Abstract
Aux/IAA genes are early auxin response genes that encode short-lived nuclear proteins with four conserved domains, referred to as I, II, III, and IV. Arabidopsis Aux/IAA proteins repressed transcription on auxin-responsive reporter genes in protoplast transfection assays. Mutations in domain II resulted in increased repression, whereas mutations in domains I and III partially relieved repression. Aux/IAA proteins fused to a heterologous DNA binding domain were targeted to promoters of constitutively expressed reporter genes and actively repressed transcription in an auxin-responsive and dose-dependent manner. In comparison with an unfused luciferase protein, luciferase fused to Aux/IAA proteins displayed less luciferase activity, which further decreased in the presence of auxin in transfected protoplasts. Domain II mutations increased and domain I mutations decreased luciferase activity with the fusion proteins. These results suggested that Aux/IAA proteins function as active repressors by dimerizing with auxin response factors bound to auxin response elements and that early auxin response genes are regulated by auxin-modulated stabilities of Aux/IAA proteins.
INTRODUCTION
Aux/IAA genes are, in general, rapidly induced by exogenous auxin treatment and encode 25- to 35-kD proteins that are short-lived and localized to the nucleus (reviewed by Abel and Theologis, 1996; Hagen and Guilfoyle, 2001). Arabidopsis contains 29 Aux/IAA genes (Liscum and Reed, 2001). Most Aux/IAA proteins contain four conserved motifs (referred to as domains I, II, III, and IV). Domain II plays a role in destabilizing Aux/IAA proteins and may be a target for ubiquitination (Colon-Carmona et al., 2000; Worley et al., 2000; Ouellet et al., 2001). Domain III is part of a motif predicted to resemble the amphipathic βαα-fold found in the β-ribbon multimerization and DNA binding domains of Arc and MetJ repressor proteins (Abel et al., 1994). The predicted βαα motif has been shown to play a role in dimerization/multimerization of Aux/IAA proteins and in heterodimerization among Aux/IAA and auxin response factor (ARF) proteins (Kim et al., 1997; Ulmasov et al., 1997b; Morgan et al., 1999; Ouellet et al., 2001). The function of domains I and IV in Aux/IAA proteins is not clear, but recent experiments suggest that domain I may play a role in homodimerization of Aux/IAA proteins (Ouellet et al., 2001).
The function of Aux/IAA proteins has not been unequivally established, but their nuclear localization and resemblance to Arc and MetJ DNA binding proteins led to the suggestion that Aux/IAA proteins are transcription factors (Abel et al., 1994). Although DNA binding by Aux/IAA proteins has not been demonstrated, their ability to heterodimerize with ARF DNA binding proteins suggests that Aux/IAA proteins could function as transcription factors in the absence of their binding DNA directly (Ulmasov et al., 1997a, 1997b). Results of Ulmasov et al. (1997b) indicate that at least some Aux/IAA proteins repress transcription of auxin-responsive reporter genes when they are expressed from effector plasmids in protoplast transfection assays.
A number of mutations in Aux/IAA genes have been identified in Arabidopsis that provide insight into the role played by these proteins in auxin responses. The Arabidopsis shy2, axr2, axr3, and iaa28 mutant proteins contain amino acid substitutions within domain II of IAA3, IAA7, IAA17, and IAA28, respectively (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). The mutant plants display abnormalities in a variety of auxin responses and abnormal morphologies as seedlings and adults. Intergenic revertant alleles of iaa7 and iaa17 have also been identified, and, interestingly, the two revertant alleles contain a second site mutation, resulting in an identical amino acid substitution in domain I (Rouse et al., 1998; Nagpal et al., 2000). Two other revertant alleles of iaa3 contain second site mutations that result in amino acid substitutions in domain III (Rouse et al., 1998).
When domain II of the pea PS-IAA4/5 protein was fused in-frame to the luciferase (LUC) reporter gene, the lifetime of the LUC protein in transient assays decreased substantially (Worley et al., 2000), suggesting that domain II of Aux/IAA proteins may be involved in regulating the short lifetime of these proteins. That mutations in domain II result in the stabilization of Aux/IAA proteins is supported by the increased level of the SHY2 protein detected immunologically in the shy2 mutant compared with the level in the wild type (Colon-Carmona et al., 2000). Immunological detection of IAA17/AXR3 in pulse-chase experiments with 35S-labeled proteins revealed that the lifetime of this Aux/IAA protein was sevenfold greater in the axr3 mutant compared with that in the wild type (Ouellet et al., 2001).
Here, we have used protoplast transfection assays to test whether Aux/IAA proteins, in general, function as transcriptional repressors on auxin-responsive reporter genes. In addition, we have tested the effects of site-directed mutations in the conserved domains of Aux/IAA proteins on their ability to alter auxin-responsive reporter gene activity. In addition, we have tested whether Aux/IAA proteins tethered to a GAL4 DNA binding domain can alter the activity of constitutive promoters containing GAL4 DNA binding sites. We show that repression by both untethered and tethered Aux/IAA is auxin responsive, and that the level of repression correlates with the relative stabilities of wild-type and mutant versions of Aux/IAA proteins.
RESULTS
Arabidopsis Aux/IAA Proteins Are, in General, Repressors of Auxin-Responsive Reporter Genes
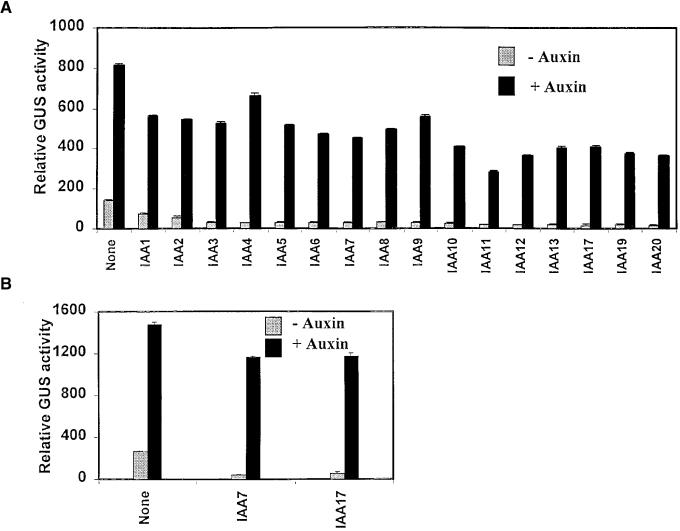
We have previously shown that the Aux/IAA proteins, soybean Aux22 and GH1 and pea PS-IAA4/5 and PS-IAA6, repressed expression of auxin-responsive reporter genes when they were expressed from effector plasmids in carrot protoplast transfection assays (Ulmasov et al., 1997b). Because only four Aux/IAA proteins were tested in this analysis, the possibility remained that some Aux/IAA proteins might function as transcriptional repressors, whereas others might be activators (see the discussion by Ulmasov et al. [1999a]). To examine this possibility, we tested whether overexpression of sixteen different Arabidopsis Aux/IAA proteins in carrot protoplasts resulted in repression or activation of the auxin-responsive P3(4X):GUS (β-glucuronidase) reporter gene (Ulmasov et al., 1997a). Effector plasmids, that consisted of a Cauliflower mosaic virus (CaMV) 35S promoter driving expression of full-length Aux/IAA proteins with an N-terminal hemagglutinin (HA) epitope-tag, were cotransfected into carrot protoplasts along with the GUS reporter gene (see Figure 1 and Methods; Ulmasov et al., 1997b). Figure 2A shows that overexpression of each Aux/IAA protein tested resulted in repression of the reporter gene in either the presence (ranging from 20 to 66% repression) or absence (ranging from 51 to 89% repression) of the auxin 1-naphthalene acetic acid (1-NAA). When IAA7 and IAA17 effector plasmids were tested with a GUS reporter gene containing the soybean GH3 promoter, repression was also observed with this auxin-responsive reporter gene (Figure 1B). These results are consistent with Aux/IAA proteins functioning exclusively as transcriptional repressors on TGTCTC-type auxin-responsive reporter genes, and they suggest it is unlikely that subsets of Aux/IAA proteins function as transcriptional activators on genes containing TGTCTC auxin response elements (AuxREs).
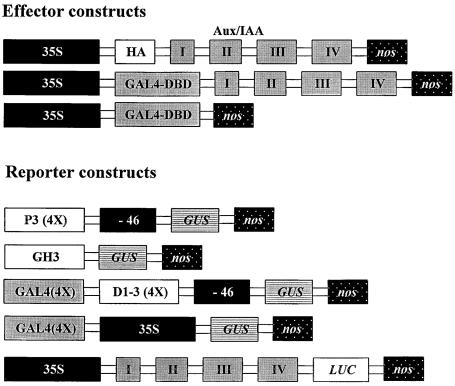
Figure 1.
Schematic Diagrams of Effector and Reporter Constructs.
Expression of all effector constructs was driven by the CaMV 35S promoter, and each effector construct contained a nopaline synthase (nos) 3′ untranslated sequence (Ulmasov et al., 1997b). Effector plasmids encoding Aux/IAA proteins contained a HA epitope tag (see Methods) fused in-frame to their N termini and are diagrammed with conserved domains I, II, III, and IV. Effector plasmids encoding GAL4 DBD–Aux/IAA fusion proteins contained a yeast GAL4 DBD (amino acids 1 to 147) fused in-frame at the N termini of Aux/IAA proteins. An effector gene expressing only the GAL4 DBD is also shown. The P3(4X):GUS reporter gene consisted of the auxin responsive P3(4X) sequence (four tandem copies of the P3 AuxRE) fused to a CaMV minimal −46 promoter driving expression of the GUS reporter gene (Ulmasov et al., 1997a). The GH3:GUS reporter gene contained a 592-bp auxin-responsive soybean GH3 promoter-driving expression of the GUS reporter gene (Liu et al., 1994). The GAL4(4X)-D1-3(4X):GUS reporter gene consisted of four tandem copies of the GAL4 DNA binding site (see Methods) fused immediately upstream of four tandem copies of the constitutive D1-3 element, which in turn was fused to a CaMV minimal −46 promoter-driving expression of the GUS reporter gene. The GAL4(4X)-35S:GUS reporter gene consisted of four tandem copies of the GAL4 DNA binding site fused immediately upstream of the CaMV 35S promoter-driving expression of the GUS reporter gene. Reporter plasmids encoding Aux/IAA-LUC fusion proteins contained a LUC protein fused in-frame at the C termini of Aux/IAA proteins.
Figure 2.
Repression of Auxin-Responsive Reporter Genes by Arabidopsis Aux/IAA Effector Plasmids.
(A) Repression with the P3(4X):GUS reporter gene. Effector plasmids encoding different Aux/IAA proteins were cotransfected with the auxin-responsive P3(4X):GUS reporter gene into carrot protoplasts, and protoplasts were incubated with or without 25 μM auxin (1-NAA). IAA effector plasmids are indicated using the nomenclature of Abel et al. (1995) and Kim et al. (1997).
(B) Repression of the soybean GH3:GUS reporter gene with IAA7 and IAA17 effector plasmids. Effector plasmids encoding either IAA7 or IAA17 were cotransfected into carrot protoplasts and assayed as described in (A).
GUS activities were measured 24 hr after transfections. Standard errors are indicated. None indicates GUS activities with the reporter gene in the absence of an effector gene.
Mutations in Domain II Increase the Repressor Activity of Aux/IAA Proteins
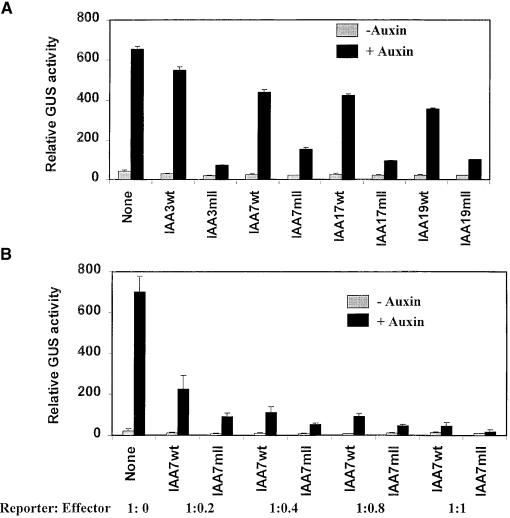
Because mutations in domain II of Aux/IAA proteins are semidominant and result in increased lifetimes of these proteins in Arabidopsis plants (reviewed by Liscum and Reed, 2001; Ouellet et al., 2001), we tested whether the repressor activities of effector plasmids encoding Aux/IAA proteins with motif II mutations were altered compared with repressor activities of effector plasmids encoding wild-type Aux/IAA proteins. Aux/IAA effector genes with domain II mutations identical to those found in shy2-2/iaa3 (IAA3mII), axr2-1/iaa7 (IAA7mII), and axr3-1/iaa17 (IAA17mII) were constructed (see Table 1). An IAA19 effector gene containing a domain II mutation (IAA19mII) was also constructed. The effector plasmids were cotransfected with the P3(4X):GUS reporter gene and tested for GUS activity in carrot protoplasts with or without auxin. Figure 3A shows that each effector plasmid harboring a domain II mutation increased repression by severalfold compared with the wild-type effectors when tested in the presence of auxin (i.e., eightfold for IAA3wt versus IAA3mII, threefold for IAA7wt versus IAA7mII, fivefold for IAA17wt versus IAA17mII, and fourfold for IAA19wt versus IAA19mII). Because GUS expression was low in the absence of auxin treatment, it was difficult to assess the level of repression in these cases. In carrot protoplasts, the soybean GH3:GUS reporter gene has higher activity than does the P3(4X):GUS reporter gene in the absence of auxin treatment, making it possible to observe severalfold repression of the reporter gene by wild-type and domain II mutant effectors but greater repression by the mutant effector (data not shown).
Table 1.
Wild-Type and Mutant Proteins Used in This Study a
| Aux/IAA Proteins | Mutations | Domain I | Domain II | Domain III |
|---|---|---|---|---|
| IAA3 | Wild type (wt) | ETELRLGLPG | GWPPVRSY | PYLRKIDL |
| mII | —b | GWSPVRSY | — | |
| IAA7 | Wild type (wt) | ATELCLGLPG | GWPPVRNY | PYLRKVDL |
| mIa | ATELCFGLPG | — | — | |
| mIb | ATVRCLGLPG | — | — | |
| mII | — | GWSPVRNY | — | |
| mIa/mII | ATELCFGLPG | GWSPVRNY | — | |
| mIb/mII | ATVRCLGLPG | GWSPVRNY | — | |
| mIII | — | — | PYLKKVDL | |
| mII/mIII | — | GWSPVRNY | PYLKKVDL | |
| IAA17 | Wild type (wt) | ETELCLGLPG | GWPPVRSY | PYLRKIDL |
| mI | ETVRCLGLPG | — | — | |
| mII | — | GWPLVRSY | — | |
| mI/mII | ETVRCLGLPG | GWPLVRSY | — | |
| mIII | — | — | PYLRKIES | |
| IAA19 | Wild type (wt) | ITELRLGLPG | GWPPVCSY | DKLFGFRGI |
| dI | Deletion of box I (1 to 34 amino acids) | — | — | |
| mII | — | GWSPVCSY | — | |
| dI/mII | Deletion of box I (1 to 34 amino acids) | GWSPVCSY | — | |
| mII/mIII | — | GWSPVCSY | DKESGFRGI |
The amino acids mutated within a conserved region of each domain are indicated in boldface.
No change from wild type.
Figure 3.
Repression of Auxin-Responsive Reporter Genes by Wild-Type and Domain II Mutant Versions of Arabidopsis Aux/IAA Proteins.
(A) Repression by IAA effector plasmids in transfected carrot protoplasts with the P3(4X):GUS reporter gene. Effector genes encoding wild-type and domain II mutated versions (see Table 1) of IAA3/SHY2, IAA7/AXR2, IAA17/AXR3, and IAA19 were assayed for GUS activity with the reporter gene in the presence and absence of 25 μM auxin (1-NAA).
(B) Repression by IAA7 effector plasmids in Arabidopsis protoplasts with the soybean GH3:GUS reporter gene. Effector genes encoding the wild type and a domain II mutated version of IAA7/AXR2 were assayed for GUS activity with the reporter gene in the presence and absence of 1 μM auxin (1-NAA). The ratios of reporter gene to effector gene in the assays are indicated below the effector lanes. Because of the relatively large standard errors for GUS assays with Arabidopsis protoplasts, t tests were used to confirm that reporter gene activity was significantly different for wild-type and domain II mutant effector genes at each reporter:effector ratio; 1:0.2 (P < 0.05), 1:0.4 (P < 0.05), 1:0.8 (P < 0.05), and 1:1 (P < 0.1).
GUS activities were measured 24 hr after transfections. Standard errors are indicated. None indicates GUS activities with the reporter gene in the absence of an effector gene.
As a second system to test repressor activity of Aux/IAA proteins, protoplasts derived from Arabidopsis leaf mesophyll cells were used in transfection assays (Kovtun et al., 2000). With Arabidopsis protoplasts, the soybean GH3:GUS reporter gene was superior to the P3(4X):GUS reporter gene for assessing auxin-responsive gene expression (X.-J. Wang, unpublished results). IAA7 wild-type (IAA7wt) and domain II mutant (IAA7mII) effector plasmids were cotransfected into Arabidopsis protoplasts at increasing amounts relative to the reporter plasmid. Figure 3B shows that as the ratio of either wild-type or domain II mutant effector plasmid to reporter plasmid increased, the level of repression also increased. Increased repression by domain II mutant effectors relative to wild-type effectors was not as great as that observed with the P3(4X):GUS reporter gene in carrot protoplasts; in all cases, however, the domain II mutant effectors brought about stronger repression than did wild-type effectors. We found that similar to assays described in Figure 3A, it was not possible to assess levels of repression in protoplasts assayed without auxin, because the reporter gene activities were very low. Our results with two different protoplast systems indicate that mutations in domain II, which appear to increase the stability of Aux/IAA proteins in Arabidopsis plants (Colon-Carmona et al., 2000; Ouellet et al., 2001), have the outcome of substantially increasing their repressor activity on auxin-responsive reporter genes in transfection assays.
Mutations in Domains I and III of Aux/IAA Proteins Partially Suppress the Increased Repressor Activity of Mutations in Domain II
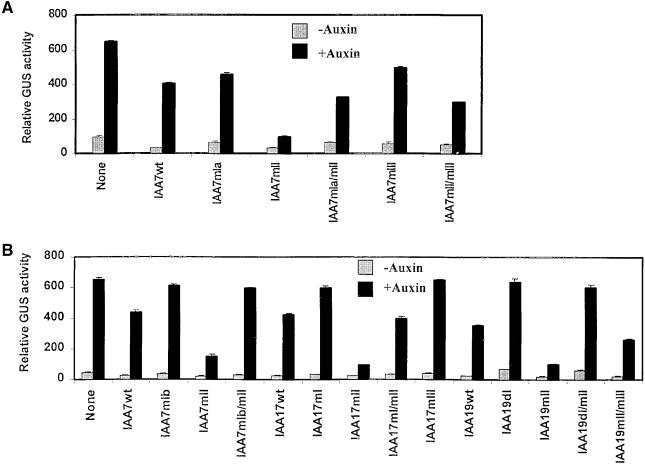
On the basis of the intragenic suppressor alleles described for axr2-1/iaa7 (Nagpal et al., 2000), we constructed effector plasmids with mutations in domain I, domains I plus II, domain III, and domains II plus III. Identical amino acids were substituted for wild-type amino acids in IAA7 as were those found in iaa7/axr2-1-r4 (mIa) and iaa7/axr2-1-r3 (mIII) revertant alleles (Nagpal et al., 2000; see Table 1). Effector constructs were cotransfected into carrot protoplasts along with the P3(4X):GUS reporter gene, and GUS activities were assayed in the presence and absence of auxin. With IAA7 effector constructs corresponding to iaa7/axr2-1-r4 (mIa) and iaa7/axr2-1-r3 (mIII), repression of reporter gene activity was reduced compared with that in which the effector contained only a mutation in domain II (i.e., corresponding to iaa7/axr2) and approached the repressor activity observed with wild-type IAA7 effector gene (Figure 4A; cf. none or no effector, IAA7wt, IAA7mII, IAA7mIa/mII, and IAA7mII/mIII). The amino acid substitutions in domains I and III (i.e., corresponding to those found in iaa7/axr2-1-r4 and iaa7/axr2-1-r3) were also partially effective in relieving the repressor activity of a IAA7 effector gene containing a wild-type domain II (cf. none or no effector, IAA7wt, IAA7mIa, and IAA7mIII).
Figure 4.
Reversal of Repression by Mutations in Domains I and III of Aux/IAA Proteins.
(A) Natural mutations in domains I and III partially reverse repression conferred by wild-type and domain II mutant IAA7/AXR2 proteins. The natural mutations correspond to those found in axr2-1, axr2-1-r3, and axr2-1-r4. Domain I, II, and III mutations in IAA7/AXR2 are described in Table 1.
(B) Additional mutations in domains I and III, that have not been found in mutant screens, partially reverse repression by wild-type and domain II mutant Aux/IAA proteins.
P3(4x):GUS was used as the reporter gene in carrot protoplasts. GUS activities were measured 24 hr after transfections. Standard errors are indicated. None indicates GUS activities with the reporter gene in the absence of an effector gene.
To determine if there was anything special about the specific amino acid substitutions found in domains II and I or III in iaa7/axr2-1-r4 and iaa7/axr2-1-r3, we made additional amino acid substitutions in domains I and III that differed from those found in the revertant alleles (see Table 1). For domain I, the sequence TELCLGL was converted to TVRCLGL in IAA7, and the sequence TELCLGL was converted to TVRCLGL in IAA17. For IAA19, the entire domain I was deleted (amino acids 1 to 34) from a domain II mutant construct. The IAA7 and IAA17 domain I mutant effectors showed reduced repressor activity compared with that of wild-type IAA7 and IAA17 effectors (Figure 4B; cf. none or no effector, IAA7wt and IAA7mIb, and cf. none or no effector, IAA17wt and IAA17mI). When a domain II mutation was introduced into the IAA7 and IAA17 mutant domain I effector constructs, the repressor activity was strongly reduced compared with effectors that contained only domain II mutations (cf. IAA7mIb/mII with IAA7mII, and IAA17mI/mII with IAA17mII). An effector gene encoding an IAA19 domain I deletion in a domain II mutant also showed reduced expression compared with that of the IAA19 domain II mutant (cf. IAA19wt, IAA19dI/mII, and IAA19mI). For domain III mutations, the sequence RKIDL was converted to RKIES in a wild-type IAA17, and the sequence DKLFG was converted to DKESG in a IAA19 domain II mutant. The IAA17 mutant domain III effector gene showed reduced repressor activity compared with that of the wild-type effector (Figure 4B; cf. IAA17wt with IAA17mIII). With the IAA19 domain II and III double mutant effector genes, the repressor activity was again strongly reduced compared with that of the IAA19 effector that contained only a domain II mutation (cf. IAA19mII with IAA19mII/mIII). The above results indicate that amino acid substitutions (mutations) in domains I and III suppress the repressor activity of wild-type and domain II mutant Aux/IAA proteins.
Aux/IAA Proteins Function as Auxin-Responsive Repressors When Targeted to Constitutive Reporter Genes
To determine if Aux/IAA proteins can repress reporter genes that are not auxin responsive, we constructed two constitutively expressed reporter genes that contained four tandem copies of the yeast GAL4 DNA binding site upstream of the promoters (see Figure 1). The reporter genes consisted of the CaMV 35S promoter (GAL4CaMV35S:GUS) or the D1-3(4X) element plus the −46 CaMV 35S minimal promoter fused to GUS (GAL4D1-3:GUS; Ulmasov et al., 1995; Ulmasov et al., 1997b). The D1-3 element was derived from the D1 composite AuxRE in the soybean GH3 promoter by mutating the C-terminal half of the TGTCTC element, thus creating a constitutive element unresponsive to auxin (Ulmasov et al., 1995). Wild-type and mutant versions of Aux/IAA proteins fused to the GAL4 DNA binding domain (GAL4 DBD) were cotransfected with the reporter genes into carrot protoplasts and tested for GUS activity in the absence and presence of auxin.
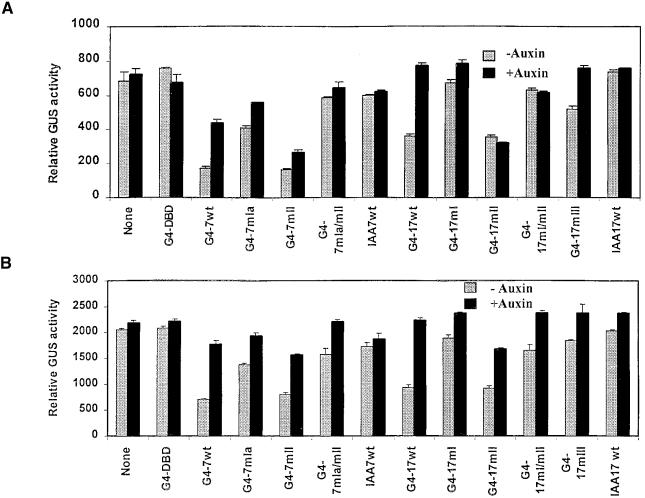
Figure 5A shows GUS activities for the GAL4D1-3:GUS reporter gene with GAL4 DBD-IAA7 and GAL4 DBD-IAA17 fusion effector genes, respectively. In the absence of effectors, the reporter genes showed no response to auxin. An effector encoding only the GAL4 DBD (G4-DBD) with a nuclear localization signal had little or no effect on reporter gene expression and showed no auxin response. An effector consisting of the GAL4 DBD fused to the herpes simplex virion protein-16 activation domain resulted in strong activation of the reporter genes (i.e., ∼1700 units of GUS activity with or without auxin treatment, compared with 700 GUS units with G4-DBD), indicating that the GAL4 DBD targets the effector plasmids to the reporter genes (data not shown). Effector genes encoding Aux/IAA proteins with no GAL4 DBD (IAA7wt and IAA17wt) also had little or no effect on reporter gene expression and showed no auxin response. With both reporter genes, greater than twofold repression was observed with an effector plasmid encoding the GAL4 DBD fused to the full-length IAA7/AXR2 (G4-7wt) or IAA17/AXR3 (G4-17wt) protein; however, this repression was partially relieved by the addition of auxin. These results indicate that when wild-type Aux/IAA proteins are targeted to reporter genes by a heterologous DBD, they function as active repressors, and that the repression they confer is sensitive to auxin. The most likely explanation for relief of repression observed with auxin could be the result of increased instability of the GAL4 DBD–Aux/IAA fusion proteins (i.e., instability conferred by the Aux/IAA part of the fusion).
Figure 5.
Aux/IAA Proteins Repress Constitutive Reporter Genes in an Auxin-Responsive Manner When Targeted to DNA Binding Sites in Promoters.
Effector genes encoding GAL4 DBD and GAL4 DBD–Aux/IAA fusion proteins were cotransfected into carrot protoplasts along with a constitutively expressed GUS reporter gene containing four tandem GAL4 DNA binding sites (see Methods for details on effector and reporter genes). GUS activities were measured 24 hr after transfections. Transfected protoplasts were incubated in the presence or absence of 25 μM auxin (1-NAA). Transfections were performed with effectors encoding GAL4 DBD fused to wild type (wt) and domain I, II, and III mutant versions (denoted with as mI, mII, and mIII; see Table 1) of IAA7 (indicated with a 7) and IAA17 proteins (indicated with a 17). G4 indicates GAL4 DBD; G4-DBD indicates an effector encoding the unfused GAL4 DBD. IAA7wt and IAA17wt refer to effector genes encoding IAA7 and IAA17 proteins that were not fused to a GAL4 DBD. None, no response to auxin. The effector:reporter ratio was 1:1.
(A) Transfections with a GAL4D1-3:GUS reporter gene.
(B) Transfections with a GAL4CaMV35S:GUS reporter gene.
Standard errors are indicated.
Effector genes encoding proteins with mutations (see Table 1) in domain I of IAA7 (G4-7mIa and G4-7mIb) and domain I of IAA17 (G4-17mI) showed decreased repression compared with wild-type effectors, especially when auxin was withheld from the transfection assays. Likewise, an effector gene encoding a protein with a mutation in domain III (G4-17mIII) of IAA17 showed decreased repression compared with that of a wild-type effector in the absence of auxin treatment. With effectors encoding proteins with mutations in domain II (G4-7mII and G4-17mII), repression was equivalent to that of the wild-type IAA17 in the absence of auxin, but derepression in the presence of auxin was reduced (G4-7mII) or eliminated (G4-17mII). Effectors encoding Aux/IAA proteins with double mutations in domains I and II (G4-7mI/mII and G4-17mI/mII) showed reduced repression and little or no auxin response compared with the effectors with only domain II mutations (cf. G4-7mI/mII with G4-7mII, and G4-17mI/mII with G4-17mII). Results with effectors encoding Aux/IAA proteins with mutations in domains I and II indicate that mutations in domain II reduce or eliminate the auxin response and that mutations in domain I partially reverse the repression conferred by wild-type or domain II mutant proteins. These results further suggest that a wild-type domain I may be required for or strongly contribute to repression by Aux/IAA proteins.
Figure 5B shows GUS activities for the GAL4CaMV-35S:GUS reporter gene with GAL4 DBD-IAA7 and GAL4 DBD-IAA17 fusion effector genes, respectively. Like the GAL4D1-3:GUS reporter gene, the GAL4CaMV35S:GUS reporter gene was unresponsive to auxin when tested with no effector, the GAL4 DBD effector (G4-DBD), and IAA effector plasmids lacking a GAL4 DBD (IAA7wt and IAA17wt). Although the GAL4CaMV35S:GUS reporter gene displayed threefold higher activity than did the GAL4D1-3:GUS reporter gene in carrot protoplasts, the relative amount of repression, derepression, and auxin responsiveness conferred by the wild-type and mutant GAL4-IAA fusion protein effectors was similar to results summarized for the GAL4D1-3:GUS reporter gene in Figure 5A, except that the G4-7mII and G4-17mII activities are less responsive to auxin than shown in Figure 5A.
Level of Repression Conferred by Aux/IAA Proteins Is Dependent on Their Auxin-Responsive Instability
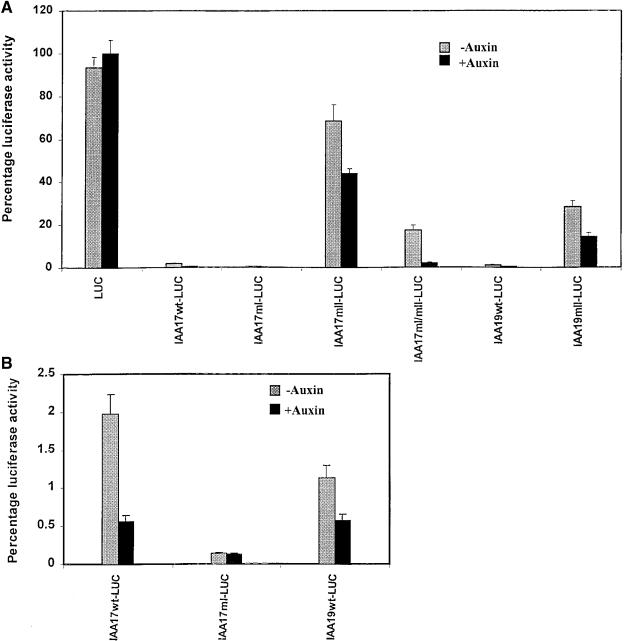
The amount of repression observed with wild-type and mutant Aux/IAA effectors documented above could result wholly or partially from the relative stabilities of the wild-type and mutant Aux/IAA proteins in transfected protoplasts. We have attempted to directly assess whether the domain II mutations, which increase the repressive nature of Aux/IAA proteins in transfection assays (see Figures 3 and 4), result in increased levels of Aux/IAA proteins in transfected protoplasts. To perform this analysis, we HA-tagged Aux/IAA proteins at their N termini, and then used anti–HA epitope antibodies in attempts to detect the epitope-tagged proteins in cell-free extracts from protoplasts by protein gel blotting. Detection of the proteins was, however, inconsistent and unreliable. Similar problems were encountered using anti–GAL4 DBD antibodies for protein gel blotting of extracts from protoplasts transfected with an effector gene encoding GAL4 DBD–Aux/IAA fusion proteins. To circumvent problems with antibody detection of proteins expressed from effector plasmids, we fused wild-type and mutant versions of IAA17 and IAA19 to the N terminus of LUC and assayed transfected protoplasts for luciferase activity. Expression of all luciferase constructs were under control of the CaMV 35S promoter. These types of fusion proteins have been used previously in plants to demonstrate that an Aux/IAA–LUC fusion protein is much less stable than is an unfused LUC protein and that Aux/IAA domain II plays a role in destabilizing the fusion protein (Worley et al., 2000).
Figure 6A shows the relative luciferase activities obtained with genes expressing LUC alone, wild-type Aux/IAA proteins fused to LUC, and mutant Aux/IAA proteins fused to LUC. The 35S:LUC gene (LUC) showed the highest level of expression for all genes tested, and this gene showed no response to auxin. Reporter genes encoding wild-type IAA17 (IAA17wt-LUC) and IAA19 (IAA19wt-LUC) proteins fused to LUC had very low luciferase activities even without addition of auxin (2.0% and 1.1% of 35S:LUC gene activity for IAA17 and IAA19, respectively); however, activities were further decreased in the presence of auxin (0.56% and 0.57% of 35S:LUC gene activity for IAA 17 and IAA 19, respectively; Figure 6B). Mutations in domain II have been reported to stabilize Aux/IAA proteins (Ouellet et al., 2001) and Aux/IAA-LUC fusion proteins in plants (Worley et al., 2000). Without auxin treatment, protoplasts transfected with genes expressing IAA17 (IAA17mII-LUC) and IAA19 (IAA19mII-LUC) domain II mutant proteins (see Table 1) fused to LUC had 35-fold and 26-fold higher luciferase activities, respectively, than did protoplasts transfected with genes expressing wild-type IAA17 and IAA19 fusion proteins. In the presence of auxin, luciferase activities were 78-fold and 30-fold higher for IAA17mII-LUC and IAA19mII-LUC, respectively, compared with those of wild-type reporters. Protoplasts expressing an IAA17 domain I mutant protein fused to LUC (IAA17mI-LUC) displayed even less luceriferase activity than did the wild-type fusion protein with or without auxin, suggesting that mutations in domain I may further decrease the stability of Aux/IAA proteins. Protoplasts expressing an IAA17 domain I and II double mutant (IAA17mI/mII-LUC) had luciferase activity higher than that of the wild type but lower than that of domain II mutant fusion proteins (cf. IAA17mI/mII-LUC with IAA17-LUC and IAA17mII-LUC) in the presence or absence of auxin. The IAA17 domain I and II double mutant fusion protein appears to be considerably less stable than the IAA17 domain II mutant fusion protein. Taken together, these results suggest that wild-type Aux/IAA proteins are highly unstable in transfected protoplasts, that their stability is increased by mutations in domain II, and that their stability is decreased by mutations in domain I. Furthermore, the Aux/IAA proteins are less stable in the presence of auxin than in the absence of auxin.
Figure 6.
Auxin Decreases the Stability of Wild-Type and Mutant Aux/IAA-LUC Fusion Proteins in Transfected Carrot Protoplasts.
(A) Relative luciferase activities with effector genes encoding luciferase alone (LUC) and luciferase fused to wild-type and domain I and II mutant versions of IAA17 and IAA19. Reporter genes encoding fusion proteins consisted of wild-type (wt) and mutant versions (mI and mII; see Table 1) of IAA17 and IAA19 fused to the N terminus of LUC (see Methods). Activities are relative to the reporter gene encoding an unfused LUC protein (100%).
(B) An expanded scale of data from (A) showing results with reporter genes encoding IAA–LUC fusion proteins that displayed low luciferase activities.
Standard errors are indicated.
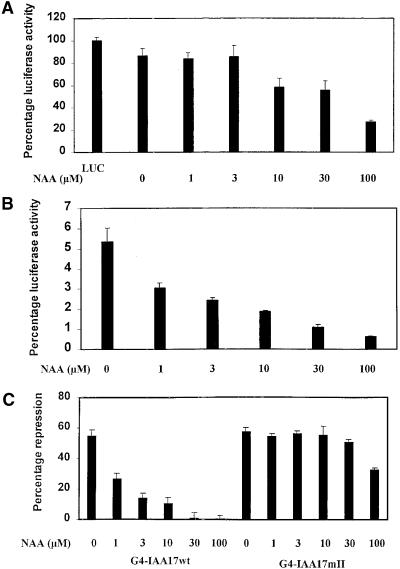
To determine if the auxin-modulated stability of Aux/IAA proteins was dependent on the dose of auxin, we tested wild-type 35S:IAA17wt-LUC and domain II mutant 35S: IAA17mII-LUC reporter genes in transfected carrot protoplasts with a range of 1-NAA concentrations. Figures 7A and 7B show auxin dose–responses for luciferase activity in carrot cells transfected with 35S:IAA17mII-LUC and 35S:IAA17wt-LUC, respectively. Results in Figure 7B show that with the 35S:IAA17wt-LUC reporter gene, auxin concentrations as low as 1 μM cause a nearly twofold reduction in luciferase activity, and as the concentration of auxin is increased from 1 to 100 μM 1-NAA, the luciferase activity declines in a dose-dependent manner. Figure 7A shows that with the 35S:IAA17mII-LUC reporter gene, auxin concentrations in the range of 1 to 3 μM had little or no effect on reporter gene activity, and only when auxin concentrations were elevated to 10 μM and above did luciferase activity decline. These results suggest that the stability of the wild-type IAA17 protein is highly sensitive to auxin in a dose-dependent manner and that a domain II mutant IAA17 protein is much more stable to auxin over a concentration range of 1 to 100 μM.
Figure 7.
Auxin Dose Response for Reporter Genes Encoding the Wild Type and a Domain II Mutant Version of Arabidopsis IAA17.
The auxin (1-NAA) was applied at the concentration indicated below each bar. Luciferase activity is relative to the reporter gene expressing the unfused LUC protein in transfected carrot protoplasts (100%).
(A) Dose response for the 35S:IAA17mII-LUC reporter plasmid encoding IAA17 with a domain II mutation fused to LUC.
(B) Dose response for the 35S:IAA17-LUC reporter plasmid encoding wild-type IAA17 fused to LUC.
(C) Dose response for repression by effector genes encoding wild-type IAA17 (G4-IAA17wt) and domain II mutant IAA17 (G4-IAA17mII) with the GAL4D1-3:GUS reporter gene. Transfection assays were conducted as described in Figure 5 with a effector:reporter ratio of 1:1 at different NAA concentrations. GUS activity with the reporter gene in the absence of effector was considered 100% activity and 0% repression. Percentage of repression was calculated by subtracting the percentage of activity of the reporter gene observed with the effector gene from the percentage of activity of the reporter gene without the effector gene at each NAA concentration. There was no response of the reporter gene to auxin in the absence of effector.
Standard errors are indicated.
Effector genes encoding G4-IAA17wt and G4-IAA17mII proteins were tested with the GAL4D1-3:GUS reporter gene (see Figure 5A) over the same auxin concentration range used in Figures 7A and 7B to determine if the derepression by auxin was also dose dependent. The GAL4D1-3:GUS reporter gene was used for the auxin dose response because this reporter gene does not respond to auxin in the absence of effector genes that encode GAL4 DBD–IAA fusion proteins. Figure 7C shows that repression of the reporter gene with G4-IAA17wt was relieved as auxin concentrations increased from 1 to 100 μM. In contrast, repression of the reporter gene with G4-IAA17mII showed little response to auxin until the concentration exceeded 30 μM. These results indicate that the auxin dose responses for luciferase activity (see Figures 7A and 7B) are mirrored by the dose responses for derepression of the GAL4D1-3:GUS reporter gene when assayed with wild-type and domain II mutant effector genes.
DISCUSSION
Our results showed that sixteen different Arabidopsis Aux/IAA proteins repressed transcription of an auxin-responsive reporter gene to varying degrees when transfected into protoplasts as effector plasmids. This included IAA17/AXR3, which might have been predicted to be an activator based upon ectopic expression of auxin-responsive reporter genes in iaa17/axr3-1 mutant plants (Leyser et al., 1996; Oono et al., 1998). These results, along with results reported previously with four other Aux/IAA proteins from pea and soybean (Ulmasov et al., 1997b), are consistent with a repressive role for Aux/IAA proteins in regulating expression of early response genes containing TGTCTC AuxREs. Because our assays with Aux/IAA effector genes were conducted only with carrot and Arabidopsis protoplasts, it remains possible that some Aux/IAA proteins might not function as repressors in all cell types, and it remains possible that some Aux/IAA proteins might function as activators in cells that differ from the protoplasts examined in our experiments.
Selected Aux/IAA proteins from pea and Arabidopsis have been shown to have short lifetimes and to be nuclear localized (Abel et al., 1995; Ouellet et al., 2001). Several semi-dominant mutants have been identified that have mutations in domain II of Aux/IAA proteins (reviewed by Liscum and Reed, 2001). For the first of these identified (i.e., iaa17/axr3), it was predicted that this gain-of-function (i.e., in terms of auxin response) mutation might result in stabilizing/increasing the lifetime of the IAA17 protein (Rouse et al., 1998). This indeed appears to be the case based on recent pulse-chase experiments that measured the lifetimes of IAA17 in wild-type and axr3 mutant plants (Ouellet et al., 2001). It is likely that similar mutations in domain II of other Aux/IAA proteins result in increased lifetimes of the proteins; immunological evidence for this has been reported for the IAA3 protein in the iaa3/shy2-2 mutant (Colon-Carmona et al., 2000). On the basis of transfection results presented here, the outcome of this increased lifetime of Aux/IAA proteins in the domain II mutants is enhanced repression of early auxin-response genes.
Our results further indicate that mutations in domains I and III partially reverse the repression observed with both wild-type Aux/IAA proteins and Aux/IAA proteins that contain a mutation in domain II. A likely explanation for reversal of repression by domain III mutations is partial disruption of the dimerization domain in Aux/IAA proteins, which would reduce their ability to interact with other Aux/IAA proteins and with ARF proteins that bind to the TGTCTC AuxREs in the reporter genes (Ulmasov et al., 1997a, 1997b, 1999b; Kim et al. 1997; Ouellet et al., 2001). The mechanism for reversal of repression by domain I mutations is less clear, but our results suggest that this domain may also play a role in protein stability and might be required for repression. Ouellet et al. (2001) have reported the iaa17/axr3-1R3 protein (Rouse et al. 1998), which contains a mutation in both domains I and II, fails to interact with itself in a yeast two-hybrid system. The same mutation in domain I of AXR3 did not prevent homodimerization or heterodimerization with other Aux/IAA or ARF proteins in the absence of a domain II mutation (Ouellet et al., 2001). On the basis of these results, Ouellet et al. (2001) suggested that a second site mutation in domain I may provide an interaction domain important for homodimerization, or it may alter the structure of the Aux/IAA protein in such a way that it becomes susceptible to degradation even with a domain II mutation. Regardless of the mechanism, reversal of repression by the domain I mutations in transfection assays is generally greater than that observed with domain III mutations with or without a domain II mutation.
Aux/IAA proteins do not appear to be DNA binding proteins that have affinity for TGTCTC AuxREs in auxin-responsive promoters (Ulmasov et al., 1997b). Thus, repression of auxin-responsive genes by Aux/IAA proteins is thought to involve their interaction with ARF transcriptional activators that bind to TGTCTC AuxREs (reviewed by Guilfoyle et al., 1998; Guilfoyle and Hagen, 2001). Interactions between Aux/IAA and ARF proteins have been shown to occur through conserved C-terminal dimerization domains that are found in both Aux/IAA and ARF proteins (Kim et al., 1997; Ulmasov et al., 1997a, 1997b; Ouellet et al., 2001). At this point, there is no direct evidence that the repressive effects of Aux/IAA proteins occur as a result of their being targeted to AuxREs indirectly by interacting with DNA-bound ARFs. It is possible that repression by Aux/IAA proteins occurs because they prevent ARF transcriptional activators from reaching their AuxRE target sites. Results presented here show, however, that when Aux/IAA proteins are targeted to DNA binding sites by a heterologous DBD on constitutive promoters, reporter gene expression is repressed. Thus, Aux/IAA proteins can function as active transcriptional repressors. Whether the Aux/IAA proteins recruit a co-repressor complex to bring about this repression or repress by some other mechanism remains to be investigated. In any case, our results indicate that the GAL4 DBD–Aux/IAA fusion proteins can repress expression at a considerable distance from the transcription start site on a complex CaMV 35S promoter, as well as at a short distance from the transcription start site on a simple, multimerized constitutive element fused to a −46 minimal 35S promoter.
Results presented here also indicate that auxin plays a dose-dependent role in modulating the lifetimes of Aux/IAA proteins and thus their capacity to repress early auxin response genes. There appears to be a direct correlation between stability of Aux/IAA proteins and ability of Aux/IAA proteins to repress transcription on early auxin response genes. On the basis of our transfection assays with Aux/IAA–LUC fusion proteins, Aux/IAA proteins become less stable as auxin concentrations increase. This instability may result from Aux/IAA proteins being more rapidly targeted to the proteasome for degradation at high auxin concentrations. The ultimate consequence of increased instability of the Aux/IAA proteins is a decline in Aux/IAA protein levels, which, in turn, results in a decline in their repressive activity on early response genes.
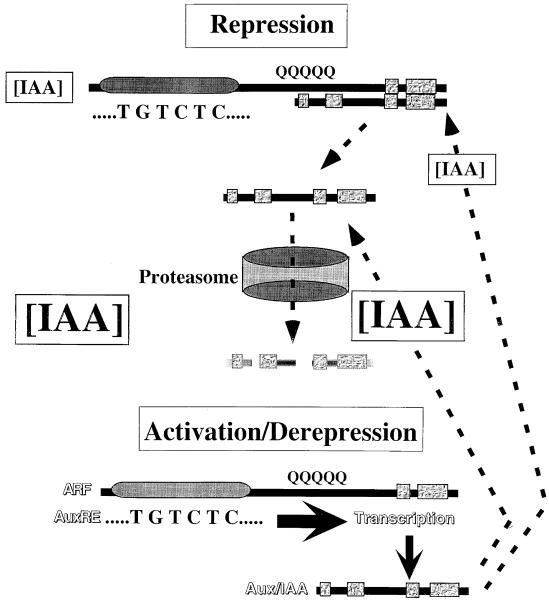
On the basis of previous reports and results presented here, we have formulated the following model for the role of Aux/IAA proteins in regulating early auxin response genes (see Figure 8). When auxin concentrations are low within cells or tissues, primary/early auxin response genes are actively repressed by Aux/IAA proteins, which are more stable at low auxin concentrations. Repression is likely to result from dimerization or multimerizaton between Aux/IAA repressors with ARF transcriptional activators that are bound to AuxRE target sites on early auxin response genes. Active repression by the interacting Aux/IAA proteins would somehow mask the activator function of the ARF protein. When auxin concentrations are elevated, early auxin response genes are rapidly derepressed/activated (reviewed by Hagen and Guilfoyle, 2001). An early event in derepression might be the dissociation of Aux/IAA repressors from their ARF counterparts bound to AuxREs, concomitant with or prior to degradation of the Aux/IAA proteins by the ubiquitin/proteasome pathway (reviewed by Gray and Estelle, 2000). Early gene activation might be further potentiated by the binding of additional ARF transcriptional activators (i.e., by dimerization/multimerization through domains III and IV) to the DNA-bound ARF (Ulmasov et al., 1999a).
Figure 8.
Model for the Auxin-Responsive Repression of Early Auxin Response Genes by Aux/IAA Proteins.
The model predicts that ARF transcriptional activators (e.g., ARF5, -6, -7, -8) with glutamine-rich (Q) middle regions are bound via their DBDs (gray oval; Ulmasov et al., 1997a, 1999b) to AuxRE target sites in the promoters of early auxin response genes independent of auxin levels within cells (see Ulmasov et al., 1999a). When auxin levels are low (small IAA in brackets), primary/early auxin response genes are actively repressed by Aux/IAA proteins that dimerize with ARF transcriptional activators. Dimerization occurs via interactions between domains III and IV, which are found in both Aux/IAA and ARF proteins. When auxin levels increase (large IAA in brackets), Aux/IAA proteins dissociate from the DNA-bound ARF proteins and are rapidly degraded through the proteasome pathway (reviewed by Gray and Estelle, 2000). Dissociation of the Aux/IAA proteins from the ARF DNA complexes results in rapid transcriptional derepression/activation of early response genes, some of which encode Aux/IAA proteins. The transcription of these Aux/IAA and other early response genes continues at high levels for up to several hours as long as auxin concentrations remain high (Hagen et al., 1984; Theologis et al., 1985; Abel et al., 1995). The Aux/IAA proteins synthesized at high auxin concentrations are rapidly degraded and only accumulate to levels sufficient for repression when auxin levels decline, ultimately resulting in feedback inhibition of their own transcription.
At least some Aux/IAA genes contain TGTCTC AuxREs, and these genes might be regulated by auxin as described above, resulting in increased abundance of the Aux/IAA mRNAs at high auxin concentrations. The mRNAs would be translated into Aux/IAA proteins, but the Aux/IAA proteins would be unstable and subject to rapid degradation via the ubiquitin/proteasome pathway as long as auxin concentrations remained high. As auxin concentrations eventually decline, the accumulated Aux/IAA mRNAs would continue to be translated and Aux/IAA protein concentration would eventually reach sufficient levels that the accumulated Aux/IAA proteins could feed back on their own genes as well as other early response genes containing TGTCTC AuxREs to downregulate their expression.
METHODS
Reporter Gene Constructs
The auxin-responsive P3(4X):GUS (β-glucuronidase) and GH3:GUS reporter genes and the constitutive D1-3(4x):GUS reporter genes have been previously described (Liu et al., 1994; Ulmasov et al., 1995, 1997a). Reporter gene constructs containing yeast GAL4 DNA binding sites were constructed by cloning four tandem copies of the GAL4 DNA binding sequence, 5′-AGGAAGACTCTCCTCCG-3′, immediately upstream of the CaMV 35S:GUS reporter gene (Skuzeski et al., 1990), which is referred to as GAL4CaMV35S:GUS, and the D1-3(4X):GUS reporter gene, which is referred to as GAL4D1-3:GUS. For Aux/IAA–LUC fusion reporter constructs, the luciferase open reading frame (ORF) (pGL2 vector; Promega, Madison WI) was cloned in-frame to the C terminus of IAA17 and IAA19 ORFs. Mutations into domains I and III IAA17 and IAA19 proteins were introduced by polymerase chain reaction (PCR) using standard methods (Ausubel et al., 1998), and all mutant constructs were verified for accuracy by DNA sequencing. Expression of the LUC and Aux/IAA–LUC fusion constructs was driven by the CaMV 35S promoter.
Effector Gene Constructs
All effector gene constructs were placed under the control of the CaMV 35S double enhancer promoter followed by a translational enhancer from the Tobacco mosaic virus 5′ leader (Skuzeski et al., 1990) and contained a 3′ nopaline synthase untranslated region (Liu et al., 1994; Ulmasov et al., 1995). Full-length ORFs for Aux/IAA proteins either were obtained from cDNA clones (provided by A. Theologis, Plant Gene Expression Center, Albany, CA) or were PCR amplified directly from an Arabidopsis cDNA library (Clontech, Palo Alto, CA). For Aux/IAA effector constructs, wild-type and mutated versions of Aux/IAA ORFs were fused in-frame with a sequence encoding the hemagglutinin (HA) epitope tag, MGYPYDVPYAH. For GAL4 DBD–Aux/IAA fusion effector constructs, the yeast GAL4 DBD (amino acids 1 to 147) with its nuclear localization signal was fused in-frame with the N terminus of wild-type and mutated versions of IAA7/AXR2 and IAA17/AXR3 ORFs. Mutations into domains I, II, and III of Aux/IAA proteins were introduced by PCR using standard methods (Ausubel et al., 1998), and all mutant constructs were verified for accuracy by DNA sequencing.
Protoplast Transfection Assays
Carrot protoplasts were isolated from suspension culture cells, and transfections, GUS assays, and luciferase assays were performed as previously described (Liu et al., 1994; Ulmasov et al., 1995, 1997b). Arabidopsis protoplasts were isolated from young leaves, and transfection assays were performed as described by Kovtun et al. (2000). Transfection assays in carrot and Arabidopsis protoplasts conducted with auxin contained 25 and 1 μM auxin 1-naphthalene acetic acid (1-NAA), respectively. All reporter and effector plasmids used in transfection assays were prepared using the EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA). Effector plasmids (5 μg) were cotransfected with reporter plasmids (10 μg) at a ratio of 1:2 unless indicated otherwise. When applicable, the amount of DNA in each transfection was equalized by adding the appropriate amount of a CaMV 35S:chloramphenicol acetyl transferase reporter plasmid (35S:CAT; Ulmasov et al., 1997b). The efficiency of transfection was standardized by adding 100 ng of a CaMV 35S:LUC reporter gene, which showed no response to auxin (Liu et al., 1994). For transfections with Aux/IAA-LUC fusion constructs, 2.5 μg of plasmid was transfected into carrot protoplasts, and extracts from equivalent numbers of protoplasts were assayed for luciferase activity. All transfection assays were performed in triplicate, and at least two independent preparations of protoplasts were used in transfection assays to ensure the reproducibility of the results.
Acknowledgments
We thank Sean Doke for technical assistance, Dr. Jen Sheen for advice on transfection of Arabidopsis protoplasts, and Dr. Athanasios Theologis for providing Aux/IAA cDNA clones. This work was supported by National Science Foundation Grant No. MCB 0080096 to T.J.G. and G.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010289.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds (1998). Current Protocols in Molecular Biology. (New York: Wiley and Sons).
- Colon-Carmona, A., Chen, D.L., Yeh, K.C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Reg., in press.
- Hagen, G., and Guilfoyle, T.J. (2001). Auxin-responsive gene expression: Genes, promoters, and regulatory factors. Plant Mol. Biol., in press. [PubMed]
- Hagen, G., Kleinschmidt, A.J., and Guilfoyle, T.J. (1984). Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162, 147–153. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress–activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Liscum, M., and Reed, J. (2001). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol., in press. [PubMed]
- Liu, Z.-B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). The soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, K.E., Zarembinski, T.I., Theologis, A., and Abel, S. (1999). Biochemical characterization of recombinant polypeptides corresponding to the predicted beta alpha alpha fold in Aux IAA proteins. FEBS Lett. 454, 283–287. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono, Y., Chen, Q.G., Overvoorde, P.J., Kohler, C., and Theologis, A. (1998). age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10, 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Skuzeski, J.M., Nichols, L.M., and Gesteland, R.F. (1990). Analysis of leaky viral translation termination codons in vivo by transient expression of improved β-glucuronidase vectors. Plant Mol. Biol. 15, 5–69. [DOI] [PubMed] [Google Scholar]
- Theologis, A., Huynh, T.V., and Davis, R.W. (1985). Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J. Mol. Biol. 183, 53–68. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z.-B., Hagen, G., and Guilfoyle, T.J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Activation and repression of transcription by auxin response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]