Abstract
Previous analysis of aerobic, glucose-limited continuous cultures of Escherichia coli revealed that G:C-to-T:A (G:C→T:A) transversions were the most commonly occurring type of spontaneous mutation. One possible explanation for the preponderance of these mutations was that nutrient limitation repressed MutY-dependent DNA repair, resulting in increased proportions of G:C→T:A transversions. The regulation of the mutY-dependent DNA repair system was therefore studied with a transcriptional mutY-lacZ fusion recombined into the chromosome. Expression from the mutY promoter was fourfold higher under aerobic conditions than under anaerobic conditions. But mutY expression was higher in glucose- or ammonia-limited chemostats than in nutrient-excess batch culture, so mutY was not downregulated by nutrient limitation. An alternative explanation for the frequency of G:C→T:A transversions was the common appearance of mutY mutator mutations in the chemostat populations. Of 11 chemostat populations screened in detail, six contained mutators, and the mutator mutation in four cultures was located in the region of mutY at 66 min on the chromosome. The spectrum of mutations and rate of mutation in these isolates were fully consistent with a mutY-deficiency in each strain. Based on PCR analysis of the region within and around mutY, isolates from three individual populations contained deletions extending at least 2 kb upstream of mutY and more than 5 kb downstream. In the fourth population, the deletion was even longer, extending at least 5 kb upstream and 5 kb downstream of mutY. The isolation of mutY mutator strains from four independent populations with extensive chromosomal rearrangements suggests that mutY inactivation by deletion is a means of increasing mutation rates under nutrient limitation and explains the observed frequency of G:C→T:A mutations in glucose-limited chemostats.
There is an increasing awareness that mutational processes in bacteria are under environmental control or at least susceptible to stress responses (3, 9, 12, 13, 35, 37). Most of the studies on these influences used batch cultured bacteria or colonies on plates with their constantly changing or spatially nonuniform environments. The complexity of the stress, be it starvation, slow growth or external challenge, can be simplified by using a steady-state application of particular stresses as can be achieved in chemostat culture (14). As an initial study in this direction, the mutational changes in three genes were analyzed in glucose-limited chemostats of Escherichia coli (23, 24). Mutations in malT, mlc, and mgl genes were identified and consisted of mainly base changes under steady-state glucose limitation. The majority of nucleotide substitutions were G:C-to-T:A (G:C→T:A) transversions in all three genes. This communication addresses possible causes of the mutational spectrum observed under glucose limitation, since it differs markedly from the generally observed spectrum of spontaneous mutations in E. coli (29, 34).
A clue as to the origin of malT, mlc, and mgl mutations was that the spectrum is characteristic of mutator mutants lacking the oxodG repair system (20). The genes mutM and mutY are involved in this process, but mutM changes are only weakly mutagenic (2). Hence the mutational spectrum under glucose limitation was most likely a function of mutY regulation or mutation. A question addressed here is whether mutY regulation is changed by growth conditions to reduce DNA repair. Alternatively, was a mutation in mutY or, less likely, mutM commonly found in populations growing under glucose-limited conditions?
Both mutM and mutY genes are part of complex operons (11), but no physiological data were available on expression of mutY until recently (10), and few data on mutM were available (16). mutY was shown to be the first gene in an operon and to be regulated by aerobiosis, but not by oxygen stress regulators (10, 11). Also, the effect of mutM and mutY on stationary-phase, starvation-associated mutation indicated that mutY, but not mutM, mutants increase mutation rates under these conditions (2). The regulation of these genes by steady-state nutrient limitation was not previously studied, and to observe whether control of mutY was responsible for the spectrum of mutations in chemostat cultures, the expression of mutY needed to be studied under steady-state nutrient limitation. Given the lack of information on mutY regulation, other growth conditions were also investigated. In particular, mutY repair is potentially important in repair of oxidative DNA damage (20), so the effect of O2 stress on mutY expression was also assessed.
The alternative explanation for the mutational spectra was that mutY mutator mutations spread in glucose-limited populations and bias the type of DNA changes in these populations. The advantage of mutators in adapting to chemostat conditions is well-known (5). Mutator mutations have been observed in long-term recycled batch cultures (31, 32) and occur at a finite level in natural bacterial populations (15). Mutator mutations are coenriched by selection pressure for advantageous mutations (18), which are common in nutrient-limited populations (23, 24). There are both advantages and disadvantages in maintaining elevated mutation rates, and it was proposed that mutators may appear transiently in populations (32). Also, there was no obvious reason why mutY as opposed to other mutators, such as mutS, should accumulate in glucose-limited populations; previous analysis of mutator appearances in bacterial culture generally resulted in mismatch-repair mutants (1). Hence, it became important to establish whether mutY mutations were indeed prevalent under glucose limitation. We addressed this question by directly looking for mutators as described below.
MATERIALS AND METHODS
Bacterial strains.
All bacterial strains used in this study are derivatives of E. coli K-12 and are shown in Table 1. P1 transduction was carried out using P1 cml clr1000 according to the method of Miller (19). A P1 lysate made on strain JM2053, containing galP::Tn10, was used to transduce selected chemostat isolates with increased mutation rates. For each chemostat isolate, a P1 lysate was made of a tetracycline-resistant transductant that still had an elevated mutation rate. The mutation causing the elevated mutation rate was then cotransduced with galP::Tn10 into a clean background (MC4100 or BW3500). P1 lysates of these chemostat isolates were also used to transduce strain AT2446 to Met+, selecting for growth on minimal glucose plates to map mutator activity by cotransduction with metC.
TABLE 1.
Strains and plasmids used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| AT2446 | Hfr relA metC69 thi | Coli genetic stock center |
| BW2952 | MC4100 Φ(malG-lacZ) | 22 |
| BW3143 | BW2952 mglA::Tn10 | 23 |
| BW3185 | MC4100 mutS srl::Tn10 (from chemostat isolate) | 25 |
| 21Qa1 (BW3490) | BW2952 mutY 3490 + unknown | Chemostat 21 isolate |
| L3Oa1 (BW3489) | BW3143 mutY 3489 + unknown | Chemostat L3 isolate |
| 10U6 (BW3488) | BW2952 mutY 3488 + unknown | Chemostat 10 isolate |
| C7F1 (BW3487) | BW2952 mutY 3487 + unknown | Chemostat C7 isolate |
| BW3500 | MC4100 λ att[Φ(mutY′-′lacZYA)] | This study |
| BW3509 | BW3500 galP::Tn10 | This study |
| BW3510 | BW3500 galP::Tn10 mutY 3490 | This study |
| BW3507 | MC4100 galP::Tn10 | This study |
| BW3508 | MC4100 galP::Tn10 mutY 3490 | This study |
| BW3517 | MC4100 galP::Tn10 mutY 3488 | This study |
| BW3518 | MC4100 galP::Tn10 mutY 3487 | This study |
| BW3523 | DY330 mutY::Tn10 | This study |
| BW3526 | BW2952 mutY::Tn10 | This study |
| BW3527 | BW3500 mutY::Tn10 | This study |
| DY330 | W3110 ΔlacU169 gal490 λ cI857 Δ(cro-bioA) | 38 |
| JM2053 | HfrC galP::Tn10 mglP ptsM ptsG-con ptsF his gndR thyA ilv nag | P. J. F. Henderson |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 deoC1 relA ptsF25 flbB5301 rbsR | 4 |
| NM522 | F−supE thi Δ(lac-proAB) hsd5 (r− m−) [F′ proAB+lacIqZΔM15] | Promega Corp. |
| pAM2555 | pRS415 cut with SmaI containing 825-bp PCR fragment amplified from MC4100 and primers mutYF1 and mutYR1 (Apr) | This study |
| pTTY | Intact mutY cloned in pTrc99A | 2 |
| pRS415 | Vector for gene fusions; tet′ ori bla T14 ′trp-lacZYA (Apr) | 30 |
Construction of a mutY-lacZ transcriptional fusion.
A region encompassing 300 bp upstream of mutY as well as 300 bp of the mutY gene was amplified by PCR using two primers: mutYF1 and mutYR1 (see below). The PCR-generated fragment was end polished and blunt-end ligated into vector pRS415 (30) cut with SmaI to form a transcriptional fusion to lacZ. Plasmids were transformed into strain NM522, and those forming blue colonies on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) plates were digested with EcoRI to test whether the insert was ligated in the correct orientation. The fusion construct was then recombined into the complementary phage λ RS45 (30) before insertion at the lambda attachment site in strain MC4100. Insertion in single copy in the chosen recombinant was verified by the method of Powell et al. (27) and designated strain BW3500. The promoter insert was fully sequenced to ensure no errors were generated during cloning or amplification.
Construction of a mutY::Tn10 insertion.
A mutY::Tn10 insertion was constructed by the method of Yu et al. (38). Primers F1Tn10F (5′-GCACAAAACTACGGATACGGCGCAGTGGGCACTCGACATCTTGGTTACCG-3′) and R2Tn10R (5′-ATCCTGACCTTCTGCTTCACGTTGCAGGAACAAGAGGGTCATTATATTTCG-3′) were designed with tet homology at the 3′ ends to amplify the tetracycline resistance cassette from strain BW3143 (mglA::Tn10) and with sequences homologous to the mutY region at the 5′ ends. PCR was carried out under standard conditions with primer annealing at 58°C and extension for 2 min. The 2,075-bp product was purified using Wizard PCR preps DNA purification kit (Promega Corp., Sydney, Australia) and electroporated into DY330 as per the method of Yu et al. (38). Recombination in the DY330 background resulted in deletion of the entire mutY gene, including 203 bp upstream and 54 bp downstream, and replacement with the tet resistance cassette to form strain BW3523. A P1 lysate was then made of this strain and used to transduce BW2952 and BW3500, selecting for tetracycline resistance, to give strains BW3526 and BW3527, respectively.
Culture conditions.
The basal salts medium used in all experiments was minimal medium A (MMA) (19) supplemented with glucose, lactate, or glycerol as specified for each experiment. Batch cultures contained 0.4% (wt/vol) sugar, unless otherwise specified, and were harvested during mid-exponential growth as defined in the individual experiments. TA plates contained NaCl (5 g/liter), tryptone (10 g/liter), and 1.5% (wt/vol) agar. The expression of the mutY-lacZ fusion in strain BW3500 was measured during growth in minimal medium containing 0.02% (wt/vol) glucose with vigorous shaking at 37°C. Paraquat (5 μM), H2O2 (0.0025% [vol/vol]), and novobiocin (20 μg/ml) were added to the culture medium, and expression was measured at early exponential phase. Anaerobic cultures were grown in filled bottles without shaking.
Glucose-limited chemostats (80 ml) were set up as previously described (6). Monitoring of mutY-lacZ expression in strain BW3500 was studied in chemostats run for several days, limited for both glucose and nitrogen. Glucose-limiting chemostats had 0.02% (wt/vol) glucose in the feed medium and were run at dilution rates of 0.1, 0.3, and 0.6 h−1. Nitrogen-limiting chemostats at a dilution rate of 0.3 h−1 had reduced ammonium sulfate levels from 1 to 0.04 g/liter, and the glucose concentration increased to 0.2% (wt/vol).
Chemostats containing mutators were part of an ongoing study (17, 23–25) and varied with respect to dilution rates, glucose concentrations in the feed medium, and the starting strains. Population 21 and population L3 were cultured with a concentration of 0.02% (wt/vol) glucose and a dilution rate of 0.3 h−1. Population 10 was cultured with a glucose concentration of 0.02% (wt/vol) and a dilution rate of 0.6 h−1. Population C7 had a glucose concentration of 0.006% (wt/vol) and a dilution rate of 0.3 h−1. All were started with strain BW2952 except population L3, which was started with strain BW3143. The chemostats were maintained for up to 4 weeks. For sampling, 10 ml of culture was collected daily aseptically from the overflow port and was either directly subjected to the T5 resistance assay or stored in 40% (wt/vol) glycerol at −70°C for later use. Samples were streaked on nonselective nutrient agar, and randomly separated colonies were purified and labeled (e.g., sampling day 1 = A, day 2 = B, etc.) for further testing.
β-Galactosidase assays.
The β-galactosidase activity of lacZ fusions of bacteria grown in batch culture or the chemostat for 3 days was assayed by the method of Miller (19) and expressed in Miller units. The β-galactosidase activities presented in Tables 1 and 2 were the averages of three to five separate culture results.
TABLE 2.
Expression of mutY-lacZ fusion under various growth conditions
| Culture methoda | Limiting substrate | Dilution rate (h−1) | Growth conditions
|
β-Galacto-sidaseb (Miller units) | |||
|---|---|---|---|---|---|---|---|
| O2 | Para-quat | H2O2 | Novo-biocin | ||||
| Batch | + | − | − | − | 65 ± 10 | ||
| Batch | + | + | − | − | 78 ± 2 | ||
| Batch | + | − | + | − | 86 ± 5 | ||
| Batch | + | − | − | + | 108 ± 5 | ||
| Batch | − | − | − | − | 17 ± 1 | ||
| Batch | − | − | − | + | 36 ± 4 | ||
| Chemostat | Glucose | 0.1 | + | − | − | − | 159 ± 5 |
| Chemostat | Glucose | 0.3 | + | − | − | − | 131 ± 3 |
| Chemostat | Glucose | 0.6 | + | − | − | − | 148 ± 8 |
| Chemostat | Nitrogen | 0.3 | + | − | − | − | 178 ± 7 |
Batch-grown cells were grown in 0.02% glucose-MMA until exponential phase (OD580 between 0.3 and 0.4) either aerobically by shaking vigorously at 300 rpm or anaerobically in filled bottles without shaking. Paraquat was added to cultures at a concentration of 5 μM and H2O2 at a concentration of 0.0025% (vol/vol). Novobiocin was added at a concentration of 20 μg/ml. Chemostat cultures were grown aerobically under nitrogen or glucose limitation at various dilution rates.
The values reported are means and standard deviations measured from at least three independent cultures.
Phenotypic characterization of chemostat isolates.
The spread of mutations in the chemostat populations were monitored using T5 phage resistance assays (for neutral mutations), mutator assays, and phenotypic tests for mgl, ptsG, mlc, and malT mutations as previously described (17, 23–25). For comparison of mutation frequencies, equal numbers (5 × 108 bacteria) of four independent cultures of each isolate as well as the mutS and mutY control strains were cultured on rifampin plates as described previously (25).
PCR protocols and mutation analysis.
PCR amplification of the galS (also known as mglD) sequence and the mgl operator used primers GalSF1 (5′-GCAATCTCATAACAGGTAGTG-3′) and GalSR2 (5′-ATGACGCTGTTACCTCGGC-3′). The reaction profile consisted of 34 cycles of the following in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.): denaturation at 94°C for 30s, annealing at 58°C for 30s, and extension at 72°C for 1.5 min. PCR amplification of the ptsG gene used two primers: ptsGF1 (5′-CTGTTTCACATCGATCGACGCTTCC-3′) and ptsGR1 (5′-CCAGCGCGGATACGCCATCG-3′). The reaction profile was similar to that for mgl except primers were annealed at 60°C. The mutY gene and surrounding region were amplified using an extensive set of primers and with an annealing temperature of 70°C. Primers were as follows: mutYF1 (5′-CTACGGA TACGGCGCAGTGGGC-3′), mutYF2 (5′-CATGCGCAGAAGCCAGGCACGC-3′), mutYF3 (5′-GCTGCTATGCTGTAAGCGGCTGG-3′), mutYF5 (5′-GCTATCTTCCAGAGTCACAACGCG-3′), mutYF7 (5′-CCACAGTTAGACGAGGCGATTGC-3′), mutYF9 (5′-GGATACCAGTTCACTACTGGCGGC-3′), mutYF10 (5′-CTGCAACGTGAAGCAGAAGGTCAGG-3′), mutYR1 (5′-CGAACATTTCGGTTTCGAGCGCGT-3′), mutYR2 (5′-CCTGACCTT CTGCTTCACGTTGCAG-3′), mutYR5 (5′-CCGGCAGTATGTTGTACCACCTGC-3′), mutYR8 (5′-CCCAGTAAGACTGCACTAGCGTTG-3′), and mutYR11 (5′-GCGTGCCTGGCTTCTGCGCATG-3′). PCR products were purified directly with Wizard PCR preps DNA purification system (Promega Corp.). The nucleotide sequence was determined using the above primers and dye-terminator sequencing reactions and were analyzed on a Catalyst Robotic Workstation. Mutations were located in mutant sequences by aligning with the known galS (mglD), ptsG, and mutY sequences in the E. coli genome database using software available in the Australian National Genomic Information Service (Sydney, Australia) database.
DNA extraction and blotting analysis.
Chromosomal DNA of the starter strain (BW2952), the mutY::Tn10-containing strain (BW3526), and the four chemostat isolates was prepared as follows. A 10-ml aliquot of overnight culture grown on Luria broth was centrifuged at 3,120 × g for 10 min and resuspended in 5 ml of TE buffer, pH 8 (50 mM Tris, pH 8; 50 mM EDTA, pH 8). The suspension was frozen at −20°C for 30 min and then thawed at room temperature in the presence of lysozyme (1 mg/ml). Once thawed, the culture was placed on ice for 45 min. One milliliter of STE buffer (0.5% [wt/vol] sodium dodecyl sulfate; 50 mM Tris, pH 7.5; 0.4 M EDTA, pH 8; proteinase K, 1 mg/ml) was added before heating at 50°C for 60 min. DNA was extracted with an equal volume of Tris-buffered phenol (pH 8) and precipitated with 3 M sodium acetate and ethanol. The DNA precipitate was spooled out and dissolved in 5 ml of TE buffer, pH 7.5 (50 mM Tris, pH 7.5; 1 mM EDTA, pH 8) containing 100 μg of RNase per ml. The DNA was extracted once with an equal volume of chloroform before precipitation with sodium acetate and ethanol and then finally was dissolved in 2 ml of TE buffer, pH 7.5, and stored at 4°C. This DNA was used both in PCRs and for dot blot analysis. Equivalent loadings of chromosomal DNA (as determined by staining in an agarose gel) were boiled for 10 min before blotting directly onto a positively charged nylon membrane (Hybond-N+; Amersham Int. Plc.) using a Bio-Dot Slot format apparatus (Bio-Rad Laboratories, Richmond, Calif.). The membrane was air dried for 30 min and then baked at 120°C for 30 min. Prehybridization and hybridization reactions were carried out overnight at 50°C with digoxigenin (DIG)-dUTP-tailed probes including mutYF2, mutYF3, and mutYF9. DIG-labeled dUTP was added to the 3′ end of each oligonucleotide probe using a DIG oligonucleotide labeling kit (Roche Diagnostics Australia, Pty. Ltd.) and the recommended protocol. As a positive control, the strains were probed with an oligonucleotide complementary to the housekeeping gene encoding enolase (eno: 5′-TGACGGAGCAGCTGCCATACCGAC-3′). A DIG luminescence detection kit for nucleic acids (Roche Diagnostics Australia, Pty. Ltd.) with the recommended protocol was used to detect DIG-labeled DNA.
RESULTS
Regulation of mutY expression.
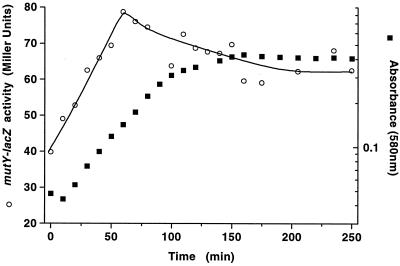
A transcriptional mutY-lacZ fusion was constructed, recombined in single copy into the chromosome, and grown under a variety of physiological conditions to test the range of variation in mutY expression. As shown in Fig. 1, growth phase controlled the mutY promoter. Expression was maximal approaching stationary phase in aerobic cultures but decreased in stationary phase. In confirmation of recent transcript data, aerobiosis was another factor in mutY regulation (10). As shown in Table 2, the presence of O2 resulted in a four- to fivefold elevation of mutY fusion activity. The stimulation by O2 did not appear to be part of a stress response, given that neither superoxide (from added paraquat) or H2O2 affecting stress responses (36) stimulated mutY expression much above aerobic levels. The lack of target sequences in the mutY promoter for global stress regulators (11) and recent transcript data (10) were also consistent with the lack of superoxide or H2O2 effects. The aerobic-to-anaerobic transition is regulated by multiple transcriptional factors as well as DNA-structural changes in E. coli (28). This left open the possibility that mutY was regulated by the level of DNA supercoiling. Indeed, the extent of regulation of mutY in Fig. 1 and Table 2 resembled that of tonB, which is also influenced by aerobiosis and growth phase in a similar manner (7). To test the possible influence of supercoiling, the effect of the DNA gyrase inhibitor novobiocin was tested as shown in Table 2. Reduced negative supercoiling associated with novobiocin resulted in increased mutY expression in both aerobic and anaerobic cultures, also by a similar margin as with tonB (7).
FIG. 1.
Expression of a mutY-lacZ fusion in strain BW3500 was monitored by measuring β-galactosidase activity during growth in batch culture in minimal medium containing 0.02% (wt/vol) glucose. Growth was monitored by measuring the absorbance at 580 nm.
Aside from batch cultures, mutY expression was also monitored in continuous cultures limited in growth rate either by glucose or ammonia (N) limitation. As shown in Table 2, there was no difference between glucose and N limitation at a particular growth rate and no difference between different growth rates. These results point to lack of regulation by RpoS, cAMP, or ppGpp, all of which markedly change in concentration under the range of conditions analyzed (8). Importantly, the level of expression in nutrient-limited cultures was actually higher than that in nutrient-excess exponential batch cultures. The level of supercoiling in nutrient-limited cultures may be lower and potentially explains the higher expression of mutY under nutrient limitation. However, an obvious conclusion is that glucose-limited continuous cultures are unlikely to have a particular mutational spectrum due to increased mutation rates caused by downregulation of mutY.
Appearance of mutY mutator mutations in glucose-limited populations.
In the absence of mutY downregulation, continuous cultures were analyzed for mutational changes affecting mutY. Eleven populations growing under glucose limitation at dilution rates of 0.3 or 0.6 h−1 were tested for mutators in daily samples of the cultures. Random colonies from each day sample (between 8 and 20 colonies tested) were screened for increased mutation frequencies on rifampin plates for Rifr. Of the 11 independent chemostat populations growing and assessed daily for mutators in this way, 6 cultures had isolates with increased mutation rates. In these cultures, isolates with elevated mutation frequencies comprised 30 to 100% of the population in daily samples after approximately 100 to 150 generations of continuous culture. The appearance of mutators coincided with a 60- to 70-fold increase in the frequency of neutral T5-resistant mutants as well as the emergence of selectively favorable mutations (in mgl, malT, mlc, and ptsG [17, 23, 24]) in a high percentage of the population (results not shown).
Of the six populations with mutators, two contained mutS mutations as determined previously (25). The mutS isolates exhibited an increase in mutation frequencies of more than 100-fold when cultured on Rif plates, whereas the other four populations contained isolates with lower mutation frequencies. The lower increase (approximately 40-fold) in mutation frequency in members of populations 10, 21, C7, and L3 was comparable with the increase in a mutY deletion strain (BW3526) and that found in mutY mutators in other studies (references 21 and 33 and results not shown). Mapping of the mutator mutation in an isolate of each of the four populations by P1 transduction located the mutations in the galP-metC region at 66 min on the chromosome of E. coli, consistent with mutY as the mutated locus. Sequencing of mutations in mgl and ptsG arising in these populations showed that the DNA changes were indeed transversions, characteristic of the mutY mutator genotype (Table 3) (21). Transfer of the mutation by P1 transduction with galP::Tn10 into a strain containing a mutY-lacZ fusion (BW3500) at the lambda attachment site (to create BW3510), showed there was no influence on mutY transcription in trans (Table 4); therefore, the mutation affected mutY directly and was not in a nearby regulator of mutY. A similar result was obtained with the mutY::Tn10 insertion on mutY transcription (Table 4), so mutY does not autoregulate its own expression. Finally, introduction of intact mutY on a plasmid, pTTY (2), reduced mutation rates back to the wild-type (WT) level for each of the four presumptive mutY isolates, but not for the mutS isolates (result not shown).
TABLE 3.
Spectrum of DNA changes in mql and ptsG
| Mutation | Base changea | No. of isolates sequencedb |
|---|---|---|
| Population L3 (ptsG) | 15 | |
| V12F | G→T | |
| G13C | G→T | |
| Population 21 (mglDO) | 30 | |
| A11D | C→A | |
| H122Q | C→A | |
| W198C | G→T | |
| A252E | C→A | |
| D246Y | G→T | |
| A296E | C→A | |
| mglO | G→T |
Changes within the codon position or promoter specified for genes ptsG or mglD.
The number of independent isolates sequenced from each of the populations subsequent to the appearance of mutators in the populations.
TABLE 4.
Effect of mutY mutation on mutY-lacZ expression
| Growth conditionsa | β-Galactosidase activity (Miller units)b in λ att [Φ(mutY-lacZ)] strain:
|
|||
|---|---|---|---|---|
| BW3509 (MutY+) | BW3510 (MutY−) | BW3500 (MutY+) | BW3527 (mutY::Tn10) | |
| MMA-glucose, aerobic, exponential | 60 ± 5 | 62 ± 10 | 58 ± 7 | 60 ± 10 |
| MMA-glucose, aerobic, stationary | 74 ± 3 | 73 ± 10 | 74 ± 6 | 65 ± 15 |
| MMA-glucose, anaerobic, stationary | 16 ± 8 | 17 ± 5 | 23 ± 6 | 24 ± 6 |
Batch-grown cells were grown in 0.02% glucose-MMA until early exponential phase (OD580 = 0.1) aerobically by shaking vigorously at 300 rpm. Stationary-phase bacteria were left for 24 h before sampling.
The values reported are means and standard deviations measured from at least four independent cultures.
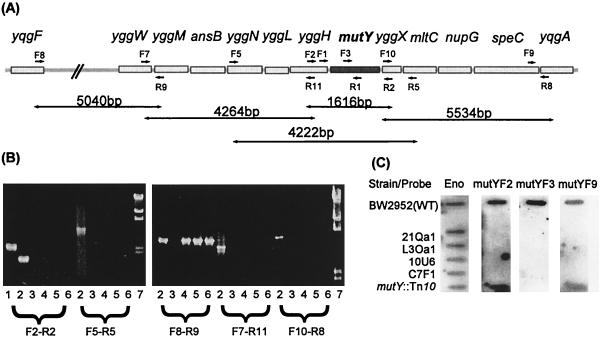
The nature of the mutator mutation in an example of each of the chemostat populations (isolate 21Qa1 from population 21; isolate L3Oa1 from population L3; isolate 10U6 from population 10; and isolate C7F1 from population C7) was investigated by a PCR analysis using mutY-flanking primers. Initially, a primer pair surrounding the mutY gene was used (mutYF2 and mutYR2) to amplify the region (Fig. 2A) in both the WT and the four mutant strains. The fragment could be amplified in the WT strain (BW2952), but no PCR fragment was amplified in any of the four mutator mutants. As there may have been a loss of one or both of the primer sites, more primer pairs were designed (as shown in Fig. 2A). A 4,222-bp fragment extending from primers mutYF5 to mutYR5 also gave no PCR product in each of the mutants (Fig. 2B). Primers were then used in pairs that amplified a wider region surrounding mutY.
FIG. 2.
(A) Locations of primers used in PCR, as probes in dot blots, and for construction of mutY-lacZ transcriptional fusion. (B) Primer pairs (specified below gels) were used to amplify the mutY gene and surrounding region in several chemostat mutator mutants. Agarose concentrations in the gels were 1 and 0.8%, respectively. The gel lane numbered 1 corresponded to strain BW3526 carrying the mutY::Tn10 insertion. Lanes 2, WT strain BW2952; lanes 3, isolate 21Qa1; lanes 4, isolate L3Oa1; lanes 5, isolate 10U6; lanes 6, isolate C7F1; lanes 7, DNA markers with sizes of 23,130, 9,416, 6,557, 4,361, 2,322, and 2,027 bp. (C) Results of slot blotting of DNA extracted from mutator strains. Hybridization was at 50°C with DIG-dUTP-tailed probes mutYF2, mutYF3, and mutYF9 as well as the positive control probe for an unlinked housekeeping gene, eno.
Upstream of mutY, primers mutYF8 to mutYR9 amplified a 5,040-bp region in the WT and in three of the chemostat isolates: L3Oa1, 10U6, and C7F1. This suggests this region is still intact in these isolates. However, PCR failed to amplify any fragment in isolate 21Qa1, suggesting a deletion that extends at least beyond the mutYR9 site 4.5 kb upstream of mutY.
Other pairs of primers included mutYF7 to mutYR11 (4,264-bp fragment), and mutYF10 to mutYR8 (5,534-bp fragment). There was no amplification using these primers in any of the isolates, suggesting rearrangement or loss of DNA downstream of mutY as well. As a control to ensure that the DNA prepared was intact, all isolates were subject to PCR using primers galSF1 and galSR2 amplifying the galS gene. All gave the correct amplification product (results not shown).
Dot blotting using DIG-dUTP-labeled probes was able to confirm the deletion results. Primers mutYF2 (300 bp upstream of mutY), mutYF3 (mid-mutY), and mutYF9 (approximately 5 kb downstream of mutY) were chosen and hybridized to DNA on membranes at 50°C (Fig. 2C). All three probes hybridized to the WT strain but not to any of the mutator mutants, while mutYF2 and mutYF9 hybridized to the second control strain (BW3526) containing the mutY::Tn10 insertion. The mutYF3 probe did not react in this strain as this region is deleted and replaced with the Tn10 insertion. The positive control probe, eno, specific for the housekeeping gene enolase, located at 60 min on the chromosome, hybridized with all strains.
DISCUSSION
The role of mutY in E. coli is to contribute to repair of DNA damaged by reactive O2 species (20). Hence, it is not surprising that there was a higher level of mutY expression under aerobic conditions than under anaerobic growth conditions. The extent of the increase with the mutY fusion was in line with recent results with transcript analysis (10). Also confirming the results of Gifford et al., the lack of stimulation by the major O2 stresses through superoxide and peroxide is consistent with the lack of obvious binding sites for global regulators in the mutY promoter (11). Much of the mutY regulation described in Fig. 1 and Table 2 and in reference 10 can be rationalized on the basis of the aerobic-to-anaerobic transition affecting mutY promoter structure. The results with the gyrase inhibitor novobiocin and the influence of aerobiosis on DNA supercoiling (7) suggest that the pattern of regulation of mutY has many similarities to the supercoiling-mediated control of tonB expression by aerobiosis and growth phase (7). The absence of regulation by growth rate under nutrient limitation also rules out mutY as being part of the RpoS-mediated general stress response or ppGpp-dependent regulation (8).
The expression of mutY under nutrient limitation ruled out the notion that the high frequency of G:C→T:A transversions under glucose limitation was due to reduced mutY expression. The earlier mutation data were more likely to be due to the repeated occurrence of mutator mutants in long-term continuous culture populations. Evidence for the occurrence of mutators was obtained in the populations 21, C7, 10, and L3 analyzed in detail. The appearance of mutators was accompanied by major population shifts as well as biased mutation rates. The appearance of mutY mutations in two of the chemostat populations coincided with the appearance of mgl and ptsG changes, with transversions being the predominant class of mutation. Unfortunately, reanalysis of the earlier populations (23, 24) failed to find mutators in the stored weekly samples, but the 7-day intervals previously used could easily miss a transient mutator sweep that can appear and disappear within days (reference 25 and results not shown).
The mutY mutation present in isolates from each of the chemostat populations was stable and maintained for multiple generations after subculture. The PCR analysis of the mutations showed the mutations were caused by a substantial deletion in each case. The rearrangement extended at least 2 to 5 kb on one or more sides of the gene. Since the endpoints of the deletion were not sequenced, there is no clue as to the cause of the mutY mutations. Given the similarity of events in four independent populations, it would be interesting to determine if particular features of the surrounding genome contribute to these events. An inspection of the chromosomal region for insertion sequences, BIME sites, chi sites, or repeats did not reveal obvious features contributing to the results.
Our observations with experimental populations may well be mirrored in evolving natural populations. Loss of the mutY gene was evident in evolving Pseudomonas aeruginosa populations in cystic fibrosis patients, and two individual isolates from two individual patients both failed to amplify mutY (26). The extent of the deletions was not determined, but PCR primers surrounding mutY failed to amplify the gene. Perhaps the major mutator gene rearrangements we observe occur in Pseudomonas as well.
There is a curious similarity between these mutY deletions and the extensive rearrangement and/or deletion of the mutS region previously observed (15). In chemostat populations, the two independent mutS mutators (populations 20 and 26 [25]) are not fully characterized yet, but preliminary evidence also suggests major deletions. This leaves open the possibility that mutator deletions are significant in evolving populations, but the source of these deletions requires further definition. In any case, the mutY inactivation by a chromosomal rearrangement is a means of increasing mutation rates in times of nutrient limitation and results in the prevalence of G:C→T:A mutations in glucose-limited populations.
Acknowledgments
We thank B. Bridges for supplying the mutY plasmid and P. J. F. Henderson for strains.
We also thank the Australian Research Council for grant support.
REFERENCES
- 1.Bregeon, D., I. Matic, M. Radman, and F. Taddei. 1999. Inefficient mismatch repair: genetic defects and down regulation. J. Genet. 78:21–28. [Google Scholar]
- 2.Bridges, B. A., M. Sekiguchi, and T. Tajiri. 1996. Effect of mutY and mutM/fpg-1 mutations on starvation-associated mutation in Escherichia coli: implications for the role of 7,8-dihydro-8-oxoguanine. Mol. Gen. Genet. 251:352–357. [DOI] [PubMed] [Google Scholar]
- 3.Bridges, B. A., and A. Timms. 1998. Effect of endogenous carotenoids and defective RpoS sigma factor on spontaneous mutation under starvation conditions in Escherichia coli: evidence for the possible involvement of singlet oxygen. Mutat. Res. 403:21–28. [DOI] [PubMed] [Google Scholar]
- 4.Casabadan, M. J. 1976. Transposition and fusion of the lac operon to selected promoters in E. coli using bacteriophage Lambda and Mu. J. Mol. Biol. 104:541–555. [DOI] [PubMed] [Google Scholar]
- 5.Chao, L., and E. C. Cox. 1983. Competition between high and low mutating strains of Escherichia coli. Evolution 37:125–134. [DOI] [PubMed] [Google Scholar]
- 6.Death, A., L. Notley, and T. Ferenci. 1993. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J. Bacteriol. 175:1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorman, C. J., G. C. Barr, N. N. Bhriain, and C. F. Higgins. 1988. DNA supercoiling and the anaerobic and growth phase regulation of the tonB gene. J. Bacteriol. 170:2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferenci, T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208–213. [DOI] [PubMed] [Google Scholar]
- 9.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gifford, C. M., J. O. Blaisdell, and S. S. Wallace. 2000. Multiprobe RNase protection assay analysis of mRNA levels for the Escherichia coli oxidative DNA glycosylase genes under conditions of oxidative stress. J. Bacteriol. 182:5416–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gifford, C. M., and S. S. Wallace. 1999. The genes encoding formamidopyrimidine and MutY DNA glycosylases in Escherichia coli are transcribed as part of complex operons. J. Bacteriol. 181:4223–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, P. J. Hastings, M. E. Winkler, and S. M. Rosenberg. 1999. Mismatch repair is diminished during stationary-phase mutation. Mutat. Res. Rev. Mutat. Res. 437:51–60. [PubMed] [Google Scholar]
- 13.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, M. E. Winkler, and S. M. Rosenberg. 1997. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 11:2426–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubitschek, H. E. 1970. Introduction to research with continuous cultures. Prentice-Hall Inc., Englewood Cliffs, N.J.
- 15.Leclerc, J. E., B. G. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211. [DOI] [PubMed] [Google Scholar]
- 16.Lee, H. S., Y. S. Lee, H. S. Kim, J. Y. Choi, H. M. Hassan, and M. H. Chung. 1998. Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: roles of fnr, arcA, and fur. Free Radical Biol. Med. 24:1193–1201. [DOI] [PubMed] [Google Scholar]
- 17.Manche, K., L. Notley-McRobb, and T. Ferenci. 1999. Mutational adaptation of Escherichia coli to glucose limitation involves distinct evolutionary pathways in aerobic and oxygen-limited environments. Genetics 153:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao, E. F., L. Lane, J. Lee, and J. H. Miller. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Miller, J. H. 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50:625–643. [DOI] [PubMed] [Google Scholar]
- 21.Nghiem, Y., M. Cabrera, C. G. Cupples, and J. H. Miller. 1988. The mutY gene: a mutator locus in Escherichia coli that generates GC-TA transversions. Proc. Natl. Acad. Sci. USA 85:2709–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notley, L., and T. Ferenci. 1995. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient stressed Escherichia coli. Mol. Microbiol. 160:121–129. [DOI] [PubMed] [Google Scholar]
- 23.Notley-McRobb, L., and T. Ferenci. 1999. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ. Microbiol. 1:33–43. [DOI] [PubMed] [Google Scholar]
- 24.Notley-McRobb, L., and T. Ferenci. 1999. The generation of multiple coexisting mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1:45–52. [DOI] [PubMed] [Google Scholar]
- 25.Notley-McRobb, L., and T. Ferenci. 2000. Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics 156:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1253. [DOI] [PubMed] [Google Scholar]
- 27.Powell, B. S., D. L. Court, Y. Nakamura, M. P. Rivas, and C. L. Turnbough. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawers, G. 1999. The aerobic/anaerobic interface. Curr. Opin. Microbiol. 2:181–187. [DOI] [PubMed] [Google Scholar]
- 29.Schaaper, R. M., B. N. Danforth, and B. W. Glickman. 1986. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J. Mol. Biol. 189:273–284. [DOI] [PubMed] [Google Scholar]
- 30.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. [DOI] [PubMed] [Google Scholar]
- 31.Sniegowski, P. D., P. J. Gerrish, and R. E. Lenski. 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387:703–705. [DOI] [PubMed] [Google Scholar]
- 32.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godelle. 1997. Role of mutator alleles in adaptive evolution. Nature 387:700–702. [DOI] [PubMed] [Google Scholar]
- 33.Tajiri, T., H. Maki, and M. Sekiguchi. 1995. Functional cooperation of mutT, mutM and mutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. DNA Repair 336:257–267. [DOI] [PubMed] [Google Scholar]
- 34.Takimoto, K., A. Tachibana, H. Ayaki, and K. Yamamoto. 1997. Spectrum of spontaneous mutations in the cyclic AMP receptor protein gene on chromosomal DNA of Escherichia coli. J. Radiation Res. 38:27–36. [DOI] [PubMed] [Google Scholar]
- 35.Tsui, H. C. T., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tweeddale, H., L. Notley-McRobb, and T. Ferenci. 1999. Assessing the effect of reactive oxygen species on Escherichia coli using a metabolome approach. Redox Rep. 4:237–241. [DOI] [PubMed] [Google Scholar]
- 37.Wright, B. E. 1996. The effect of the stringent response on mutation rates in Escherichia coli K-12. Mol. Microbiol. 19:213–219. [DOI] [PubMed] [Google Scholar]
- 38.Yu, D. G., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]