Abstract
The reaction center core of photosystem II is composed of two chlorophyll binding proteins, D1 and D2, that are encoded by the chloroplast genes psbA and psbD. These chlorophyll binding proteins are damaged during photochemistry, especially under high irradiance. Photosystem II function is maintained under these conditions through turnover and resynthesis of D1 and D2. Blue light–activated transcription of psbD from a special light-responsive promoter is part of the repair system. In this study, light-activated chloroplast and psbD transcription were studied after dark adaptation of 21-day-old light-grown Arabidopsis plants. Illumination of dark-adapted plants with red light increased chloroplast transcription activity and transcription from the psbD light-responsive promoter. Blue light further increased chloroplast transcription activity and stimulated differential transcription from the psbD light-responsive promoter. Photoreceptor mutants showed that blue light–specific activation of chloroplast transcription and the psbD light-responsive promoter involve cryptochrome 1 (cry1) or cryptochrome 2 (cry2) and phytochrome A (phyA). Blue light–induced activation of the psbD light-responsive promoter was normal in det2-1 and hy5-1 but attenuated in det3-1. Therefore, cry1/cry2/phyA–mediated blue light activation of the psbD light-responsive promoter in 21-day-old Arabidopsis plants does not involve hy5, a transcription factor that mediates other phyA and blue light–induced responses.
INTRODUCTION
Light plays a central regulatory role in plant and chloroplast development in addition to being the source of energy for plant life (for reviews, see Mullet, 1988; Link, 1991; Chory, 1997). Information about light quality, intensity, and duration is measured by a large number of different plant photoreceptors (reviewed by Briggs and Liscum, 1997; Chory, 1997; Fankhauser and Chory, 1997; Briggs and Huala, 1999; Neff et al., 2000). The selective blue light/UVA/UVB photoreceptors include the cryptochromes 1 and 2 (cry1 and cry2), phototropin (nph1), carotenoids (i.e., zeaxanthin), and other less well characterized blue light photoreceptors (reviewed by Briggs and Huala, 1999). The red light photoreceptors include the phytochromes, protochlorophyllide holochrome, and chlorophyll. Although the red light photoreceptors are so named because they absorb red light, these pigments also absorb and respond to blue light via their soret absorption bands (Schafer and Haupt, 1983). The plant photoreceptors modulate germination, hypocotyl and leaf elongation, phototropism, leaf and chloroplast development, stomatal conductance, onset of flowering, circadian rhythms, and nuclear and chloroplast gene expression (reviewed by Briggs and Liscum, 1997; Chory, 1997; Fankhauser and Chory, 1997; Briggs and Huala, 1999; Neff et al., 2000). The extent of each photoreceptor's influence varies with plant species, stage of development, and gene examined.
Light drives primary photochemistry that generates ATP and reducing power for carbon fixation and all other aspects of plant growth and development. The primary photochemical reactions occur in photosystem I and photosystem II, large protein complexes that provide a precise scaffold for the pigments and cofactors that mediate vectorial primary charge separation and electron transfer steps. Exposure of plants to high light, especially under conditions of abiotic stress, limited cardon dioxide availability, or excess carbon, often results in photodamage to the photosynthetic apparatus (Barber and Andersson, 1992; Melis et al., 1992; Aro et al., 1993; Melis, 1999). Photosystem II, the site of oxygen evolution, is particularly susceptible to damage when plants are subjected to high light intensities (Barber and Andersson, 1992; Melis et al., 1992; Aro et al., 1993). The photosystem II reaction center chlorophyll binding proteins D1 and D2 are most often damaged under these conditions (Mattoo et al., 1984; Ohad et al., 1985). Plants are able to repair damaged photosystem II complexes through disassembly of photosystem II complexes, synthesis of new D1 and D2 subunits, and reassembly of the proteins and cofactors into functional complexes (Melis, 1999). In mature leaves, synthesis of D1 and D2 is primarily needed to repair photosystem II complexes after subunit turnover. At this stage of development, the genes encoding D1 and D2 are differentially expressed relative to other plastid genes (Baumgartner et al., 1993; Christopher and Mullet, 1994). Differential expression of psbA involves light-activated transcription (Klein and Mullet, 1990; Chun et al., 2001), extraordinary RNA stability (Deng and Gruissem, 1987; Mullet and Klein, 1987; Rapp et al., 1992; Baumgartner et al., 1993; Kim et al., 1993), and light-regulated translation (Klaff and Gruissem, 1991; Danon and Mayfield, 1994; Kim and Mayfield, 1997). In contrast, psbD transcripts are differentially maintained in mature chloroplasts primarily because of the activity of an unusual blue light–activated promoter (Gamble and Mullet, 1989; Sexton et al., 1990a).
Higher plant psbD genes are located in a complex chloroplast operon that, in some plant species, also contains psbC, psbK, orf62, and trnG (Sexton et al., 1990b). In barley, this operon is transcribed from at least three different promoters. One of these promoters, the psbD light-responsive promoter (psbD-LRP), is selectively activated by illumination of plants with high-fluence blue/UVA light (Gamble and Mullet, 1989; Christopher and Mullet, 1994). Transcription from the psbD-LRP is low early in leaf development and in cotyledons but increases during leaf and plant development (Christopher, 1996; Christopher and Hoffer, 1998). In mature light-grown plants, transcripts derived from the psbD-LRP are the most abundant psbD RNAs (Christopher and Mullet, 1994). The psbD-LRP is conserved among cereals, dicots, and black pine (Christopher et al., 1992). However, this promoter is not present in the liverwort Marchantia polymorpha (Ohyama et al., 1986) and three non-green algae that grow in low light or underwater environments (Kowallik et al., 1995; Reith and Munholland, 1995; Stirewalt et al., 1995). This is consistent with a role for the psbD-LRP in high-light, UV-A–rich environments, conditions that promote PSII photo damage. Accumulation of psbD-LRP transcripts is also regulated by circadian cycling, providing yet another level of control over transcription from this promoter (Nakahira et al., 1998; Thum et al., 2001).
Over the past several years, the architecture of the psbD-LRP has been intensively investigated in vitro (Kim and Mullet, 1995; To et al., 1996; Nakahira et al., 1998; Kim et al., 1999b) and in transgenic plants (Allison and Maliga, 1995; Thum et al., 2001). The plastid-encoded RNA polymerase (PEP), which is similar in structure and function to bacterial RNA polymerases, transcribes the psbD-LRP (Hajdukiewicz et al., 1997; Nakahira et al., 1998; Hess and Börner, 1999; Kim et al., 1999b). A −10 sequence (TATTCT) immediately upstream of the site of transcription initiation is essential for psbD-LRP activity (To et al., 1996; Nakahira et al., 1998; Kim et al., 1999b; Thum et al., 2001). Unlike most PEP promoters, the psbD-LRP does not require a −35 prokaryotic-like transcription element (TTGACA) for activity (To et al., 1996; Nakahira et al., 1998; Kim et al., 1999b; Thum et al., 2001). Instead, this function is replaced by a sequence immediately upstream from −35, named the AAG-box, which binds an activating factor, termed AGF, that is required for transcription from this promoter. A basic helix-loop-helix DNA binding protein encoded by the nucleus is a component of the AGF complex (Baba et al., 2001). The AGF is thought to act like bacterial transcription activators by binding and positioning the chloroplast RNA polymerase on the psbD-LRP (Allison and Maliga, 1995; Kim et al., 1999b). A second complex, termed PGTF, binds upstream of the AGF (Kim and Mullet, 1995). The binding of this complex to DNA is decreased by ADP-dependent phosphorylation, leading to the suggestion that PGTF binding modulates the activity of the psbD-LRP in response to light/dark cycles (Kim et al., 1999a).
Our understanding of how light regulates chloroplast transcription and differential transcription from the psbD-LRP is still rudimentary. Early analysis of light induction demonstrated that high-fluence blue light activates the psbD-LRP (Gamble and Mullet, 1989; Christopher and Mullet, 1994). However, the blue light photoreceptor involved has not been identified. During leaf development, red and far-red light also modulate psbD-LRP activity, presumably through photosynthetic electron transport and phytochrome, respectively (Christopher, 1996). Recent advances in our understanding of blue and red light photoreceptors and the availability of photoreceptor and light-signaling mutants provide an opportunity to clarify the photobiology of blue light activation of the psbD-LRP. In this study, analysis of Arabidopsis photobiology mutants showed that blue light activation of the psbD-LRP is mediated by cry1 or cry2 and phytochrome A (phyA). To our knowledge, the psbD-LRP is the first plastid-encoded gene that shows this type of photoreceptor dependence.
RESULTS
Light Modulates psbD-LRP Transcript Levels in 21-Day-Old Arabidopsis Plants
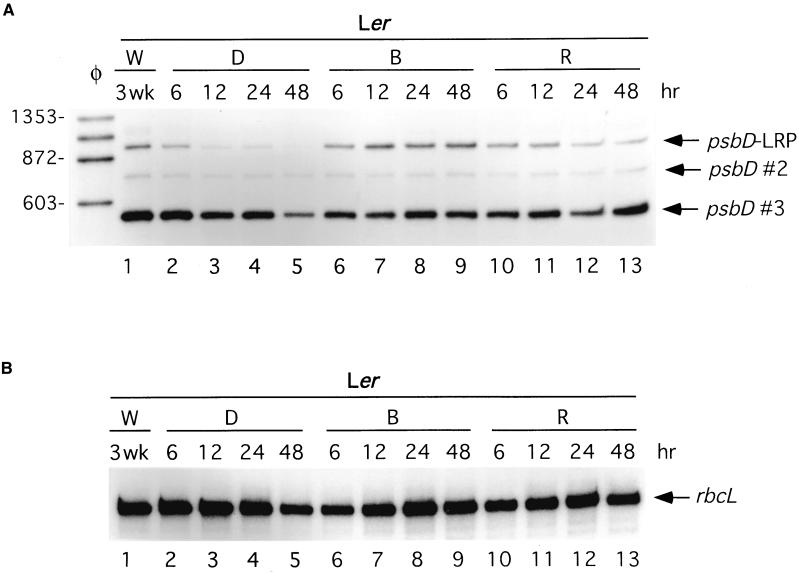
Primer extension assays shown in Figure 1 were used to characterize light-modulated changes in psbD and rbcL transcript abundance in 21-day-old Arabidopsis thaliana plants of the ecotype Landsberg erecta (Ler). Consistent with previous analysis (Hoffer and Christopher, 1997), transcripts derived from the psbD-LRP were located 950 bp upstream of the psbD open reading frame (Figure 1, transcripts marked psbD-LRP). The two smaller psbD transcripts shown in Figure 1 are derived either from processing the 950-nucleotide transcript or from uncharacterized promoters downstream of the psbD-LRP (Figure 1, psbD #2 and psbD #3). Transcripts derived from rbcL were also analyzed using primer extension assays (Figure 1B). Light-induced changes in psbD and rbcL transcript abundance were characterized in plants grown for 21 days in continuous white light and then transferred to darkness for 48 hr. After dark adaptation, plants were illuminated with red light or blue light at 30 μE·m−2·sec−1 for an additional 48 hr. Primer extension assays showed that transcripts arising from the psbD-LRP decreased in abundance within 6 hr after the transfer of plants to darkness and declined further during the 48 hr of dark treatment (Figure 1A, lanes 2 to 5). Dark treatment also caused a decrease in rbcL RNA level; however, the decrease in transcript abundance occurred slowly (Figure 1B, lanes 2 to 5). When 48-hr dark-adapted plants were illuminated with blue light, psbD-LRP transcript levels increased within 6 hr, reached a maximum between 12 and 24 hr, and remained elevated in plants illuminated for 48 hr (Figure 1A, lanes 6 to 9). Illumination of dark-adapted plants with red light also increased psbD-LRP transcript abundance within 6 hr (Figure 1, lane 5 versus 10). However, psbD-LRP transcript levels declined when red light illumination was continued for 24 to 48 hr (Figure 1A, lanes 10 to 13). The abundance of other psbD transcripts changed to a lesser extent during the dark-adaptation and illumination treatments (Figure 1, psbD #2 and psbD #3).
Figure 1.
Light-Induced Changes in psbD and rbcL Transcript Levels in 21-Day-Old Arabidopsis Plants.
The abundance of psbD (A) and rbcL (B) transcripts was monitored using primer extension analysis in response to a 48-hr dark adaptation (D) and subsequent illumination with blue (B) or red (R) light (both at 30 μE·m−2·sec−1) for an additional 48 hr. The lanes labeled W represent tissue from plants grown for 3 weeks (wk) under continuous white light. Tissue samples were taken at the time points indicated above each of the lanes.
(A) psbD transcripts derived from the LRP (950 nucleotides) are indicated by an arrow and labeled as such. Two other psbD transcripts are designated by an arrow and labeled psbD #2 and psbD #3. Lane φ shows some of the φX174-HaeIII restriction fragments used as length markers.
(B) rbcL transcripts are indicated by an arrow and labeled.
Cry1 or Cry2 Can Mediate Blue Light Activation of Chloroplast Transcription and Differential Accumulation of psbD-LRP Transcripts
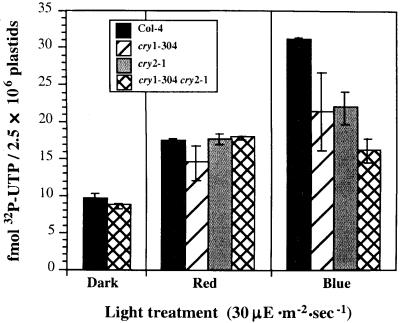
The results in Figure 1 show that both red light and blue light can stimulate accumulation of transcripts from the psbD-LRP, although the extent and kinetics of transcript accumulation differ in the two light treatments. Figure 2 shows that illumination of dark-adapted plants for 8 hr with red light increased chloroplast transcription twofold, whereas illumination with blue light activated transcription fourfold (Figure 2, Col-4). Red light could stimulate chloroplast transcription through chlorophyll-driven photosynthetic electron transport and/or phytochrome-induced changes in gene expression. Blue light–induced increases in chloroplast transcription could be mediated by chlorophyll, phytochrome, cryptochrome, and/or phototropin because these photoreceptors have absorption bands in the blue. As a starting point, we analyzed blue light–induced activation of chloroplast transcription in a CRY1 mutant (cry1-304), CRY2 mutant (cry2-1), and a CRY1/CRY2 double mutant (cry1-304 cry2-1). Mutant plants were dark-adapted and then illuminated with red or blue light for 8 hr before chloroplast isolation and run-on transcription assays. Figure 2 shows that illumination of wild-type plants with blue light increased chloroplast transcription fourfold; however, illumination of the cry1-304 cry2-1 double mutant with blue light increased chloroplast transcription only twofold, the same as did red light. This result indicates that cryptochromes help mediate the blue light–induced increase in overall chloroplast transcription activity in older dark-adapted plants. Interestingly, partial blue light activation of chloroplast transcription occurred in cry1-304 and cry2-1, the single CRY mutants. This suggests that either cry1 or cry2 can mediate blue light activation of chloroplast transcription after dark adaptation. A fluence response study is needed to determine if cry1 and cry2 can independently induce full activation of chloroplast transcription.
Figure 2.
Analysis of Light-Induced Chloroplast Transcription in Cryptochrome Mutants.
Chloroplast transcription activity in the wild type (Col-4) and the cryptochrome single and double mutants cry1-304, cry2-1, and cry1-304 cry2-1 was monitored after a 24-hr dark adaptation followed by illumination with red or blue light at 30 μE·m−2·sec−1. Total chloroplast transcription activity is expressed as fmol 32P-UTP incorporation per 2.5 × 106 plastids in a 5-min run-on transcription assay. Light treatments are indicated below each bar in the figure. Error bars indicate ±sd.
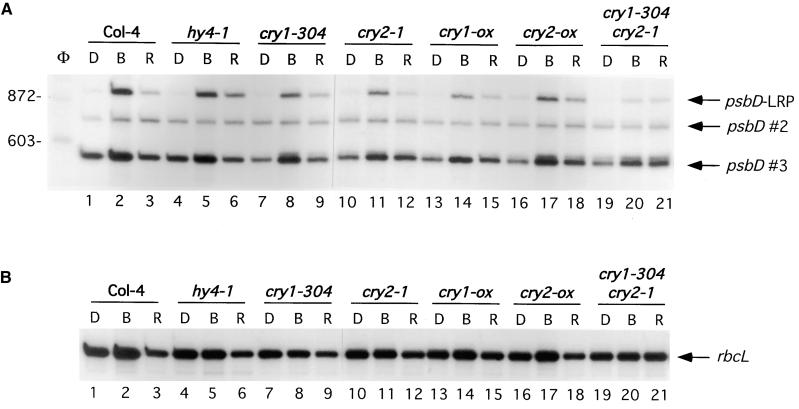
The involvement of cryptochrome in blue light–induced psbD-LRP transcript accumulation was tested using several single and double CRY mutants (Figure 3). As previously reported (Christopher and Hoffer, 1998), blue light was able to induce accumulation of psbD-LRP transcripts in CRY1 mutants (Figure 3, lanes 4 to 9, hy4-1 and cry1-304). A similar analysis was performed on cry2-1, a mutant that accumulates no CRY2 apoprotein because of a complete deletion of the CRY2 gene (Guo et al., 1998). Illumination of dark-adapted cry2-1 plants with red and blue light showed that cry2 was not essential for selective blue light–activated transcription from the psbD-LRP (Figure 3A; cf. lanes 1 to 3 with 10 to 12). Arabidopsis plants overexpressing CRY1 or CRY2, cry1-ox, and cry2-ox, respectively, also had similar levels of blue and red light–induced accumulation of psbD and rbcL transcripts compared with that of wild-type plants of the Columbia ecotype (Col-4) (Figures 3A and 3B; cf. lanes 1 to 3 with 13 to 15 and 16 to 18). In contrast, illumination of a CRY1/CRY2 double mutant (cry1-304 cry2-1) with red or blue light induced only a small increase in psbD-LRP transcript abundance compared with that in wild-type plants (Figure 3A, lanes 19 to 21). Taken together, these results indicate that blue light activation of the psbD-LRP can be mediated by either cry1 or cry2.
Figure 3.
Analysis of psbD-LRP Transcript Accumulation in Cryptochrome Mutants.
Primer extension analysis of psbD (A) and rbcL (B) transcript levels in the wild type (Col-4), single cryptochrome mutants hy4-1, cry1-304, and cry2-1, cryptochrome-overexpressing plants cry1-ox, cry2-ox, and the double mutant cry1-304 cry2-1. Dark-adapted plants (D) were illuminated for 48 hr with blue (B) or red (R) light at 30 μE·m−2·sec−1.
(A) psbD transcripts derived from the LRP (950 nucleotides) are indicated by an arrow and labeled as such. Other psbD transcripts are designated by an arrow and labeled as psbD #2 and psbD #3. Lane φ shows several φX174-HaeIII restriction fragments that were used as size markers.
(B) rbcL transcripts are designated by arrows and labeled.
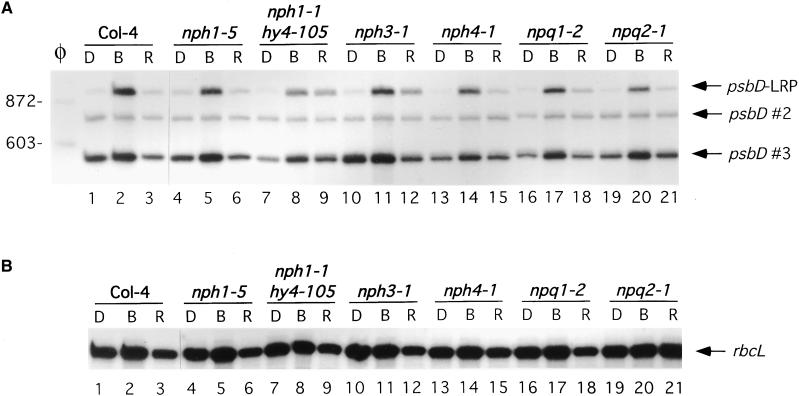
The possible involvement of phototropin and zeaxanthin in blue light–induced accumulation of psbD-LRP transcripts was tested in the phototropin minus NPH1 null mutant nph1-5 (Huala et al., 1997) and in nph3-1 and nph4-1 plants that are defective in phototropism (Liscum and Briggs, 1995). Additionally, a phototropin/cry1 double mutant, nph1-5 hy4-105 (Liscum and Briggs, 1995), was also analyzed. Blue light–induced accumulation of psbD-LRP transcripts was not significantly altered in any of these mutants (Figure 4, lanes 4 to 15). Similarly, blue light–induced accumulation of transcripts from the psbD-LRP and rbcL was not altered in npq1-2 and npq2-1, mutants blocked in zeaxanthin biosynthesis (Figure 4, lanes 16 to 21) (Niyogi et al., 1998).
Figure 4.
Light-Induced Accumulation of psbD-LRP Transcripts Is Not Altered in Phototropin (nph) and Zeaxanthin (npq) Blue Light Photoreceptor Mutants.
Primer extension analysis was used to monitor psbD-LRP (A) and rbcL (B) transcript levels in the wild type (Col-4), in the single mutants nph1-5, nph3-1, nph4-1, npq1-2, and npq2-1, and in the double mutant nph1-1 hy4-105. Dark-adapted plants (D) were illuminated for 48 hr with blue (B) or red (R) light at 30 μE·m−2·sec−1.
(A) psbD transcripts derived from the psbD-LRP (950 nucleotides) are indicated by an arrow and labeled as such. Other psbD transcripts are designated by an arrow and labeled as psbD #2 and psbD #3. Lane φ shows φX174-HaeIII restriction fragments used as size markers.
(B) rbcL transcripts are indicated by an arrow and labeled.
PhyA Is Required for Blue Light Induction of the psbD-LRP
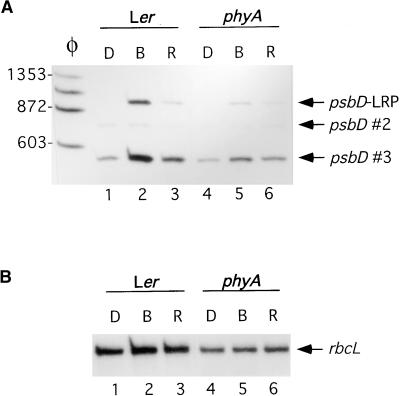
Blue light–induced increases in chloroplast transcription in dark-adapted Arabidopsis are attenuated in plants lacking functional phyA (Chun et al., 2001). Figure 5 shows that blue light–induced accumulation of psbD-LRP transcripts was also attenuated in phyA plants (Figure 5, lane 2 versus lane 5). Plants lacking phytochrome B showed normal blue light–induced accumulation of the psbD-LRP transcripts (data not shown). Run-on transcription assays were used to further analyze the influence of phyA on light-induced activation of psbD-LRP transcription (Table 1). Chloroplasts from dark-adapted wild-type and phyA plants transcribed psbD-LRP and rbcL at low rates (ratio of 0.5). Illumination of wild-type and phyA plants with red light for 7 hr increased transcription from both promoters, although activation of transcription from the psbD-LRP was greater than it was from rbcL (ratio of 2.3) (Table 1). In wild-type plants, blue light increased transcription from the psbD-LRP and rbcL promoters more than did red light and increased the ratio of transcription of psbD-LRP to rbcL from 2.3 to 5.4 (Table 1). In contrast, illumination of phyA plants with blue light increased transcription from rbcL and psbD-LRP to the same extent (ratio of 1.8). These results show that phyA, in addition to cry1 or cry2, is required for differential blue light–induced transcription from the psbD-LRP.
Figure 5.
Light-Induced Accumulation of psbD-LRP Transcripts Is Reduced in phyA Mutants.
Primer extension analysis was used to monitor psbD-LRP (A) and rbcL (B) transcript levels in the wild type (Ler) and phyA mutants. Plants were dark-adapted (D) and then illuminated for 48 hr with blue (B) or red (R) light at 30 μE·m−2·sec−1.
(A) psbD transcripts derived from the psbD-LRP (950 nucleotides) are indicated by an arrow and labeled as such. Other psbD transcripts are designated by an arrow and labeled as psbD #2 and psbD #3. Lane φ shows φX174-HaeIII restriction fragments used as size markers.
(B) rbcL transcripts are indicated by an arrow and labeled.
Table 1.
Blue Light–Induced Transcription of the psbD-LRP Is Reduced in PhyA-Deficient Plantsa
| Light Treatment
|
||||||
|---|---|---|---|---|---|---|
| Wild Type
|
phyA
|
|||||
| Gene | Dark | Red Light | Blue Light | Dark | Red Light | Blue Light |
| psbD-LRP | 0.1 ± 0.2 | 0.9 ± 0.2 | 8.1 ± 0.7 | 0.2 ± 0.2 | 0.8 ± 0.3 | 2.8 ± 0.4 |
| rbcL | 0.2 ± 0.1 | 0.4 ± 0.1 | 1.5 ± 0.1 | 0.3 ± 0.1 | 0.4 ± 0.1 | 1.6 ± 0.2 |
| Ratio of LRP:rbcL | 0.5 | 2.3 | 5.4 | 0.7 | 2.0 | 1.8 |
Wild-type and phyA plants were dark-adapted for 24 hrs and then exposed to 8 hrs of additional darkness, red or blue light (15 μE·m−2·sec−1). Gene- and promoter-specific transcription activities were measured using run-on transcription assays (mean ±sd). The ratio of transcription from the psbD-LRP versus the rbcL promoter is shown in the lower portion of the Table. Units: 1 = 0.1 fmols UMP incorporated (5 × 107 plastids·kb·10 min)−1.
Blue Light–Induced Accumulation of psbD-LRP Transcripts in DET/HY5 Mutants
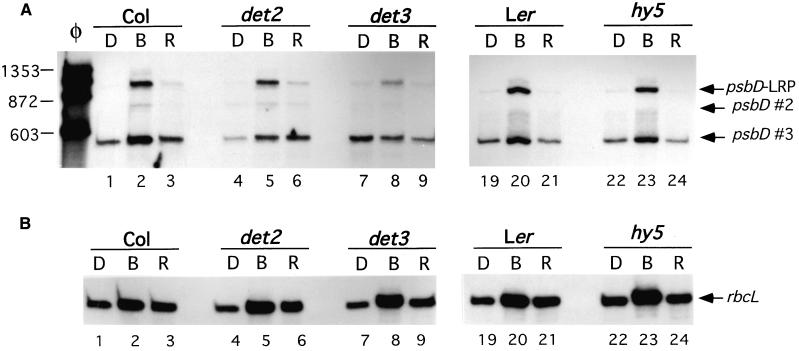
The COP/DET/HY5 genes play important roles in light-modulated plant growth, development, and gene expression (Kwok et al., 1996; Wei and Deng, 1996; Deng and Quail, 1999). The action of these genes is regulated by the phytochrome and cryptochrome signaling pathways (Whitelam and Devlin, 1998). Therefore, the COP/DET/HY5 genes could be involved in cryptochrome- and phytochrome-mediated activation of psbD-LRP transcript accumulation in older plants after dark adaptation. To test this possibility the mutants det2-1, det3-1, and hy5-1 and their corresponding wild-type plants were grown in continuous light for 21 days, dark-adapted for 48 hr, and then illuminated with blue or red light for an additional 48 hr. Figure 6 shows changes in rbcL and psbD-LRP RNA levels in mutant and wild-type plants when dark-adapted plants are illuminated with either red or blue light. Most of the mutants showed blue light–induced changes in psbD-LRP and rbcL transcript abundance that were similar to those in wild-type plants. However, blue light–induced accumulation of psbD-LRP transcripts in det3-1 plants was attenuated compared with that of the controls (Figure 6A, lanes 1 to 3 versus lanes 7 to 9).
Figure 6.
Analysis of psbD-LRP Transcript Accumulation in det2, det3, and hy5 Mutants.
Primer extension analysis of the blue and red light effects on psbD (A) and rbcL (B) transcript levels in det2-1, det3-1, and hy5-1 mutants and their wild-type Arabidopsis ecotypes such as Columbia (Col) and Landsberg erecta (Ler). All plants were grown for 3 weeks in continuous light, dark-adapted (D) for 2 days, and then illuminated for 48 hr with blue (B) or red (R) light at 30 μE·m−2·sec−1.
(A) Transcripts derived from the psbD-LRP are indicated by an arrow and labeled as such. Other psbD transcripts are designated by an arrow and labeled as psbD #2 and psbD #3. Lane φ shows φX174-HaeIII restriction fragments used as size markers.
(B) rbcL transcripts are indicated by an arrow and labeled.
DISCUSSION
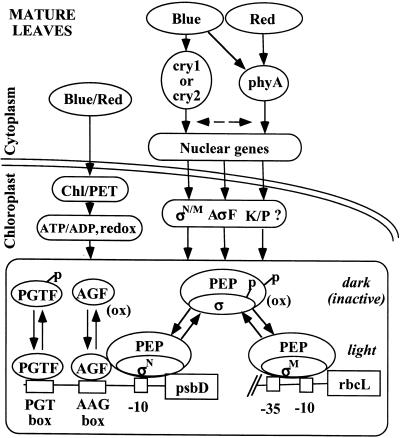
Light regulates numerous aspects of chloroplast transcription through the action of chlorophyll, cryptochrome, and phytochrome, as shown in Figure 7. Light-induced increases in chloroplast transcription are partly the result of photosynthetic electron transport and associated changes in stromal pH, redox state, and the ratio of ATP/ADP and NADPH/NADP in chloroplasts. In addition, as shown in this study, photoreceptors such as cry1, cry2, and phyA that are located in the cytoplasm/nucleus modulate chloroplast transcription activity and differential use of plastid promoters such as the psbD-LRP. The combination of photosynthesis, photoregulation, and circadian cycling (Krupinska, 1992; Nakahira et al., 1998; Thum et al., 2001) helps plants activate and modulate chloroplast transcription during the light phase and in response to varying light environments.
Figure 7.
Diagram of Pathways and Components Involved in Light-Regulated Transcription from the psbD-LRP in Light-Grown Plants.
Chloroplast transcription mediated by PEP is modulated by light through the action of chlorophyll (Chl), cryptochrome (cry1, cry2), and phytochrome (phyA). Light-driven photosynthetic electron transport (PET) can modulate chloroplast transcription by changing stromal pH, ATP/ADP and NADPH/NADP ratios, and redox state. Blue light is shown modulating nuclear gene expression through cry1, cry2, and phyA, and red light through phyA. Nuclear genes encoding plastid-localized sigma factors (σ), putative anti-sigma factors (AσF), kinases (K), and phosphatases (P) are shown as possible targets for light regulation. The rbcL promoter contains −10 and −35 promoter elements, whereas the psbD-LRP consists of a −10 element and upstream protein binding sequences (AAG box, PGT box). The activity of PEP, the DNA binding complexes AGF and PGTF, is modulated by phosphorylation, redox state, and possibly the relative abundance of different sigma factors.
Sigma factors (σM, σN/M, σN) designate the involvement of different unspecified sigma factors. Phosphorylated (p) and oxidized (ox) protein complexes are noted.
In this study, transfer of 21-day-old Arabidopsis plants grown in continuous light to darkness caused the abundance of psbD-LRP transcripts to decrease significantly within 6 hr. In contrast, the abundance of rbcL transcripts changed more slowly and to a smaller extent in response to light/dark treatment. The slow change in rbcL mRNA level after light/dark treatment is likely due in part to the stability of rbcL transcripts. This interpretation is consistent with a half-life of rbcL transcripts of 15 to 33 hr in the expanded portion of barley leaves (Kim et al., 1993) and increased stability of rbcL transcripts in dark-treated plants (Shiina et al., 1998). No direct measurement of the stability of psbD-LRP transcripts has been reported. However, the relatively rapid decline of these transcripts upon transfer of plants to darkness suggests a half-life of <6 hr. Therefore, changes in the rate of transcription from the psbD-LRP during dark/light cycles and in response to various light conditions have a rapid and significant impact on the potential for chloroplasts to synthesize D2 and CP43 in vivo.
The overall rate of chloroplast transcription is higher in illuminated plants compared with that in dark-adapted plants. Low transcription activity in dark-grown plants has been correlated with inactivation of the PEP by increased phosphorylation and the redox state of dark-adapted chloroplasts (Figure 7) (Tiller and Link, 1993; Baginsky et al., 1999; Tullberg et al., 2000). Activation of chloroplast transcription after illumination of dark-adapted plants is consistent with the influence of chlorophyll-driven photosynthetic electron transport. In the current study, illumination of dark-adapted plants with red light caused an increase in transcription from both psbD-LRP and rbcL (Table 1). Red light also increased the abundance of psbD-LRP transcripts to a small extent in wild-type plants, CRY1/CRY2 double mutants, and in plants lacking phytochrome (Figures 3 and 5, data not shown). These results suggest that illumination of 21-day-old dark-adapted plants with red (or blue) light activates overall chloroplast transcription and transcription from rbcL and the psbD-LRP in part through chlorophyll-driven photosynthetic electron transport (Figure 7, Chl/PET). Further analysis is needed to characterize the specific mechanisms involved in this activation.
Cry1 or Cry 2 Can Mediate Blue Light Activation of Chloroplast Transcription and Differential Transcription of the psbD-LRP
Illumination of dark-adapted plants with blue/UVA light activates overall chloroplast transcription and differentially increases transcription from the psbD-LRP (Christopher and Mullet, 1994; Christopher, 1996; Hoffer and Christopher, 1997; Chun et al., 2001). In this study, illumination of dark-adapted plants with blue light increased overall chloroplast transcription fourfold, whereas illumination with red light caused a twofold increase (Figure 2). However, illumination of CRY1/CRY2 double mutants with blue or red light resulted in a similar twofold increase in chloroplast transcription. This result shows that the cryptochrome contributes to the reactivation of chloroplast transcription after dark adaptation. In the absence of cry1 and cry2 function, red light and blue light probably activate chloroplast transcription through chlorophyll-driven electron transport and/or the phytochrome signaling pathway. Plants containing either cry1 or cry2 show a partial blue light–specific increase in overall chloroplast transcription. This result suggests that cry1 and cry2 can both mediate blue light activation of overall chloroplast transcription. Further study is required to determine if cry1 or cry2 is sufficient for full induction of this response and to elucidate the mechanism of activation.
Illumination of plants with blue light caused a greater increase in transcription from both rbcL and psbD-LRP relative to plants illuminated with red light. In addition, blue light–activated transcription from psbD-LRP to a greater extent than did rbcL (Table 1). The differential blue light activation of psbD-LRP transcription was paralleled by differential accumulation of psbD-LRP transcripts in blue light (Figure 3). Mutants lacking either cry1 or cry2 still showed differential accumulation of psbD-LRP transcripts in blue light (Figure 3, lanes 4 to 12). However, differential blue light–induced accumulation of psbD-LRP transcripts was not observed in a CRY1/CRY2 double mutant (Figure 3, lanes 19 to 21). This result indicates that cryptochrome is required for differential blue light activation of psbD-LRP and that both cry1 and cry2 can transduce the blue light signal involved. Analysis of phototropin and zeaxanthin mutants showed that these potential photoreceptors were not involved in blue light–induced transcription from psbD-LRP (Figure 4).
The cry1 and cry2 photoreceptors have related amino acid sequences, and both photoreceptors contain a pterin antennae and an FAD chromophore that mediate light-dependent responses either through changes in protein conformation or by electron transfer (Ahmad and Cashmore, 1993; Lin et al., 1996b; Cashmore et al., 1999). Some responses mediated by the cryptochromes can be induced by either cry1 or cry2, whereas others are induced primarily by one of these photoreceptors (Ahmad et al., 1998; Briggs and Huala, 1999; Cashmore et al., 1999). For example, cry1 or cry2 can accelerate flowering, but cry2 selectively blocks phytochrome B (phyB) inhibition of flowering (Lin et al., 1998; Mockler et al., 1999; Lin, 2000a, 2000b). Cry1 has a primary role in induction of CHS, anthocyanin synthesis, and inhibition of hypocotyl elongation, whereas cry2 is primarily responsible for blue light–mediated cotyledon expansion (Lin, 2000a, 2000b). The selective involvement of one of the two photoreceptors may be related to differences in the stability, expression, or signaling pathways used by cry1 and cry2. For example, cry1 is stable in illuminated plants, whereas cry2 shows light-induced turnover (Ahmad et al., 1998; Lin, 2000a, 2000b). CRY1 and CRY2 have different expression patterns and interact with different downstream factors (Ahmad et al., 1998; Cashmore et al., 1999; Lin, 2000a, 2000b). The activation of chloroplast and psbD-LRP transcription in mature leaves by both cry1 and cry2 could be explained if both photoreceptors induce a common cellular state (i.e., calcium flux) that is required for activation of the psbD-LRP. Alternatively, signaling may occur selectively through cry1 and cry2, but both pathways activate transcription from the psbD-LRP. The involvement of cry1 in psbD-LRP activation is consistent with the need for induction of this promoter in plants exposed to high-light irradiance where cry2 levels may be low as a result of light-induced turnover of this photoreceptor. On the other hand, plants, or leaves of plants grown in low light, may benefit from psbD-LRP activation by cry2 that accumulates under these conditions. Plants grown in low light increase the amount of chlorophyll antennae associated with photosystem II, making them susceptible to damage by moderate light intensity and light flecks. In contrast, CHS gene activation and anthocyanin synthesis induced primarily through cry1 are required for screening under high-light conditions but may be less important in plants grown in low light, thereby reducing the need for cry2 activation of this response.
PhyA Is Required for Blue Light Activation of psbD-LRP Transciption
Blue light–induced transcription from the psbD-LRP and accumulation of psbD-LRP transcripts were attenuated in plants lacking phyA (Figure 5, Table 1) but not phyB (data not shown). In contrast, blue light–induced rbcL transcription was not altered by mutation of phyA (Table 1). Therefore, photoactivation of cry1 or cry2 and phyA is required for differential light-induced stimulation of the psbD-LRP in 21-day- old dark-adapted Arabidopsis. PhyA was recently reported to be required for full blue light activation of chloroplast transcription in dark-adapted Arabidopsis plants and to increase transcription of psbA and rrn16 transcription in these plants (Chun et al., 2001). It is possible that the phyA-dependent blue light activation of psbA and rrn16 also involves cry1 and cry2, as observed for the psbD-LRP in this study. Experiments are under way to test this possibility to understand the full complement of plastid genes under cry1/2 and phyA regulation.
This study and the results of Chun et al. (2001) show that illumination of plants with blue light is sufficient to stimulate responses mediated by cry1/2 and phyA as expected based on the absorption spectra of these photoreceptors (Schafer and Haupt, 1983). Other phyA-mediated responses such as cotyledon expansion, hypocotyl growth inhibition, and LHCB gene expression can also be activated by blue light (Whitelam et al., 1993; Hamazato et al., 1997; Neff and Chory, 1998). Christopher and Mullet (1994) showed that light-induced accumulation of psbD-LRP transcripts required blue light with a fluence threshold of 1 μE·m−2·sec−1 and saturation at 100 μE·m−2·sec−1. The finding that blue light induction of this response requires both cry1/2 and phyA implies that the earlier fluence response data represent a combination of the requirements for both photoreceptors. It is possible, therefore, that low-fluence blue light saturates the response mediated by cry1/2, whereas high-fluence blue light is required for phyA-mediated signaling (or vice versa). Separate analysis of each photoreceptor's fluence requirement may help clarify the reason why plants use cry1/2 and phyA to regulate transcription from the psbD-LRP.
PhyA plays a central role in chloroplast development during early seedling development in dicots. For example, leaf development, plastid DNA synthesis, and transcription are activated in pea seedlings by continuous far-red light treatment mediated by phyA (Dubell and Mullet, 1995). In seedlings, phyA probably activates chloroplast development and transcription by derepressing leaf and chloroplast developmental program through its action on a subset of the HY5/COP/DET/FUS gene products. During light-induced plant development, the levels of phyA and PHYA gene expression decrease dramatically (Quail, 1994; Reed, 1999) and blue light becomes an important regulator of chloroplast gene expression and nuclear genes such as RBCS and CHS (Mohr, 1994). However, phyA is present and functional in light-grown plants (Clack et al., 1994), and dark adaptation of light-grown plants leads to the reaccumulation of phyA (Hunt and Pratt, 1980; Smith, 1995). Therefore, the role of phyA observed here is consistent with the activity associated with this photoreceptor in light-grown plants.
Mechanism of Blue Light–Induced Transcription from the psbD-LRP in Green Leaves
Light-regulated transcription of the psbD-LRP is controlled at several levels and by multiple photoreceptors as shown in Figure 7. Cry1/2 and phyA are required to fully activate transcription from the psbD-LRP, but the molecular basis of co-regulation is not known. PhyA has been shown to phosphorylate cry1 and cry2 (Ahmad et al., 1998); therefore, co-regulation could occur through direct interaction. Alternatively, output from the phyA signaling pathway could regulate the cry1/2 signaling pathway (or vice versa), or both pathways could modulate a common downstream regulator of psbD-LRP activity. The COP/DET/HY5 genes regulate many light-induced events in leaf and chloroplast development, and the activity of these genes is modulated by the cryptochrome and phytochrome signaling pathways (Whitelam and Devlin, 1998). For example, hy5, a bZIP transcription factor, acts as a positive component in the transduction of light signals perceived by both phytochromes and blue/UV-A photoreceptors, and cop1 inhibits the activity of hy5 (Ang et al., 1998; Chattopadhyay et al., 1998; Khurana et al., 1998; Whitelam and Devlin, 1998). However, cry1/2- and phyA-mediated activation of chloroplast transcription and psbD-LRP transcription was not altered in 21-day-old det2 or hy5 plants (Figure 6). Therefore, the cry/phyA signaling pathways involved in psbD-LRP activation do not act through hy5 or det2 at this stage of plant development. However, blue light–mediated activation of psbD-LRP was attenuated in det3-1 plants. Dark-grown det3-1 has short hypocotyls, expanded cotyledons, and differentiated leaves, but chloroplast development and expression of photosynthetic genes are inhibited (Cabrera y Poch et al., 1993). Light-grown det3-1 plants have reduced stature and apical dominance, which may be explained by impaired sugar uptake into vacuoles and modified carbohydrate levels in the cytoplasm (Schumacher et al., 1999). Sugars are known to inhibit expression of photosynthetic genes, and this may also explain the attenuated response of the psbD-LRP to blue light in this mutant.
The cry1/2 and phyA signaling pathways most likely regulate the synthesis of one or more nuclear-encoded proteins that selectively increase the affinity of the PEP for the psbD-LRP. The psbD-LRP lacks a functional −35 element found in promoters of many other plastid genes such as rbcL and requires an upstream activating complex (AGF) for transcription (Figure 7). In bacteria, special sigma factors are involved in the recognition of promoters lacking −35 promoter elements (Helmann and Chamberlin, 1988). Moreover, six different nuclear genes encoding putative plastid sigma factors have been identified in Arabidopsis (Tanaka et al., 1997; Allison, 2000; Fujiwara et al., 2000). Therefore, it is possible that blue light activates the synthesis of a plastid sigma factor that enhances transcription from the psbD-LRP. Alternatively, blue light could modulate synthesis of anti-sigma factors or a kinase/phosphatase (Christopher et al., 1997) that modulates activity of the psbD-LRP. For example, T4 bacteriophage encode an anti-σ70 factor that binds to and modifies σ70 recognition of −35 promoter elements (Hughes and Mathee, 1998). This activity enhances transcription from promoters that lack a −35 element but bind the activator, MotA, in the −30 region (Hughes and Mathee, 1998). Further analysis is required to clarify the specific mechanism involved in blue light induction of the chloroplast psbD-LRP.
METHODS
Plant Material and Growth
Seed stocks of Arabidopsis thaliana photomorphogenic mutants hy5-1, det2-1, det3-1, npq1-2, npq2-1, phyA, and hy4-1 were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Dr. Winslow Briggs (Stanford University, Palo Alto, CA) provided seeds for the nph1-5 (Huala et al., 1997), nph3-1, and nph4-1 (Liscum and Briggs, 1995) mutants. Seed stocks of cry1-304, cry2-1 (Guo et al., 1998), H3 (cry2-ox) (Lin et al., 1998), and cry1-304 cry2-1 (Mockler et al., 1999) were provided by Dr. Chentao Lin (University of California, Los Angeles). Dr. Margaret Ahmad (Université Paris) provided the seed stock for the cry1-ox mutant, overexpressing the cry1 apoprotein (Lin et al., 1996a). Seed stock for the nph1-1 hy4-105 double mutant (Liscum and Briggs, 1995) was provided by Dr. Emmanuel Liscum (University of Missouri, Columbia).
To investigate light-regulated transcription from the psbD-LRP, all Arabidopsis photomorphogenic mutant seeds and their respective wild- type ecotypes were sterilized and plated on half-strength Murashige and Skoog phytagar plates, pH 5.7 (Thum et al., 2001). After cold treatment at 8°C for 24 hr, the seeds on the plates were germinated and grown under continuous white light (120 μE·m−2·sec−1) at 23°C for 7 days. Seedlings were then transplanted to pots containing Metro-Mix 360 (Scotts-Sierra Horticultural Products Company, Maryland, OH) and watered with half-strength Hoagland solution. Plants were grown for another 2 weeks in continuous white light (120 μE·m−2·sec−1) and then dark-adapted for 48 hr at 23°C. After dark adaptation, plants were either harvested or illuminated for 48 hr with red or blue light (30 μE·m−2·sec−1) at 23°C before harvesting for RNA analysis. To determine the effect of blue and red light on plastid transcription in wild-type Col-4 and the cryptochrome mutants cry1-304, cry2-1, and cry1-304 cry2-1, seeds were planted in large flats containing Metro-Mix 360 and watered with half-strength Hoagland solution. After cold treatment at 8°C for 24 hr, plants were grown for 21 days under continuous white light illumination (120 μE·m−2·sec−1). Plants were then dark-adapted for 24 hr and illuminated with red (30 μE·m−2·sec−1) or blue (30 μE·m−2·sec−1) light for an additional 8 hr before chloroplast isolation and run-on transcription assays.
Light Sources
Photon fluence rates of white, blue, and red light mentioned above were measured using a quantum photometer (model LI-1800; LI-COR Inc., Lincoln, NE). White light was obtained from fluorescent light tubes (model F72T12/CW; Philips Lighting Company, Somerset, NJ) plus incandescent bulbs (60 W; General Electric). Actinic Blue 7100k light tubes (peak at 420 nm; CORALIFE, Pembroke Pines, FL) were used as the blue light source. Red light was obtained by passing fluorescent light (model F48T12/CW; Philips Lighting Company) plus incandescent light through a red Plexiglas filter (3.0 mm thick, for above 600 nm, peak at 650 nm; Acme Glass Co., Bryan, TX). Far-red light was obtained by passing incandescent bulbs (60 W) through a far-red filter. All light experiments were performed in light-tight temperature-controlled growth chambers.
RNA Isolation and Primer Extension Analyses
All experiments, from seedling growth to light treatments to primer extension analyses, were performed at least two times. RNA was isolated from cotyledons and leaves as described by Kim et al. (1993). Primer extension analysis was performed according to Kim and Mullet (1995). Primers used in this study were as follows: (1) for psbD transcripts: 5′-GTCATAGTGATCCTCCTATTC-3′, complementary to nucleotide positions 34,445 to 34,465 of the RNA-like strand of the tobacco psbD gene (Shinozaki et al., 1986) (previously, this primer was used to detect the psbD blue light–induced transcript produced from various dicot and monocot plants; Christopher et al., 1992); and (2) for rbcL transcripts: 5′-GTAGGGAGGGACTTATGTC-3′, complementary to positions 57,573 to 57,591 of the RNA-like strand of tobacco rbcL (Shinozaki et al., 1986), which is a conserved site in several rbcL genes (Crossland et al., 1984) (previously, this primer was used to detect the rbcL transcript produced from various monocots [Christopher et al., 1992] and dicot plants).
Plastid Run-on Transcription Assays
Intact plastids were isolated from wild-type Col-4 and the cryptochrome mutants cry1-304, cry2-1, and cry1-304 cry2-1, which were grown and treated as described above. Plastid isolation was performed according to Mullet and Klein (1987), in which all manipulations were performed in a light-tight cold room (4 to 8°C) under red or blue light, depending on the light treatment of the plants. Plastid concentration was determined by counting in a hemacytometer. Plastid run-on transcription assays were performed according to Mullet and Klein (1987), in which a plastid concentration of 105 per μL of reaction mixture and 15 μCi of 32P-UTP (specific activity 800 Ci/mmol) were used per reaction. Plastid transcription was performed for 5 min at 25°C in red light (plastids isolated from red light–illuminated plants) or blue light (plastids isolated from blue light–illuminated plants). Reactions were stopped by spotting aliquots on DE-81 paper (Hallick et al., 1976). Incorporation of 32P-UTP was quantitated by washing the spotted DE-81 paper extensively in 5% Na2HPO4, followed by washing in water and ethanol (Hallick et al., 1976). Filters were air dried and counted in a scintillation counter. Femtomoles of 32P-UTP incorporated per 2.5 × 106 plastids was determined from the specific activity of 32P-UTP. These experiments were performed twice; each individual reaction within the experiment was performed in triplicate. Isolation of Arabidopsis chloroplasts and run-on transcription assays for psbD-LRP and rbcL promoter activities was as previously described (Hoffer and Christopher, 1997). Chloroplasts were isolated from Arabidopsis plants grown for 27 days at a photoperiod of 11.5 hr of light (white light, 90 to 110 μE·m−2·sec−1) and 12.5 hr of dark. Plants were dark-adapted for 24 hr and then placed in 8 hr of red or blue light or kept in the dark. Red and blue light fluences were 15 μE·m−2·sec−1 (±3 μE·m−2·sec−1).
Acknowledgments
This research was supported by National Institutes of Health Grant GM 37987, the Perry Adkisson Chair in Agricultural Biology, and the Texas Agricultural Experiment Station. M.K. was partially supported by the Seoul National University Research Fund, and United States Department of Energy Biosciences Program Grant No. DE-F603-97ER20273 to D.A.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010345.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J., and Cashmore, A. (1998). Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison, L.A. (2000). The role of sigma factors in plastid transcription. Biochimie 82, 537–548. [DOI] [PubMed] [Google Scholar]
- Allison, L.A., and Maliga, P. (1995). Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 14, 3721–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, L.-H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Aro, E.-M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage, and turnover. Biochim. Biophys. Acta 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Baba, K., Nakano, T., Yamagishi, K., and Yoshida, S. (2001). Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol. 125, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky, S., Tiller, K., Pfannschmidt, T., and Link, G. (1999). PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol. Biol. 39, 1013–1023. [DOI] [PubMed] [Google Scholar]
- Barber, J., and Andersson, B. (1992). Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 17, 61–66. [DOI] [PubMed] [Google Scholar]
- Baumgartner, B.J., Rapp, J.C., and Mullet, J.E. (1993). Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol. 101, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., and Huala, E. (1999). Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol. 15, 33–62. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Liscum, E. (1997). Blue light–activated signal transduction in higher plants. In Signal Transduction in Plants, P. Aducci, ed (Basel, Switzerland: Birkhauser Verlag), pp. 107–135.
- Cabrera y Poch, H., Peto, C., and Chory, J. (1993). A mutation in the Arabidopsis DET3 gene uncouples photoregulated leaf development from gene expression and chloroplast biogenesis. Plant J. 4, 6671–6682. [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.-J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.-H., Puente, P., Deng, X.-W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J. (1997). Light modulation of vegetative development. Plant Cell 9, 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher, D.A. (1996). Leaf development and phytochrome modulate the activation of psbD-psbC transcription by high-fluence blue light in barley chloroplasts. Photosynth. Res. 47, 239–251. [DOI] [PubMed] [Google Scholar]
- Christopher, D.A., and Hoffer, P.H. (1998). DET1 represses a chloroplast blue light–responsive promoter in a developmental and tissue-specific manner in Arabidopsis thaliana. Plant J. 14, 1–11. [DOI] [PubMed] [Google Scholar]
- Christopher, D.A., and Mullet, J.E. (1994). Separate photosensory pathways co-regulate blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 104, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher, D.A., Kim, M., and Mullet, J.E. (1992). A novel light-regulated promoter is conserved in cereal and dicot chloroplasts. Plant Cell 4, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher, D.A., Xinli, L., Kim, M., and Mullet, J.E. (1997). Involvement of protein kinase and extraplastidic serine/threonine protein phosphatases in signaling pathways regulating plastid transcription and the psbD blue light–responsive promoter in barley. Plant Physiol. 113, 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, L., Kawakami, A., and Christopher, D.A.. (2001). Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol. 125, 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack, E., Matthews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Crossland, L.D., Rodermel, S.R., and Bogorad, L. (1984). Single gene for the large subunit of ribulosebisphosphate carboxylase in maize yields two differentially regulated mRNAs. Proc. Natl. Acad. Sci. USA 81, 4060–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon, A., and Mayfield, S.P. (1994). Light-regulated translation of chloroplast messenger RNAs through redox potential. Science 266, 1717–1719. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., and Gruissem, W. (1987). Control of plastid gene expression during development: The limited role of transcriptional regulation. Cell 49, 379–387. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., and Quail, P.H. (1999). Signaling in light-controlled development. Sem. Cell Dev. Biol. 10, 121–129. [DOI] [PubMed] [Google Scholar]
- DuBell, A.N., and Mullet, J.E. (1995). Continuous far-red light activates plastid DNA synthesis in Pea leaves but not full cell enlargement or an increase in plastid number per cell. Plant Physiol. 109, 95–103. [Google Scholar]
- Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229. [DOI] [PubMed] [Google Scholar]
- Fujiwara, M., Nagashima, A., Kanamaru, K., Tanaka, K., and Takahashi, H. (2000). Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase σ factors in Arabidopsis thaliana. FEBS Lett. 481, 47–52. [DOI] [PubMed] [Google Scholar]
- Gamble, P.E., and Mullet, J.E. (1989). Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J. 8, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T.J., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick, R.B., Lipper, C., Richards, O.C., and Rutter, W.J. (1976). Isolation of a transcriptionally active chromosome from chloroplasts of Euglena gracilis. Biochemistry 15, 3039–3045. [DOI] [PubMed] [Google Scholar]
- Hamazato, F., Shinomura, T., Hanazawa, H., Chory, J., and Furuya, M. (1997). Fluence and wavelength requirements for Arabidopsis Cab gene induction by different phytochromes. Plant Physiol. 115, 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann, J.D., and Chamberlin, M.J. (1988). Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 57, 839–872. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., and Börner, T. (1999). Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 190, 1–59. [DOI] [PubMed] [Google Scholar]
- Hoffer, P.H., and Christopher, D.A. (1997). Structure and blue-light-responsive transcription of a chloroplast psbD promoter from Arabidopsis thaliana. Plant Physiol. 115, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala, E., Oeller, P.W., Liscum, E., Han, I., Larsen, E., and Briggs, W.R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278, 2120–2123. [DOI] [PubMed] [Google Scholar]
- Hughes, K.T., and Mathee, K. (1998). The anti-sigma factors. Annu. Rev. Microbiol. 52, 231–286. [DOI] [PubMed] [Google Scholar]
- Hunt, R.E., and Pratt, L.H. (1980). Radioimmunoassay of phytochrome content in green, light-grown oats. Plant Cell Environ. 3, 91–95. [Google Scholar]
- Khurana, J.P., Kochhar, A., and Tyagi, A.K. (1998). Photosensory perception and signal transduction in higher plants: Molecular genetic analysis. Crit. Rev. Plant Sci. 17, 465–539. [Google Scholar]
- Kim, J., and Mayfield, S.P. (1997). Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278, 1954–1957. [DOI] [PubMed] [Google Scholar]
- Kim, M., and Mullet, J.E. (1995). Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light–responsive psbD-psbC promoter. Plant Cell 7, 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Christopher, D.A., and Mullet, J.E. (1993). Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol. Biol. 22, 447–463. [DOI] [PubMed] [Google Scholar]
- Kim, M., Christopher, D.A., and Mullet, J.E. (1999. a). ADP-dependent phosphorylation regulates association of a DNA-binding complex with the barley chloroplast psbD blue-light-responsive promoter. Plant Physiol. 119, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Thum, K.E., Morishige, D.T., and Mullet, J.E. (1999. b). Detailed architecture of the barley chloroplast psbD-psbC blue light–responsive promoter. J. Biol. Chem. 274, 4684–4692. [DOI] [PubMed] [Google Scholar]
- Klaff, P., and Gruissem, W. (1991). Changes in chloroplast mRNA stability during leaf development. Plant Cell 3, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, R.R., and Mullet, J.E. (1990). Light-induced transcription of chloroplast genes. J. Biol. Chem. 265, 1895–1902. [PubMed] [Google Scholar]
- Kowallik, K.V., Stobe, B., Schaffran, I., and Freier, U. (1995). The chloroplast genome of a chlorophyll a+c containing alga, Odontella sinensis. Plant Mol. Biol. Rep. 13, 336–342. [Google Scholar]
- Krupinska, K. (1992). Transcriptional control of plastid gene expression during development of barley primary foliage leaves under a daily light–dark regime. Planta 186, 294–303. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Misera, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. (2000. a). Photoreceptors and regulation of flowering time. Plant Physiol. 123, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. (2000. b). Plant blue-light receptors. Trends Plant Sci. 5, 337–342. [DOI] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., and Cashmore, A.R. (1996. a). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10, 893–902. [DOI] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., Chan, J., and Cashmore, A.R. (1996. b). CRY2: A second member of the Arabidopsis cryptochrome gene family. Plant Physiol. 110, 1047.8819875 [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, G. (1991). Photoregulated development of chloroplasts. In The Photosynthetic Apparatus: Molecular Biology and Operation, L. Bogorad and I.K. Vasil, eds (San Diego, CA: Academic Press), pp. 365–394.
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo, A.K., Hoffman-Falk, H., Marder, J.B., and Edelman, M. (1984). Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc. Natl. Acad. Sci. USA 81, 1380–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis, A. (1999). Photosystem-II damage and repair cycle in chloroplasts: What modulates the rate of photodamage in vivo? Trends Plant Sci. 4, 130–135. [DOI] [PubMed] [Google Scholar]
- Melis, A., Nemson, J.A., and Harrison, M.A. (1992). Damage to functional components and partial degradation of photosystem II reaction center proteins upon chloroplast exposure to ultraviolet-B radiation. Biochim. Biophs. Acta 1100, 312–320. [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Mohr, H. (1994). Coaction between pigment systems. In Photomorphogenesis in Plants, R.E. Kendrick and G.H.M. Kronengerg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 353–376.
- Mullet, J.E. (1988). Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 475–502. [Google Scholar]
- Mullet, J.E., and Klein, R.R. (1987). Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 6, 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira, Y., Baba, K., Yoneda, A., Shiina, T., and Toyoshima, Y. (1998). Circadian-regulated transcription of the psbD light-responsive promoter in wheat chloroplasts. Plant Physiol. 118, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytocrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Björkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, I., Kyle, D.J., and Hirschberg, J. (1985). Light-dependent degradation of the QB-protein in isolated pea thylakoids. EMBO J. 4, 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama, K., et al. (1986). Chloroplast gene organization deducted from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574. [Google Scholar]
- Quail, P.H. (1994). Phytochrome genes and their expression. In Photomorphogenesis in Plants, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 71–104.
- Rapp, J.C., Baumgartner, B.J., and Mullet, J. (1992). Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes: Transcription rates and mRNA levels vary over 300-fold; predicted mRNA stabilities vary 30-fold. J. Biol. Chem. 267, 21404–21411. [PubMed] [Google Scholar]
- Reed, J.W. (1999). Phytochromes are Pr-ipatetic kinases. Curr. Opin. Plant Biol. 2, 393–397. [DOI] [PubMed] [Google Scholar]
- Reith, M., and Munholland, J. (1995). Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 13, 333–335. [Google Scholar]
- Schafer, E., and Haupt, W. (1983). Blue light effects in phytocrome-mediated responses. In Encyclopedia of Plant Physiology, W. Shropshire Jr. and H. Mohr, eds (New York: Springer-Verlag), p. 16A.
- Schumacher, K., Vafeados, D., McCarthy, M., Sze, H., Wilkins, T., and Chory, J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H+ATPast in plant growth and development. Genes Dev. 13, 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, T.B., Christopher, D.A., and Mullet, J.E. (1990. a). Light-induced switch in barley psbD-psbC promoter utilization: A novel mechanism regulating chloroplast gene expression. EMBO J. 9, 4485–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, T.B., Jones, J.T., and Mullet, J.E. (1990. b). Sequence and transcriptional analysis of the barley ctDNA region upstream of psbD-psbC encoding trnK(UUU), rps16, trnQ(UUG), psbK, psbI and trnS(GCU). Curr. Genet. 17, 445–454. [DOI] [PubMed] [Google Scholar]
- Shiina, T., Allison, L., and Maliga, P. (1998). rbcL transcript levels in tobacco plastids are independent of light: Reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell 10, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. (1995). Physiological and ecological function within the phytochrome family. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 289–315. [Google Scholar]
- Stirewalt, V.L., Michalowski, C.B., Löfflehardt, W., Bohnert, H.J., and Bryant, D.A. (1995). Nucleotide sequence of the cyanelle genome from Cyanophora paradoxa. Plant Mol. Biol. Rep. 14, 327–332. [Google Scholar]
- Tanaka, K., Tozawa, Y., Mochizuki, N., Shinozaki, K., Nagatani, A., Wakasa, K., and Takahashi, H. (1997). Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: Evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 413, 309–313. [DOI] [PubMed] [Google Scholar]
- Thum, K.E., Kim, M., Morishige, D.T., Eibl, C., Koop, H.-U., and Mullet, J.E. (2001). Analysis of barley chloroplast psbD light responsive promoter elements in transplastomic tobacco. Plant. Mol. Biol. 47, 353–366. [DOI] [PubMed] [Google Scholar]
- Tiller, K., and Link, G. (1993). Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapsis alba L.). EMBO J. 12, 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, K., Cheng, M., Suen, D., Mon, D., Chen, L.O., and Chen, S.G. (1996). Characterization of the light-responsive promoter of rice chloroplast psbD-C operon and the sequence-specific DNA binding factor. Plant Cell Physiol. 37, 660–666. [DOI] [PubMed] [Google Scholar]
- Tullberg, A., Alexciev, K., Pfannschmidt, T., and Allen, J.F. (2000). Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol. 41, 1045–1054. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1996). The role of the COP/DET/FUS genes in light control of arabidopsis seedling development. Plant Physiol. 112, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., and Devlin, P.F. (1998). Light signalling in Arabidopsis. Plant Physiol. Biochem. 36, 125–133. [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]