Abstract
The interaction of human immunodeficiency virus type 1 (HIV-1) Nef with p21-activated kinase 2 (Pak2) has been proposed to play an important role in T-cell activation and disease progression during viral infection. However, the mechanism by which Nef activates Pak2 is poorly understood. Mutations in most Nef motifs previously reported to be required for Pak2 activation (G2, PxxP72, and RR105) also affect other Nef functions, such as CD4 or major histocompatibility complex class I (MHC-I) downregulation. To better understand Nef interactions with Pak2, we performed mutational analysis of three primary HIV-1 Nef clones that exhibited similar capacities for downregulation of CD4 and MHC-I but variable abilities to associate with activated Pak2. Our results demonstrate that Nef amino acids at positions 85, 89, 187, 188, and 191 (L, H, S, R, and F in the clade B consensus, respectively) are critical for Pak2 association. Mutation of these Nef residues dramatically altered association with Pak2 without affecting Nef expression levels or CD4 and MHC-I downregulation. Furthermore, compensation occurred at positions 89 and 191 when both amino acids were substituted. Since residues 85, 89, 187, 188, and 191 cluster on the surface of the Nef core domain in a region distinct from the dimerization and SH3-binding domains, we propose that these Nef residues form part of a unique binding surface specifically involved in association with Pak2. This binding surface includes exposed and recessed hydrophobic residues and may participate in an as-yet-unidentified protein-protein interaction to facilitate Pak2 activation.
The Nef protein of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV) is an important determinant of progression to AIDS. Nef is required for maintenance of high viral load and disease induction in SIV-infected rhesus monkeys (29). Additionally, Nef-defective viruses have been associated with long-term nonprogression in HIV-1-infected individuals (31). These findings together with the demonstration that a transgenic mouse expressing the HIV-1 nef gene exhibits AIDS-like disease (23) suggest that Nef is important for viral replication and pathogenicity in vivo.
HIV-1 Nef is a 27-kDa, membrane-associated cytoplasmic protein that is posttranslationally myristoylated and phosphorylated. Many functions of Nef have been demonstrated in cell culture, although their relative contributions to AIDS pathogenesis are unclear (reviewed in references 3, 7, 14, 17, and 62). Functions of Nef include CD4 downregulation, major histocompatibility complex class I (MHC-I) downregulation, enhancement of viral infectivity, and modulation of cellular signaling pathways. CD4 and MHC-I downregulation are well-described Nef functions (45). To achieve CD4 downregulation, Nef bridges CD4 with the adaptor protein complex of clathrin-coated pits (8, 19) and then transfers CD4 to COP-I for transport to lysosomes (27, 44). Nef motifs required for CD4 downregulation include the LL164 motif of the C-terminal flexible loop, which is required for Nef interaction with the adaptor protein complex. For MHC-I downregulation, Nef acts to link MHC-I and the endosome-to-Golgi PACS-1 sorting pathway. This function is dependent on the binding of PACS-1 to the acidic EEEE62-65 motif of Nef (46). The proline-rich SH3-binding domain (PxxP domain) is also important for MHC downregulation (20, 38). The mechanism by which Nef enhances viral infectivity has not been elucidated.
Nef expression enhances HIV replication in resting peripheral blood mononuclear cells (PBMC) but not in most cultured cell lines (40, 60). Enhancement of HIV replication most likely results from activation of resting T cells (1, 6, 13, 57, 58, 61, 67). Nef interaction with p21-activated kinase 2 (Pak2), a cellular serine/threonine kinase, has been proposed to play an important role in T-cell activation (37). Additionally, Nef interacts via its SH3-binding PxxP domain with several signaling molecules that could potentially contribute to T-cell activation, including Vav and Src tyrosine kinases such as Lck, Fyn, and Hck (reviewed in reference 51). However, the biological significance of these interactions remains unclear.
Nef-associated kinase (NAK) was initially detected as a 62-kDa serine kinase in in vitro kinase assays (IVKAs) of anti-Nef immunoprecipitates from infected T cells (53). NAK was shown to be activated by Nef, and this activation was blocked by dominant-negative forms of Pak and the p21 GTPases Rac1 and Cdc42 (37). Subsequently, NAK was identified as Pak2, a member of the Pak family of serine/threonine kinases (4, 42, 49, 50). Pak2 is involved in the regulation of cellular processes such as cytoskeleton rearrangement, cell morphology, motility, apoptosis, and gene transcription and is activated in response to a variety of cellular stresses (reviewed in references 5 and 12). Endogenous Pak2 is activated by the binding of GTP-bound forms of p21 GTPase Rac1 or Cdc42, which triggers a cascade of autophosphorylation events that culminate in full phosphorylation and activation (66).
The mechanism by which Nef activates Pak2 is poorly understood. Nef is thought to activate Pak2 through a multiprotein complex, but the low abundance and transient nature of this complex have made it difficult to identify its components and the nature of their interaction with Nef (4, 26, 33, 47). Motifs of Nef reported to be required for Pak2 association and activation include the N-terminal myristoylation signal, the PxxP domain, R106, and F191, but mutations of most of these motifs have pleiotropic effects (16, 30, 39, 43, 55, 64). Myristoylation is required for membrane association and for nearly all Nef functions (17). The PxxP motif of HIV Nef is required for interaction with the SH3 domains of Src tyrosine kinases and other signaling molecules in addition to Pak2 (38, 51). However, in SIV Nef the effects of mutation of the PxxP motif may be more specific for Pak2 activation (10, 30, 34). The RR105LL mutation in Nef abolishes Pak2 association and activation (30, 54) but is also multiply defective (16, 56, 64). R106 is likely a structural element at which even conservative changes deleteriously affect multiple functions (43). Only the F191I mutation has been demonstrated to specifically knock out Nef association with Pak2 without reducing other Nef functions (16).
To better understand Nef interactions with Pak2, we performed mutational analysis of three primary HIV-1 Nef clones that exhibited similar capacities for downregulation of CD4 and MHC-I but varied in their capacities for association with activated Pak2. Mutations at Nef residues 85, 89, 187, 188, and 191 reduced or abolished Pak2 association but did not affect CD4 or MHC-I downregulation. Furthermore, Pak2 association was restored by compensatory covariation at some of these Nef residues. Structural analysis showed that these residues cluster on the surface of the core domain in a region distant from R106 and distinct from domains involved in dimerization and interaction with SH3 domains. These findings suggest that residues 85, 89, 187, 188, and 191 form part of a unique binding surface specifically involved in Nef association with activated Pak2.
MATERIALS AND METHODS
Viral isolates.
The primary HIV-1 viruses MACS2-LN, MACS3-Br, and MACS3-LN were isolated from two late-stage AIDS patients (MACS2 and MACS3) and characterized as described previously (18). Briefly, autopsy lymph node and brain tissue samples were homogenized and cocultured with CD8-depleted PBMC. Culture supernatants testing positive for reverse transcriptase (RT) activity were filtered and stored at −80°C. Although MACS3-Br was isolated from brain tissue, phylogenetic analysis of nef and env sequences indicates that it is most closely related to lymphoid-derived rather than brain-derived viruses (data not shown).
Nef amplification and plasmids.
Genomic DNA was extracted, using a QIAGEN DNeasy kit, from pellets of cultured PBMC infected with the primary virus isolates. HIV-1 nef genes were amplified by nested PCR performed with 1 μl (∼100 ng) total genomic DNA. Maximally conserved primer binding sites were selected. Nucleotide positions of the primer sequences in the HIV-1 HXB2 genome are given in parentheses. Outer primers were Nef1 (5′-GCCCGAAGGAATAGAAGAAG-3′) (8411 to 8430) and Nef2 (5′-GCACTCAAGGCAAGCTTTATTGAGGC-3′) (9631 to 9605). The Nef2 primer was previously published (25). Inner primers were Nef3b.5 (5′-TTAGTGAACGGATCCTTAGCACTTATCTGGG-3′) (8466 to 8496) and Nef4.5 (5′-GCGGAAAGTCCCTTGTAGCAACATCGATGTCA-3′) (9446 to 9415). The inner primers included the restriction sites BamHI and ClaI (underlined). All reactions were performed with Pfu polymerase (Stratagene). The cycling conditions for the 50-μl, first-round reaction were 94°C (2 min), followed by 30 cycles of 94°C (15 s), 57°C (15 s), and 68°C (3 min) and then the final elongation step at 72°C (7 min). One microliter of the first-round PCR product was used as the template for the second-round reaction. The cycling conditions for the second-round primer set were the same except that hybridization was performed at 55°C. PCR products were cloned into the pCR3.1 expression vector containing BamHI and ClaI restriction sites. From each isolate, three to five individual clones as well as the bulk PCR product were sequenced. The SF2 nef allele was cloned into pCDNA3.1. The pCR3.1 vector lacking an insert was used for mock transfections. pCDNA3.1-FLAG-Pak2-K278R was created from pCDNA1-HA-Pak2 (where HA is hemagglutinin) (4) by site-directed mutagenesis followed by addition of the FLAG tag by PCR amplification and subcloning with EcoRV-XhoI into pCDNA3.1. pCDNA3.1-Cdc42-V12 was created by subcloning the HindIII-NotI fragment from pCS2+-Cdc42V12 (obtained from M. Kirschner) into pCDNA3.1.
Site-directed mutagenesis.
Mutations were created using a QuikChange site-directed mutagenesis kit (Stratagene), and all mutants were confirmed by sequencing.
Cell culture.
293T cells and HIJ cells (HeLa cells retrovirally transduced to express high levels of CD4), kindly provided by David Kabat (28), were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 100 μg/ml penicillin-streptomycin. Jurkat T-antigen cells (41), a kind gift of Heinrich Gottlinger, were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 100 μg/ml penicillin-streptomycin.
IVKAs.
IVKAs were performed as previously described (16). Briefly, 293T cells transfected (Lipofectamine 2000; Invitrogen) with Nef and pN1GFP plasmids were lysed in IVKA lysis buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 10% glycerol, 0.5% IGEPAL CA-630 [Sigma], 1 mM EDTA, 25 mM NaF, 2 mM Na3VO4, 20 mM beta-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 25 mM benzamidine, and Complete protease inhibitors [Roche]). Cleared whole-cell lysates (600 μg) were immunoprecipitated with sheep anti-Nef serum. Kinase reactions were performed with immunoprecipitates in the presence of [γ-32P]ATP and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Dried gels were exposed to a phosphorimager screen (Packard).
CD4 and MHC-I downregulation assays.
For CD4 downregulation assays, HIJ cells were cotransfected with nef and pCDNA3-EGFP plasmids (where EGFP is enhanced green fluorescent protein) by use of Lipofectamine 2000 (Invitrogen). Cells were harvested after 24 to 28 h and stained for 30 min on ice with saturating amounts of phycoerythrin (PE)-conjugated mouse anti-CD4 antibody and 7AAD (7-amino-actinomycin D) (BD Pharmingen). 7AAD staining was used to identify nonviable cells. Stained cells were fixed in 2% paraformaldehyde and analyzed by flow cytometry (FACScan; Becton Dickinson). Events were plotted on a log scale, and the geometric mean of CD4-PE fluorescence intensity was calculated for transfected (GFP+) and untransfected (GFP−) cell populations. To calculate the relative mean fluorescence intensity (relMFI) of each transfected sample, we used the formula relMFIsample = [(MFIGFP+)/(MFIGFP−)]sample/[(MFIGFP+)/(MFIGFP−)]Mock, where MFI is the mean intensity. This formula corrects for slight staining variation between stained samples and expresses the CD4 surface levels relative to those for the mock-transfected negative control. MHC-I downregulation assays were performed by cotransfecting Jurkat T-antigen cells with nef and pCDNA3-EGFP plasmids by using Lipofectamine 2000. Cells were activated 4 h later with 1 μg/ml phytohemagglutinin L and 50 ng/ml phorbol myristate acetate. Cells were harvested after 24 to 36 h and stained with 7AAD and a saturating amount of PE-conjugated mouse anti-HLA-ABC antibody (BD Pharmingen). Stained cells were fixed and analyzed by flow cytometry as described above. The relMFI of HLA-ABC-PE fluorescence was calculated for high GFP expression by use of the formula described above.
Western blotting.
Aliquots of cell lysates used for the IVKAs were tested for Nef expression by Western blotting with sheep polyclonal anti-Nef SF2 serum (1:2,000 dilution) as described previously (16), rabbit anti-Nef SF2, or rabbit anti-Nef no. 2949 (1:2,000 dilution) (AIDS Research and Reference Reagent Program). GFP expression was detected with a mouse anti-GFP antibody (Zymed). In other experiments, 293T cells were transfected using the calcium phosphate method and harvested 48 h after washing. Cells were lysed in either IVKA lysis buffer or cold lysis buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, and Complete protease inhibitors [Roche]). Lysates were centrifuged at 14,000 rpm for 20 min at 4°C to remove cell debris. Equal amounts of total protein were run on 12% SDS-PAGE gels and analyzed by Western blotting with rabbit anti-Nef no. 2949.
Coimmunoprecipitations.
293T cells were transfected using the calcium phosphate method and harvested 48 h after washing. Cells were lysed in cold lysis buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, and Complete protease inhibitors [Roche]) for 20 min at 4°C, followed by centrifugation at 14,000 rpm for 20 min at 4°C to remove cell debris. Lysates were normalized by amount of total protein, precleared with protein A Sepharose beads (Amersham Biosciences), and immunoprecipitated with sheep anti-Nef serum (2 h, 4°C) followed by protein A Sepharose beads. Immunoprecipitated proteins were eluted by boiling in 2× SDS-PAGE sample buffer, followed by separation on 10% SDS-PAGE gels and immunoblotting with M2 anti-FLAG-horseradish peroxidase (Sigma) or rabbit anti-Nef no. 2949. To confirm consistent levels of protein expression, aliquots of initial cell lysates were run on 10% SDS-PAGE gels and submitted to Western blotting.
Structural modeling.
DeepView (Swiss PDB viewer) was used to analyze the crystal structure of the Nef dimer bound to the SH3 domain of Fyn kinase (Protein Data Base accession number 1AVZ) (2). The solution nuclear magnetic resonance (NMR) structure of the core domain of Nef BH10 (Protein Data Base accession number 2NEF) (22) was also viewed with DeepView and analyzed for possible F191 rotomers and residues within 8 or 10 Å of F191.
Phylogenetic analysis.
Nef protein sequence alignments were made using Clustal W and contained no gaps. Bootstrapped phylogenetic trees were created by the neighbor-joining method using Clustal X. Percent conservation of specific Nef amino acids was determined using the programs Epilign and SeqPublish and downloadable alignments from the Los Alamos National Laboratory sequence database.
Nucleotide sequence accession numbers.
The nucleotide and amino acid sequences of Nef clones MACS2LN-5C, MACS3Br-6I, and MACS3LN-7D and nef clones amplified directly from MACS2 lymph node and spleen have been submitted to the GenBank sequence database and have been assigned the accession numbers DQ357219 to DQ357221 and DQ358012 to DQ358047.
RESULTS
Identification of primary nef clones that associate with activated Pak2.
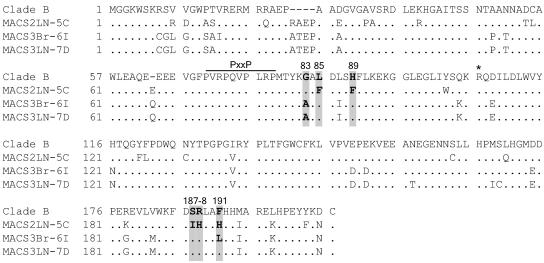
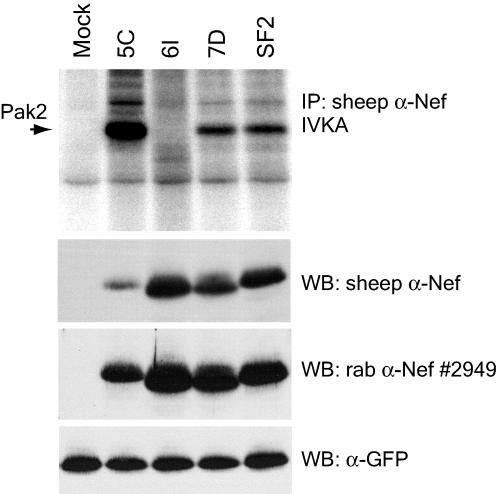
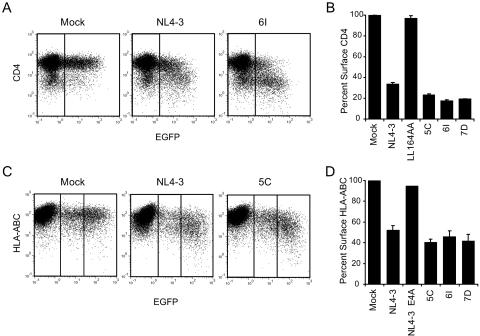
Three nef alleles, MACS2LN-5C (5C), MACS3Br-6I (6I), and MACS3LN-7D (7D), were cloned from primary HIV-1 isolates previously characterized by Gorry et al. (18). Each allele represents the consensus of nef sequences from the corresponding viral isolate. Phylogenetic analysis of the nucleotide sequences of nef alleles 5C, 6I, and 7D showed that they represent clade B nef alleles (data not shown). Figure 1 shows the amino acid sequences of each primary Nef aligned with the clade B consensus Nef. All three nef alleles expressed high levels of full-length 27-kDa protein (Fig. 2), and each Nef strongly downregulated both cell surface CD4 and MHC-I relative to the positive control, NL4-3 Nef (Fig. 3).
FIG. 1.
Amino acid alignment of primary HIV-1 Nef clones compared to the clade B consensus. Amino acids G83, L85, H89, S187, R188, and F191, numbered relative to the NL4-3 Nef sequence, are shaded. Boldface type indicates positions that vary from the consensus. Also labeled are R106, previously proposed to be important for Pak2 activation (51, 54), and the PxxP SH3-binding domain. The clade B consensus is from the LANL sequence database (August 2004). Alanine is the consensus amino acid at position 83 of an earlier LANL clade B Nef consensus sequence.
FIG. 2.
Associations of primary HIV-1 Nefs with activated Pak2 are variable. 293T cells transiently expressing GFP and the indicated Nefs were immunoprecipitated with sheep anti-Nef (α-Nef) (raised to SF2 Nef) and assayed by IVKA (top panel) as described in Materials and Methods. SF2 Nef, previously demonstrated to strongly activate Pak2 (4), is included as a positive control. An arrow indicates the 62-kDa band corresponding to Pak2. Whole-cell lysates used for IVKA immunoprecipitations (IP) were immunoblotted with sheep anti-Nef (second panel) or rabbit (rab) anti-Nef antibody no. 2949. As a control for transfection efficiency, lysates were immunoblotted for GFP expression (bottom panel). WB, Western blot.
FIG. 3.
Downregulation of cell surface CD4 and MHC-I by primary Nef alleles is conserved. Flow cytometric analysis of CD4 downregulation (A and B) and MHC-I downregulation (C and D). (A) Flow cytometric analysis of HIJ cells cotransfected with pCDNA3-EGFP and either pCR3.1 (Mock), pCR3.1-Nef-NL4-3 (NL4-3), or pCR3.1-Nef-MACS3Br-6I (6I). Vertical lines indicate the threshold for GFP expression. Data shown are representative of two independent experiments. (B) Quantitation of CD4 downregulation data. Percent surface CD4 was calculated from the geometric mean fluorescence in GFP-positive cells as described in Materials and Methods and is shown relative to that for GFP-positive mock-transfected cells. The LL164AA mutation, previously shown to abolish CD4 downregulation, was made in the background of NL4-3 Nef and was included as a negative control. Averages are from two independent experiments. Error bars represent standard deviations. (C) Flow cytometric analysis of Jurkat T-antigen cells cotransfected with EGFP and either pCR3.1 (Mock), NL4-3 Nef, or 5C Nef. Surface MHC-I levels were determined by staining with anti-HLA-ABC-PE. Vertical lines indicate gates for GFP-negative, low-GFP-expressing, and high-GFP-expressing cells. Data are representative of four independent experiments. (D) Quantitation of MHC-I downregulation. Percent surface MHC-I was calculated as described above but includes only the high-GFP-expressing cells (right panels in dot plots). E4A is the EEEE65AAAA mutation of NL4-3 Nef, a mutation that has been shown to abolish MHC-I downregulation (46) and was included as a negative control. Averages are from one to four independent experiments. Error bars represent standard deviations.
To assess the ability of these Nefs to associate with Pak2 activity, IVKAs were performed with anti-Nef immunoprecipitates (Fig. 2). SF2 Nef, previously demonstrated to strongly activate Pak2, was used as a positive control (4). IVKAs with anti-Nef SF2 immunoprecipitates produced a 62-kDa phosphorylated band (Fig. 2) previously identified as Pak2 (4, 50). IVKA is a sensitive method of visualizing Nef-associated Pak2, which is of too-low abundance to be seen in Western blots (47). As shown in Fig. 2 (top panel), 7D Nef associated with Pak2 activity, while 6I Nef was defective for association. 6I and 7D Nefs (both from patient MACS3) have a high level of sequence similarity, are expressed at similar levels, and are similarly active in CD4 and MHC-I downregulation assays, suggesting that the Pak2 association defect of 6I Nef is specific.
5C Nef strongly associated with Pak2 (Fig. 2, top panel). Surprisingly, the sheep anti-Nef SF2 antibody, used to immunoprecipitate the Nef complexes for the IVKAs, recognized 5C Nef poorly by Western blotting, perhaps due to the high level of sequence divergence in 5C Nef. Comparison of Nef protein levels in lysates before and after immunoprecipitation confirmed that the sheep anti-Nef antibody efficiently immunoprecipitated all Nefs, including 5C Nef (data not shown). To better visualize 5C Nef expression, we also used rabbit anti-Nef antibody no. 2949 to measure Nef expression by immunoblotting. In these blots, 5C Nef appears to express well but at a somewhat lower level than 6I and 7D Nefs. Therefore, we conclude that 5C Nef strongly associates with Pak2 and immunoprecipitates a high amount of Pak2 activity despite having a lower expression level.
F191 is critical for association of 6I and 7D Nefs with Pak2.
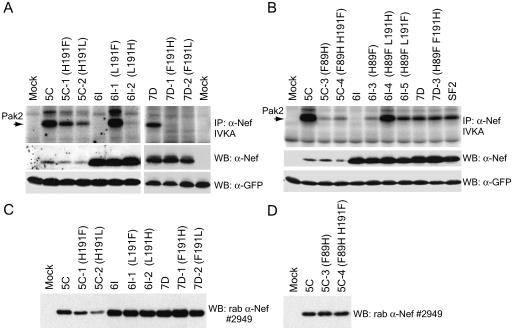
We next investigated the amino acid variation that could account for reduced association of 6I Nef with Pak2. Arginine 106 has been proposed to be involved in Pak2 association and activation (51, 54, 64). However, R106 is conserved in all three primary Nefs (Fig. 1). Thus, other amino acid variations must account for differences in Pak2 association with 5C, 6I, and 7D Nefs. Mutation of phenylalanine 191 to isoleucine was previously shown to abolish Pak2 association and activation (16). Among clade B Nef proteins in the LANL database (n = 455), F191 is 91% conserved. F191 is one of only five amino acids that differ between the 6I and 7D Nefs derived from patient MACS3 (Fig. 1). The conserved F191 is present in 7D Nef, whereas 6I Nef has a leucine at this position. Only 4% of clade B Nef proteins in the LANL database have a leucine at position 191. To investigate whether this single-amino-acid change in 6I Nef accounts for the loss of Pak2 association, reciprocal mutants of 6I and 7D were constructed. As expected, mutant 7D-2 (F191L) Nef was defective for Pak2 association while Pak2 association was fully restored for 6I-1 (L191F) Nef (Fig. 4A). Both mutants were similar to their parental Nefs in expression levels (Fig. 4A and C). Mutant Nef 6I-1 was also similar to its parental Nef in CD4 and MHC-I downregulation activity (data not shown). These results demonstrate the importance of F191 for Nef association with Pak2 in divergent nef alleles and show that mutation of this residue alone can specifically attenuate Pak2 association without affecting stability, expression, or other functions of Nef.
FIG. 4.
Amino acids 89 and 191 of Nef are critical for Pak2 association, and mutations at these positions are compensatory in the 6I and 7D Nef backgrounds. (A and B) IVKAs. Whole-cell lysates of 293T cells transiently expressing GFP and single and double mutants of 5C, 6I, and 7D Nefs were immunoprecipitated with sheep anti-Nef (α-Nef) and assayed by IVKA (top panel) as described in Materials and Methods. An arrow indicates the 62-kDa band corresponding to Pak2. Whole-cell lysates used for IVKA immunoprecipitations (IP) were immunoblotted with rabbit (rab) or sheep anti-Nef antibodies (raised to SF2) (middle panel) or anti-GFP (bottom panel). (C and D) Independent transfection experiments to confirm relative expression levels of Nef mutants. 293T cells were transfected with GFP and Nef as described for panels A and B. Whole-cell lysates were immunoblotted with rabbit anti-Nef antibody no. 2949. WB, Western blot.
5C Nef, with a histidine at position 191, strongly associates with Pak2, while the F191H mutation in 6I and 7D Nefs abolishes Pak2 association.
Although 5C Nef differs from 6I, 7D, and ConB Nefs by several amino acids (Fig. 1), the most striking variation of 5C Nef is the histidine at position 191. Because F191 has been shown to be critical for Pak2 association, the strong association of 5C Nef with Pak2 was surprising. To determine if histidine and phenylalanine are functionally equivalent at position 191 or if histidine 191 acts as a gain-of-function mutation, reciprocal mutants were made. 6I-2 (L191H) was severely impaired for Pak2 association relative to 6I-1 (L191F), and 7D-1 (F191H) was completely defective for this activity (Fig. 4A). Both 5C-1 (H191F) and 5C-2 (H191L) had reduced Pak2 association (Fig. 4A), but these mutants also appeared to have lower expression levels (Fig. 4C). Densitometry revealed that the reduction in amount of Pak2 activity associated with mutants 5C-1 and 5C-2 was equivalent to the reduction in expression of these mutants (data not shown). While it is possible that the mutations in 5C-1 and 5C-2 Nefs altered their recognition by anti-Nef antibodies, these data suggest that mutations H191F and H191L in the 5C Nef background do not specifically reduce Pak2 association but instead reduce 5C Nef stability and/or expression.
Pak2 association with 6I and 7D Nefs was highest when residue 191 was phenylalanine and was severely reduced or abolished when it was histidine. In contrast, 5C Nef was expressed at higher levels and associated with more Pak2 activity when residue 191 was a histidine. Together, these data show that histidine 191 is neither a gain-of-function variation nor equivalent to phenylalanine 191. Furthermore, the results suggest that variant amino acids in the background of 5C Nef create a requirement for H191 for optimal expression and Pak2 association with 5C Nef.
Variation H89F compensates for F191H in 6I and 7D Nefs to restore association with Pak2.
In addition to the F191H variation, 5C Nef has a phenylalanine at position 89 instead of the histidine common to clade B Nefs. In Nefs from non-B clades of HIV-1, amino acids arginine, leucine, and histidine are more frequently found at position 191. In clades C and E, these amino acids are often correlated with a phenylalanine at position 89 (43). Therefore, we hypothesized that F89 might compensate for the H191 variation in 5C Nef and might even contribute to the requirement for H191 for optimal expression and Pak2 association.
To determine if H89F and F191H are indeed compensatory changes, double mutants were made in the 5C (F89 H191), 6I (H89 L191), and 7D (H89 F191) Nef backgrounds. As discussed above, single mutants 6I-2 (H89 H191) and 7D-1 (H89 H191) were defective for Pak2 association (Fig. 4A). Introduction of secondary H89F mutations fully rescued the ability of the 6I and 7D Nef F191H mutants to associate with Pak2 activity (Fig. 4B). Double mutants 6I-4 (F89 H191) and 7D-3 (F89 H191) associated with equivalent amounts of active Pak2 compared to the fully functional 6I-1 (H89 F191) Nef and 7D (H89 F191) Nef. In addition, the double mutant 6I-5 Nef (F89 F191) had reduced Pak2 association relative to the fully functional 6I-1 (H89 F191) Nef. Therefore, while 6I and 7D Nefs have minimal tolerance for phenylalanine at both positions (F89 F191) or histidine at both positions (H89 H191), they tolerate F89 H191 and H89 F191 equally well. All mutants were expressed at levels equivalent to those of their parental clones and were fully functional for CD4 and MHC-I downregulation (data not shown), suggesting that these mutations did not affect the overall structure or stability of 6I and 7D Nefs. These results demonstrate that Nef H89 and F191 are both critical for Pak2 association and that variations at positions 89 and 191 are compensatory.
F89H cannot compensate for H191F in the 5C Nef background.
Surprisingly, the double mutant 5C-4 (H89 F191) had severely reduced ability to associate with Pak2 relative to 5C, even though it has the clade B consensus amino acids at positions 89 and 191 (Fig. 4B). In fact, double mutant 5C-4 was less able to pull down active Pak2 than the single mutant 5C-1 (F89 F191). Mutant 5C-4 (H89 F191) and the parental 5C Nef (F89 H191) were expressed at similar levels (Fig. 4B and D) and had similar activities by use of CD4 and MHC-I downregulation assays (data not shown). Therefore, in contrast to the secondary H89F mutations in the 6I and 7D Nef backgrounds, the secondary mutation F89H did not compensate for the deleterious single mutation H191F in the 5C Nef background. Instead, F89 and H191 are required for optimal Pak2 association with 5C Nef. This finding suggests that 5C Nef contains other variant amino acids that restrict tolerance for variation at positions 89 and 191.
Amino acids F85, I187, and H188 in 5C Nef cluster near F89 and H191 and are specifically required for Pak2 association.
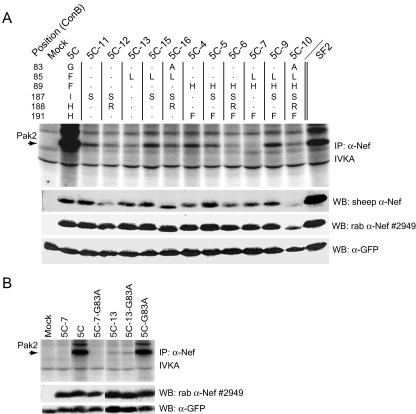
To identify additional amino acids of Nef potentially involved in Pak2 association, we examined published crystal and NMR structures of Nef (2, 22). Three divergent amino acids in 5C Nef, residues 85, 187, and 188, clustered adjacent to H89 and F191 on the surface of the Nef core (see Fig. 7A). To test the involvement of Nef residues 85, 187, and 188 in Pak2 association, a series of mutations was made in the 5C Nef background that replaced these residues with amino acids found in the clade B consensus Nef. All mutations in 5C Nef at residues 85, 187, and 188 significantly reduced Pak2 association, and each mutation exhibited a similar phenotype, whether in the context of 5C Nef (F89 H191) or 5C-4 Nef (H89 F191) (Fig. 5A). Both I187S mutants (5C-11 and 5C-5) had similarly low levels of Pak2 association. The F85L mutants (5C-13 and 5C-7) had severely reduced Pak2 association, and mutant 5C-7 appeared to be completely defective for Pak2 association. Double mutants with both F85L and I187S mutations (5C-15 and 5C-9) had slightly increased Pak2 association compared to Nefs with either mutation alone, indicating that these mutations might have compensatory effects. The H188R mutation reduced Pak2 association in the context of 5C Nef, 5C-4 Nef, and all other mutants tested. Double mutant 5C-12 (I187S/H188R) had reduced Pak2 association compared to 5C-11 (I187S). Similarly, 5C-6, 5C-10, and 5C-16 had reduced association with Pak2 compared to 5C-5, 5C-9, and 5C-15, respectively. Importantly, mutations I187S, F85L, and H188R did not affect 5C Nef expression levels (Fig. 5A, middle panels) or reduce CD4 or MHC-I downregulation activity (data not shown), indicating that these mutations specifically reduced Pak2 association.
FIG. 7.
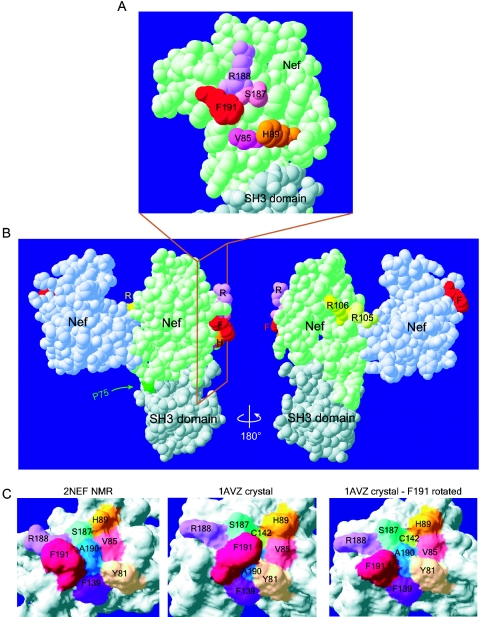
Residues 85, 89, 187, 188, and 191 cluster together on the surface of the Nef core domain in a region distinct from the dimerization and SH3-binding domains. Computer modeling of Nef core domain structures demonstrates the proximity of amino acids important for Pak2 association. Structures were obtained from the Protein Data Bank and were visualized with Swiss PDB viewer DeepView. (A and B) van der Waals surfaces of the 1AVZ Nef crystal structure (2). The Nef core domain (light green) is bound to a second Nef core (light blue) and to the Fyn kinase SH3 domain (gray). The Nef core domain comprises amino acids 71 to 148 and 179 to 203. (A) Residues 85 (fuchsia), 89 (orange), 187 (pink), 188 (violet), and 191 (red) cluster together on the surface of the Nef core domain, with phenylalanine 191 protruding from the surface. The SH3 domain of Fyn is shown binding to the PxxP domain of Nef. Panel A is an enlargement, rotated 90°, of the boxed region in panel B. (B) F191 (red) is distant from the dimerization surface and arginines 105 and 106 (yellow). P75 (dark green) is part of the PxxP SH3-binding domain. (C) F191 of Nef can adopt two different confomers that either expose or hide the recessed A190 (dark blue). (Left) The molecular surface of the 2NEF NMR model of the Nef core domain was computed with DeepView (22). The confomer adopted by F191 in this model occurs in 50% of the 40 published NMR models, while the other 50% are similar to that shown in the middle panel. (Middle) Molecular surface of the Nef core domain of the 1AVZ crystal structure (2). Here, contact with a neighboring molecule in the crystal lattice sterically hinders F191 and constrains it into adopting the confomer shown. (Right) Molecular surface of the 1AVZ crystal structure with the side chain of F191 rotated to adopt a confomer, as in 2NEF. No steric hindrances occurred with this confomer.
FIG. 5.
Amino acids F85, I187, and H188 are specifically required for association of 5C Nef with Pak2. (A and B) IVKAs. Whole-cell lysates of 293T cells transiently expressing GFP and expressing 5C Nef mutants were immunoprecipitated with sheep anti-Nef and assayed by IVKA (top panels) as described in Materials and Methods. An arrow indicates the 62-kDa band corresponding to Pak2. Whole-cell lysates used for IVKA immunoprecipitations (IP) were immunoblotted with sheep anti-Nef (α-Nef) (raised to SF2), rabbit (rab) anti-Nef antibody no. 2949 (middle panels), or anti-GFP (bottom panels). The chart at the top of panel A indicates the amino acid changes made for each mutant. All lanes shown in panel B are from the same experiment. WB, Western blot.
Mutation A83G has no effect on Pak2 association with 5C Nef.
Analysis of the 1AAV crystal structure (2) revealed that A83 participates in an alpha helix that also contains H89 and V85 (data not shown). The G83 variation found in 5C Nef is relatively common among clade B Nefs. However, glycines are capable of disrupting alpha helices. Therefore, we hypothesized that the G83 variation might restrict the tolerated amino acids at position 85, 191, and 89 by disrupting the alpha helix. Contrary to our prediction, however, mutation G83A had no effect on the ability of 5C to associate with Pak2 (Fig. 5B). Like the parental 5C Nef, mutant 5C-19 Nef (G83A) associated with very high levels of active Pak2. Furthermore, single mutant 5C-13 Nef (F89L) and double mutant 5C-14 Nef (F89L/G83A) had equally low levels of Pak2 association, and mutant 5C-7 Nef (F89H/H191F/F89L) and 5C-8 Nef (F89H/H191F/F89L/G83A) were equally defective for Pak2 association.
Mutation F89H abolishes association of 5C Nef with exogenous Pak2.
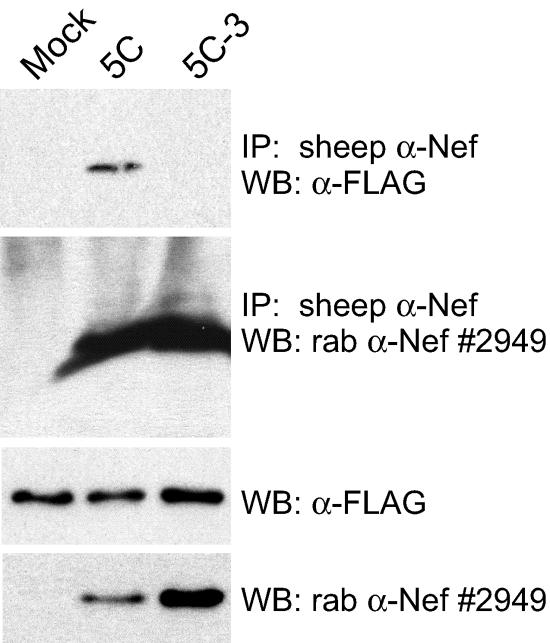
To demonstrate that the mutants discussed here disrupt Pak2 association rather than activation of Pak2 or another 62-kDa protein, we performed immunoprecipitations of lysates expressing exogenous FLAG-tagged Pak2-K278R, Cdc42-V12, and either vector alone or primary or mutant Nefs. Kinase-dead Pak2-K278R was shown (47) to stabilize the Nef/Pak2 complex and facilitate visualization of Nef-associated Pak2 by Western blotting. Dominant active Cdc42-V12 is thought to stabilize or catalyze the formation of the Nef/Pak2 complex. Pak2 coimmunoprecipitated with 5C Nef but not with 5C-3 (F89H) Nef (Fig. 6). Both Pak2-K278R and Cdc42-V12 were required to visualize 5C Nef-associated Pak2 by Western blotting. These results show that mutation F89H disrupts the specific association of Nef with Pak2.
FIG. 6.
Mutation F89H abolishes association of 5C Nef with exogenous Pak2. 293T cells were cotransfected with FLAG-tagged, kinase-dead Pak2-K278R, dominant active Cdc42-V12, and either vector alone (Mock), 5C, or mutant 5C-3. Cell lysates were immunoprecipitated with sheep anti-Nef (α-Nef) antibody. Nef-associated FLAG-Pak2-K278R was detected by anti-FLAG antibody (top panel), and immunoprecipitated Nef was detected by rabbit (rab) anti-Nef antibody no. 2949 (second panel). Expression of FLAG-Pak2-K278R and Nef in cell lysates was assayed by anti-FLAG and anti-Nef Western blotting (third and fourth panels). Results are representative of two independent experiments. IP, immunoprecipitation; WB, Western blot.
Residues 85, 89, 187, 188, and 191 are unlikely to bind SH3 domains of interacting proteins or participate in Nef dimerization.
Our data suggest that residues 85, 89, 187, 188, and 191 may form an important binding surface of Nef specifically involved in association with active Pak2. Structural modeling shows that these residues are within 10 Å of each other on the surface of the Nef core, include two exposed hydrophobic amino acids, and surround a third, recessed hydrophobic amino acid, A190 (Fig. 7A and C). To confirm that these residues are not participating in any known protein interactions, we examined the 1AVZ crystal structure of a dimerized Nef core domain bound to the SH3 domain of Fyn kinase (Fig. 7B) (2). From this structure, it is evident that the residues mutated in this study cannot participate in the dimerization of Nef. Although F191 and residues 85, 89, 187, and 188 are near the PxxP domain of Nef, they do not participate in binding to the SH3 domain of Fyn. These residues are distant from R106, a residue previously proposed to be involved in Pak2 association (51). Therefore, Nef residues 85, 89, 187, 188, and 191 may participate in a novel protein-protein interaction required for association with Pak2.
Covariation of residues 89 and 191 occurs naturally within the viral populations of patient MACS2.
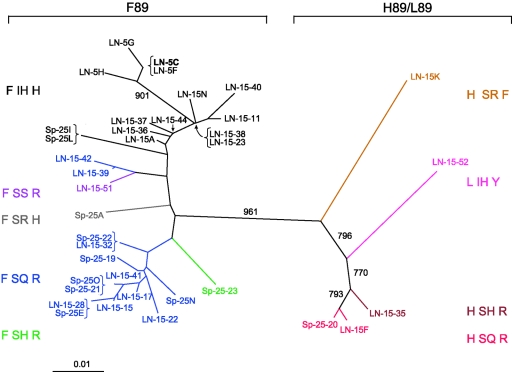
To investigate how well clone 5C represents Nef proteins from its patient source, 3 additional nef alleles were cloned from isolate MACS2LN and 33 nef alleles were directly amplified from the lymph node and spleen of patient MACS2. Sequencing revealed that a large number of Nefs from MACS2 share the amino acid sequence of 5C: F85, F89, I187, H188, and H191 (labeled in black text in Fig. 8). Therefore, Nef clone 5C is not a rare allele in this patient. For reference, the sequence of ConB Nef is L H S R F at these positions. Most strikingly, phylogenetic analysis of the protein sequences revealed that Nefs from MACS2 clustered into two groups, with H89 corresponding to one branch and F89 corresponding to the other. The level of divergence between the two groups of clones is striking. The bootstrap value of 961 (out of 1,000) indicates a high level of confidence in this branching. This branching could reflect negative selection against intermediate clones, distinct archival populations, or a founder effect. However, the high level of variation and high viral RNA copy numbers in these tissues argues against a bottleneck or founder effect (data not shown). Alignment with other amplified nef clones and BLAST searches indicated that the MACS2 clones were most closely related to each other and that none represented a possible contaminant.
FIG. 8.
Phylogenetic analysis of 37 full-length Nef clones from patient MACS2 reveals distinct populations that segregate according to variation at residue 89. Full-length Nef amino acid sequences amplified by PCR from the MACS2LN viral isolate (4 sequences) or directly from autopsy lymph node (22 sequences) or spleen tissue (11 sequences) were aligned with Clustal W. Clones are colored according to their amino acid sequence at residues 89, 187, 188, and 191, as indicated to the left and right of the tree. Clustal X was used to create the bootstrapped, neighbor-joining tree. The numbers associated with each branch represent the bootstrap values, which indicate the reliability of the branching order and were derived from 1,000 replicates.
Unlike most clade B Nefs, MACS2 Nefs had a very high frequency of histidines and arginines at position 191. H191 was found in 16 out of 37 total clones. R191 was found in 19 out of 37 clones from MACS2. There was evidence for covariation at positions 191 and 89 among Nef clones from MACS2. All 16 H191-containing clones also had F89. Clones with R191 had either F89 (16 of 19) or H89 (3 of 19). The only clone from this patient with F191 was similar to ConB at residues 89, 187, and 188 (see clone LN-15K in orange in Fig. 8). Interestingly, all Nefs cloned from this patient had a phenylalanine at position 85, which is uncommon with clade B Nefs (7.3%). Some sequences at position 89, 187, and 188 were highly represented (colored in black and blue in Fig. 8). Fifteen out of 37 total sequences had F89 I187 H188 H191, and 14 out of 37 total sequences had F89 S187 Q188 R191. This finding may reflect collective conservation of these amino acids in vivo or the fact that these Nefs are very fit alleles subjected to positive selection. Together, these results support our mutagenesis data and suggest that covariation at HIV-1 Nef positions 89 and 191 occurs in vivo.
DISCUSSION
In this study, we demonstrate that HIV-1 Nef residues 85, 89, 187, 188, and 191 (L, H, S, R, and F in the Nef clade B consensus, respectively) are critical for Nef-associated Pak2 activity. These residues cluster on the surface of the Nef core domain in a region distinct from the dimerization region and the SH3-binding domain. Mutations at these positions reduced or abolished association with endogenous Pak2 activity but did not affect CD4 and MHC-I downregulation. Furthermore, mutation of 5C Nef residue 89 specifically abolished association with exogenous Pak2, suggesting that these mutations influence association with rather than activation of Pak2. Compensatory covariation occurs at positions 89 and 191, indicating that these residues are subject to selection pressure. Based on these findings, we propose that residues 85, 89, 187, 188, and 191 form part of a unique binding surface specifically required for association with active Pak2.
Our studies provide evidence that Nef residues 85, 89, 187, 188, and 191 play a critical role in Pak2 association and activation. F191L and F191H mutations abolished association of 6I and 7D Nefs with Pak2 activity, in agreement with previous work reporting a similar defect for the D.con F193I mutant (corresponding to 191 in NL4-3 Nef) (16). Analyses of residues 85 and 188 and point mutations of residue 89 have not been reported. Deletion of residues 89 and 90 (delta 89/90) was shown to reduce binding to Hck and abolish binding to Lck but was not tested for Pak2 association or activation (11). Since F90 interacts with the RT loop of SH3 domains (35) and the F90R point mutation reduces binding to Hck (39), the Lck- and Hck-binding defects of the delta 89/90 mutant can be attributed to deletion of residue 90 alone.
Importantly, we show that point mutation of residue 89 can specifically abolish Pak2 association or rescue Nefs with deleterious mutations at position 191 (Fig. 4 and 6). The I187S mutation in 5C Nef significantly reduced Pak2 association without affecting CD4 and MHC downregulation. In contrast, mutation S187R in D.con and D88-11 Nefs reduced both Pak2 activation and MHC-I downregulation (16). The S187R mutation, which replaces a small, uncharged polar amino acid with a large basic amino acid, may be less stable or less well tolerated in the context of D.con, since S187A had no effect on D.con function. The structure of 5C may provide a more plastic environment with greater tolerance for variations at this position. None of the point mutations that reduced Pak2 activation in this study reduced other functions of Nef. Therefore, our analysis of primary HIV-1 nef alleles has identified amino acid determinants specifically important for association with active Pak2.
We have shown that within 6I and 7D Nefs, variations H89F and F191H are fully compensatory for association with Pak2. Additionally, mutations F85L and I187S were slightly compensatory in the 5C Nef background. Structural analysis showed that these positions are within 10 Å of each other. Compensatory mutations in amino acids that are spatial neighbors often indicate interaction. The effects of compensatory mutations can be either structural or functional, but the finding that most of our mutants appear to be stably expressed argues against broad structural effects. Together, our data suggest that these residues form a binding surface that participates in a protein-protein interaction important for Pak2 association.
Residues 85, 89, 187, 188, and 191 cluster on the surface of the Nef core domain in a region distant from the SH3-binding domain, the Nef dimerization region, and R106. Positions 85, 187, and 188 are within 6 to 8 Å of F191, while H89 is within 10 Å from F191 (Fig. 7A), so these residues are in close proximity. Residues 89 and 90 are on opposite faces of a ridge-like alpha helix that separates our cluster of residues from the RT loop interaction domain. This observation, together with the fact that MHC-I downregulation requires functional SH3 domain-mediated interactions, makes participation in RT loop or SH3 domain interactions unlikely. Therefore, H89, F191, and nearby residues are likely to participate in an as-yet-unidentified protein-protein interaction. Further inspection of this binding surface reveals both protruding and recessed hydrophobic molecules. F191 protrudes from the surface of Nef and has the freedom to move between two energetically favorable confomers (Fig. 7C). Surface exposure of highly hydrophobic molecules such as phenylalanine is unusual and often results in a high degree of entropy, suggesting it would be energetically favorable for this amino acid to participate in a binding interaction. The two confomers of F191 either expose or obscure a recessed A190. Exposure of A190 is of particular interest since it appears to form the bottom of a pocket at the center of hydrophobic amino acids F191, H89, and V85. A190 is conserved in 98% of 1,031 sequences from all clades of HIV-1 and SIV. Although A190 is likely to be important for structural integrity of the core domain, its position at the bottom of a hydrophobic pocket is suggestive of a binding site. Natural variation at position 85 is limited mostly to hydrophobic amino acids; in clade B, the most common are L (46.8%), V (37.6%), F (7.3%), and I (3.3%). The clustering of the protruding hydrophobic F191, the recessed A190, and L/V/F85 in a surface region important for association with Pak2 suggests that this surface could be a hydrophobic binding pocket involved in interaction with a critical member of the Nef-associated Pak2 activation complex.
Our findings, together with other studies, suggest there are at least two distinct regions of Nef critical for stability of the Nef-Pak2 activation complex: (i) the PxxP domain and (ii) a binding surface formed by residues 85, 89, 187, 189, and 191. Recent work demonstrated that R106, a residue previously proposed to be important for Pak2 activation, is not specifically involved in Pak2 activation, since even conservative mutations at this position impact multiple Nef functions (43). We favor a model in which Pak2, an SH3 domain-containing protein, and possibly another protein(s) simultaneously and/or cooperatively bind to Nef at these regions to facilitate activation of Pak2, consistent with previous work suggesting that Nef activates Pak2 through a multiprotein complex (4, 26, 33, 47). Pulkkinen et al. demonstrated that Pak2 activation occurs in two mechanistically distinct steps, association and activation, that are nearly concurrent within the Nef-Pak2 multiprotein complex (47). Association is required for Pak2 activation, but Pak2 activity promotes dissociation of the complex, rendering the complex transient. Evidence from studies of diverse HIV-1 and SIV Nefs indicates that Pak2 association may be a more conserved function than Pak2 activation (32, 47). Since the residues discussed in this study are important for Nef association with Pak2, their mutation would be expected to abolish activation of Pak2 as well. It will be important to identify the cellular factor that interacts with our proposed binding surface and determine what role that factor plays in Nef-Pak2 association and activation.
Of several proposed cellular interactors, only Pak2 and an SH3 domain-containing protein are known to be critical members of the Nef-Pak2 complex. At least one SH3 domain-containing protein must stabilize the association of Pak2 with Nef, and yet Pak2 itself does not contain an SH3 domain (24, 39). Previous work reported that the guanine exchange factor Vav, binding via the PxxP domain, stabilizes Nef-Pak2 association (15). However, other studies did not detect Vav in anti-Nef immune complexes (26). Other interacting molecules potentially important for Nef-mediated Pak2 activation include Rac, Cdc42, PIX, phosphatidylinositol-3-kinase (PI3K), DOCK2, and ELMO1 (9, 21, 26, 36, 37, 48, 50, 52, 65, 66). Further studies are necessary to elucidate which cellular proteins are critical for Nef-mediated Pak2 activation and their interactions within the Nef-Pak2 complex. Our identification of a potential hydrophobic binding surface on the Nef core specific for Pak2 association will facilitate these studies.
There has been considerable debate about the biological importance of Nef-mediated Pak2 activation in HIV infection. One study showed that HIV-1 Nef interaction with Pak2 correlates with enhanced virion infectivity in vitro, but interpretation of these results is confounded by the pleiotropic effects of the mutations used (myristoylation site, PxxP domain, and RR105LL mutations) (43, 64). Studies of chronically infected rhesus macaques infected with SIV mutated in the PxxP domain of Nef found that Nef-mediated activation of Pak2 correlated with induction of high viral loads and disease progression (30, 55). In contrast, SIVmac239 containing Nef mutation PxxP/AxxA caused AIDS and rapid death in two rhesus macaques without acquiring reversions (34). These findings raise the possibility that Pak2 activation may be more important during chronic infection, when infected T cells are more likely to be resting, than during rapid progression, when T cells have higher levels of activation. Wei et al. showed that Nef induces phosphorylation of merlin in primary human T cells (63). Merlin is a key protein in the Rac signaling pathway that is phosphorylated by Pak2. Nef mutations at F191 and S187 inhibited this effect, supporting a functional role for these residues in Pak2 activation and downstream signaling. Simmons et al. recently demonstrated Nef-mediated activation of Cdc42 and formation of a complex between Cdc42, PIX, and c-Cbl that displaces UbcH7 from lipid rafts (59). UbcH7 is involved in Cbl-mediated ubiquitination and negative regulation of T-cell signaling, so Nef-mediated disruption of Cbl activity is predicted to positively regulate T-cell signaling. Because PIX binds to Pak2 and Pak2 is activated by Cdc42, these findings shed light on possible roles of the Nef-Pak2 complex in T-cell activation. Nef mutants specifically defective for Pak2 activation, such as those analyzed in this study, will help to elucidate the contribution of Nef-mediated Pak2 activation to T-cell activation and HIV infection and pathogenesis, as well as the potential importance of the Nef-Pak2 interaction as a therapeutic target.
Acknowledgments
We thank J. Foster, R. Desrosiers, H. Gottlinger, M. Farzan, J. Sodroski, B. Appleton, A. Mehle, P. Ancuta, J. Wang, and P. Gorry for helpful discussions and technical advice. We thank J. Foster and E. O'Neill for sharing data prior to publication. We also thank D. Kabat for providing HIJ cells, H. Gottlinger for Jurkat T-antigen cells, M. Kirschner for pCS2-Cdc42-V12, and the NIH AIDS Reagent Program for Nef antibody no. 2949 (donated by R. Swanstrom).
This work was supported by NIH grant NS35734 to D.G. K.A. was supported in part by a fellowship from the Cancer Research Institute. B.L.W. and J.V.G. were supported by NIH grant AI33331 to J.V.G. Core facilities were supported by Center for AIDS Research and DFCI/Harvard Center for Cancer Research grants.
REFERENCES
- 1.Alexander, L., Z. Du, M. Rosenzweig, J. U. Jung, and R. C. Desrosiers. 1997. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 Nef alleles in lymphocyte activation. J. Virol. 71:6094-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arold, S., P. Franken, M. P. Strub, F. Hoh, S. Benichou, R. Benarous, and C. Dumas. 1997. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5:1361-1372. [DOI] [PubMed] [Google Scholar]
- 3.Arold, S. T., and A. S. Baur. 2001. Dynamic Nef and Nef dynamics: how structure could explain the complex activities of this small HIV protein. Trends Biochem. Sci. 26:356-363. [DOI] [PubMed] [Google Scholar]
- 4.Arora, V. K., R. P. Molina, J. L. Foster, J. L. Blakemore, J. Chernoff, B. L. Fredericksen, and J. V. Garcia. 2000. Lentivirus Nef specifically activates Pak2. J. Virol. 74:11081-11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagrodia, S., and R. A. Cerione. 1999. Pak to the future. Trends Cell Biol. 9:350-355. [DOI] [PubMed] [Google Scholar]
- 6.Baur, A. S., E. T. Sawai, P. Dazin, W. J. Fantl, C. Cheng-Mayer, and B. M. Peterlin. 1994. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity 1:373-384. [DOI] [PubMed] [Google Scholar]
- 7.Bour, S., and K. Strebel. 2000. HIV accessory proteins: multifunctional components of a complex system. Adv. Pharmacol. 48:75-120. [DOI] [PubMed] [Google Scholar]
- 8.Bresnahan, P. A., W. Yonemoto, S. Ferrell, D. Williams-Herman, R. Geleziunas, and W. C. Greene. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 8:1235-1238. [DOI] [PubMed] [Google Scholar]
- 9.Brown, A., X. Wang, E. Sawai, and C. Cheng-Mayer. 1999. Activation of the PAK-related kinase by human immunodeficiency virus type 1 Nef in primary human peripheral blood lymphocytes and macrophages leads to phosphorylation of a PIX-p95 complex. J. Virol. 73:9899-9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carl, S., A. J. Iafrate, S. M. Lang, N. Stolte, C. Stahl-Hennig, K. Mätz-Rensing, D. Fuchs, J. Skowronski, and F. Kirchhoff. 2000. Simian immunodeficiency virus containing mutations in N-terminal tyrosine residues and in the PxxP motif in Nef replicates efficiently in rhesus macaques. J. Virol. 74:4155-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, H., J. P. Hoxie, and W. P. Parks. 1999. The conserved core of human immunodeficiency virus type 1 Nef is essential for association with Lck and for enhanced viral replication in T-lymphocytes. Virology 264:5-15. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, R. H., and G. M. Bokoch. 1999. p21-activated protein kinase: a crucial component of morphological signaling? Trends Biochem. Sci. 24:350-355. [DOI] [PubMed] [Google Scholar]
- 13.Du, Z., P. O. Ilyinskii, V. G. Sasseville, M. Newstein, A. A. Lackner, and R. C. Desrosiers. 1996. Requirements for lymphocyte activation by unusual strains of simian immunodeficiency virus. J. Virol. 70:4157-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fackler, O. T., and A. S. Baur. 2002. Live and let die: Nef functions beyond HIV replication. Immunity 16:493-497. [DOI] [PubMed] [Google Scholar]
- 15.Fackler, O. T., W. Luo, M. Geyer, A. S. Alberts, and B. M. Peterlin. 1999. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol. Cell 3:729-739. [DOI] [PubMed] [Google Scholar]
- 16.Foster, J. L., R. P. Molina, T. Luo, V. K. Arora, Y. Huang, D. D. Ho, and J. V. Garcia. 2001. Genetic and functional diversity of human immunodeficiency virus type 1 subtype B Nef primary isolates. J. Virol. 75:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer, M., O. T. Fackler, and B. M. Peterlin. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorry, P. R., G. Bristol, J. A. Zack, K. Ritola, R. Swanstrom, C. J. Birch, J. E. Bell, N. Bannert, K. Crawford, H. Wang, D. Schols, E. De Clercq, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg, M., L. DeTulleo, I. Rapoport, J. Skowronski, and T. Kirchhausen. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8:1239-1242. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg, M. E., A. J. Iafrate, and J. Skowronski. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimsley, C. M., J. M. Kinchen, A. C. Tosello-Trampont, E. Brugnera, L. B. Haney, M. Lu, Q. Chen, D. Klingele, M. O. Hengartner, and K. S. Ravichandran. 2004. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 279:6087-6097. [DOI] [PubMed] [Google Scholar]
- 22.Grzesiek, S., A. Bax, J. S. Hu, J. Kaufman, I. Palmer, S. J. Stahl, N. Tjandra, and P. T. Wingfield. 1997. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 6:1248-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanna, Z., D. G. Kay, N. Rebai, A. Guimond, S. Jothy, and P. Jolicoeur. 1998. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell 95:163-175. [DOI] [PubMed] [Google Scholar]
- 24.Hiipakka, M., P. Huotari, A. Manninen, G. H. Renkema, and K. Saksela. 2001. Inhibition of cellular functions of HIV-1 Nef by artificial SH3 domains. Virology 286:152-159. [DOI] [PubMed] [Google Scholar]
- 25.Huang, Y., L. Zhang, and D. D. Ho. 1995. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 69:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janardhan, A., T. Swigut, B. Hill, M. P. Myers, and J. Skowronski. 2004. HIV-1 Nef binds the DOCK2-ELMO1 complex to activate Rac and inhibit lymphocyte chemotaxis. PLoS Biol. 2:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janvier, K., H. Craig, S. Le Gall, R. Benarous, J. Guatelli, O. Schwartz, and S. Benichou. 2001. Nef-induced CD4 downregulation: a diacidic sequence in human immunodeficiency virus type 1 Nef does not function as a protein sorting motif through direct binding to β-COP. J. Virol. 75:3971-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabat, D., S. L. Kozak, K. Wehrly, and B. Chesebro. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 68:2570-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kestler, H. W., III, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 30.Khan, I. H., E. T. Sawai, E. Antonio, C. J. Weber, C. P. Mandell, P. Montbriand, and P. A. Luciw. 1998. Role of the SH3-ligand domain of simian immunodeficiency virus Nef in interaction with Nef-associated kinase and simian AIDS in rhesus macaques. J. Virol. 72:5820-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 32.Kirchhoff, F., M. Schindler, N. Bailer, G. H. Renkema, K. Saksela, V. Knoop, M. C. Muller-Trutwin, M. L. Santiago, F. Bibollet-Ruche, M. T. Dittmar, J. L. Heeney, B. H. Hahn, and J. Munch. 2004. Nef proteins from simian immunodeficiency virus-infected chimpanzees interact with p21-activated kinase 2 and modulate cell surface expression of various human receptors. J. Virol. 78:6864-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krautkramer, E., S. I. Giese, J. E. Gasteier, W. Muranyi, and O. T. Fackler. 2004. Human immunodeficiency virus type 1 Nef activates p21-activated kinase via recruitment into lipid rafts. J. Virol. 78:4085-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang, S. M., A. J. Iafrate, C. Stahl-Hennig, E. M. Kuhn, T. Nisslein, F. J. Kaup, M. Haupt, G. Hunsmann, J. Skowronski, and F. Kirchhoff. 1997. Association of simian immunodeficiency virus Nef with cellular serine/threonine kinases is dispensable for the development of AIDS in rhesus macaques. Nat. Med. 3:860-865. [DOI] [PubMed] [Google Scholar]
- 35.Lee, C. H., K. Saksela, U. A. Mirza, B. T. Chait, and J. Kuriyan. 1996. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell 85:931-942. [DOI] [PubMed] [Google Scholar]
- 36.Linnemann, T., Y. H. Zheng, R. Mandic, and B. M. Peterlin. 2002. Interaction between Nef and phosphatidylinositol-3-kinase leads to activation of p21-activated kinase and increased production of HIV. Virology 294:246-255. [DOI] [PubMed] [Google Scholar]
- 37.Lu, X., X. Wu, A. Plemenitas, H. Yu, E. T. Sawai, A. Abo, and B. M. Peterlin. 1996. CDC42 and Rac1 are implicated in the activation of the Nef-associated kinase and replication of HIV-1. Curr. Biol. 6:1677-1684. [DOI] [PubMed] [Google Scholar]
- 38.Mangasarian, A., V. Piguet, J. K. Wang, Y. L. Chen, and D. Trono. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J. Virol. 73:1964-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manninen, A., M. Hiipakka, M. Vihinen, W. Lu, B. J. Mayer, and K. Saksela. 1998. SH3-domain binding function of HIV-1 Nef is required for association with a PAK-related kinase. Virology 250:273-282. [DOI] [PubMed] [Google Scholar]
- 40.Miller, M. D., M. T. Warmerdam, K. A. Page, M. B. Feinberg, and W. C. Greene. 1995. Expression of the human immunodeficiency virus type 1 (HIV-1) nef gene during HIV-1 production increases progeny particle infectivity independently of gp160 or viral entry. J. Virol. 69:579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Northrop, J. P., K. S. Ullman, and G. R. Crabtree. 1993. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J. Biol. Chem. 268:2917-2923. [PubMed] [Google Scholar]
- 42.Nunn, M. F., and J. W. Marsh. 1996. Human immunodeficiency virus type 1 Nef associates with a member of the p21-activated kinase family. J. Virol. 70:6157-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill, E., L. S. Kuo, J. F. Krisko, D. R. Tomchick, J. V. Garcia, and J. L. Foster. 2006. Dynamic evolution of the human immunodeficiency virus type 1 pathogenic factor, Nef. J. Virol. 80:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piguet, V., F. Gu, M. Foti, N. Demaurex, J. Gruenberg, J. L. Carpentier, and D. Trono. 1999. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell 97:63-73. [DOI] [PubMed] [Google Scholar]
- 45.Piguet, V., O. Schwartz, S. Le Gall, and D. Trono. 1999. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol. Rev. 168:51-63. [DOI] [PubMed] [Google Scholar]
- 46.Piguet, V., L. Wan, C. Borel, A. Mangasarian, N. Demaurex, G. Thomas, and D. Trono. 2000. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulkkinen, K., G. H. Renkema, F. Kirchhoff, and K. Saksela. 2004. Nef associates with p21-activated kinase 2 in a p21-GTPase-dependent dynamic activation complex within lipid rafts. J. Virol. 78:12773-12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reiser, J., G. Harmison, S. Kluepfel-Stahl, R. O. Brady, S. Karlsson, and M. Schubert. 1996. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc. Natl. Acad. Sci. USA 93:15266-15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renkema, G. H., A. Manninen, D. A. Mann, M. Harris, and K. Saksela. 1999. Identification of the Nef-associated kinase as p21-activated kinase 2. Curr. Biol. 9:1407-1410. [DOI] [PubMed] [Google Scholar]
- 50.Renkema, G. H., A. Manninen, and K. Saksela. 2001. Human immunodeficiency virus type 1 Nef selectively associates with a catalytically active subpopulation of p21-activated kinase 2 (PAK2) independently of PAK2 binding to Nck or β-PIX. J. Virol. 75:2154-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renkema, G. H., and K. Saksela. 2000. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front. Biosci. 5:D268-D283. [DOI] [PubMed] [Google Scholar]
- 52.Sanui, T., A. Inayoshi, M. Noda, E. Iwata, J. V. Stein, T. Sasazuki, and Y. Fukui. 2003. DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood 102:2948-2950. [DOI] [PubMed] [Google Scholar]
- 53.Sawai, E. T., A. Baur, H. Struble, B. M. Peterlin, J. A. Levy, and C. Cheng-Mayer. 1994. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc. Natl. Acad. Sci. USA 91:1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawai, E. T., A. S. Baur, B. M. Peterlin, J. A. Levy, and C. Cheng-Mayer. 1995. A conserved domain and membrane targeting of Nef from HIV and SIV are required for association with a cellular serine kinase activity. J. Biol. Chem. 270:15307-15314. [DOI] [PubMed] [Google Scholar]
- 55.Sawai, E. T., I. H. Khan, P. M. Montbriand, B. M. Peterlin, C. Cheng-Mayer, and P. A. Luciw. 1996. Activation of PAK by HIV and SIV Nef: importance for AIDS in rhesus macaques. Curr. Biol. 6:1519-1527. [DOI] [PubMed] [Google Scholar]
- 56.Schindler, M., J. Münch, M. Brenner, C. Stahl-Hennig, J. Skowronski, and F. Kirchhoff. 2004. Comprehensive analysis of Nef functions selected in simian immunodeficiency virus-infected macaques. J. Virol. 78:10588-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrager, J. A., and J. W. Marsh. 1999. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc. Natl. Acad. Sci. USA 96:8167-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simmons, A., V. Aluvihare, and A. McMichael. 2001. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 14:763-777. [DOI] [PubMed] [Google Scholar]
- 59.Simmons, A., B. Gangadharan, A. Hodges, K. Sharrocks, S. Prabhakar, A. Garcia, R. Dwek, N. Zitzmann, and A. McMichael. 2005. Nef-mediated lipid raft exclusion of UbcH7 inhibits Cbl activity in T cells to positively regulate signaling. Immunity 23:621-634. [DOI] [PubMed] [Google Scholar]
- 60.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, J. K., E. Kiyokawa, E. Verdin, and D. Trono. 2000. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc. Natl. Acad. Sci. USA 97:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, B. L., V. K. Arora, J. L. Foster, D. L. Sodora, and J. V. Garcia. 2003. In vivo analysis of Nef function. Curr. HIV Res. 1:41-50. [DOI] [PubMed] [Google Scholar]
- 63.Wei, B. L., V. K. Arora, A. Raney, L. S. Kuo, G.-H. Xiao, E. O'Neill, J. R. Testa, J. L. Foster, and J. V. Garcia. 2005. Activation of p21-activated kinase 2 by human immunodeficiency virus type 1 Nef induces merlin phosphorylation. J. Virol. 79:14976-14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiskerchen, M., and C. Cheng-Mayer. 1996. HIV-1 Nef association with cellular serine kinase correlates with enhanced virion infectivity and efficient proviral DNA synthesis. Virology 224:292-301. [DOI] [PubMed] [Google Scholar]
- 65.Wolf, D., V. Witte, B. Laffert, K. Blume, E. Stromer, S. Trapp, P. d'Aloja, A. Schürmann, and A. S. Baur. 2001. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 7:1217-1224. [DOI] [PubMed] [Google Scholar]
- 66.Wu, H., and Z. X. Wang. 2003. The mechanism of p21-activated kinase 2 autoactivation. J. Biol. Chem. 278:41768-41778. [DOI] [PubMed] [Google Scholar]
- 67.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]