic channeling prevents the formation of daidzein.
When daidzein is fed to uninfected leaves of transgenic alfalfa ectopically expressing IOMT, it is converted to isoformononetin (He and Dixon, 2000). In contrast, isoformononetin was not produced from daidzein fed to elicited alfalfa cell cultures in the present work. This finding can be explained by the fact that, in the elicited cell cultures, endogenous 2-HIS and IOMT are coinduced and coupled closely according to the model in Figure 9, precluding daidzein from entry to the active site of IOMT. In contrast, unelicited transgenic alfalfa leaves overexpressing IOMT do not express 2-HIS, and the product of the IOMT transgene would be localized to the cytosol and available for the 7-O-methylation of daidzein.
Isoformononetin has been reported as a minor isoflavonoid component of soybean by Ingham et al. (1981), although the major constitutive and inducible isoflavonoids in this species are not O-methylated and daidzein is a major metabolite. Interestingly, isoflavone 2′-hydroxylase (I2′H) from soybean can act on daidzein to form 2′-hydroxydaidzein, the substrate for isoflavone reductase from that species and an intermediate in the biosynthesis of the phytoalexin glyceollin (Fischer et al., 1990). In contrast, I2′H from legume species in which the B-ring is methylated requires the 4′-methoxy group for activity (Dixon, 1999). Thus, the subcellular colocalization of 2-HIS and IOMT, together with the substrate specificity of I2′H, another microsomal cytochrome P450, determine the metabolic fate of liquiritigenin in isoflavonoid biosynthesis.
When IOMT is overexpressed in transgenic alfalfa, increased production of formononetin and medicarpin is observed after elicitation (He and Dixon, 2000), suggesting that IOMT is a rate-determining enzyme for the production of 4′-O-methylated isoflavonoids. At first, this appears to be incompatible with our data that suggest tight coupling of IOMT with 2-HIS on the microsomes but that also reveal a pool of IOMT, in excess of the requirements for coupling, in the cytosol. However, it is possible that all of the enzyme is localized microsomally in unperturbed cells that do not overexpress IOMT from an introduced transgene. Furthermore, transgenic overexpression of IOMT results in the increased expression of 2-HIS and several other enzymes of the isoflavonoid pathway in elicited alfalfa leaves (He and Dixon, 2000), which may be the explanation for the apparent rate-determining role of IOMT. Detailed flux control analysis will be necessary to resolve this question.
It remains to be determined how IOMT locates to the ER after elicitation. We have yet to address whether IOMT is localized via direct interactions with the 2-HIS P450 or whether it is associated with the endomembrane system by some other mechanism, such as membrane targeting after post-translational modification that brings it into close, but not direct, contact with 2-HIS. Resolution of this question may require the generation of transgenic alfalfa plants with epitope-tagged IOMT and 2-HIS for fluorescence resonance energy transfer studies or chemical cross-linking approaches. Post-translational modifications to IOMT associated with membrane targeting may lead to reduced resistance to proteolytic cleavage, as suggested by the presence of the 36-kD form of IOMT in microsomes from elicited leaves. The determination of exactly how IOMT is modified in response to elicitation will be important for a fuller understanding of the assembly of metabolic complexes on the ER. Arabidopsis chalcone isomerase has been proposed to undergo lipid modification that then directs the enzyme to a metabolic complex with chalcone synthase and perhaps other enzymes of flavonoid biosynthesis (Burbulis and Winkel-Shirley, 1999). In this case, however, the modified enzyme has reduced electrophoretic mobility (Burbulis and Winkel-Shirley, 1999). There are no obvious sites for lipid modification in the IOMT protein sequence. Nevertheless, other proteins, such as the soybean β-glucan elicitor binding protein, are known to exhibit membrane localization without possessing any obvious membrane localization domains (Umemoto et al., 1997).
Metabolic Channeling Involving Cytochrome P450s
The very complexity of metabolism itself argues for the need for physical ordering of sets of reactions, particularly in cases in which metabolites may participate in more than one reaction, as in the dehydration or O-methylation of 2,4′,7-trihydroxyisoflavanone in the present work, or cases in which different isoenzymic forms might participate in pathways leading to different end products from common intermediates. Membrane-associated cytochrome P450 enzymes occur in many stages of plant natural product biosynthesis (Chapple, 1998) and might serve as “anchoring points” for the assembly of associated enzymes on the outer surface of the ER (Hrazdina and Wagner, 1985). In the case of the phenylpropanoid pathway, such enzymes include cinnamate 4-hydroxylase in the core phenylpropanoid pathway, ferulate (coniferaldehyde) 5-hydroxylase in the lignin branch pathway, flavonoid 3′,5′-hydroxylase in flavonoid biosynthesis, and 2-HIS at the entry point into isoflavonoid biosynthesis. Metabolic channeling has been either demonstrated or proposed for sets of consecutive reactions involving each of these cytochrome P450s (Burbulis and Winkel-Shirley, 1999; Rasmussen and Dixon, 1999; Dixon et al., 2001; Winkel-Shirley, 2001). However, it is not known in any of these cases how the interactions between operationally soluble and integral membrane-bound enzymes occur. Understanding the cellular mechanism of isoflavone methylation may provide a model for understanding a more widespread form of metabolic control.
METHODS
Plant Materials
Transgenic alfalfa (Medicago sativa cv Regen SY) plants were grown under greenhouse conditions. Details of the lines, which harbored either empty vector or vector containing alfalfa isoflavone O-methyltransferase 8 (IOMT8) under the control of the 35S promoter of cauliflower mosaic virus, have been given elsewhere (He and Dixon, 2000). Young seedlings (4 to 5 days old) were cut at the mid stem, and the upper portions were allowed to stand in aqueous 0.3 mM copper chloride for 8 hr. The leaves were either used for particle bombardment or frozen in liquid N2 and stored at −80°C before enzyme or metabolite analyses.
Alfalfa cell suspension cultures were initiated and maintained in Schenk and Hildebrandt medium at 24°C in the dark, as described previously (Dalkin et al., 1990). The cultures were subcultured every 14 days. Cells were treated with yeast elicitor (Schumacher et al., 1987) (final concentration, 50 μg glucose equivalent/mL) 7 days after subculture. Cells were collected after 24 hr (for enzyme assay) or 48 hr (for metabolite analysis) by vacuum filtration through a nylon mesh and frozen in liquid N2.
Chemicals
S-Adenosyl-l-methyl-3H-methionine (SAM; 3.4 TBq/mmol) was purchased from Amersham Pharmacia Biotech (Buckinghamshire, UK). 3H-Liquiritigenin (740 GBq/mmol) was from Sibtech, Inc. (Newington, CT). Isoformononetin and 2′-hydroxyformononetin were from Apin (Abingdon, UK). Other isoflavonoids were from Indofine Chemical Co. (Somerville, NJ). All other chemicals were from Sigma (St. Louis, MO).
Precursor Dilution Experiments
Alfalfa suspension cell cultures (20-mL batches, 5 days after subculture) were treated with yeast elicitor as described above. A mixture of 1 μmol of 3H-liquiritigenin (2 μCi/μmol) with or without 2 μmol of unlabeled daidzein or formononetin was fed to the cells. After incubation for 48 hr, the cells were collected, washed four times with 20 mL of water, and frozen in liquid N2. Two grams (fresh weight) of cells was homogenized and extracted three times with 10 mL of cold acetone (−20°C). The extracts were pooled and concentrated under a stream of N2. The residues were dissolved in 25 mM citrate phosphate buffer, pH 5.2, digested with 100 units of β-glucosidase at 37°C for 24 hr, and then extracted three times with 4 mL of ethyl acetate. The extracts were combined and concentrated again, and the residues were dissolved in 100 μL of methanol for HPLC analysis. Aliquots of medium were adjusted to pH 2.0 to 3.0 by treatment with acetic acid and extracted twice with an equal volume of ethyl acetate. The extracts were pooled, concentrated, dissolved in methanol, and subjected to HPLC for determination of the amount of daidzein and formononetin remaining in the medium. Standard curves for isoflavonoids were constructed from HPLC diode array determinations using authentic compounds.
Preparation of Microsomes
Suspension-cultured cells or leaves (2 to 3 g fresh weight) were homogenized in 5 mL of buffer A (0.1 M potassium phosphate, pH 7.5, containing 0.4 M sucrose and 14 mM 2-mercaptoethanol) with 1% (w/w) polyvinylpolypyrrolidone and 0.3 g of Dowex-1 × 2 (preequilibrated with 0.1 M phosphate buffer, pH 7.5) per gram of cells. The homogenate was centrifuged at 2,300g for 15 min at 4°C, and the supernatant was filtered through glass wool and centrifuged at 135,000g for 1 hr at 4°C. The supernatant was decanted for enzyme assay, and the resultant pellet was washed carefully two times using buffer B (0.1 M potassium phosphate, pH 8.0, containing 0.4 M sucrose and 3.5 mM 2-mercaptoethanol). The pellet was resuspended in 300 μL of buffer B with a rubber spatula, and the microsome preparation was assayed directly. An alternative method for microsome preparation used buffer C (0.2 M Tris-HCl, pH 7.5, 0.4 M sucrose, 28 mM 2-mercaptoethanol, and 10 mM EDTA) for extraction and resuspension of the microsomes.
In Vitro Channeling Assays
The assay mixture contained assay buffer (0.1 M potassium phosphate, pH 8.5, containing 0.4 M sucrose and 3.5 mM 2-mercaptoethanol), 2 mM Pefablock (Boehringer Mannheim), 20 μM 3H-liquiritigenin (7.4 GBq/mmol), 2.5 mM SAM, 1 mM NADPH, and 50 μL of microsome preparation (50 to 70 μg of protein) in a final volume of 100 μL. Reactions were started by adding microsomes and incubated at 15°C for 2 hr with gentle shaking. NADPH and/or SAM were omitted from controls. Reactions were stopped by adding 30 μL of acetic acid and extracted three times with 300 μL of ethyl acetate. The extracts were concentrated under a stream of N2 and dissolved in 100 μL of methanol. After mixing with authentic standards of daidzein, isoformononetin, formononetin, and 2′-hydroxyformononetin, the samples were resolved by HPLC with diode array detection. Corresponding fractions were collected, and the radioactivity was determined by scintillation counting (LS1701; Beckman). Protein was determined using a protein assay kit from Bio-Rad (Hercules, CA).
Assay of IOMT Activity
Soluble and microsomal fractions for IOMT assay were prepared according to Edwards and Kessmann (1992) in 0.2 M Tris-HCl buffer, pH 7.5, containing 14 mM 2-mercaptoethanol and 5 mM EDTA. Reaction mixtures (60 μL) contained 50 nmol of daidzein, 150 nmol of SAM (246 MBq/mmol), and 30 μL of soluble fraction or 20 μL of microsomal fraction. Mixtures were incubated at 30°C for 30 min, and the reactions were stopped by the addition of 10 μg of formononetin. The mixtures were extracted twice with ethyl acetate, the extracts were concentrated to dryness, and the residues were resuspended in 50 μL of methanol and separated by HPLC or thin-layer chromatography.
Analysis of Isoflavonoids by HPLC and Thin-Layer Chromatography
Reaction products from IOMT enzyme assays (20 μL) and extracts from precursor feeding experiments with suspension-cultured cells (30 μL) were analyzed by HPLC. Samples were applied to an ODS2 reverse phase column (Metachem Technologies Inc., Torrance, CA) (5 μm particle size, 4.6 × 250 mm) and eluted in 1% phosphoric acid with an increasing concentration gradient of acetonitrile. Four different gradients were used. Gradient I (for IOMT assays and nondigested cell extracts): 0 to 45 min, 20 to 54% acetonitrile at a constant flow rate of 0.8 mL/min. Gradient II (for in vitro channeling assays and analysis of glucosidase-treated extracts after precursor feeding): 0 to 5 min, 30 to 31% acetonitrile; 5 to 15 min, 31%; 15 to 19 min, 31 to 32%; 19 to 42 min, 32%; 42 to 59 min, 32 to 43%, at a constant flow rate of 0.8 mL/min. Gradient III (for resolution of daidzein from liquiritigenin): 0 to 22 min, 18% acetonitrile; 22 to 34 min, 18 to 22%, 34 to 38 min, 22%, at a constant flow rate of 2 mL/min. Gradient IV (for analysis of isoflavone conjugates after feeding daidzein): 0 to 5 min, 5% acetonitrile; 5 to 10 min, 5 to 10%; 10 to 15 min, 10 to 15%; 15 to 20 min, 15%; 20 to 25 min, 15 to 17%; 25 to 30 min, 17 to 23%; 30 to 65 min, 23 to 50%, at a constant flow rate of 1 mL/min. UV light absorption was monitored at 254 and 287 nm with a photodiode array detector. The corresponding fractions were collected, and radioactivity was determined by scintillation counting.
Further details of the identification of alfalfa isoflavonoids by HPLC have been presented elsewhere (Sumner et al., 1996; He and Dixon, 2000). The identifications in the present work were based on comparisons of chromatographic behavior and UV light spectra with our previous reports and parallel analysis of authentic samples.
Thin-layer chromatography analysis of reaction products from IOMT assays and in vivo labeling studies on Si250 silica gel (J.T. Baker, Phillipsburg, NJ) was performed as described previously (Edwards and Dixon, 1991). Spots corresponding to positions of authentic standards were observed under UV light at 324 nm, and the areas of silica gel were scraped from the plates for determination of radioactivity by scintillation counting.
SDS-PAGE and Protein Gel Blot Analysis
Based on the leaf homogenate volume, equivalent amounts of soluble and microsomal proteins were resolved on precast 12% Tris-glycine gels (Novex, San Diego, CA) and transferred to nitrocellulose membranes using a Bio-Rad Trans-Blot apparatus. The membranes were blocked with 3% (w/v) BSA and probed with a primary antibody against recombinant alfalfa IOMT8 (He and Dixon, 2000) at a dilution of 1:5000 in 5% fat-free milk powder in phosphate-buffered saline plus Tween 20 (PBST). Goat anti-rabbit IgG horseradish peroxidase conjugate was used as the secondary antibody at a dilution of 1:10,000 in PBST, with visualization using a chemiluminescence assay kit (Amersham). Kaleidoscope prestained protein standards (Bio-Rad) were used as molecular mass markers.
Construction of Chimeric Genes with Enhanced Green Fluorescent Protein and Red Fluorescent Protein
To generate a fusion protein of IOMT8 with the enhanced green fluorescent protein (EGFP; Clontech, Palo Alto, CA), a two-step recombinant polymerase chain reaction (PCR) strategy was applied (Higuchi, 1990). In the first step, the IOMT8 open reading frame was amplified using pfu DNA polymerase (Stratagene, La Jolla, CA) with a forward primer to introduce an EcoRI restriction site (IOMT forward, 5′-ACGGAATTCAAATGGCTTCATCAATTAA-3′) and a reverse primer containing reverse complementary sequences from the end of the IOMT8 and the start of the EGFP open reading frames (IOMT reverse, 5′-GCCCTTGCTCACCATTGGATAGATCTCAATAA-3′). Similarly, EGFP was amplified using a forward primer complementary to the IOMT reverse primer (EGFP forward, 5′-ATTGAGATCTATCCAATGGTGAGC-AAGGGCGA-3′) and a reverse primer with an XbaI restriction site (EGFP reverse, 5′-AGTTATCTAGAGTCGCGGCC-3′). The resulting IOMT8 and EGFP open reading frame fragments were recovered from an agarose gel and served as templates in a second PCR using the IOMT forward and EGFP reverse primers. After digestion with EcoRI and XbaI, the resulting chimeric cDNA was inserted into the corresponding sites of the shuttle vector pRTL2 (Restrepo et al., 1990) under the control of a double 35S promoter.
A 2-hydroxyisoflavanone synthase (2-HIS) cDNA was isolated from a Medicago truncatula root cDNA library with a heterologous cDNA probe of CYP93C1 from soybean (Steele et al., 1999). By using the same PCR strategy described above, the open reading frame of 2-HIS was amplified using pfu polymerase, with the primers 2-HIS forward (5′-AAGGAATTCCATGTTGGTGGA-3′) and 2-HISGFP reverse (5′-GCCCTTGCTCACCATGGAGGAAAGAAGTTTA-3′). EGFP was amplified using the primers EGFP2-HIS forward (5′-ACTTCTTTCCTCCATGGTGAGCAAGGGCGA-3′) and EGFP reverse. A 1.6-kb 2-HIS open reading frame fused in frame at the N terminus of EGFP was generated, and the chimeric DNA was digested with EcoRI and XbaI and inserted into pRTL2.
The DNA fragment encoding free EGFP was amplified with a forward primer (5′-ACGGAATTCCATGGTGAGCAA-3′) containing an EcoRI restriction site and the EGFP reverse primer and also inserted in pRTL2. Red fluorescent protein (RFP)–HDEL was made from DsRed (Clontech) using the protocol described by Haseloff et al. (1997) but replacing modified green fluorescent protein (mGFP) with RFP. The resulting chimeric DNA cassette was ligated into pRTL101.
Particle Bombardment and Confocal Microscopy
Plasmid DNAs (∼5 μg) harboring the IOMT8–EGFP, 2-HIS–EGFP, mGFP4–HDEL (Haseloff et al., 1997), RFP–HDEL, or free EGFP gene under the control of the double 35S promoter was mixed with 50 μL of an aqueous suspension containing 7.5 mg of 1.0-μm gold particles (Bio-Rad). The gold–DNA suspension was dispersed with moderate vortexing and sonication in the presence of 1.25 M CaCl2 and 17 mM spermidine and kept on ice for 5 to 30 min. The DNA-coated gold particles then were collected by brief centrifugation, washed, resuspended in ethanol, and spread onto plastic carrier discs for biolistic bombardment using the particle delivery system 1000/He (Bio-Rad).
Alfalfa seedlings were treated with 0.3 mM copper chloride or distilled water for 8 hr. Young leaves were excised and placed on moist filter paper in Petri dishes. The gold particles were fired at 1100 p.s.i., and bombarded leaves were maintained in the dark at 24°C before examination. Leaves with EGFP-expressing cells were examined 12 hr after bombardment using a Bio-Rad 1024ES confocal imaging system attached to a Zeiss Axioskop microscope (Carl Zeiss, Thornwood, NY). Cells expressing EGFP were imaged with a 40× water immersion objective by excitation with the 488 line of a krypton/argon laser and use of a 522 DF35 emission filter. Serial optical sections were obtained at 0.36-μm intervals, and projections of optical sections were accomplished using LaserSharp image-processing software (Bio-Rad). Optical sections were processed and assembled digitally using Photoshop 5.0 L.E. (Adobe Systems, Mountain View, CA).
Cells cotransfected with IOMT8–EGFP and RFP–HDEL were imaged by simultaneous excitation at 488 and 568 nm, and detection of emitted light between 522 and 535 nm for EGFP and at 596 nm for RFP from a single 0.36-μm optical section. Alternatively, dual-color images were acquired by sequentially scanning with settings optimal for EGFP followed by scanning the same optical section with settings optimal for RFP and then merging projections of the individual channels using LaserSharp imaging software.
Acknowledgments
We thank Dr. Elison Blancaflor for advice and assistance with the confocal microscopy, Dr. Xian-Zhi He for useful suggestions, Jack Blount for help with HPLC analysis, Drs. Yiming Bao and Rouf Mian for critical reading of the manuscript, and Cuc Ly for artwork. This work was funded by the Samuel Roberts Noble Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010382.
References
- Akashi, T., Aoki, T., and Ayabe, S. (1999). Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 121, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, T., Sawada, Y., Aoki, T., and Ayabe, S.-I. (2000). New scheme of the biosynthesis of formononetin involving 2,7,4′-trihydroxyisoflavanone but not daidzein as the methyl acceptor. Biosci. Biotechnol. Biochem. 64, 2276–2279. [DOI] [PubMed] [Google Scholar]
- Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2000). Biochemistry, mutagenesis and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97, 11984–11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis, I.E., and Winkel-Shirley, B. (1999). Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA 96, 12929–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell, J. (1995). Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 521–547. [Google Scholar]
- Chapple, C. (1998). Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 311–343. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griggitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czichi, U., and Kindl, H. (1975). Formation of p-coumaric acid and o-coumaric acid from l-phenylalanine by microsomal membrane fractions from potato: Evidence of membrane-bound enzyme complexes. Planta 125, 115–125. [DOI] [PubMed] [Google Scholar]
- Czichi, U., and Kindl, H. (1977). Phenylalanine ammonia-lyase and cinnamic acid hydroxylase as assembled consecutive enzymes on microsomal membranes of cucumber cotyledons: Cooperation and subcellular distribution. Planta 134, 133–143. [DOI] [PubMed] [Google Scholar]
- Dalkin, K., Edwards, R., Edington, B., and Dixon, R.A. (1990). Stress responses in alfalfa (Medicago sativa L.). I. Elicitor-induction of phenylpropanoid biosynthesis and hydrolytic enzymes in cell suspension cultures. Plant Physiol. 92, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewick, P.M., and Martin, M. (1979). Biosynthesis of pterocarpan, isoflavan and coumestan metabolites of Medicago sativa: Chalcone, isoflavone and isoflavanone precursors. Phytochemistry 18, 597–602. [Google Scholar]
- Dixon, R.A. (1999). Isoflavonoids: Biochemistry, molecular biology and biological functions. In Comprehensive Natural Products Chemistry, Vol. 1, U. Sankawa, ed (New York: Elsevier), pp. 773–823.
- Dixon, R.A. (2000). Metabolic channeling in the phenylpropanoid pathway: Implications for pathway engineering in transgenic plants. Polyphenols Actualite 20, 14–18. [Google Scholar]
- Dixon, R.A., Chen, F., Guo, D., and Parvathi, K. (2001). The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57, 1069–1084. [DOI] [PubMed] [Google Scholar]
- Edwards, R., and Dixon, R.A. (1991). Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30, 2597–2606. [Google Scholar]
- Edwards, R., and Kessmann, H. (1992). Isoflavonoid phytoalexins and their biosynthetic enzymes. In Molecular Plant Pathology: A Practical Approach, Vol. 2, S.J. Gurr, M.J. McPherson, and D.J. Bowles, eds (Oxford, UK: IRL Press), pp. 45–62.
- Fischer, D., Ebenau-Jehle, C., and Grisebach, H. (1990). Phytoalexin synthesis in soybean: Purification and characterization of NADPH:2′- hydroxydaidzein oxidoreductase from elicitor-challenged soybean cell cultures. Arch. Biochem. Biophys. 276, 390–395. [DOI] [PubMed] [Google Scholar]
- Hagmann, M., and Grisebach, H. (1984). Enzymatic rearrangement of flavanone to isoflavone. FEBS Lett. 175, 199–202. [Google Scholar]
- Hakamatsuka, T., Mori, K., Ishida, S., Ebizuka, Y., and Sankawa, U. (1998). Purification of 2-hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata. Phytochemistry 49, 497–505. [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodges, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.-Z., and Dixon, R.A. (1996). Affinity chromatography, substrate/product specificity and amino acid sequence analysis of an isoflavone O-methyltransferase from alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 336, 121–129. [DOI] [PubMed] [Google Scholar]
- He, X.-Z., and Dixon, R.A. (1997). A cDNA of an additional member of the isoflavone O-methyltransferase (IOMT2) gene family in Medicago sativa. Plant Physiol. 115, 1289. [Google Scholar]
- He, X.-Z., and Dixon, R.A. (2000). Genetic manipulation of isoflavone 7-O-methyltransferase enhances the biosynthesis of 4′-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12, 1689–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.-Z., Reddy, J.T., and Dixon, R.A. (1998). Stress responses in alfalfa (Medicago sativa L.). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol. Biol. 36, 43–54. [DOI] [PubMed] [Google Scholar]
- Higuchi, R. (1990). Recombinant PCR. In PCR Protocols, M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, eds (New York: Academic Press), pp. 177–182.
- Hrazdina, G., and Wagner, G.J. (1985). Compartmentation of plant phenolic compounds: Sites of synthesis and accumulation. Annu. Proc. Phytochem. Soc. Eur. 25, 119–133. [Google Scholar]
- Ingham, J.L., Keen, N.T., Mulheirn, L.J., and Lyne, R.L. (1981). Inducibly-formed isoflavonoids from leaves of soybean. Phytochemistry 20, 795–798. [Google Scholar]
- Jackobs, S., Subramaniam, V., Schonle, A., Jovin, T.M., and Hell, S.W. (2000). EGFP and DsRed expressing cultures of Escherichia coli imaged by confocal, two-photon and fluorescence lifetime microscopy. FEBS Lett. 479, 131–135. [DOI] [PubMed] [Google Scholar]
- Jung, W., Yu, O., Lau, S.-M.C., O'Keefe, D.P., Odell, J., Fader, G., and McGonigle, B. (2000). Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18, 208–212. [DOI] [PubMed] [Google Scholar]
- Kahn, R.A., Bak, S., Svendsen, I., Halkier, B.A., and Moller, B.L. (1997). Isolation and reconstitution of cytochrome P450ox and in vitro reconstitution of the entire biosynthetic pathway of the cyanogenic glucoside dhurrin from sorghum. Plant Physiol. 115, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessmann, H., Choudhary, A.D., and Dixon, R.A. (1990). Stress responses in alfalfa (Medicago sativa L.). III. Induction of medicarpin and cytochrome P450 enzyme activities in elicitor-treated cell suspension cultures and protoplasts. Plant Cell Rep. 9, 38–41. [DOI] [PubMed] [Google Scholar]
- Kochs, G., and Grisebach, H. (1986). Enzymic synthesis of isoflavones. Eur. J. Biochem. 155, 311–318. [DOI] [PubMed] [Google Scholar]
- Rasmussen, S., and Dixon, R.A. (1999). Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, H.-M., Gundlach, H., Fiedler, F., and Zenk, M.H. (1987). Elicitation of benzophenanthridine alkaloid synthesis in Eschscholtzia cell cultures. Plant Cell Rep. 6, 410–413. [DOI] [PubMed] [Google Scholar]
- Singh, S., McCallum, J., Gruber, M.Y., Towersw, G.H.N., Muir, A.D., Bohm, B.A., Koupai-Abyazani, M.R., and Glass, A.D.M. (1997). Biosynthesis of flavan-3-ols by leaf extracts of Onobrychis viciifolia. Phytochemistry 44, 425–432. [Google Scholar]
- Srere, P.A. (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124. [DOI] [PubMed] [Google Scholar]
- Steele, C.L., Gijzen, M., Qutob, D., and Dixon, R.A. (1999). Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367, 147–150. [DOI] [PubMed] [Google Scholar]
- Sumner, L., Paiva, N.L., Dixon, R.A., and Geno, P.W. (1996). High-performance liquid chromatography/continuous-flow liquid secondary ion mass spectrometry of flavonoid glucosides in leguminous plant extracts. J. Mass Spectrom. 31, 472–485. [DOI] [PubMed] [Google Scholar]
- Umemoto, N., Kakitani, M., Iwamatsu, A., Yoshikawa, M., Yamaoka, N., and Ishida, I. (1997). The structure and function of a soybean β-glucan-elicitor-binding protein. Proc. Natl. Acad. Sci. USA 94, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (1999). Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107, 142–149. [Google Scholar]
- Winkel-Shirley, B. (2001). Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E., and Francis, C.M. (1968). Flavonoids in genotypes of Trifolium subterraneum. II. Mutants of the Geraldton variety. Phytochemistry 7, 2131–2137. [Google Scholar]
- Zubieta, C., Dixon, R.A., and Noel, J.P. (2001). Crystal structures of chalcone O-methyltransferase and isoflavone O-methyltransferase reveal the structural basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8, 271–279. [DOI] [PubMed] [Google Scholar]