Abstract
Herpesviruses replicate their double stranded DNA genomes as high-molecular-weight concatemers which are subsequently cleaved into unit-length genomes by a complex mechanism that is tightly coupled to DNA insertion into a preformed capsid structure, the procapsid. The herpes simplex virus type 1 UL25 protein is incorporated into the capsid during DNA packaging, and previous studies of a null mutant have demonstrated that its function is essential at the late stages of the head-filling process, either to allow packaging to proceed to completion or for retention of the viral genome within the capsid. We have expressed and purified an N-terminally truncated form of the 580-residue UL25 protein and have determined the crystallographic structure of the region corresponding to amino acids 134 to 580 at 2.1-Å resolution. This structure, the first for any herpesvirus protein involved in processing and packaging of viral DNA, reveals a novel fold, a distinctive electrostatic distribution, and a unique “flexible” architecture in which numerous flexible loops emanate from a stable core. Evolutionary trace analysis of UL25 and its homologues in other herpesviruses was used to locate potentially important amino acids on the surface of the protein, leading to the identification of four putative docking regions for protein partners.
The Herpesviridae constitute a large family of animal viruses, several members of which are pathogenic to humans, causing such diseases as chicken pox, cold sores, infectious mononucleosis, and genital lesions (45). Herpes simplex virus type 1 (HSV-1), the prototypical member of this family, is a large (∼2,000 Å in diameter) complex virus particle composed of an outer glycoprotein-containing envelope surrounding an amorphous protein layer or tegument. The tegument layer in turn surrounds an icosahedral capsid (1,250 Å in diameter) which encloses a linear double-stranded DNA (dsDNA) genome of ∼150 kb (50).
Assembly of the herpesvirus virion is a complicated process that proceeds along an ordered morphogenetic pathway. Initially the capsid shell proteins “polymerize” in a scaffold-mediated fashion to form an intermediate particle or procapsid (35, 36). The procapsids are closed spherical structures that have a diameter similar to that of the mature capsid but are much more fragile, with open, elongated capsomeres that share very few connections between them (56). During maturation, the internal protein scaffold is cleaved by the viral protease and is displaced from inside the capsid as the DNA genome is packaged (10, 15, 28, 44). This process results in the conversion of the spherical procapsid into a more angularized form. Three types of angularized capsids have been identified: A capsids, which are empty with no scaffolding protein or DNA; B capsids, which contain scaffolding protein but no DNA; and C capsids, which contain the DNA genome but no scaffolding protein (16, 19, 43). C capsids represent the precursors of infectious particles, in contrast to the A and B capsids, which are dead-end products most likely arising from abortive DNA-packaging events and cleavage of the internal scaffold protein in the absence of DNA packaging, respectively.
Many aspects of capsid assembly and DNA packaging in the herpesviruses are similar to those in dsDNA bacteriophages such as λ, T4, and P22 (7, 42). The replicated viral DNA genomes are synthesized as concatemers that are cleaved by the terminase complex at specific DNA sequences to generate unit-length genomes. DNA cleavage appears to be tightly coupled to insertion into the capsid, and in the absence of DNA packaging, no unit-length genomes are generated. Analysis of temperature-sensitive and null mutants of HSV-1 has demonstrated that six viral proteins (UL6, UL15, UL17, UL28, UL32, and UL33) are essential for initiation of the cleavage/packaging process (7). The terminase is likely to be a complex of UL15 with UL28 and possibly UL33 (2, 5, 12, 60), which actively pumps the DNA into the procapsid through a portal complex (comprised of 12 copies of UL6) located at a unique vertex (11, 37). The portal complex is an integral part of the capsid structure and harbors the binding sites for the terminase complex (58). The functions of UL17 and UL32 are not well defined, although both proteins have been implicated in targeting of capsids or capsid proteins to the sites of DNA packaging (25, 53).
A seventh gene product essential for HSV-1 DNA packaging, UL25, differs from the proteins described above in being required at a later stage of the DNA-packaging process. A null mutant of UL25 (KUL25NS) was isolated and has been characterized in two separate studies. McNab et al. (33) reported that A capsids and DNase I-sensitive unit-length genomes accumulated in the nuclei of cells infected with KUL25NS, suggesting that, although complete genomes could be packaged, the viral DNA was only transiently associated with the capsid and that UL25 was necessary for its retention. In contrast, Stow (51) reported that DNase-resistant (packaged) DNA was detectable in cells infected with the mutant virus and that although the amount of full-length DNA encapsidated was greatly reduced compared to that of wild-type virus, the mutant was impaired to a much lesser extent for molecules of up to about 100 kb in size. This suggested that UL25 might perform an important function during the final stages of the head-filling process. Despite these differences, the two reports agree that UL25 is essential at a stage subsequent to DNA cleavage and the initiation of DNA packaging. Such a role is consistent with the observation that procapsids contain little to no UL25, while B capsids contain some UL25 but much less than the DNA-containing C capsids (47, 55). It has been postulated that the UL25 protein acquired by the capsid during DNA packaging may function to seal the capsid after the viral DNA has been inserted (33) or to stabilize its structure as the DNA content increases towards that of a full-length genome (51). These hypotheses are also consistent with the observation that the association of other cleavage and packaging proteins with capsids is independent of UL25 (61).
There has been some work characterizing the binding partners of UL25 (38). In the absence of virus infection UL25 localized in the cytoplasm, but in the presence of the capsid triplex protein VP19C it was transported into the cell nuclei, suggesting that the two proteins interact. Direct interactions with the major capsid protein, VP5, from which the pentons and hexons are assembled, and the second triplex protein, VP23, were also detected. Immunogold labeling studies and capsid stripping experiments with guanidine-HCl suggested that UL25 was associated with the pentons and possibly also the hexons. C capsids are estimated to contain approximately 100 UL25 molecules (38). UL25 is retained in the mature virion, and its location on the outside of the C capsid suggests that it may possibly also play roles in capsid egress from the nucleus or interaction with components of the tegument layer (3, 38).
In order to advance our understanding of the structural basis of UL25 function, we have determined the crystallographic structure of an N-terminal deleted form (Δ1-133) of the 580-residue protein (UL25nt) to 2.1-Å resolution. The structure revealed a novel fold in which the unique flexible architecture appears well suited for participation in the dynamic processes of DNA encapsidation. UL25 and its homologues in other herpesviruses were additionally analyzed using the evolutionary trace (ET) (20, 27) to locate residues of likely functional importance on the surface of the molecule. This information should provide a useful starting point for future site-directed mutagenesis experiments.
MATERIALS AND METHODS
UL25 expression, purification, and crystallization.
HSV-1 nucleotides 48945 to 50555 (29), encoding residues R45 to V580 of UL25, were amplified by PCR and cloned into the pET41 expression vector (the vector contains an N-terminal glutathione S-transferase [GST] tag) using ligation independent cloning (Novagen). It was found that the removal of the N-terminal 44 amino acids increased expression of the UL25 protein severalfold. The truncated protein was expressed in Escherichia coli BL21(DE3) cells at 16°C for 24 h following induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were lysed using a Cell Buster (Microfluidics), and the protein was purified to near homogeneity using a glutathione-Sepharose column. Proteolysis to remove the GST tag was performed at 25°C for 24 h with 2 U/ml of enterokinase. Secondary cleavage by the enterokinase at a nonspecific site within the polyglycine loop between residues 116 to 130 was observed, resulting in a further truncation of the UL25 protein. This secondary cleavage resulted in a nearly 100% conversion from the expected 57-kDa protein to a new, 48-kDa product. To remove minor impurities, the 48-kDa fragment of UL25 was further purified on a Pharmacia S75 gel filtration column equilibrated in 10 mM Tris (pH 8.0), 300 mM NaCl, and 2 mM EDTA. Selected fractions were pooled and concentrated to 7 to 8 mg/ml. The protein was crystallized at 25°C using the vapor diffusion method, with the drop consisting of a 1:1 ratio of the stock protein solution and a reservoir solution of 15 to 25% polyethylene glycol monomethyl ether 2000, 200 mM trimethylanime N-oxide dehydrate, and 100 mM Tris (pH 8.0 to 8.6). Prior to data collection, the crystals were placed in increasing percentages of glycerol in the crystallization solution to a final concentration of 20% glycerol and then flash frozen in liquid nitrogen.
Structure determination.
A two-wavelength data set from a crystal of selenomethione-substituted protein was collected on beamline 19ID at the SBC-CAT at the Advanced Photon Source (APS) at the Argonne National Laboratory. Data were processed with HKL2000 and merged with SCALEPACK (40). The crystals have one molecule in the asymmetric unit and belong to the orthogonal space group P212121 with unit-cell dimensions a = 52.6 Å, b = 67.6 Å, c = 119.6 Å, and α = β = γ = 90°. The positions of six of the nine selenomethionine sites in the molecule were determined using the direct methods program SnBv2.1 (17) with data collected at the selenomethionine peak wavelength. An additional two sites were found from residual maps calculated in SHARP (13). Multiwavelength anomalous dispersion (MAD) phases were calculated to 2.1-Å resolution using SHARP and the peak and inflection point wavelength data sets. The calculated electron density map was solvent flattened with DM (4) and used to build ∼85% of the total residues using O (23). The model was refined in CNS (8), interspersed with model building and fitting of water molecules. A total of 410 out of 446 residues (92%) were ordered within the asymmetric unit. Within the molecule, residues 249 to 254, 335 to 345, 417 to 425, 479 to 483, 511 to 513, and 578 to 580 were disordered. The current model, refined at 2.1-Å resolution, has an Rcryst of 22.7 and an Rfree of 25.1. The model has good geometry, with all but one of the backbone dihedral angles in the most favored or allowed regions.
Sequence alignment and evolutionary trace.
The multiple-sequence alignment for the full-length proteins encoded by the human herpesviruses was generated using the CLUSTALW (54) and BOXSHADE programs from the San Diego Supercomputer Center biology workbench at http://workbench.sdsc.edu/index.html. A multiple-sequence alignment for all the UL25nt domains from all known herpesvirus members was generated using CLUSTALW at http://www.ebi.ac.uk/clustalw/ and manually edited to remove residues corresponding to the unresolved portions of theUL25 structure. A probable frameshift error near the C terminus of the gallidherpesvirus 1 sequence was also corrected. This alignment was submitted totheEvolutionary Trace Server (TraceSuite II) (20) at http://www.cryst.bioc.cam.ac.uk/∼jiye/evoltrace/evoltrace.html. Secondary-structure predictions were done usingPsiPred (22).
Illustrations.
Electrostatic calculations were performed using GRASP. All ribbon diagrams and space-filling models were computed using PyMol (http://www.pymol.org) as a renderer.
Protein structure accession number.
Coordinates have been deposited under PDB accession code 2F5U.
RESULTS AND DISCUSSION
Identification of a stable fragment and structure determination.
Full-length UL25 was expressed rather poorly as a GST fusion protein and was difficult to purify in the amounts necessary for crystallography. Analysis of the secondary-structure prediction revealed the presence of a long, unstructured N-terminal loop comprising the first 47 residues (Fig. 1). Removal of this loop resulted in a construct (R45 to V580; see Materials and Methods) that was expressed robustly and could be purified easily. During purification, removal of the fusion tag by protease digestion resulted in a secondary cleavage event at an unidentified site within a predicted glycine-rich loop (Fig. 1) between residues G116 and G130, resulting in a 48-kDa stable fragment, referred to as UL25nt. UL25nt exists as a monomer in solution (as determined by gel filtration) as well as in the crystal (data not shown).
FIG. 1.
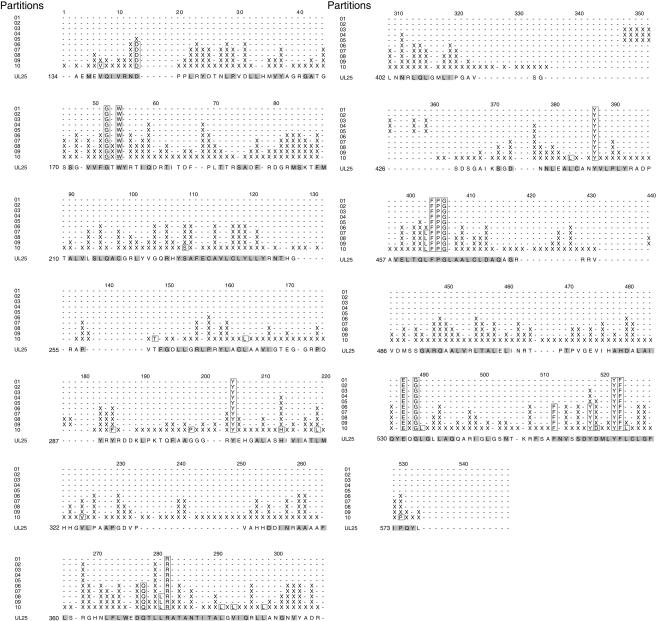
Sequence alignment of UL25 homologues from the eight human herpesviruses. The residues are colored green (completely conserved), yellow (identical residues), and cyan (similar residues). The secondary-structure elements are indicated above the sequence in red (helix), blue (strand), black (loop), and dotted line (disordered). The arrow indicates the position of the first ordered N-terminal residue (A134), and the helix and loop regions predicted for residues 1 to 133 are shown as gray wavy and straight lines, respectively.
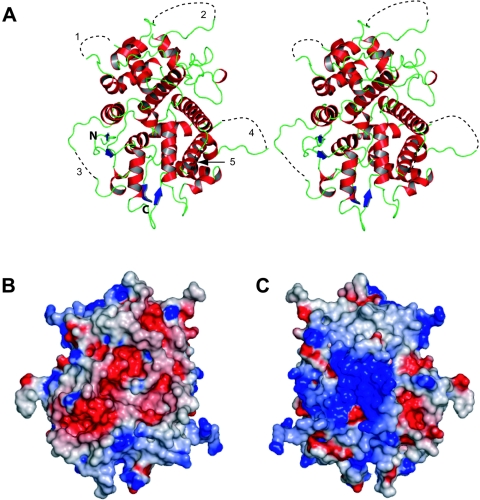
The crystal structure of UL25nt (first ordered N-terminal residue A134) was determined by MAD phasing and refined to 2.1 Å resolution (Tables 1 and 2). The structure has a box-like-shaped core with dimensions of roughly 54 by 37 by 40 Å, consisting mostly of α-helices and a few minor β-sheets (Fig. 2A). Emanating from the UL25 core are numerous loops, which in some cases stretch quite a distance from the core. Five such loops are quite flexible, as many of the residues that comprise them are disordered in the UL25nt crystals (Fig. 2A).
TABLE 1.
Crystallographic analysis statistics: data collection and phasing
| Parameter (unit) | Selenomethionine MAD, APS beamline 19IDa
|
|
|---|---|---|
| Inflection (λ1) | Peak (λ2) | |
| Wavelength (Å) | 0.97948 | 0.97937 |
| Resolution maximum (Å) | 2.1 | 2.0 |
| Total reflections | 182,995 | 191,657 |
| Unique reflections | 48,018 | 54,508 |
| Completeness (%) | 100 (100) | 97.9 (81.3) |
| Redundancy | 3.8 (3.5) | 3.5 (2.5) |
| 〈I〉/〈σ〉 | 18.9 (3.5) | 17.8 (2.5) |
| Rsymb | 4.8 (27.6) | 4.8 (29.6) |
| Figure of merit | 0.64 | |
Values in parentheses are for outer resolution shell.
Rsym = Σhkl|Ihkl − 〈Ihkl〉|/Σhkl Ihkl.
TABLE 2.
Crystallographic analysis statistics: refinement
| Parameter (unit) | Value |
|---|---|
| Resolution (Å) | 50-2.1 |
| Rcrysta (%) | 22.7 |
| Rfreea (%) | 25.1 |
| Avg B values (Å2) | 32.6 |
| Root mean square deviations from ideal | |
| Bond length (Å) | 0.009 |
| Bond angles (°) | 1.2 |
| Dihedral angles (°) | 21.2 |
| Improper torsion angles (°) | 0.71 |
R factor = Σhkl‖Fo|−|Fc‖/Σhkl|Fo| where |Fo| and |Fc| are the observed and calculated structure factor amplitudes for reflection hkl, applied to the work (Rcryst) and test (Rfree) (5% omitted from refinement) sets, respectively.
FIG. 2.
The UL25nt structure. (A) Stereo view of the ribbon representation of the backbone structure. The secondary-structure elements are colored red (α-helices), blue (β-strands), and green (loops). The dotted black lines (1 to 4) represent the disordered loops, labeled for orientation reference. N and C labels identify the amino- and carboxy-terminal ends, respectively. (B) Electrostatic surface representation of the electronegative face of UL25nt. (C) A 180° rotation of the view in panel B, showing the electropositive face of UL25nt.
An analysis of the electrostatic surface potential of UL25nt reveals an interesting distribution of surface charges (Fig. 2B and C). One face of the protein contains a large cluster of negatively charged residues, while the face 180° opposite contains a large number of positively charged residues. The significance of this unusual charge distribution is unknown. It has been proposed that UL25 binds DNA (38), and it is possible that the basic patch could mediate this interaction. Additionally, these oppositely charged faces may play a role in self-oligomerization or in interactions with other partner proteins during capsid assembly and/or in the subsequent process of tegumentation.
The UL25 protein has a novel α plus β protein-folding motif. A search for overall structural similarities against the DALI (18) database failed to reveal any significant matches. The lack of structural similarity to a known protein makes it difficult to infer structure/function relationships from the structure of UL25 alone. From the previous biochemical studies of this protein it has been suggested that UL25 binds to different partner proteins at particular steps within the capsid assembly and DNA-packaging process. The molecular architecture of UL25nt (a stable core of compact helices surrounded by long flexible loops) is consistent with the switching between conformational states as it interacts with its various protein partners during capsid assembly.
Evolutionary trace and sequence analysis.
UL25 is one of the approximately 40 core genes conserved among all members of the alpha-, beta-, and gammaherpesvirus subfamilies (12). An initial alignment of the sequences of the homologous proteins encoded by the eight human herpesviruses, representing all three subfamilies, was made to locate regions of highest sequence conservation and likely functional importance.
Although the crystallographic structure does not include the N-terminal 133 residues of UL25, the secondary structure predicted for this region comprises a long alpha helix (residues 48 to 110), possibly involved in oligomerization via a coiled-coil interaction, preceded by a 45-residue unstructured N-terminal loop. The sequence alignment reveals this area, and in particular the N-terminal loop (residues 1 to 45), to be the least conserved portion of the protein (Fig. 1). Moreover, in the human cytomegalovirus (human herpesvirus 5 [HHV-5]) protein, the region between the predicted long alpha helix and the point corresponding to the first ordered amino acid (A134) in the UL25 structure is extended by approximately 50 amino acids. However, deletion mutants of UL25 lacking the N-terminal 45 residues complement the growth of the UL25 null mutant poorly, suggesting that this region is nevertheless important for protein function (V. Preston, unpublished results).
The greatest sequence similarity lies in the UL25nt portion of the protein. This region of the HSV-1 (HHV-1) protein was, as expected, most closely related to the corresponding proteins of the other human alphaherpesviruses, i.e., HSV-2 (HHV-2) and varicella-zoster virus (HHV-3), which exhibited similarities of 93% and 64%, respectively. The proteins encoded by the human beta- and gammaherpesviruses all exhibited sequence similarities to UL25nt in the range of 37 to 39%.
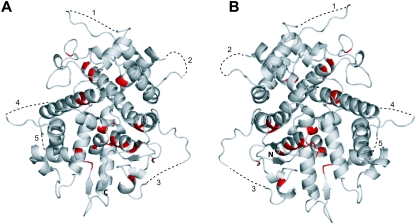
There are 24 residues within the UL25nt region which are completely conserved in the eight human herpesviruses. Mapping these amino acids onto the known structure of UL25nt revealed that they are mostly hydrophobic core residues located in the interior of the protein (Fig. 1 and 3). These residues are more likely to be critical in folding and maintaining the structural integrity of the protein than in playing a key functional role such as participating in protein-protein interactions.
FIG. 3.
Conserved residues of UL25nt. (A) Ribbon diagram of UL25nt, with the conserved residues in Fig. 1 in red. (B) A 180° rotation of the view in panel A.
In an attempt to identify and localize important functional sites on the surface of UL25nt an evolutionary trace analysis was done on a multiple-sequence alignment of UL25 and homologous protein sequences identified by a PSI-BLAST search. The ET method identifies functionally important residues from phylogenetic tree-based sequence conservation among homologous proteins and maps them onto a representative structure (27). Partitioning of the tree divides the set of proteins into a number of classes, each consisting of related sequences originating from a node within that partition. In general, residues important for protein architecture tend to exhibit the least variation among a family of related proteins, whereas residues important for function are invariant among a particular protein subgroup (i.e., a group of most closely related proteins) but vary between subgroups. Clustering of these class-specific residues on a structure is indicative of a functional site, since changes in the amino acid composition correlate with evolutionary divergence and functional specificity. This method has been used to detect functional sites in a wide variety of proteins (9, 34, 48).
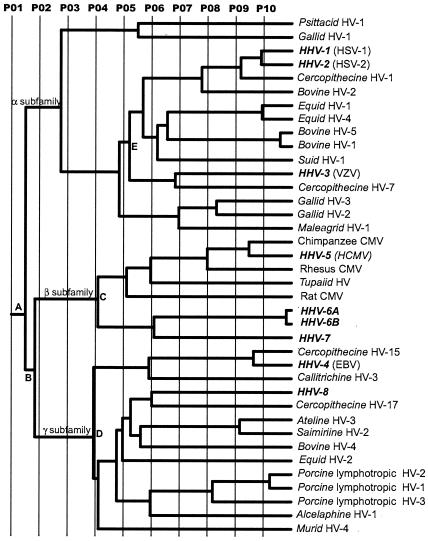
The ET phylogenetic tree for UL25 is shown in Fig. 4, and the residues identified by the evolutionary trace analysis are shown in Fig. 5. The tree is in very close agreement with results of more detailed investigations of herpesvirus phylogeny based on larger protein sets and the use of alternative approaches for tree construction (30-32). The different partitions (P01 to P10) define the protein classes within the tree. Partitioning of the phylogenetic tree at the root (P01) (Fig. 4) results in a single class of proteins, and the conserved residues correlate with the most fundamental features of the protein. Twelve absolutely conserved residues are identified at this partition (Fig. 5), representing a subset of the residues conserved among the eight human herpesviruses (Fig. 1).
FIG. 4.
Evolutionary trace of herpesvirus UL25 homologues. P01 to P10 represent the different partitions of the ET phylogenetic tree. The branches defining the three herpesvirus subfamilies are labeled α, β, and γ. Key nodes discussed in the text are identified by A to D. Virus species names in boldface represent human viruses.
FIG. 5.
Evolutionary trace residues for partitions 01 to 10. The trace residues belonging to a given partition occur in the horizontal row corresponding to the partition. The conserved residues are outlined in a box, the class-specific residues are demarcated by an X, and the dashes reflect gaps in the alignment.
At partition 02 the UL25 homologues are divided into three classes corresponding to the separate subfamilies as a result of branching at nodes A and B. Further partitioning results in the generation of increasing numbers of separate protein classes. For example the gamma herpesviruses resolve into genera Lymphocryptovirus (gamma-1) and Rhadinovirus (gamma-2) at partition 04 (bifurcation at D), and the betaherpesviruses resolve into the cytomegalo- and roseoloviruses at partition 05 (bifurcation at C).
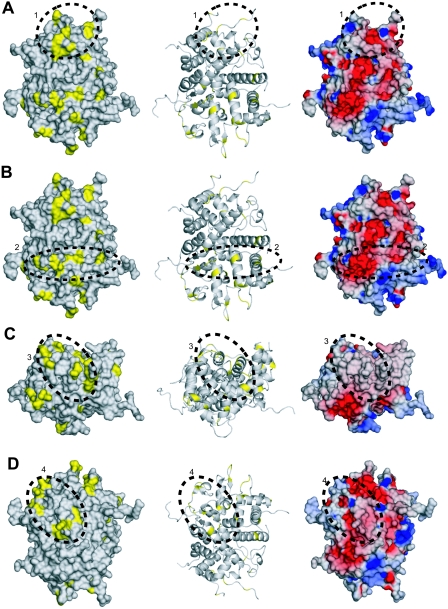
As a consequence of partitioning the phylogenetic tree, the ET analysis reveals conservation patterns that conventional sequence comparisons do not show. At P07, nine protein classes containing more than a single member are defined, which contribute to the positional information shown in Fig. 5. These classes, which are distributed among the three subfamilies, are exemplified by HSV-1, HSV-2, cercopithecine herpesvirus 1, and bovine herpesvirus 2 in the alphaherpesviruses; human, rhesus, and chimpanzee cytomegaloviruses in the betaherpesviruses; and Epstein-Barr virus (HHV-4) and cercopithecine HV-15 in the gammaherpesviruses. Mapping of the class-specific residues identified at this level to the surface of UL25nt revealed four clusters of particular interest (Fig. 5 and 6). The surface residues and the cluster to which each is classified are summarized in Table 3.
FIG. 6.
Evolutionary trace analysis. (A) The left and middle panels are a space-filling model and a ribbon trace, respectively, of UL25nt, with the class-specific exterior residues identified by the evolutionary trace shown in yellow. Cluster 1 is circled and identified with a 1. This cluster is also depicted in the electrostatic surface potential representation of the UL25nt structure (right). All three views are the same. (B, C, and D) Corresponding depictions of clusters 2, 3, and 4, respectively.
TABLE 3.
Evolutionary trace residues within the identified four clusters
| Cluster | Trace residuesa |
|---|---|
| 1 | G225, R288, R305, P327, G331, H348, R362, G363, N365 |
| 2 | R148, D150, N152, D156, L214, N508 |
| 3 | G169, S170, G172, R180, G202, R203, K206, P413 |
| 4 | R390, N396, Y398, D400, L402, R552 |
The listed residues are from partition 07. Only the external residues are listed.
Cluster 1 is located in the apical region of the protein located on the same face as the view shown in Fig. 2A near disordered loop 2. Cluster 2, located on the same face as cluster 1, traverses laterally across the entire molecule and abuts disordered loop 5. Cluster 3 is made up of loops at the base of UL25nt directly opposite cluster 1. Cluster 4 is located on the face 180° from clusters 1 and 2 and abuts disordered loop 3. The electrostatic surface potentials of the three clusters are shown in Fig. 6. All four potential binding surfaces are rather nondescript, containing both charged and uncharged residues. It is impossible to predict, based solely on electrostatics, either the likely binding partners or the mode of binding. However, it is interesting that three of the four clusters reside on or near areas of the structure where there are disordered loops. It is possible that these flexible areas might become ordered upon binding to another protein partner. The disperse distribution of binding sites on UL25nt suggests that it is capable of binding to several protein partners. The inherent elasticity of UL25nt in its apparent ability to bind multiple protein partners, its unusual charge distribution, and its structural flexibility are consistent with its key function during the dynamic processes associated with DNA encapsidation, capsid stabilization, and DNA retention.
Models for UL25 function and biological implications.
The HSV-1 UL25 protein has been proposed to be involved in the retention of packaged DNA within the capsid during the late stages of head filling (33, 51). There are phage proteins known which perform a function similar to that of UL25, as many aspects of the DNA-packaging mechanism of herpesviruses are analogous to those of dsDNA bacteriophages such as lambda, T4, and P22.
In many phages, following the packaging of the DNA, the portal is sealed to prevent the escape of the genome in a process known as head completion. Proteins that function to plug the portal during head completion include gp4, gp16, and gp26 of bacteriophage P22 and gp15 and gp16 of SPP1 (39, 41, 52). Mutants defective in any of these proteins exhibit a phenotype of unpackaged unit-length genomes similar to that described for the UL25 deletion mutant KUL25NS by McNab et al. (33), suggesting that UL25 may also be involved in the head completion process for HSV-1. The observations that HSV-1 C capsids contain more UL25 than B capsids and that only approximately half of the UL25 protein in B capsids is associated with pentons (38, 47, 55) are consistent with a role for UL25 in plugging the portal. However, no direct interaction with the UL6 portal protein has been reported, and it has not been determined whether UL25 is present at the unique portal vertex of capsids.
In the electron cryomicroscopy reconstructions of the entire capsid (62) the location of UL25 was not identified, suggesting that it does not conform to the icosahedral symmetry of the capsid and therefore its density is averaged out. However, as mentioned above, UL25 appears to associate with pentons (38), and such an interaction could be envisaged to have a role in either DNA retention or capsid stabilization. The penton channel is rather large, with a minimum diameter of ∼40 Å, and is electropositive in character (6), suggesting that escape of packaged DNA is more likely through one or more of the penton vertices than through the hexon channels, which are quite narrow (∼20 Å in diameter) and are very electronegative. The dimensions of UL25 certainly allow for it or an oligomer to fit within the penton channel and act to potentially seal the DNA in the capsid.
The gpD protein of bacteriophage lambda and gpShp of bacteriophage 21 function as head stabilization proteins (57, 59). DNA packaging in bacteriophage lambda is accompanied by expansion of the prohead to an icosahedral shape and the binding of gpD. Interestingly, gpD is necessary for the packaging of full-length genomes, whereas gpD deletion mutants with genomes <82% of wild-type length are viable (49). This deficiency at the late stages of DNA packaging is similar to the phenotype of KUL25NS described by Stow (51), suggesting that rather than physically blocking the penton or portal channel, UL25 might act to stabilize the capsid against the increasing internal pressures generated during the packaging process. As is the case with lambda gpD, new UL25 binding sites might be revealed as the shape of the HSV-1 capsid is reconfigured during DNA entry. In addition to hexons and pentons, these sites could involve two other reported partners of UL25, the capsid triplex proteins VP19C and VP23 (38). Binding sites for the gpSoc and gpHoc proteins of bacteriophage T4 are similarly exposed on the phage head following the structural transitions associated with DNA packaging (26). Although neither protein is required for DNA encapsidation, gpSoc does have a role in stabilizing the capsid against extremes of pH and temperature (21, 46). Like UL25, lambda gpD is characterized by a surface of irregular secondary structure with the majority of residues in coils and loops (59).
A possible parallel to UL25 is seen in the outer scaffold protein D from FX174, in which a rigid core structure is surrounded by flexible regions that are capable of existing in different conformations depending on their local environment (14). It is possible that UL25 could act in a scaffold-like manner, but being added rather than removed from the capsid during the maturation process. This could facilitate subsequent stages of capsid maturation such as nuclear egress or acquisition of the tegument layer. Although the structures of phage proteins involved in DNA retention or head stabilization have been determined (39, 59) no significant similarity to UL25 in their folding pattern is apparent.
In addition to its role in DNA packaging, UL25 may also perform other important functions. It is likely that UL25 incorporated into C capsids establishes interactions that are important for capsid egress from the nucleus and the acquisition of the tegument layer by the virion. Studies on a UL25 ts mutant also indicate an essential early function for the virion-contained protein (1; V. Preston, unpublished results), and in this context it is interesting to note that UL25 has been implicated in the association of incoming capsids with microtubules (24). Although our structure does not include the N-terminal 133 amino acids, the predicted coiled coil (amino acids 49 to 110) is likely to contribute to oligomerization or other protein-protein interactions, and genetic studies on N-terminally truncated proteins confirm that residues in this region are important for virus replication (V. Preston, unpublished results). It remains to be determined whether DNA packaging and/or some other function is impaired in these truncated proteins.
Conclusion.
We have presented the first crystallographic structure of a herpesvirus protein involved in DNA encapsidation. The structure revealed a novel fold with a unique “flexible” architecture, suggesting its adaptability in changing conformational states depending on its local environment. The electrostatic calculation reveals a distinct division of surface charges in which one face is highly electronegative, while the opposite face is electropositive. Evolutionary trace analysis incorporating UL25 homologues from the three herpesvirus subfamilies identified four clusters of residues that potentially form binding sites for other interacting proteins and mapped them to the structure. The distribution of binding sites, in combination with its “flexible” architecture and distinctive electrostatic pattern, suggests an inherent elasticity for UL25, which is a necessary characteristic of a protein involved in the dynamic processes of capsid assembly and DNA encapsidation. Although the structure determination cannot presently discriminate between possible models for UL25 function, it provides a basis for future studies on the role of this protein in DNA packaging and virion morphogenesis.
Acknowledgments
We thank W. Chiu and C. M. Moure for technical assistance and useful discussions and the staff at the APS SBC-CAT for help with data collection.
This research was supported in part by grants from the NIH (R01-AI38469) to Wah Chiu and the Welch Foundation (Q0581) to F.A.Q. Initial extensive work was funded by a grant from the Howard Hughes Medical Institute to F.A.Q.
REFERENCES
- 1.Addison, C., F. J. Rixon, J. W. Palfreyman, M. O'Hara, and V. G. Preston. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali, M. A., B. Forghani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216:278-283. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, S. 1994. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, B. R., M. L. Baker, F. J. Rixon, W. Chiu, and F. A. Quiocho. 2003. Structure of the herpesvirus major capsid protein. EMBO J. 22:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. C., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-153. In A. Holzenburg and E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 8.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarty, S., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2005. Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity. J. Virol. 79:554-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasgupta, A., and D. W. Wilson. 1999. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J. Virol. 73:2006-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 13.delaFortelle, E., and G. Bricogne. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276:472-494. [DOI] [PubMed] [Google Scholar]
- 14.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowman, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral procapsid with molecular scaffolding. Nature 389:308-313. [DOI] [PubMed] [Google Scholar]
- 15.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann III, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68:3702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauptman, H. A. 1997. Shake-and-bake: an algorithm for automatic solution ab initio of crystal structures. Methods Enzymol. 277:3-13. [DOI] [PubMed] [Google Scholar]
- 18.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478-480. [DOI] [PubMed] [Google Scholar]
- 19.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 20.Innis, C. A., J. Shi, and T. L. Blundell. 2000. Evolutionary trace analysis of TGF-beta and related growth factors: implications for site-directed mutagenesis. Protein Eng. 13:839-847. [DOI] [PubMed] [Google Scholar]
- 21.Ishii, T., and M. Yanagida. 1977. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 109:487-514. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 24.Kaelin, K., S. Dezelee, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leiman, P. G., S. Kanamaru, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2003. Structure and morphogenesis of bacteriophage T4. Cell Mol. Life Sci. 60:2356-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lichtarge, O., and M. E. Sowa. 2002. Evolutionary predictions of binding surfaces and interactions. Curr. Opin. Struct. Biol. 12:21-27. [DOI] [PubMed] [Google Scholar]
- 28.McClelland, D. A., J. D. Aitken, D. Bhella, D. McNab, J. Mitchell, S. M. Kelly, N. C. Price, and F. J. Rixon. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 30.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., and D. Gatherer. 2005. Integrating reptilian herpesviruses into the family Herpesviridae. J. Virol. 79:725-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., D. Gatherer, and A. Dolan. 2005. On phylogenetic relationships among major lineages of the Gammaherpesvirinae. J. Gen. Virol. 86:307-316. [DOI] [PubMed] [Google Scholar]
- 33.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihalek, I., I. Res, H. Yao, and O. Lichtarge. 2003. Combining inference from evolution and geometric probability in protein structure evaluation. J. Mol. Biol. 331:263-279. [DOI] [PubMed] [Google Scholar]
- 35.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 36.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlova, E. V., B. Gowen, A. Droge, A. Stiege, F. Weise, R. Lurz, M. van Heel, and P. Tavares. 2003. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 22:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307-326. [DOI] [PubMed] [Google Scholar]
- 41.Poteete, A. R., and J. King. 1977. Functions of two new genes in Salmonella phage P22 assembly. Virology 76:725-739. [DOI] [PubMed] [Google Scholar]
- 42.Prevelige, P. E., Jr., and J. King. 1993. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog. Med. Virol. 40:206-221. [PubMed] [Google Scholar]
- 43.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 44.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roizman, B. N., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Strauss (ed.), Field's virology. Lippincott-Raven, Philadelphia, Pa.
- 46.Ross, P. D., L. W. Black, M. E. Bisher, and A. C. Steven. 1985. Assembly-dependent conformational changes in a viral capsid protein. Calorimetric comparison of successive conformational states of the gp23 surface lattice of bacteriophage T4. J. Mol. Biol. 183:353-364. [DOI] [PubMed] [Google Scholar]
- 47.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sowa, M. E., W. He, K. C. Slep, M. A. Kercher, O. Lichtarge, and T. G. Wensel. 2001. Prediction and confirmation of a site critical for effector regulation of RGS domain activity. Nat. Struct. Biol. 8:234-237. [DOI] [PubMed] [Google Scholar]
- 49.Sternberg, N., and R. Weisberg. 1977. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 117:733-759. [DOI] [PubMed] [Google Scholar]
- 50.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 51.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strauss, H., and J. King. 1984. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol. 172:523-543. [DOI] [PubMed] [Google Scholar]
- 53.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115-125. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 57.Wendt, J. L., and M. Feiss. 2004. A fragile lattice: replacing bacteriophage lambda's head stability gene D with the shp gene of phage 21 generates the Mg2+-dependent virus, lambda shp. Virology 326:41-46. [DOI] [PubMed] [Google Scholar]
- 58.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, F., P. Forrer, Z. Dauter, J. F. Conway, N. Cheng, M. E. Cerritelli, A. C. Steven, A. Pluckthun, and A. Wlodawer. 2000. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat. Struct. Biol. 7:230-237. [DOI] [PubMed] [Google Scholar]
- 60.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]
- 61.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]