Abstract
Herpes simplex virus type 1 packages its DNA genome into a precursor capsid, referred to as the procapsid. Of the three capsid-associated DNA-packaging proteins, UL17, UL25, and UL6, only UL17 and UL6 appear to be components of the procapsid, with UL25 being added subsequently. To determine whether the association of UL17 or UL25 with capsids was dependent on the other two packaging proteins, B capsids, which lack viral DNA but retain the cleaved internal scaffold, were purified from nonpermissive cells infected with UL17, UL25, or UL6 null mutants and compared with wild-type (wt) B capsids. In the absence of UL17, the levels of UL25 in the mutant capsids were much lower than those in wt B capsids. These results suggest that UL17 is required for efficient incorporation of UL25 into B capsids. B capsids lacking UL25 contained about twofold-less UL17 than wt capsids, raising the possibilities that UL25 is important for stabilizing UL17 in capsids and that the two proteins interact in the capsid. The distribution of UL17 and UL25 on B capsids was examined using immunogold labeling. Both proteins appeared to bind to multiple sites on the capsid. The properties of the UL17 and UL25 proteins are consistent with the idea that the two proteins are important in stabilizing capsid-DNA structures rather than having a direct role in DNA packaging.
Herpesviruses have a characteristic morphology, consisting of an icosahedral capsid containing the linear double-stranded DNA genome; an amorphous layer, referred to as the tegument, surrounding the capsid; and an outer envelope. Over 30 viral structural proteins have been identified in the mature herpes simplex virus type 1 (HSV-1) particle, eight of which are associated with the capsid (14, 35, 39). Three minor virion proteins, UL17, UL25, and UL6, which are located on the capsid, are important for packaging the viral DNA into the capsid but are not essential for capsid formation (14, 24, 25, 30, 36-39). The viral genome is inserted into a spherical intermediate capsid, referred to as the procapsid, which is the precursor of DNA-containing (C) capsids. During DNA packaging the internal protein scaffold within the procapsid is cleaved and removed (26, 28). At the same time the procapsid undergoes extensive morphological changes into a more robust, angularized capsid with a well-defined capsid floor (11).
In the absence of DNA packaging, cleavage of the protein scaffold and structural transformation of the procapsid still occur, provided that the UL26 protease is functional (1, 26). The resulting angularized capsid, which retains the scaffolding protein, is referred to as the B capsid. B capsids are relatively stable, are present in large amounts in the nuclei of wild-type (wt) virus-infected cells, and accumulate in nonpermissive cells infected with DNA packaging null mutants. Although they represent dead-end products of infection by wt HSV-1, their structure closely resembles that of C capsids, and they have proved very useful in investigating the roles of individual DNA-packaging proteins during capsid maturation (19, 43, 44). Both UL17 and UL6 are present in similar amounts in procapsids and B capsids whereas UL25 is found in much lower levels in procapsids than in B capsids (31, 39). It has therefore been proposed that UL25 is not a genuine procapsid component but is incorporated into the capsid at a later stage in the assembly process (31). UL17 and UL25 but not UL6 are present in greater levels in C capsids than in B capsids, suggesting that further binding sites for these two proteins are exposed on the capsid after the DNA has been packaged (31, 39). UL17 has also been found in L particles, aberrant structures that contain a tegument and envelope but lack a capsid (30, 39). This finding, together with the observation that virions have approximately twofold-more UL17 than do C capsids, suggests that UL17 is also a tegument protein (39).
UL6 is located at a single vertex on the capsid, forming the portal for entry of viral DNA (19). Analyses of UL6 complexes suggest that the portal has a ring structure composed of 12 copies of UL6, which is similar in morphology to the complexes produced by portal or connector proteins from double-stranded DNA bacteriophage (10, 19, 40, 41). Immunoprecipitation and immunofluorescence experiments have revealed that UL6 interacts with two other DNA-packaging proteins, UL15 and UL28, which are believed to be transiently associated with the capsid since they are not present in significant amounts in DNA-containing capsids (31, 42, 43). UL15 and UL28 are thought to form the terminase, which is responsible for recognizing packaging signals on the viral DNA and cleaving the replicated concatemeric viral DNA into monomeric units (2, 8, 43). By analogy with the mechanism of genome encapsidation of double-stranded DNA-tailed bacteriophage (4, 7, 15), it is likely that these two proteins, together with UL6, comprise the molecular motor that inserts the DNA into the capsid. Another packaging protein, UL33, interacts with UL28 and UL15 and may also be a component of the packaging motor (3). Less information is available on the distribution and function of UL17 and UL25 than on those of UL6. Preliminary experiments have suggested that at least some UL25 is exposed on the outer surface of capsids at multiple sites, indicating that its role in DNA packaging may be indirect (23).
The phenotype of UL17 null mutants is very similar to that of UL6 null mutants. In the absence of UL17 the concatemeric replicated viral DNA is not cleaved into genome-sized pieces and no DNA is encapsidated in the mutant virus-infected cells, suggesting that, like UL6, UL17 is required at an early stage in DNA packaging (30). By contrast, UL25 appears to function at a later stage (14, 36) since low levels of viral DNA are cleaved and packaged in nonpermissive cells infected with a UL25 null mutant (36). In addition, greater numbers of empty capsids, lacking the scaffolding protein, are present in the UL25 null mutant-infected cells than in cells infected with wt virus or mutants with defects in other packaging proteins (14).
In this report we have examined the amounts and distribution of UL17 and UL25 on wt and mutant B capsids to extend our understanding of the roles of these two proteins.
MATERIALS AND METHODS
Cells.
Baby hamster kidney 21 clone 13 (BHK) cells were grown in Glasgow minimal essential medium supplemented with 10% tryptose phosphate, 10% newborn calf serum, 100 units of penicillin, and 100 μg streptomycin per ml. The transformed cell lines were all grown in Dulbecco's minimal essential medium containing 10% fetal calf serum and the same antibiotics.
Viruses.
wt HSV-1 strain 17(syn+), wt HSV-1 strain KOS, and the KUL25NS, lacZ-UL6−, and ΔUL17 mutants have been described previously (6, 14, 25, 30, 36). The UL17-stop virus contains a 16-bp insertion at position 32718 in the HSV-1 strain 17 genome, with stop codons in all three reading frames, truncating the UL17 open reading frame after codon 260 (30). The UL17-stop marker rescuant, MRUL17-stop, was constructed by transfecting UL17RSC5 cells (described below) with the plasmid pGX334 (containing the HSV-1 strain 17 genomic BglII p fragment; nucleotide coordinates 31097 to 35729 [13]) cleaved with BglII, using Lipofectamine and Plus reagent (Invitrogen) according to the manufacturer's recommendations. Four hours after transfection the cells were infected with 1 PFU per cell of HSV-1 UL17-stop and the cells were incubated for a further 20 h at 37°C. The cells were harvested; marker rescuants, identified by their ability to grow and form plaques on BHK cells, were plaque purified; and a stock of virus was prepared. The yield of revertants in mock-transfected cells was 80-fold lower than the level of marker rescuants in cells transfected with the plasmid containing the BglII p fragment. KUL25NS, lacZ-UL6−, and UL17 mutants were grown in 8-1 (14), UL6RSC1 (39), and UL17RSC5 (39) cells, respectively.
Purification of capsids.
Nuclear extracts were prepared from virus-infected cells at 18 to 24 h postinfection, and capsids were purified on 20 to 50% sucrose gradients as described previously (45). C capsids were banded twice on a 20 to 50% sucrose gradient and directly used in immunoelectron microscopic experiments without any concentration by centrifugation, to minimize aggregation.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto a nitrocellulose membrane, and screened for various packaging or capsid proteins using the enhanced chemiluminescence method as outlined by Thurlow et al. (39). After each enhanced chemiluminescence reaction the blots were stripped of antibody and washed extensively prior to the addition of a different primary antibody. The protein bands in exposed film were quantified as described previously (39).
Quantification of packaging proteins in capsids.
Proteins were separated on large SDS-polyacrylamide gels and were stained for 4 h with SYPRO orange (Bio-Rad Laboratories) by diluting the stock solution of dye 5,000-fold in 7.5% (vol/vol) acetic acid. The stained gels were rinsed in 7.5% acetic acid and placed on a 302-nm UV transilluminator. A digital image was taken of the gel, and the packaging proteins were quantified using Syngene gene tools software.
Antibodies.
Mouse monoclonal antibodies (MAbs) specific for UL6 (MAb 175), UL17 (MAb 203), or UL25 (MAb 166) and a rabbit polyclonal antibody specific for VP23 (Rab186) have been described previously (12, 39). Rabbit antibody Rab335 was raised against insoluble UL25 (residues 342 to 580)-glutathione S-transferase fusion protein. Rabbit antibodies Rab992 and Rab1218 were produced against purified soluble UL6 (residues 379 to 676)-pMAL fusion protein and purified, insoluble UL17 (residues 154 to 703)-pMAL fusion protein, respectively, essentially as described by Preston et al. (27). The specificity of each of the polyclonal antibodies was initially confirmed by Western blot analysis of polypeptides from insect cells infected with a recombinant baculovirus expressing the HSV protein of interest, using the nonrecombinant baculovirus-infected cell polypeptides as a negative control (data not shown).
Treatment of capsids with urea.
wt B capsids were treated with 0, 2, 4, or 6 M urea in 10 mM Tris-HCl, 1 mM EDTA, 0.5 M NaCl, pH 7.5 (NTE), containing 50 mM dithiothreitol, in the presence of protease inhibitors for 1 h at room temperature, essentially as described by Newcomb et al. (21). The samples were overlaid on a 150-μl 25% (wt/vol) sucrose cushion in NTE and centrifuged at 48,458 × g in a Beckman TLS-55 rotor for 1 h at 4°C. The pellet of capsids was resuspended in NTE, and the supernatant was retained for Western blot analysis.
Detection of UL17- and UL25-specific antibodies on capsids by immunogold labeling.
Each Formvar-coated nickel grid was placed onto a 50-μl droplet of phosphate-buffered saline, containing 6.8 mM CaCl2 and 4.9 mM MgCl2 and supplemented with 2 mM β-octylglucoside, for 2 min and blotted dry prior to the addition of 5 μl capsid sample (0.1 mg/ml in phosphate-buffered saline) for 3 min. In subsequent manipulations the grids were floated, specimen side down, on droplets of solution. After the grids had been washed twice with 0.5× NTE for 30 s per wash, they were treated with blocking solution (0.5× NTE containing 5% goat serum, 5% bovine serum albumin, and 0.1% fish gelatin) for 1 h. Each grid was subsequently transferred to a drop of purified primary antibody diluted in blocking buffer. After incubation for 1.5 h the grids were washed eight times (about 4 min per wash) in blocking solution. The washed grids were each transferred for 1 h to a droplet of secondary antibody (goat anti-rabbit immunoglobulin G [IgG] conjugated to 10-nm colloidal gold [British Biocell International]) diluted in blocking solution. The unbound secondary antibody was removed by washing the grids eight times with 0.5× NTE followed by two brief washes with H2O. The capsids were negatively stained for 15 s with 1% ammonium molybdate in 2 mM β-octylglucoside. The grids were blotted and dried, and the capsids were visualized under a JEOL 100S transmission electron microscope. Serial dilutions of primary and secondary antibodies were made to determine the optimal concentration of each antibody giving the highest amount of gold binding to wt capsids, with the lowest level of background staining of mutant capsids lacking the relevant protein.
RESULTS
Identification and abundance of UL6, UL17, and UL25 proteins in B capsids.
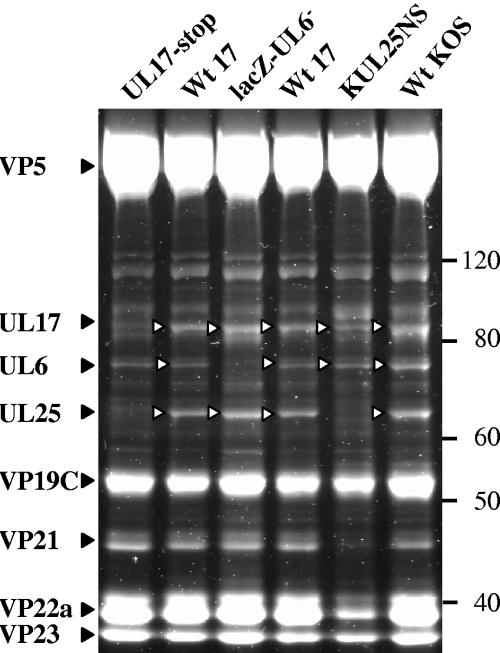
To obtain information on the identity and relative abundance of the capsid-associated packaging proteins, the proteins of B capsids of wt HSV-1 strains 17 and KOS and UL6, UL17, and UL25 null mutants (lacZ-UL6−, UL17-stop, and KUL25NS, respectively) were separated by SDS-PAGE and stained with SYPRO orange (Fig. 1). This stain was selected because of its high sensitivity and minimal protein-to-protein variation in staining, allowing quantitative comparison between proteins (33). A broad protein band with a molecular weight (MW) of about 80,000 was observed in all the capsid protein profiles except the UL17-stop insertional mutant B capsid profile, which had only a weak band in this position. A protein migrating in this region of the gel is therefore a strong candidate for the UL17 product, which has a predicted molecular mass of 74.6 kDa (13) but had previously been reported to have a slightly lower electrophoretic mobility (30). A protein band with an MW of approximately 63,000 was absent from the UL25 null mutant capsid profile, and unexpectedly this protein band was also reduced in intensity in the UL17 null mutant capsid profile. This band is likely to represent the UL25 product since it is similar in size to the calculated molecular mass of UL25 (62.7 kDa). The UL6 null mutant capsid lacked a protein with an MW of approximately 74,000, the size of UL6, which was present in all the other capsid protein profiles. The identities of the UL17, UL6, and UL25 proteins were confirmed by Western blot analysis of proteins separated on the same gel (data not shown, but see also Fig. 2).
FIG. 1.
Identification of packaging proteins UL6, UL17, and UL25 in wt virus and mutant B capsids. Purified B capsid proteins were separated by SDS-PAGE, and the proteins were detected by SYPRO orange staining. Capsid proteins are indicated on the left, and the positions of molecular weight markers (MagicMarkXP; in thousands) are shown on the right.
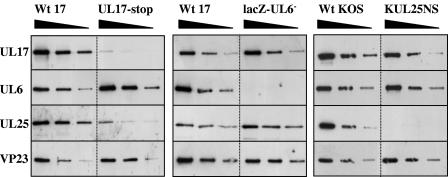
FIG. 2.
Analysis of UL6, UL17, and UL25 in wt HSV-1 strain 17, wt strain KOS, lacZ-UL6−, KUL25NS, and UL17-stop capsids by Western blotting. Serial twofold dilutions of wt and mutant capsids that had been equalized on the basis of their VP23 content were prepared. The polypeptides were resolved by SDS-PAGE and blotted onto a nitrocellulose membrane. The blots were probed sequentially with antibodies UL6 MAb 175, UL17 MAb 203, UL25 MAb 166, and VP23 antibody Rab186 as indicated on the left side. In this experiment the UL25NS B capsid preparation was banded twice on a sucrose gradient to ensure minimal contamination with UL25NS empty capsids, lacking the scaffolding proteins.
To determine the relative amounts of the three packaging proteins in B capsids, a digital image of the gel illuminated by UV light was made and analyzed by densitometry. The copy numbers of UL17 and UL25 in wt B capsids were estimated by comparing the amounts of protein present relative to UL6 and assuming that there were 12 copies of UL6 per B capsid (19). Background levels of protein in the same position in the relevant null mutant profile were subtracted. Mean copy numbers and standard deviations of 19.2 ± 5.0 for UL17 and 26.8 ± 6.2 for UL25 were obtained from six samples.
Western blot analysis of wt and mutant B capsids.
To confirm that UL25 was present in substantially reduced amounts in the UL17-stop mutant capsids, Western blot analysis was carried out. Three serial twofold dilutions of each capsid sample, standardized on the basis of VP23 content, were prepared, and the separated proteins were screened by Western blotting using UL6, UL17, UL25, and VP23 antibodies. The results (Fig. 2) show that the interaction of UL17 or UL25 with B capsids was not dependent on the presence of UL6 and, similarly, neither UL25 nor UL17 was required for the attachment of UL6 to capsids. UL17, however, was important for efficient binding of UL25 to B capsids. Densitometric analysis of the digital images of the exposed films of the Western blot revealed that the UL17-stop capsids contained about 10% of the level of UL25 found in wt strain 17 B capsids. The faint UL17-specific band present in the UL17-stop capsid profile was indicative of a low frequency of revertants in the mutant stock.
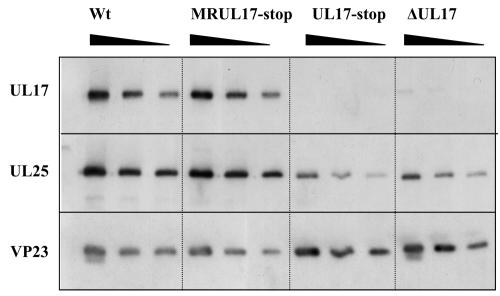
To exclude the possibility that the UL17-stop mutant had a secondary defect in UL25, the levels of UL17 and UL25 were investigated in B capsids of a marker rescuant of UL17-stop and in another UL17 mutant, the ΔUL17 UL17 null mutant (Fig. 3). The amounts of UL25 present in both ΔUL17 and UL17-stop B capsid preparations were also reduced in comparison with wt virus or the marker rescuant UL17-stop capsids. In this experiment the mutant capsids contained about 20% of the level of UL25 found in the wt or marker rescuant B capsids, confirming that UL17 is required for efficient binding of UL25 to capsids (Fig. 3). Similarly reduced amounts of UL25 (23%) were observed in the UL17-stop mutant capsids compared with wt strain 17 B capsids, calculated from densitometric analysis of capsid proteins stained with SYPRO orange (Fig. 1).
FIG. 3.
Western blot analysis of proteins of MRUL17-stop and mutant ΔUL17 B capsids. For comparison, proteins of wt strain 17 and mutant UL17-stop B capsids were also included. Serial twofold dilutions of wt and mutant B capsids that had been equalized on the basis of their VP23 content were prepared. The polypeptides were resolved by SDS-PAGE and blotted onto a nitrocellulose membrane. The blots were probed sequentially with antibodies UL17 MAb 203, UL25 MAb 166, and VP23 polyclonal antibody 186 as indicated on the left side.
The data in Fig. 2 also suggest that the KUL25NS capsids had reduced amounts of UL17 compared with wt KOS capsids, about twofold lower. Comparison of the level of UL17 in KUL25NS capsids with that of wt HSV-1 strain KOS B capsids, calculated using information from SYPRO orange-stained gels of capsid proteins, also showed that there was about a twofold reduction in the amount of UL17 in UL25NS capsids. Repeated experiments consistently revealed a reduction in the amount of UL17 in UL25NS capsids in comparison with wt B capsids. Thus, the presence of UL25 is necessary for the retention of wt amounts of UL17 in B capsids. It should be noted that in Fig. 1 the KUL25NS capsid preparation contained lower amounts of scaffolding proteins than the other capsid samples, due to the presence of empty capsids; however, similar results were obtained using this assay with other KUL25NS B capsid preparations containing higher levels of scaffolding proteins (data not shown).
The distribution of UL17 and UL25 on the surface of B capsids.
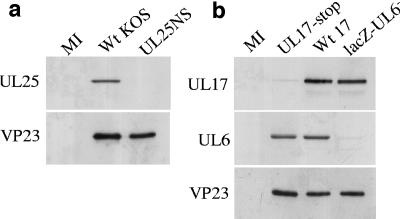
Previous work by Ogasawara et al. (23) suggested that UL25 was present at multiple sites on wt B capsids. To confirm this finding and to investigate whether UL17 had a similar pattern of distribution, immunoelectron microscopy was carried out. Since the UL25 MAb did not react with purified wt B capsids and the UL17 MAb bound to wt B capsids at low frequency, rabbit polyclonal antibodies to UL17 and UL25 were prepared as described in Materials and Methods. In addition, as a control, rabbit antibodies were raised against UL6. The specificity of each of the three polyclonal antibodies was checked by Western blot analysis of extracts of BHK cells infected with the mutant virus that did not express the relevant packaging protein or with wt virus (Fig. 4). The blots were also probed with antibodies to VP23 to show that the cells had been efficiently infected with virus. Each antibody reacted to a viral protein of the correct size that was present in the wt virus-infected cell profile but was absent in the relevant mutant-infected cell profile or in very low amounts due to the presence of revertants (Fig. 4).
FIG. 4.
Specificity of UL6, UL17, and UL25 rabbit polyclonal antibodies. BHK cells were infected with HSV-1 wt strain 17, wt strain KOS, UL17-stop mutant, lacZ-UL6−, or UL25NS or were mock infected (MI). At 24 h postinfection the cells were harvested and the proteins were separated by SDS-PAGE. Western blot analysis was carried out, probing the KOS, UL25NS, and mock-infected cell profiles sequentially with UL25 and VP23 polyclonal antibodies (a) and strain 17, lacZ-UL6−, UL17-stop, and mock-infected cell profiles sequentially with UL17, UL6, and VP23 polyclonal antibodies (b).
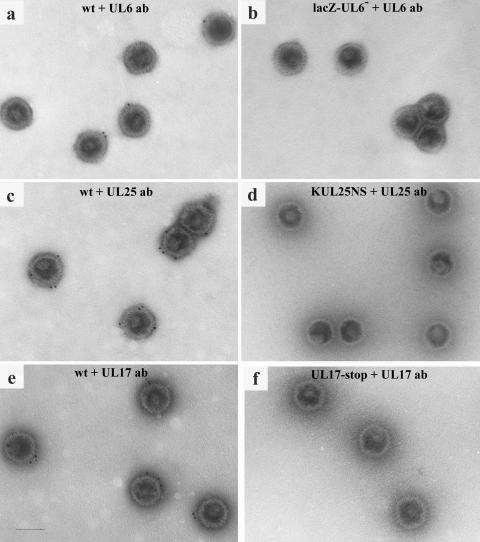
In immunoelectron microscopic experiments capsids lacking the packaging protein were included in the assay to ensure that each packaging protein antibody was attaching to wt capsids in a specific manner. wt and mutant capsids were incubated with antiserum to the packaging protein and subsequently treated with goat anti-rabbit IgG conjugated to 10-nm gold beads. The capsids were negatively stained with ammonium molybdate, visualized under the electron microscope, and photographed. The number of gold particles bound or unattached to capsids was calculated, and their pattern of distribution on the capsid was recorded (Table 1; Fig. 5). Fields of wt HSV-1 strain 17 and lacZ-UL6− B capsids treated with UL6 antibody, illustrating the pattern of gold labeling, are shown in Fig. 5a and b, respectively. About 26% of wt capsids contained gold particles whereas only 2% of capsids lacking UL6 were labeled with gold (Table 1). These results are comparable to previous findings (19). Most (97%) wt capsids and 2% of KUL25NS capsids treated with UL25 antibody were labeled with gold (Fig. 5c and d; Table 1). The majority of the wt B capsids treated with UL17 antibody were also labeled with gold (69%) whereas the UL17-stop capsids treated under the same conditions had low levels of attached gold (2%) (Fig. 5e and f; Table 1).
TABLE 1.
Immunogold staining of wt and mutant B capsids
| Antibody and B capsid type | No. of capsids analyzed | No. (%) of capsids labeled with gold | Total no. of gold particles | No. (%) of gold particles bound to capsids |
|---|---|---|---|---|
| UL6 RAba | ||||
| wt | 496 | 128 (26) | 281 | 169 (60) |
| lacZ-UL6− | 943 | 15 (2) | 76 | 20 (26) |
| UL25 RAb | ||||
| wt | 301 | 291 (97) | 1,382 | 1,299 (94) |
| KUL25NS | 170 | 3 (2) | 28 | 3 (11) |
| UL17 RAb | ||||
| wt | 370 | 257 (69) | 604 | 525 (87) |
| UL17-stop | 658 | 12 (2) | 44 | 15 (34) |
RAb, rabbit antibody.
FIG. 5.
Electron micrographs of wt and mutant capsids treated with UL6 antibody (a and b), UL25 antibody (c and d), or UL17 antibody (e and f). Capsids were incubated with the packaging protein antibody and subsequently treated with goat anti-rabbit IgG coupled to 10-nm colloidal gold. Capsids were negatively stained and examined under the electron microscope. Bar, 100 nm.
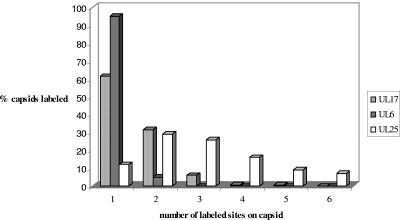
For each wt B capsid containing gold particles, the number of sites labeled with gold was recorded. Figure 6 shows a graph comparing the number of labeled sites on wt B capsids treated with UL6, UL17, or UL25 antibody. As anticipated, most of the gold on wt capsids treated with UL6 antibody was present at a single site whereas the gold particles on most of the wt capsids incubated with UL25 antibody were found at multiple sites. The pattern of distribution of UL17 was intermediate between those of UL6 and UL25. Most of the capsids were labeled with gold at a single site, but a significant proportion had gold at two and three sites on the capsid. Similar results were obtained in repeated experiments (data not shown). The numbers of UL17 and UL25 sites labeled by immunogold are likely to be an underestimate of the true number of sites because not all the sites may be accessible and the antibodies had not been raised against the full-length proteins.
FIG. 6.
Distribution of UL25, UL17, and UL6 on wt B capsids. The percentage of labeled wt B capsids (treated with UL25, UL17, or UL6 antibodies) was plotted against the number of sites labeled with gold particles on each capsid.
Distribution of UL25 and UL17 on the surface of wt C capsids.
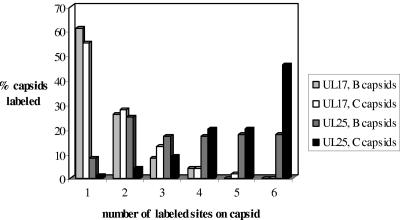
Both UL17 and UL25 are present in greater amounts in wt DNA-containing (C) capsids than in wt B capsids (31, 39). For this reason and because C capsids are intermediates in the assembly pathway whereas B capsids are dead-end products, the distributions of UL25 and UL17 in B and C capsids were compared in parallel in a separate experiment using immunoelectron microscopy. The number of sites containing gold particles on each capsid was recorded, and results were plotted as a histogram (Fig. 7). The pattern of distribution of UL17 in C capsids was similar to that of B capsids, with a slightly higher number of C capsids than B capsids labeled with UL17 antibody (69% compared with 61%). By contrast, a higher proportion of C capsids than of B capsids treated with UL25 antibodies contained gold particles at six or more different sites, suggesting that UL25 was attached to more sites on C capsids. Nearly all (98%) of the B and C capsids treated with UL25 antibody were labeled with gold particles. As a control, the distribution of UL6 in wt B and C capsids was also examined. The pattern of gold labeling and the percentage of C capsids containing gold particles were very similar to the results obtained for B capsids (data not shown), in keeping with previous findings showing that B and C capsids contain similar amounts of UL6 (31).
FIG. 7.
Comparison of the distribution of UL25 and UL17 on wt B and C capsids. The percentage of labeled B or C wt strain 17 capsids (treated with UL25 or UL17 antibodies) was plotted against the number of sites labeled with gold particles on each capsid. The last column represents all capsids with six or more sites labeled with gold particles. The capsids examined were as follows: 164 wt B capsids treated with UL25 antibody were analyzed, 96% of which were labeled with gold; 137 wt B capsids treated with UL17 antibody were analyzed, 61% of which were labeled with gold; 147 wt C capsids treated with UL25 antibody were analyzed, 99% of which were labeled with gold; and 136 wt C capsids treated with UL17 were analyzed, 69% of which were labeled with gold.
Treatment of wt virus B capsids with urea.
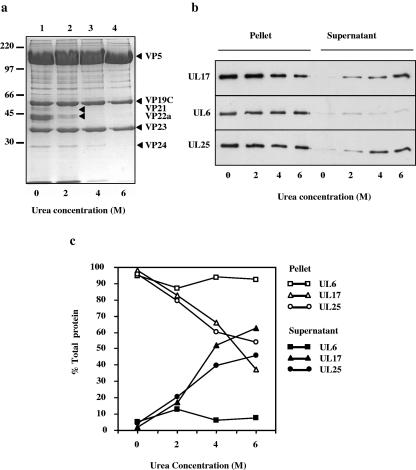
Most of the portal protein, UL6, is retained in B capsids which have been treated with 2 M guanidine HCl or 6 M urea, under conditions in which the pentons and the 10 triplexes near each penton are removed in addition to the scaffolding protein and VP26 (20, 21, 24). To determine whether the UL17 protein was strongly associated with the capsid, purified wt virus B capsids were incubated in 0, 2, 4, or 6 M urea, and the proteins in the recovered capsids, together with the proteins extracted from the capsids, were separated by SDS-PAGE in duplicate. One gel was treated with Coomassie blue stain, and the other was processed for Western blot analysis. Figure 8a shows a Coomassie blue-stained SDS-polyacrylamide gel of polypeptides from B capsids treated with urea. Consistent with previous published results, most of the scaffolding proteins VP22a and VP21 were removed from capsids treated with 2 M urea.
FIG. 8.
Treatment of capsids with urea. Purified wt HSV-1 strain 17 B capsids were treated with various concentrations of urea for 1 h and concentrated by centrifugation. The proteins from the capsid pellets were resolved by SDS-PAGE and stained with Coomassie blue (a). Molecular masses in kilodaltons are shown at the left. In addition, the separated capsid proteins from the pellets and the supernatants were blotted onto nitrocellulose and the UL6, UL17, and UL25 packaging proteins were identified by Western blotting (b). The protein bands in the exposed film from the Western blot experiment shown in Fig. 7b were quantified by densitometric analysis of digital images (c).
In Fig. 8b the separated capsid proteins and the proteins extracted with urea were blotted onto a nitrocellulose membrane and the blot was sequentially probed for UL17, UL6, and UL25. The protein bands on the exposed film were quantified by densitometric analysis, and the results are shown as a graph in Fig. 8c. The patterns of extraction of UL25 and UL17 from capsids treated with urea were similar, with increasing amounts of protein removed with increasing concentrations of urea. By contrast UL6 was less susceptible to extraction by urea.
DISCUSSION
The amounts of UL17 and UL25 in wt B capsids and capsids lacking UL17, UL6, or UL25 and the distribution of UL17 and UL25 on wt B and C capsids were determined to gain more insight about the functions of these two packaging proteins. The results of immunogold labeling experiments with wt HSV-1 strain 17 B capsids confirmed that UL25 was present at multiple sites and demonstrated that at least part of UL17 is exposed on the surface of the B capsid (Fig. 5). Greater numbers of capsids treated with UL17 antibody than of those incubated with UL6 antibody were labeled with gold (69% compared with 26%). The low level of labeling of wt B capsids treated with UL6 antibody is probably due to the fact that UL6 is present at a single site on the capsid and that much of the underside of the capsid in contact with the grid is not available for antibody binding. Similar low levels of gold binding to bacteriophage PRD1 capsids treated with antibodies specific for proteins at the unique vertex have been reported (9). Because a high proportion of the capsids incubated with UL17 antibody were labeled with gold particles and a substantial number of the capsids with UL17 antibody were labeled at two and three sites, we propose that UL17, like UL25, is present at multiple sites on the surface of the capsid.
As well as being present at multiple locations on the capsid, both UL17 and UL25 had a similar pattern of removal from B capsids treated with different concentrations of urea. UL17 and UL25 had also previously been shown to be present in elevated amounts in DNA-containing capsids compared to B capsids, unlike UL6 (39). These observations, together with the finding that stable attachment of UL25 to B capsids was dependent on the presence of UL17, suggest that these two proteins interact. This proposal is supported by the reciprocal observation that capsids lacking UL25 also had reduced amounts of UL17, suggesting that in the absence of UL25 some UL17 may be lost from the B capsid. At this stage the alternative possibility that the attachment of UL17 to capsids exposes UL25 binding sites on the capsid at positions different from those of UL17 cannot be excluded, but we consider this idea less attractive in view of the above findings.
To date, we have been unable to demonstrate an interaction between UL17 and UL25, using immunofluorescence or immunoprecipitation assays on cells transiently expressing the two proteins. Because UL17 is found at levels in procapsids similar to the amounts in B capsids whereas UL25 is present in reduced amounts in procapsids compared to the levels in B capsids, UL17 is probably inserted into capsids before UL25 (31, 39). The finding that the UL17 null mutants appear to be blocked at an earlier stage of DNA packaging than the UL25 null mutant is consistent with this suggestion (30). It is therefore probable that these two proteins associate only after UL17 has bound to the capsid.
Ogasawara et al. (23) suggested that UL25 attached to both the pentons and hexons of the B capsid since significant amounts of UL25 were retained on capsids treated with 6 M urea. If this is the case, given the low copy number of UL25 in B capsids, UL25 cannot be interacting with all the hexons. It remains possible that UL25 may be interacting with other components or features of the capsid (e.g., triplex proteins or the centers or edges of the capsid faces). Since binding of UL25 to capsids is dependent on the presence of UL17, UL25 may be interacting only indirectly with the capsid shell components via UL17. Significantly, UL17 behaved very similarly to UL25 in terms of its extraction with urea (Fig. 7), suggesting that it may be interacting, at least in part, with the pentons. Alternatively, it may be located elsewhere and, like the hexon decorating protein VP26 and the scaffolding protein, for example, be more susceptible to extraction by 6 M urea than some capsid components (17).
The estimate of 27 per capsid for the copy number of UL25 was lower than the value of 42 obtained by Ogasawara et al. (23). However, we note that their figure for the putative UL6 protein was considerably higher than subsequent published values (19). The observations that a higher proportion of B capsids was labeled with UL25 antibody than with UL17 antibody and that a greater number of these capsids than the capsids treated with UL17 antibody had gold bound to more than one site on the capsid are consistent with the finding that there were more copies of UL25 than UL17 in B capsids. However, the amount of gold bound to capsids could also be influenced by the potency of the antibody and the accessibility of the protein. The UL25 antibody recognized a greater number of sites on the surface of wt C capsids than on that of wt B capsids, consistent with the observation that there is more UL25 in C capsids than in B capsids (31). Although there are also greater amounts of UL17 in C capsids than in B capsids, the difference in binding patterns of the UL17 antibody between B and C capsids was much less marked than that of the UL25 antibody. It is possible that some UL17 is masked in C capsids.
Although UL17 is essential for DNA packaging, it is unlikely to perform a specific function at the portal because it appears to be located at multiple sites on the capsid. A possible role of UL17 could be to stabilize the procapsid. The HSV-1 procapsid is a labile structure, which, in the absence of DNA packaging, spontaneously angularizes in HSV-infected cells, provided that the scaffold is cleaved by the UL26 protease. Procapsids formed in vitro from the capsid shell and the major scaffolding protein angularize more rapidly than their in vivo counterparts, suggesting that other viral proteins may delay the transformation into angularized capsids (18, 22, 29). Since UL17 is absolutely necessary for DNA packaging and appears to be required at a slightly earlier stage than UL25, UL17 is a strong candidate for preventing premature angularization (30). The involvement of UL17 in this early process could explain the failure of UL17 null mutants to package viral DNA in nonpermissive cells.
A function of UL25 and possibly also UL17 may be to reinforce the capsid shell, stabilizing the capsid and possibly the packaging machinery at the portal during and after insertion of the genome into the capsid. It has been estimated from work on the bacteriophage φ29 that a pressure of about 6 MPa is generated inside the DNA-containing capsid (32). The HSV-1 genome, like the φ29 genome, is inserted into a very confined space. Examination of HSV-1 virions and capsids by diffraction analysis and three-dimensional reconstruction has shown that the viral DNA is arranged in concentric rings with an interlayer spacing of about 26 Å, indicative of liquid-crystalline packing (5, 44). Given the compact organization of the encapsidated genome, it is highly likely that there is also considerable pressure inside the HSV-1 DNA-containing capsid. The UL25 null mutant produces a high proportion of empty capsids lacking DNA and devoid of scaffold but only a low number of C capsids, most of which have less than a genome length of DNA (14, 36). This phenotype may be influenced not only by the absence of UL25 but also by the reduced levels of UL17 present in the UL25 null mutant capsids. The properties of the mutant are consistent with UL25 having a role in stabilizing the maturing capsid. In this respect, as pointed out previously (43), UL25 resembles the bacteriophage lambda protein gD that binds to sites on the capsid which become exposed during DNA packaging and is specifically required for packaging genomes greater than 82% of the full length of the lambda genome (16, 34). Since UL25 is located at multiple sites on the capsid, it seems much less likely that it specifically closes the portal at the unique vertex and prevents the viral DNA from being released from the capsid prematurely as previously suggested (23, 31).
Acknowledgments
We thank D. McGeoch and F. Rixon for critically reading the manuscript. We are also grateful to J. Baines, P. Desai, and A. Patel for providing the ΔUL17, KUL25NS, and lacZ-UL6− mutants, respectively, and the appropriate cell lines. Advice and help on electron microscopy from D. Bhella and J. Aitken were much appreciated.
J. Thurlow was supported by an MRC studentship.
REFERENCES
- 1.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type-1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 5.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S. M., D. A. Ritchie, and J. H. Subak-Sharpe. 1973. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18:329-346. [DOI] [PubMed] [Google Scholar]
- 7.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Gowen, B., J. K. H. Bamford, D. H. Bamford, and S. D. Fuller. 2003. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J. Virol. 77:7863-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guasch, A., J. Pous, B. Ibarra, F. X. Gomis-Rüth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa, and M. Coll. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 315:663-676. [DOI] [PubMed] [Google Scholar]
- 11.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 12.Kirkitadze, M. D., P. N. Barlow, N. C. Price, S. M. Kelly, C. Boutell, F. J. Rixon, and D. M. McClelland. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 72:10066-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 14.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, S. D., and J. P. E. Prevelige. 2002. DNA packaging: a new class of molecular motors. Curr. Biol. 12:R96-R98. [DOI] [PubMed] [Google Scholar]
- 16.Murialdo, H. 1991. Bacteriophage lambda DNA maturation and packaging. Annu. Rev. Biochem. 60:125-153. [DOI] [PubMed] [Google Scholar]
- 17.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal protein complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel, A. H., and J. B. Maclean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 25.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 26.Preston, V. G., J. A. V. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preston, V. G., F. J. Rixon, I. M. McDougall, M. McGregor, and M. F. Al-Kobaisi. 1992. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology 186:87-98. [DOI] [PubMed] [Google Scholar]
- 28.Rixon, F. J., A. M. Cross, C. Addison, and V. G. Preston. 1988. The products of herpes simplex virus type-1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J. Gen. Virol. 69:2879-2891. [DOI] [PubMed] [Google Scholar]
- 29.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413:748-752. [DOI] [PubMed] [Google Scholar]
- 33.Steinberg, T. H., L. J. Jones, R. P. Haugland, and V. L. Singer. 1996. SYPRO orange and SYPRO red protein gel stains: one step fluorescent staining of denaturing gels for the detection of nanogram levels of protein. Anal. Biochem. 239:223-237. [DOI] [PubMed] [Google Scholar]
- 34.Sternberg, N., and R. Weisberg. 1977. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 117:733-759. [DOI] [PubMed] [Google Scholar]
- 35.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 36.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliott, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75:1101-1113. [DOI] [PubMed] [Google Scholar]
- 38.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, C. A. White, I. M. McDougall, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packing protein UL17 is a virion protein, present in both the capsid and tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 42.White, C., N. D. Stow, A. H. Patel, M. Hughes, and V. Preston. 2003. The herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Z. H., B. V. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]