Abstract
Cyclin-dependent kinase inhibitors, such as the mammalian p27Kip1 protein, regulate correct cell cycle progression and the integration of developmental signals with the core cell cycle machinery. These inhibitors have been described in plants, but their function remains unresolved. We have isolated seven genes from Arabidopsis that encode proteins with distant sequence homology with p27Kip1, designated Kip-related proteins (KRPs). The KRPs were characterized by their domain organization and transcript profiles. With the exception of KRP5, all presented the same cyclin-dependent kinase binding specificity. When overproduced, KRP2 dramatically inhibited cell cycle progression in leaf primordia cells without affecting the temporal pattern of cell division and differentiation. Mature transgenic leaves were serrated and consisted of enlarged cells. Although the ploidy levels in young leaves were unaffected, endoreduplication was suppressed in older leaves. We conclude that KRP2 exerts a plant growth inhibitory activity by reducing cell proliferation in leaves, but, in contrast to its mammalian counterparts, it may not control the timing of cell cycle exit and differentiation.
INTRODUCTION
Growth is one of the most studied phenomena in multicellular organisms. It has become clear that the process of cell division plays a crucial role in the mechanisms by which higher organisms achieve appropriate development of their organs. The cell division cycle is controlled by a molecular machinery that ensures the fidelity of DNA replication and that responds to signals from both the external environment and intrinsic developmental programs. A central role in the regulation of the cell cycle is played by the cyclin-dependent kinases (CDKs). CDK activity is controlled by a variety of mechanisms, including binding to cyclins (for review, see Pines, 1994) and phosphorylation of the Thr-161 (or an equivalent) residue by the CDK-activating kinase (for review, see Dunphy, 1994).
Active cyclin/CDK complexes can be inhibited in different ways. The phosphorylation of the Thr-14 and Tyr-15 residues interferes with the correct binding of the cofactor ATP and, therefore, inhibits CDK activity (Dunphy, 1994). Indirectly, kinase activity also is inhibited by the controlled degradation of cyclin subunits (for review, see Peters, 1998). Recently, another mechanism of the negative regulation of CDK activity has become evident. A family of mainly low- molecular-weight proteins, named CDK inhibitors (CKIs), inhibit CDK activity by tight association with the cyclin/CDK complexes (for review, see Sherr and Roberts, 1995, 1999). In mammals, two different CKI families can be distinguished on the basis of their mode of action and sequence similarity: the INK4 and the Kip/Cip families.
The Kip/Cip family comprises three gene products: p21Cip1, p27Kip1, and p57Kip2. These CKIs bind to all known G1/S-specific CDKs (Toyoshima and Hunter, 1994; Lee et al., 1995). The Kip/Cip CKIs are involved in both checkpoint control and the regulation of cell cycle exit preceding differentiation. The former function is illustrated by the observed association of p21Cip1 with CDKs in a p53-dependent manner upon the occurrence of DNA damage, inhibiting replication but still allowing DNA repair (Dulić et al., 1994; Smith et al., 1994). A role of the CKIs in cell differentiation is seen during muscle development. Mice lacking both p21Cip1 and p57Kip2 display severe defects in skeletal muscle development because of prolonged proliferation and inhibited differentiation (Zhang et al., 1999). Moreover, p27Kip1 has been implicated as a mediator of various antimitogenic stimuli (Kato et al., 1994; Nourse et al., 1994; Polyak et al., 1994). Kip1 nullizygous mice are significantly larger than control mice because of an increase in the number of cells, suggesting that the absence of p27Kip1 might allow continued cell proliferation in the presence of antimitogenic signals (Fero et al., 1996; Nakayama et al., 1996).
A novel function for the Kip/Cip CKIs has been revealed by the observation that p21Cip1 and p27Kip1 associate with active cyclin D/CDK4 complexes (LaBaer et al., 1997). Not only are the cyclin D/CDK4 complexes inert toward the inhibitory function of the Kip/Cip proteins, but their activation is stimulated by the CKIs (Cheng et al., 1999). Because the Kip/Cip proteins contain interaction sites with both cyclin D and CDK subunits, they help assemble the cyclin D/CDK complexes. In addition, the CKIs direct the cyclin D/CDK complexes to the nucleus, where they are phosphorylated by the CDK-activating kinase.
In plants, two major groups of CDKs have been studied: the A-type and B-type CDKs (Mironov et al., 1999). The A-type CDKs, represented by CDKA;1 (previously designated CDC2aAt; Joubès et al., 2000) in Arabidopsis, show kinase activity during the S, G2, and M phases of the cell cycle. In contrast, the activity of B-type CDKs, represented by CDKB1;1 (previously designated CDC2bAt) in Arabidopsis, is linked prominently to mitosis (Magyar et al., 1997; our unpublished results). These data indicate that A-type CDKs regulate both the G1-to-S and G2-to-M transitions, whereas the B-type CDKs regulate the G2-to-M transition only. Down- regulation of A-type CDK activity in plants does not affect the relative duration of G1 and G2. In contrast, plants with reduced B-type CDK activity have an increased duration of G2 (Hemerly et al., 1995; our unpublished results).
To date, only two structurally related CKI-like molecules have been described for plants, ICK1 and ICK2 (Wang et al., 1997; Lui et al., 2000). Interestingly, ICK1 was demonstrated to be twofold to threefold transcriptionally induced upon abscisic acid treatment, suggesting that this CKI might be responsible for the growth inhibitory effect of abscisic acid (Wang et al., 1998). Despite their low sequence identity with the mammalian Kip/Cip inhibitors, recombinant ICK1 and ICK2 proteins inhibited CDK activity in an in vitro assay (Wang et al., 1997; Lui et al., 2000). Recently, the CDK inhibitory activity of ICK1 was demonstrated by its overproduction in Arabidopsis, resulting in dwarf plants with a reduced number of cells that were much larger than those of wild-type plants (Wang et al., 2000).
Here we report the existence of five additional CKI genes in Arabidopsis. We show that (1) the various CKI genes are expressed differentially, whereas their gene products, with the exception of the Kip-related protein 5, all bind the same CDK; (2) the reduced cell number and size of CKI transgenic plants are a consequence of inhibited cell division and leaf expansion rates, whereas the temporal pattern of development is unaffected; and (3) the rate of endoreduplication is inhibited as well.
RESULTS
Cloning of p27Kip1-Like Genes from Arabidopsis
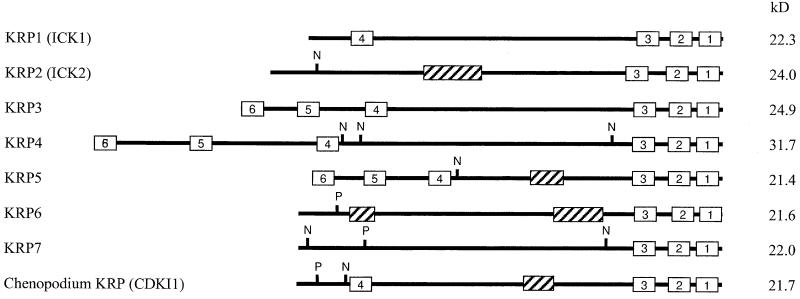
To investigate the presence and function of plant CKIs, a two-hybrid screen was performed with the Arabidopsis CDKA;1. Among the interacting clones, three putative CKIs were identified, two of which were identical to the ICK1 and ICK2 genes described previously (Wang et al., 1997; Lui et al., 2000). In parallel, we identified four related genes in the Arabidopsis genomic databases, of which the cDNAs were isolated by reverse transcriptase–mediated polymerase chain reaction (PCR). Because all proteins showed sequence identity to the mammalian p27Kip1 CKI in their C-terminal domains, we propose to name these proteins Kip-related proteins (KRPs) with the numbers 1 to 7, where KRP1 and KRP2 correspond to ICK1 and ICK2, respectively. Overall, the KRPs have no significant sequence identity to any proteins in the databases. They also display only low sequence identity to each other. However, a detailed analysis allowed us to identify small sequence elements shared by different KRP members (Figure 1, Table 1).
Figure 1.
Structural Organization of KRP1, KRP2, KRP3, KRP4, KRP5, KRP6, KRP7, and Chenopodium KRP.
Conserved sequence boxes are indicated (1 to 6). N, nuclear localization signal; P, CDK consensus phosphorylation site; striped boxes, PEST domains. The predicted molecular masses (kD) are indicated at right.
Table 1.
Conserved Motifs in the Plant KRPs
| Protein | Motif 1 | Motif 2 | Motif 3 | Motif 4 | Motif 5 | Motif 6 |
|---|---|---|---|---|---|---|
| KRP1 | 180-PLEGRYEW | 167-FKKKYNFD | 151-EIEDFFVEAE | 20-YMQLRSRR | ||
| KRP2 | 197-LGGGRYEW | 183-CSMKYNFD | 164-ELEDFFQVAE | |||
| KRP3 | 210-PLSGRYEW | 197-FMEKYNFD | 181-EMEEFFAYAE | 58-YLQLRSRR | 26-SPGVRTRA | 1-MGKYMKKSK |
| KRP4 | 274-PLPGRFEW | 261-FIEKYNFD | 245-EMDEFFSGAE | 102-YLQLRSRR | 44-SLGVLTRA | 1-MGKYIRKSK |
| KRP5 | 177-PLPGRYEW | 164-FIQKYNFD | 148-EIEDFFASAE | 54-YLQLRSRR | 24-ALGFRTRA | 1-MGKYIKKSK |
| KRP6 | 186-PLEGRYKW | 173-FIEKYNFD | 155-EIEDLFSELE | |||
| KRP7 | 183-PLEGRYQW | 170-FTEKYNYD | 154-ELDDFFSAAE | |||
| CDKI1a | 184-PLKGRYDW | 171-FSEKYNFD | 155-EIEEFFAVAE | 25-IPQLRSRR |
a Chenopodium KRP.
Three domains located at the extremity of the C-terminal part of the proteins are shared by all KRPs and are found in a CKI-like molecule from Chenopodium rubrum. Only the two last motifs are present in mammalian CKIs such as p27Kip1. The other three domains are not common among all of the KRPs but were identified in at least two different members of the KRP family. A motif 3–related sequence (ELDEFFAEAE) is shared with the erythronolide synthase from Streptomyces erythraeus, and a motif 6–related sequence (SLGFLTRA) is located in the TPR1 protein of fission yeast, which is involved in potassium transport. However, the significance of these motifs is unknown. Motifs 4 and 6 share no similarity with any protein in the databases. In addition to the motifs mentioned above, KRP2, KRP5, and KRP6 all have PEST domains (PESTfind score > +10), which are polypeptide sequences enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) residues that serve as proteolytic signals (Rogers et al., 1986) (Figure 1). Furthermore, KRP2, KRP4, KRP5, and KRP7 carry at least one nuclear localization signal (Figure 1). Finally, KRP6 and KRP7 can be distinguished from other KRPs by the presence of a consensus CDK phosphorylation site (S/TPXK/R) in their N-terminal domains (Figure 1).
KRPs Do Not Bind the Mitotic CDKB1;1
KRP1 and KRP2 have been shown to interact with CDKA;1 but not with CDKB1;1 in a two-hybrid system (Lui et al., 2000). We tested the binding specificity for all identified KRPs toward the same CDKs. Plasmids that encode the CDKA;1 or CDKB1;1 protein fused to the GAL4 DNA binding domain (GAL4-BD) were cotransformed in a yeast reporter strain with vectors that encode a fusion protein between the different KRPs and the GAL4 activation domain (GAL4-AD). Transformants were streaked on medium lacking histidine, because the yeast reporter strain would grow only in the absence of histidine when the proteins interacted. As summarized in Table 2, all KRPs, except KRP5 and KRP6, interacted with CDKA;1, and none interacted with CDKB1;1. In contrast, the CDK docking factor CKS1At interacted with both CDKs, demonstrating that CDKB1;1 was functional in our two-hybrid assay. In a reciprocal two-hybrid experiment in which the KRPs and CDKs were cloned as fusion proteins with the GAL4-BD and GAL4-AD, respectively, KRP6 also interacted with CDKA;1 (Table 2). This interaction might be attributable to the fusion between KRP6 and the GAL4-BD being more stable than that with the GAL4-AD. The KRP1, KRP2, and KRP3 proteins could not be used in this assay because the fusion of these proteins with the GAL4-BD activated the reporter gene in the absence of a prey. Thus, except for KRP5, all KRPs bound specifically with CDKA;1.
Table 2.
Interaction of KRPs with CDKA;1, CDKB1;1, and CYCD4
| CDKA;1
|
CDKB1;1
|
CYCD4
|
|||
|---|---|---|---|---|---|
| Prey/Baita | GAL-BD | GAL-AD | GAL-BD | GAL-AD | GAL-AD |
| KRP1 | + | − | |||
| KRP2 | + | − | |||
| KRP3 | + | − | |||
| KRP4 | + | + | − | − | + |
| KRP5 | − | − | − | − | + |
| KRP6 | − | + | − | − | + |
| KRP7 | + | + | − | − | + |
| CKS1At | + | + | + | + | − |
Depending on the CDK-containing construct.
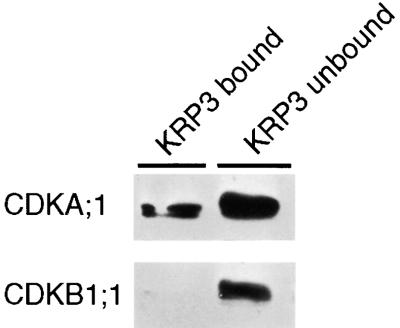
For KRP3, the binding specificity toward CDKA;1 was confirmed by an in vitro assay. The recombinant KRP3 protein was synthesized in Escherichia coli and purified to homogeneity. The purified protein was coupled to Sepharose beads, which were used to make an affinity column. To this column, total Arabidopsis cell suspension extracts were applied. Both the unbound and bound protein fractions were separated subsequently by electrophoresis and transferred to membranes. Afterward, these membranes were probed with CDKA;1- and CDKB1;1-specific antibodies. As can be seen in Figure 2, CDKA;1 was detected in the flow-through fraction, but a significant amount also was bound to the column, indicating again a stable interaction between KRP3 and CDKA;1. In contrast, the CDKB1;1 protein was detected only in the flow-through fraction.
Figure 2.
In Vitro CDKA;1 Binding by KRP3.
Protein extracts of 3-day-old cell suspensions of Arabidopsis were loaded onto a KRP3-Sepharose column, and bound and unbound fractions were tested for the presence of CDKA;1 or CDKB1;1 with specific antibodies.
The mammalian Cip/Kip family members interact with both the CDK and cyclin subunits. Accordingly, the KRP1 protein was shown to interact with the D-type cyclin CYCD3 from Arabidopsis (Wang et al., 1998). Here we tested the interaction of the KRPs with CYCD4 (De Veylder et al., 1999). As seen in Table 2, all KRPs tested, including KRP5, interacted with this particular cyclin. These results show that the inability of KRP5 to interact with CDKA;1 was not caused by the instability of the protein in yeast and suggest that KRP5 binds to a yet-unidentified CDK from Arabidopsis that can make a complex with CYCD4.
The KRP Genes Are Expressed Differentially in Plant Tissues
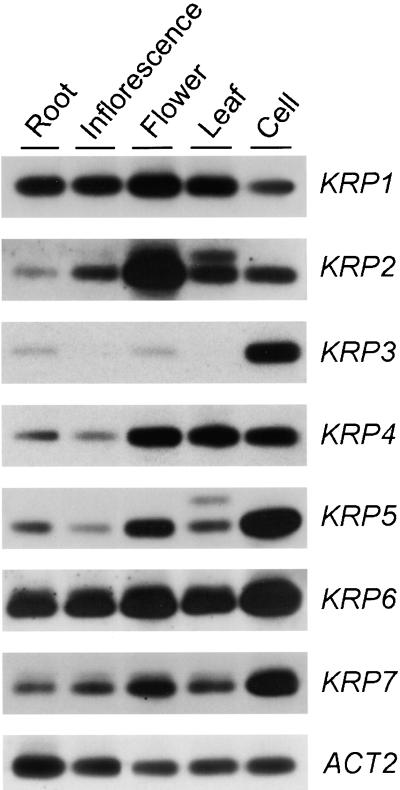
The expression of the different KRP genes in various plant organs (roots, inflorescence stems, flower buds, and 3-week-old leaves) and in a 3-day-old, actively dividing suspension culture was studied by semiquantitative reverse transcriptase–mediated PCR because not all of the genes were expressed strongly enough to be detected by classic RNA gel blotting. PCR was performed with a specific primer set for each of the KRPs (see Methods). Amplification with actin 2–specific primers was used to ensure equal input of cDNA for each sample. As can be seen in Figure 3, only KRP1 and KRP6 were expressed ubiquitously. KRP4, KRP5, and KRP7 were detected in all tissues, but mRNA clearly was more abundant in the tissues that displayed high mitotic activity (flowers and suspension cultures), with KRP4 also being abundantly present in leaves. KRP2 was most abundant in flowers. Remarkably, the expression of KRP3 was particularly high in actively dividing suspension cultures but was not, or was barely (in roots and flowers), detectable in intact plant organs. The organ-specific expression patterns of KRP1 and KRP2 corresponded with those reported previously by Lui et al. (2000), although minor differences in the amount of transcript accumulation were observed that may be explained by differences in time of harvesting the plant material. Together, the transcription profiles suggest that the various KRPs might play distinct roles in plant development.
Figure 3.
Differential Expression of KRP Genes in Various Arabidopsis Organs and a 3-Day-Old Cell Suspension Culture.
cDNA prepared from the indicated organs and the suspension cell culture were subjected to semiquantitative reverse transcriptase–mediated PCR analysis using gene-specific primers (see Methods). The actin 2 gene (ACT2) was used as a loading control.
Inhibition of CDKA;1 Activity by KRPs in Yeast
To test the CKI activity of the KRPs in vivo, a genetic experiment in fission yeast was designed. In this assay, CDK inhibition was evaluated as the potential to convert prematurely dividing cells into noncycling cells. The assay is illustrated here for the KRP2 protein, but similar results were obtained for the other KRP proteins tested (KRP1 and KRP4). As illustrated in Figure 4, yeast cells that expressed the KRP2 gene exhibited no growth abnormalities or changes in cell morphology compared with control cells (Figures 4A, 4B, and 4E), demonstrating that the expression of KRP2 did not inhibit the cell cycle of yeast. This result is not surprising, because the KRP2 protein is entirely unrelated to any of the known CKIs from yeast. In contrast, cells that expressed a dominant positive allele of the CDKA;1 gene (CDKA;1.A14F15) divided prematurely, resulting in cells being slightly smaller than control cells (Figures 4A, 4C, and 4E). It has been demonstrated previously that this reduced cell size results from an increase of the total CDK activity in the yeast cells (Porceddu et al., 1999) and that CDKA;1.A14F15 is expected to sequester the majority of the yeast cyclins. However, when the mutant CDKA;1 allele was coexpressed with KRP2, cells were much longer than those of the wild type (Figures 4D and 4E), indicating cell cycle arrest. The most probable explanation for this cell cycle arrest is the conversion by the KRP2 protein of the dominant positive form of CDKA;1 into an inactive form. In any case, these data show indirectly the potential of KRP2 to inhibit specifically plant CDK activity.
Figure 4.
KRP2 Inhibition of Advanced Mitosis in Yeast Caused by the CDKA;1.A14F15 Gene.
(A) Wild-type fission yeast cells transformed with empty control vectors.
(B) Yeast cells expressing the KRP2 gene.
(C) Yeast cells expressing the dominant positive CDKA;1.A14F15 gene.
(D) Yeast cells coexpressing the CDKA;1.A14F15 and KRP2 genes.
(E) Protein gel blot analysis of protein levels of CDKA;1 and KRP2 in the cells from (A) to (D).
Bars in (A) to (D) = 30 μm.
KRP Overexpression in Plants
To analyze the roles of the KRPs in plant development, we generated transgenic plants containing the KRP1, KRP2, KRP3, or KRP4 gene under the control of the constitutive cauliflower mosaic virus 35S promoter. For KRP4, no transgenic lines could be obtained, although several independent transformations were undertaken. In contrast, numerous independent transgenic lines were generated for KRP1, KRP2, and KRP3. Here we report in detail the phenotype of the KRP2-overproducing lines.
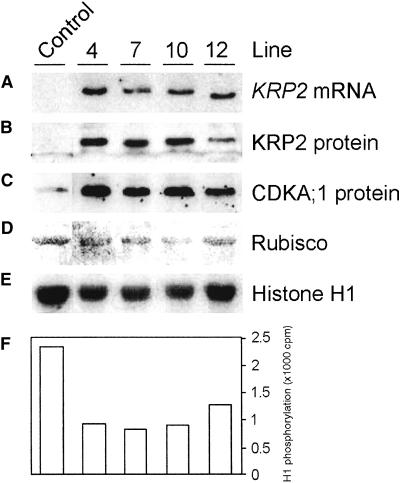
Thirty-nine lines were generated in which the levels of the KRP2 mRNA and the KRP2 protein exceeded those found in untransformed plants (Figures 5A and 5B). The presence of the KRP2 protein correlated with a decrease in extractable CDK activity (Figures 5E and 5F). Remarkably, the overproduction of KRP2 led to an increase in CDKA;1 protein level (Figure 5C). No change in the CDKA;1 mRNA level was observed (data not shown), indicating that overproduction of the KRP2 protein may stabilize the CDKA;1 protein.
Figure 5.
KRP2 Transgene Expression and CDK Histone H1 Activity in Untransformed and Four Independent Transgenic Arabidopsis Plants.
(A) KRP2 mRNA levels.
(B) KRP2 protein levels.
(C) CDKA;1 protein levels.
(D) Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) pro-tein levels visualized by Ponceau S staining (loading control).
(E) CDK histone H1 activity bound to p10CKS1At beads.
(F) Quantification of signals in (E).
KRP2-overproducing plants had narrower leaves than did wild-type plants. As reported for the overproduction of KRP1 (Wang et al., 2000), the leaves of KRP2-overproducing plants were distinctly serrated (Figures 6A to 6D). In addition, sepals and petals were modified in size, and flowers were partially male sterile. Furthermore, the number of lateral roots appeared to be reduced by ∼50%. There was no obvious effect on stem size, root elongation rate, and hypocotyl length (data not shown). Because the leaf phenotype was the most striking feature, we focused on this organ.
Figure 6.
Phenotypic Analysis of KRP2-Overproducing Lines.
(A) Four-week-old soil-grown control plant. The inflorescence was removed to visualize the rosette leaves.
(B) KRP2-overproducing plant with removed inflorescence.
(C) Fifth leaf of a control plant grown in vitro.
(D) Fifth leaf of a KRP2-overproducing plant.
(E) Adaxial epidermal cells of the fifth leaf of a control plant.
(F) Adaxial epidermal cells of a KRP2-overproducing plant.
(G) Palisade cells of the fifth leaf of a control plant.
(H) Palisade cells of a KRP2-overproducing plant.
Bars in (A) and (B) = 2 mm; bars in (C) and (D) = 5 mm; bars in (E) to (H) = 50 μm.
In the T2 population, the leaf phenotype segregated strictly with the presence and expression of the transgene. The number of leaves was unchanged (per wild-type plant, the mean was 7.25 ± 0.85 [n = 139] compared with 7.28 ± 1.06 [n = 137] and 7.54 ± 1.03 [n = 196] in two independent KRP2 transgenic plants). Microscopic analysis of mature fifth leaves revealed that all tissue layers in leaves of the KRP2-overproducing lines had much larger cells than did those of control plants, as illustrated for the adaxial epidermis (Figures 6E and 6F) and palisade parenchyma (Figures 6G and 6H). For the adaxial epidermis, the mean cell area was increased almost sixfold, from 599 ± 155 μm2 in the wild-type plants to 3334 ± 1538 μm2 in the transgenic plants. Palisade cell area was threefold larger (from 357 ± 31 to 1210 ± 572 μm2). Similarly, larger cells were observed in the spongy parenchyma and the abaxial epidermis. No difference in cell size was observed for stomata and trichomes, although occasionally enlarged stomata were seen in the cotyledons (data not shown). Transverse sections through the central part of the first leaf revealed that, in addition to having larger areas, all cell types were enlarged conspicuously in the dorsoventral direction (Figure 7). The number of cell layers, however, was unaffected by the transgene, and consequently, KRP2 leaves were somewhat thicker than wild-type leaves (112.48 ± 18.56 μm in KRP2-overproducing lines compared with 89.23 ± 27.17 μm [n = 4] in wild-type plants).
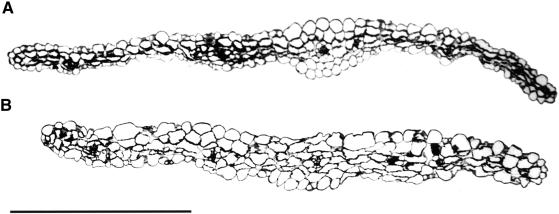
Figure 7.
Transverse Sections through the Central Part of the First Leaf.
(A) Sixteen-day-old wild-type leaf grown in vitro.
(B) Sixteen-day-old KRP2-overproducing leaf grown in vitro.
Bar = 500 μm.
To gain insight into the effect of KRP2 overexpression on leaf development and cell cycle duration, a kinematic analysis was performed on the first two initiated leaves of plants grown in vitro. We measured cell size and stomatal index on positions 25 and 75% from the base to the tip of the leaf, which, in addition to giving better estimates for the average of the leaf, also allowed detection of the presence of developmental gradients along the length of the blade. The average of the cell areas in these two positions was used in combination with the measured total leaf area to estimate total cell number per leaf. To validate this approach, we took a number of 9-day-old leaves of both wild-type and KRP2-overproducing plants that showed strong basipetal developmental gradients in cell size and stomatal complexes, indicating that they were approximately at the end of their meristematic development. The entire abaxial epidermis of these leaves was drawn on paper with the aid of a drawing tube. On these images, we manually counted the number of epidermal cells and subsequently estimated this number by extrapolating the average cell size at the two reference positions to the whole leaf area. This comparison showed no significant difference between estimated and counted numbers for either genotype (the estimates were 7 ± 4% and 1 ± 10% [average ± se; n = 5 and 3] higher than the actual counts for ecotype Columbia and KRP2-overproducing plants, respectively).
We used leaves 1 and 2 because they are nearly indistinguishable and probably are best synchronized among replicate plants. From day 5 until day 21 after sowing, leaves of transgenic and wild-type plants were harvested and leaf size and number, size of the abaxial epidermal cells, and stomatal index were determined (see Methods). As can be seen from the linear increase on the logarithmic plot, the leaf area expanded exponentially until day 11 in both wild-type and transgenic lines, after which expansion rates decreased; the mature size was reached approximately 3 weeks after sowing (Figure 8A). Clearly, the duration of expansion was unaffected by the transgene. At maturity, the areas of leaves of the transgenic line were only 25% of those of wild-type leaves (4 and 16 mm2, respectively). At day 5, however, the primordia of the transgenic lines were, if anything, slightly larger than those of wild-type lines. Thus, the differences in mature leaf size were caused entirely by differences in leaf expansion rates.
Figure 8.
Kinematic Analysis of Leaf Growth of the First Leaf Pair of Wild-Type (Col-0) and KRP2-Overproducing (35S-KRP2) Plants.
(A) Leaf blade area.
(B) Epidermal cell number on the abaxial side of the leaf.
(C) Average cell division rates of the epidermal cells on the abaxial side of the leaf.
(D) Epidermal cell size on the abaxial side of the leaf.
The number of abaxial epidermal cells in mature leaves also increased exponentially (Figure 8B). In mature leaves of KRP2-overproducing plants, 10-fold fewer cells were present than in wild-type leaves, whereas at day 5 the cell number differed only slightly. Average cell division rates for the whole leaf could be calculated as exponential increase of the cell number (Figure 8C). Analogous to expansion of the leaf, the period in which cell division occurred was not influenced by KRP2 overexpression; cell division rates were approximately constant until 9 days after sowing, then they decreased rapidly during the next 4 to 5 days in both control and transgenic lines. Average cell cycle duration, the time between one phase of mitosis and the next, can be estimated as the inverse of cell division rate. Between days 5 and 9, the average cell cycle duration more than doubled from 20.7 hr in wild-type plants to 43.3 hr in KRP2-overproducing lines.
Average cell size depends on the balance between division and expansion rates (Green, 1976). In the control line, the average cell size was approximately constant during the entire period of cell division, from day 5 to day 9 (Figure 8D), indicating that cell division and expansion rates were balanced. After day 9, cell size increased exponentially because expansion continued in the absence of division. In contrast, in the KRP2-overproducing lines, the average cell size had already increased during the period of cell division, indicating that division rates were more inhibited than were expansion rates. From the start of the analysis at day 5, there was a large difference in cell size between the transgenic and control lines, which must originate from a developmental stage before those studied here.
In Arabidopsis, exit from cell division followed by cell enlargement and differentiation starts at the tip of the leaf and progresses subsequently in a basipetal direction (Pyke et al., 1991). To verify the validity of average cell division rates for the whole leaf and to analyze the effects of KRP2 overexpression on developmental timing, we determined the evolution of the stomatal index. Divisions that give rise to stomata are the final divisions that occur at a particular position (Donnelly et al., 1999), indicating the end of proliferative activity. The stomatal index was measured in both the tip and the base of the leaves (Figure 8E). The stomatal index remained close to zero until day 9 and then increased until ∼15 days after sowing. Stomata were first observed at the tip of the leaf, and the final index was reached a few days earlier, reflecting the basipetal direction of maturation. These data indicate that some cells started leaving the cell cycle 9 days after sowing and that cell division finally stopped at approximately day 15, corresponding well with the independently determined cell division data. The final stomatal index and its evolution were very similar in wild-type and transgenic lines, showing that KRP2 did not influence the timing or the degree of cell differentiation.
KRP2 Overproduction Suppresses Endoreduplication
Arabidopsis leaves undergo extensive endoreduplication, increase of DNA content by consecutive doubling of the genomic DNA in the absence of chromatin segregation, and cytokinesis. A positive correlation between DNA level and cell size has been established (Melaragno et al., 1993; Folkers et al., 1997). Because KRP2 overexpression results in enlarged cells, the ploidy levels of both wild-type and KRP2-overproducing lines were measured. Different seed stocks often germinate at different times, resulting in small differences in ploidy levels at the moment of analysis. To avoid this problem, we used heterozygous seed stocks of KRP2-overproducing lines. These seed stocks segregated for plants showing normal leaves (1:4) and the serrated leaf phenotype (3:4). Plants were considered as control or transgenic on the basis of their phenotypes. Ploidy levels were analyzed in three different leaves of 3-week-old plants, namely, the youngest leaf (leaf 5), the third leaf, and the oldest leaf (leaf 1). At that stage of development, the youngest leaf is still actively dividing, as seen by the presence of high CDK activity (data not shown), leaf 3 has stopped dividing and is expanding, and leaf 1 is fully expanded and differentiated (Figure 9).
Figure 9.
Ploidy Distribution Diagrams of Leaves of Wild-Type (WT) and KRP2-Overexpressing Lines.
Values are means of two independent measurements. Maximum differences found between two samples were 4.0, 3.0, and 2.0% for 2C, 4C, and 8C, respectively.
For the youngest leaf, no significant difference between the control and transgenic lines was observed. In wild-type leaves, the fraction of cells with a DNA content of 4C and 8C increased with leaf age at the cost of the 2C fraction because of endoreduplication (Figure 9). This result is in agreement with earlier reports (Galbraith et al., 1991; Jacqmard et al., 1999) demonstrating that the process of endoreduplication is regulated developmentally. In KRP2-overexpressing plants, this decrease was significantly slower, showing that, in addition to proliferation, KRP2 overexpression inhibits the endoreduplication cycle. It can be concluded that the observed cell enlargement occurred in the absence of enhanced endoreduplication.
DISCUSSION
By means of two-hybrid screening and database mining, we identified seven different genes from Arabidopsis with sequence identity to the mammalian CKI p21Kip1. Sequence similarity is restricted to a region of ∼25 amino acids located at the extreme C-terminal end of each KRP protein. The remaining parts of the KRPs are not significantly similar to those of any protein in the databases. The CDK inhibitory and binding domains of p27Kip1 consist of three structural domains: a β-hairpin, a β-strand, and a 310 helix (Russo et al., 1996). The first two structures associate with the N-terminal lobe of the CDKs, resulting in the destabilization of the ATP binding site. The 310 helix binds to the catalytic cleft and occupies the ATP binding site. The KRPs share only sequence identity along the β-hairpin and β-strand and lack the 310 helix. Nevertheless, CDK activity is inhibited by at least some of the KRPs in vitro (KRP1 and KRP2; Wang et al., 1997; Lui et al., 2000), in a yeast assay (KRP1, KRP2, and KRP4; this study), and in planta (KRP1, KRP2, and KRP3; Wang et al., 2000; this study), showing that the 310 helix domain is not required for CDK inhibition. This result is consistent with the observation that a truncated form of p27Kip1 that lacks the 310 helix still retains inhibitory activity (Polyak et al., 1994; Luo et al., 1995).
Why has Arabidopsis so many CKIs? One reason might be related to the diversity of spatial patterns of transcript accumulation, suggesting that KRPs could operate in a tissue-specific manner. Furthermore, differences in the domain organization suggest that the distinct KRPs have dissimilar biochemical properties. We identified distinct sequence motifs within the KRPs, of which only three are shared by all family members. Although the major CDK and cyclin binding domain of the KRPs have been mapped at the C-terminal domain (Wang et al., 1998), the additional motifs found in the N-terminal part of the KRPs could help to determine specificity toward different cyclin/CDK complexes. Alternately, these domains might interact with other unknown proteins. Kip/Cip family members are reported to interact with many other regulatory proteins, including the proliferating cell nuclear antigen (Chen et al., 1995), the coactivator of c-Jun JAB1 (Tomoda et al., 1999), and the syntaxin-like molecule CARB (McShea et al., 2000).
In addition to the different motifs found in the KRPs, they also can be distinguished by the presence (KRP2, KRP4, KRP5, and KRP7) or absence (KRP1, KRP3, and KRP6) of a nuclear localization signal, the presence (KRP2, KRP5, and KRP6) or absence (KRP1, KRP3, KRP4, and KRP7) of PEST domains, and the presence (KRP6 and KRP7) or absence (KRP1, KRP2, KRP3, KRP4, and KRP5) of a consensus CDK phosphorylation site. A CDK phosphorylation site also has been identified in p27Kip1. Phosphorylation triggers the degradation of p27Kip1 by the ubiquitin/proteasome-dependent pathway, allowing cells to enter the S phase (Vlach et al., 1997; Tomoda et al., 1999). Ubiquitin-dependent degradation of cell cycle proteins in plants has been reported previously (Genschik et al., 1998). In addition, a ubiquitin-ligase complex has been isolated from Arabidopsis (Gray et al., 1999), opening the possibility that the phosphorylation of KRP6 and KRP7 triggers their own destruction in a manner similar to that reported for p27Kip1.
The overexpression of KRP2 affected various organs differentially, the most obvious effect being a large reduction in leaf area. The ∼75% reduction in leaf area (Figure 8) was offset by an ∼30% increase in leaf thickness (Figure 7). Hence, leaf volume was reduced by approximately two-thirds in response to the transgene. The inhibitory effect on leaf area expansion was studied in detail for the KRP2-overproducing lines with a kinematic approach. This method, which has been used previously for dicotyledonous leaves (Granier and Tardieu, 1998), was adapted to the small size of Arabidopsis leaves. With this method, we found almost constant division rates in epidermal cells during the first 9 days after sowing, after which division stopped rapidly, a pattern similar to that observed in sunflower (Granier and Tardieu, 1998). Moreover, the average cell cycle duration of approximately 20 hr corresponds very well with that found in Arabidopsis root tip meristems growing under similar conditions (Beemster and Baskin, 1998).
In the KRP2-overproducing lines, a decrease of 50 to 60% in CDK activity was measured. Because CDK activity was shown to correlate linearly with growth rates (Granier et al., 2000), KRP2-overproducing plants were expected to grow twice as slowly as controls, which is exactly what we observed: an increase of the cell cycle duration from 20 to 43.3 hr. Whereas the timing of the duration of division and expansion during development remained unaltered, mature leaves showed a more than 10-fold reduced epidermal cell number compared with wild-type plants. Because the number of cell layers did not vary (Figure 7), these data suggest that KRP2 primarily regulates the rate and, thereby, the total number of anticlinal cell divisions during leaf development. Surprisingly, KRP2 overexpression did not alter the period in which cell division occurred, nor did it influence the timing of cell differentiation, which was measured by the appearance of stomata, suggesting that KRP2, unlike its mammalian counterpart p27Kip1, does not regulate cell cycle exit and the onset of cell differentiation. Some of the other KRPs might be involved in these processes.
In the young leaves, no significant differences in ploidy levels were observed between the control and transgenic lines, indicating that both the G1-to-S and G2-to-M transitions are inhibited by KRP2. This conclusion is in agreement with the observation that KRP2 specifically binds CDKA;1, which displays kinase activity at both transition points. In the KRP2 transgenic plants, progression from 2C to 8C was slower than in wild-type plants. Apparently, not only the mitotic cell cycle but the endoreduplication cycle as well is delayed by KRP2 overproduction.
Previously, a correlation was observed between ploidy level and cell size in wild-type epidermal cells and trichomes, suggesting that endoreduplication levels could determine cell size (Melaragno et al., 1993; Folkers et al., 1997). In contrast to these results, our data on KRP2-overproducing lines showed that the leaves displayed enlarged cells, whereas DNA replication clearly was suppressed, illustrating that DNA amplification is not a prerequisite for cell enlargement.
In the KRP2-overproducing lines, an increase in cell size compensated partially for the cell division inhibition, reflecting an uncoupling of cell growth from cell division. A similar uncoupling, which has been seen in tobacco plants with reduced A-type CDK activity (Hemerly et al., 1995), illustrates the plasticity of cells in response to changes in cell division activity. In contrast to this uncoupling of division and expansion, leaf morphology was changed: in the KRP2-overproducing lines, the slightly serrated phenotype that can be observed in control plants became very pronounced. In analogy to the differential sensitivity between organs, the inhibitory effect of KRP2 in specific domains of the leaf might underlie this phenotype. It has been noted that the expression of CYCB1;1 and CYCA2;1 at the leaf teeth remains high during late foliar development, when these mitotic cyclins are already not expressed in other parts of the leaves (Van Lijsebettens and Clarke, 1998; Burssens et al., 2000). Therefore, increased KRP2 levels might be less effective at inhibiting cell division activity at the teeth. However, to understand exactly how KRP2 overexpression results in abnormal leaf morphology, it will be necessary to determine when differences in shape become apparent during leaf development and how this is correlated with changes in cell division rates.
KRP2 is not the only cell cycle gene that induces abnormal leaf morphology. Overexpression of the fission yeast cdc25 gene in tobacco results in plants in which the leaves show a twisted appearance, with the interveinal regions being pocketed (Bell et al., 1993). Because overexpression of cdc25 is supposed to stimulate CDK activity positively, this phenotype may be the opposite of the KRP2-induced phenotype. No change in leaf morphology was reported upon the overproduction of other core cell cycle proteins, such as CDKA;1, CYCD2;1, CYCB1;1, and CKS1At (Hemerly et al., 1995; Doerner et al., 1996; Cockcroft et al., 2000; De Veylder et al., 2001). Because the serrated leaf phenotype also was observed in plants that overexpressed KRP1 or KRP3, it remains unknown whether KRP2 has a true morphogenic function or whether the observed phenotype is secondary to the strong reduction in cell number. Regardless, our data show that the rate of cell division can be an important factor in determining leaf shape.
METHODS
Isolation of Kip-Related Protein Genes
A two-hybrid screen using the CDKA;1 protein as a bait was performed as described (De Veylder et al., 1999). Among the positive clones, three different p27Kip1-like genes were identified: KRP1, KRP2, and KRP3. Full-length clones were obtained by screening a flower cDNA library obtained from the Arabidopsis Biological Resources Center (library number CD4-6; Columbus, OH). By screening the Arabidopsis databases, four additional KRP genes were discovered located on the genomic clones with accession numbers AC003974 (KRP4), AB028609 (KRP5), AP000419 (KRP6), and AC011807 (KRP7). The corresponding cDNAs were isolated using RNA prepared from cell suspensions of Arabidopsis thaliana (ecotype Columbia [Col-0]) and the Superscript RT II kit (Gibco BRL, Gaithersburg, MD) according to the manufacturer's protocol. Primers used were 5′-CCGGAATTCATGGGGAAATACATAAGAAAGAGC-3′ and 5′-GGCGGATCCGTTTCTAATCATCTACCTTCGTCC-3′ for KRP4, 5′-GGGAATTCATGGGAAAATACATTAAG-3′ and 5′-GGGGATCCTCATGGCATCACTTTGACC-3′ for KRP5, 5′-GGGAATTCATGAGCGAGAGAAAGCG-3′ and 5′-GGGGATCCTTAAAGTCGATCCC-3′ for KRP6, and 5′-GGGAATTCATGAGCGAAACAAAACCC-3′ and 5′-GGGGATCCCTAAGGTTTCAGACTAACCC-3′ for KRP7. The resulting polymerase chain reaction (PCR) fragments were cut with EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pGBT9 and pGAD424 (Clontech, Palo Alto, CA), resulting in the vectors pGADKRP4, pGADKRP5, pGADKRP6, pGADKRP7, pGBTKRP4, pGBTKRP5, pGBTKRP6, and pGBTKRP7. Binding specificity of the different Kip-related proteins (KRPs) toward CDKA;1, CDKB1;1, and CYCD4 was tested as described (De Veylder et al., 1997). Motifs were identified using the GCG software package (Genetics Computer Group, Madison, WI).
Reverse Transcriptase–Mediated PCR Analysis
RNA was isolated from flowers, roots, inflorescence stems, and 3-day-old cell suspensions of Arabidopsis (ecotype Col-0) using Trizol reagent (Amersham Pharmacia Biotech, Little Chalfont, UK). The RNA was treated with DNase for 30 min and was column purified (Chromaspin 400; Clontech). First-strand cDNA synthesis was performed on 3 μg of total RNA with the Superscript RT II kit (Gibco BRL) and oligo(dT)18 according to the manufacturer's instructions. A 1-μL aliquot of the total reverse transcription reaction volume (20 μL) was used as a template in semiquantitative reverse transcriptase–mediated PCR amplification, ensuring that the amount of amplified product remained in linear proportion to the initial template present in the reaction. Ten microliters from the PCR reaction was separated on a 0.8% agarose gel and transferred onto Hybond N+ membranes (Amersham Pharmacia Biotech). The membranes were hybridized at 65°C with fluorescein-labeled probes (Gene Images random prime module; Amersham Pharmacia Biotech). The hybridized bands were detected using the CDP Star detection module (Amersham Pharmacia Biotech). Primers used were 5′-GCAGCTACGGAGCCGGAGAATTGT-3′ and 5′-TCTCCTTCTCGAAATCGAAATTGTACT-3′ for KRP1, 5′-CGGCTCGAGGAGAACCACAAACACGC-3′ and 5′-CGAAACTAGTTAATTACCTCAAGGAAG-3′ for KRP2, 5′-GATCCCGGGCGATATCAGCGTCATGG-3′ and 5′-GATCCCGGGTTAGTCTGT-TAACTCC-3′ for KRP3, 5′-GGCGGATCCGTTTCTAATCATCTACCTTCGTCC-3′ and 5′-GAATCCATGGGGTACATAAG-3′ for KRP4, 5′-GGCCATGGGAAAATACATTAAGAA-3′ and 5′-GGGGATCCTCATGGCATCACTTTGACC-3′ for KRP5, 5′-GGCCATGGGCGAGAG-AAAGCGAGAGC-3′ and 5′-GGGGATCCTTAAAGTCGATCCC-3′ for KRP6, 5′-GGCCATGGGCGAAACAAAACCCAAGAG-3′ and 5′-GGG-GATCCCTAAGGTTTCAGACTAACCC-3′ for KRP7, and 5′-CTAAGCTCTCAAGATCAAAGGCTTA-3′ and 5′-TTAACATTGCAAAGAGTT-TCAAGGT-3′ for ACT2.
Generation of KRP2 and KRP3 Proteins and Antibodies
The coding regions of KRP2 and KRP3 were amplified by PCR. The KRP3-amplified coding sequence contained a protein starting at Met-11. Primer pairs used to amplify KRP2 and KRP3 were 5′-TAGGAGCATATGGCGGCGG-3′ and 5′-ATCATCGAATTCTTCATGGAT-TC-3′ and 5′-ATATCAGCGCCATGGAAGTC-3′ and 5′-GGAGCT-GGATCCTTTTGGAATTCATGG-3′, respectively. The obtained KRP2 PCR fragment was cut with NdeI and EcoRI and cloned into the NdeI and EcoRI sites of pRK172 (McLeod et al., 1987). The obtained KRP3 PCR fragment was cut with NcoI and BamHI and cloned into the NcoI and BamHI sites of pET21d. KRP3pET21d was transformed into Escherichia coli BL21(DE3). KRP2pRK172 was cotransformed into E. coli BL21(DE3) with pSBETa (Schenk et al., 1995). pSBETa encoded tRNAUCU, which is a low-abundance tRNA in E. coli corresponding to codons AGG and AGA (Arg). Because of the presence of an AGGAGAAGA sequence (Arg-5, Arg-6, and Arg-7) at the beginning of the KRP2 coding sequence, an increase of the tRNAUCU pool of E. coli was necessary for the translation of KRP2.
The KRP3pET21d/BL21(DE3) and KRP2pRK172[pSBETa/BL21(DE3)] E. coli recombinant strains were grown on selective Luria-Bertani medium. The cells were grown at 37°C until the density of the culture reached A600 = 0.7. At this time, 0.4 mM isopropyl-β-d-thiogalactopyranoside was added to induce production of the recombinant protein. Cells were collected 3 hr later by centrifugation. The bacterial pellet from 250 mL of culture was suspended in 10 mL of lysis buffer (Tris-HCl, pH 7.5, 1 mM DTT, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 0.1% Triton X-100) and subjected to three freeze/thaw cycles before sonication. Cell lysate was clarified by centrifugation for 20 min at 8000 rpm. The pellet was collected and resuspended in extraction buffer. Third and fourth washes were performed in the same way with Tris extraction buffer with and without 1 M NaCl, respectively.
After the different washing steps, the pellet contained KRP3 or KRP2 proteins at 90% homogeneity. The pellets were suspended in Laemmli loading buffer (Laemmli, 1970), and KRP3 and KRP2 were further purified by SDS-12% PAGE. The gel was stained in 0.025% Coomassie Brilliant Blue R 250 in water and destained in water. The strong band comigrating at the 31-kD marker position was cut from the gel with a scalpel. The polyacrylamide fragments containing KRP3 or KRP2 were lyophilized and reduced to powder. The rabbit immunization was performed subcutaneously with complete Freund's adjuvant with this antigen preparation. One injection corresponded to 100 μg of protein. The boosting injections were performed subcutaneously with incomplete Freund's adjuvant. With the obtained sera, bands of the expected size were found in protein extracts prepared from 2-day-old actively dividing cell cultures. No signals were observed with the preimmune serum.
In Vitro KRP3 Binding Assay
Purified recombinant KRP3 protein was coupled to cyanogen bromide–activated Sepharose 4B (Amersham Pharmacia Biotech) at a concentration of 5 mg/mL according to the manufacturer's instructions. Protein extracts were prepared from a 2-day-old cell suspension culture of Arabidopsis Col-0 in homogenization buffer (HB) containing 50 mM Tris-HCl, pH 7.2, 60 mM β-glycerophosphate, 15 mM nitrophenyl phosphate, 15 mM EGTA, 15 mM MgCl2, 2 mM DTT, 0.1 mM vanadate, 50 mM NaF, 20 μg/mL leupeptin, 20 μg/mL aprotinin, 20 μg/mL soybean trypsin inhibitor, 100 μM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 0.1% Triton X-100. Protein extract (200 μg) in a total volume of 100 μL of HB was loaded onto 50 μL of 50% (v/v) KRP3-Sepharose or control Sepharose beads and incubated on a rotating wheel for 2 hr at 4°C. The unbound proteins were collected for later analysis. The bead-bound fractions were washed three times with HB. The beads were resuspended in 30 μL of SDS loading buffer and boiled. The supernatants (bead-bound fractions) and 10 μL of the unbound fractions were separated on a 12.5% SDS-polyacrylamide gel and electroblotted onto nitrocellulose membranes (Hybond C+; Amersham Pharmacia Biotech). Filters were blocked overnight with 2% milk in PBS, washed three times with PBS, probed for 2 hr with specific antibodies for CDKA;1 (1:5000 dilution) or CDKB1;1 (1:2500 dilution) in PBS containing 0.5% Tween 20 and 1% albumin, washed for 1 hr with PBS containing 0.5% Tween 20, incubated for 2 hr with peroxidase-conjugated secondary antibody (Amersham Pharmacia Biotech), and washed for 1 hr with PBS containing 0.5% Tween 20. Proteins were detected by the chemiluminescence procedure (Pierce Chemical Co., Rockford, IL).
Regeneration and Molecular Analysis of KRP2-Overexpressing Plants
The full length KRP2 coding region was amplified by PCR with the 5′-AGACCATGGCGGCGGTTAGGAG-3′ and 5′-GGCGGATCCCGTCTTCTTCATGGATTC-3′ primers, introducing NcoI and BamHI restriction sites. The amplified fragment was cut with NcoI and BamHI and cloned between the NcoI and BamHI sites of PH35S (Hemerly et al., 1995), resulting in the 35SKRP2 vector. The cauliflower mosaic virus 35S/KRP2/nopaline synthase cassette was released by EcoRI and XbaI and cloned blunt into the SmaI site of PGSV4 (Hérouart et al., 1994). The resulting vector, PGSKRP2, was mobilized by the helper plasmid pRK2013 into Agrobacterium tumefaciens C58C1RifR harboring the plasmid pMP90. Arabidopsis plants (ecotype Col-0) were transformed by the floral dip method (Clough and Bent, 1998). Transgenic plants were obtained on kanamycin-containing medium and later transferred to soil for optimal seed production. For all analyses, plants were grown in vitro under a 16-hr-light/8-hr-dark photoperiod at 22°C on germination medium (Valvekens et al., 1988). Molecular analysis of the obtained transformants was performed by RNA and protein gel blotting, and cyclin-dependent kinase (CDK) activity measurements were as described (De Veylder et al., 1997, 1999). Leaves were sectioned according to Beeckman and Viane (2000).
Kinematic Analysis of Leaf Growth
For the kinematic analysis of leaf growth, eight plants of the wild type and three KRP2-overproducing lines were sown in quarter sections of round 12-cm Petri dishes filled with 100 mL of 1 × Murashige and Skoog (1962) medium (Duchefa, Haarlem, The Netherlands) and 0.6% plant tissue culture agar (Lab M, Bury, UK). The plates were placed horizontally on cooled benches in a growth chamber (temperature, 22°C; irradiation, 65 μE·m−2·sec−1 photosynthetically active radiation; photoperiod, 16-hr-light/8-hr-dark). From day 5 (when cotyledons started to expand) until day 21 after sowing (when leaves were fully expanded), a single plate was harvested daily (except on days 17, 18, and 20). All healthy plants were placed in methanol overnight to remove chlorophyll, and subsequently they were cleared and stored in lactic acid for microscopy.
A strong KRP2-overexpressing line was selected for analysis. The youngest plants were mounted whole on a slide and covered. Older primordia that had visible petioles were dissected, whereas younger primordia were left on the plant. The primordia were observed with differential interference contrast optics on a microscope (DMLB; Leica, Wetzlar, Germany). The total leaf (blade) area of the oldest two primordia (leaves 1 and 2) of each seedling was determined from drawing tube images that were scanned with a flatbed scanner using the public domain image analysis program ImageJ (version 1.17y; http://rsb.info.nih.gov/ij/). At older stages, the primordia were digitized directly with a charge-coupled device camera mounted on a binocular (Stemi SV11; Zeiss, Jena, Germany), which was connected to a personal computer fitted with a frame-grabber board LG3 (Scion Corp., Frederick, MD) running the image analysis program Scion Image (version 3b for Windows NT).
Cell density and stomatal index were determined from scanned drawing tube images of outlines of at least 20 cells of the abaxial epidermis located 25 and 75% from the distance between the tip and the base of the leaf primordium (or blade once the petiole was present), halfway between the midrib and the leaf margin. In the youngest primordia (up to day 6), a single group of cells was drawn. The following parameters were determined: total area of all cells in the drawing, total number of cells, and number of guard cells. From these data, we calculated the average cell area and the stomatal index (i.e., the fraction of guard cells in the total population of epidermal cells). We estimated the total number of cells per leaf by dividing the leaf area by the average cell area (averaged between the apical and basal positions). Finally, average cell division rates for the whole leaf were determined as the slope of the log 2–transformed number of cells per leaf, which was done using five-point differentiation formulas (Erickson, 1976).
Flow Cytometric Analysis of Leaves
Leaves were chopped with a razor blade in 300 μL of buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-[N-morpholino]propanesulfonic acid, pH 7, and 1% Triton X-100) (Galbraith et al., 1991). To the supernatants, 1 μL of 4′,6-diamidino-2-phenylindole from a stock of 1 mg/mL was added, which was filtered over a 30-μm mesh. The nuclei were analyzed with the BRYTE HS flow cytometer and WinBryte software (Bio-Rad, Hercules, CA).
KRP2 Analysis in Yeast
The CDKA;1.A14F15 and KRP2 genes were fused transcriptionally to the nmt1 promoter in the fission yeast vectors pREP41 (with a leucine marker) and pREP4X (with a uracil marker), respectively (Maundrell, 1993). The pREPCDKA;1.A14F15 vector has been described (Porceddu et al., 1999). The pREP4KRP2 vector was obtained by cloning the KRP2 gene into the XhoI and BamHI sites of the pREP4X vector after the XhoI and BamHI sites had been introduced by PCR with the primers 5′-GGCATATGGCGGCGGTTAGGAG-3′ and 5′-GGCGGATCCCGTCTTCTTCATCATGGATTC-3′. Plasmids were transformed into the fission yeast strain 972 leu1-32 ura4-D18 h− using the lithium acetate method (Gietz et al., 1992). All yeast manipulations were performed as described (Porceddu et al., 1999).
GenBank Accession Numbers
The GenBank accession numbers are as follows: KRP1, U94772; KRP2, AJ251851; KRP3, AJ301554; KRP4, AJ301555; KRP5, AJ301556; KRP6, AJ301557; KRP7, AJ301558; Chenopodium KRP, AJ002173.
Acknowledgments
The authors thank the members of the cell cycle group for fruitful discussions and useful suggestions, Stefaan Rombauts for analyzing the sequences, Martine De Cock for help in preparing the manuscript, and Peter Chaerle, Rebecca Verbank, Stijn Debruyne, and Karel Spruyt for illustrations. This work was supported by grants from the Interuniversity Poles of Attraction Program (Belgian State, Prime Minister's Office, Federal Office for Scientific, Technical and Cultural Affairs; P4/15) and the European Union (ECCO QLG2-CT1999-00454). L.D.V. is indebted to the “Onderzoeksfonds van de Universiteit Gent” for a postdoctoral fellowship.
References
- Beeckman, T., and Viane, R. (2000). Embedding thin plant specimens for oriented sectioning. Biotech. Histochem. 75, 23–26. [DOI] [PubMed] [Google Scholar]
- Beemster, G.T.S., and Baskin, T.I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, M.H., Halford, N.G., Ormrod, J.C., and Francis, D. (1993). Tobacco plants transformed with cdc25, a mitotic inducer gene from fission yeast. Plant Mol. Biol. 23, 445–451. [DOI] [PubMed] [Google Scholar]
- Burssens, S., de Almeida Engler, J., Beeckman, T., Richard, C., Shaul, O., Ferreira, P., Van Montagu, M., and Inzé, D. (2000). Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211, 623–631. [DOI] [PubMed] [Google Scholar]
- Chen, J., Jackson, P.K., Kirschner, M.W., and Dutta, A. (1995). Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature 374, 386–388. [DOI] [PubMed] [Google Scholar]
- Cheng, M., Olivier, P., Diehl, J.A., Fero, M., Roussel, M.F., Roberts, J.M., and Sherr, C.J. (1999). The p21Cip1 and p27Kip1 CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18, 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G.W., Healy, J.M.S., and Murray, J.A.H. (2000). Cyclin D control of growth rate in plants. Nature 405, 575–579. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Segers, G., Glab, N., Casteels, P., Van Montagu, M., and Inzé, D. (1997). The Arabidopsis Cks1At protein binds to the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 412, 446–452. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., De Almeida Engler, J., Burssens, S., Manevski, A., Lescure, B., Van Montagu, M., Engler, G., and Inzé, D. (1999). A new D-type cyclin of Arabidopsis thaliana expressed during lateral root primordia formation. Planta 208, 453–462. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beemster, G.T.S., Beeckman, T., and Inzé, D. (2001). CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell cycle progression. Plant J. 25, 617–626. [DOI] [PubMed] [Google Scholar]
- Doerner, P., Jørgensen, J.-E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380, 520–523. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Dulić, V., Kaufmann, W.K., Wilson, S.J., Tlsty, T.D., Lees, E., Harper, J.W., Elledge, S.J., and Reed, S.I. (1994). p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell 76, 1013–1023. [DOI] [PubMed] [Google Scholar]
- Dunphy, W.G. (1994). The decision to enter mitosis. Trends Cell Biol. 4, 202–207. [DOI] [PubMed] [Google Scholar]
- Erickson, R.O. (1976). Modeling of plant growth. Annu. Rev. Plant Physiol. 27, 407–434. [Google Scholar]
- Fero, M.L., Rivkin, M., Tasch, M., Porter, P., Carow, C.E., Firpo, E., Polyak, K., Tsai, L.-H., Broudy, V., Perlmutter, R.M., Kaushansky, K., and Roberts, J.M. (1996). A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27Kip1-deficient mice. Cell 85, 733–744. [DOI] [PubMed] [Google Scholar]
- Folkers, U., Berger, J., and Hülskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779–3786. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle–dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the protease inhibitor MG132. Plant Cell 10, 2063–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier, C., and Tardieu, F. (1998). Spatial and temporal analyses of expansion and cell cycle in sunflower leaves: A common pattern of development for all zones of a leaf and different leaves of a plant. Plant Physiol. 116, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier, C., Inzé, D., and Tardieu, F. (2000). Spatial distribution of cell division rate can be deduced from that of p34cdc2 kinase activity in maize leaves grown at contrasting temperatures and soil water conditions. Plant Physiol. 124, 1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P.B. (1976). Growth and cell pattern formation on an axis: Critique of concepts, terminology, and modes of study. Bot. Gaz. 137, 187–202. [Google Scholar]
- Hemerly, A., de Almeida Engler, J., Bergounioux, C., Van Montagu, M., Engler, G., Inzé, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14, 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérouart, D., Van Montagu, M., and Inzé, D. (1994). Developmental and environmental regulation of the Nicotiana plumbaginifolia cytosolic Cu/Zn-superoxide dismutase promoter in transgenic tobacco. Plant Physiol. 104, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacqmard, A., De Veylder, L., Segers, G., de Almeida Engler, J., Bernier, G., Van Montagu, M., and Inzé, D. (1999). CKS1At expression in Arabidopsis thaliana suggests a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inzé, D., Umeda, M., and Renaudin, J.-P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43, 607–620. [DOI] [PubMed] [Google Scholar]
- Kato, J.-y., Matsuoka, M., Polyak, K., Massagué, J., and Sherr, C.J. (1994). Cyclic AMP-induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell 79, 487–496. [DOI] [PubMed] [Google Scholar]
- LaBaer, J., Garrett, M.D., Stevenson, L.F., Slingerland, J.M., Sandhu, C., Chou, H.S., and Harlow, E. (1997). New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11, 847–862. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, M.-H., Reynisdóttir, I., and Massagué, J. (1995). Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 9, 639–649. [DOI] [PubMed] [Google Scholar]
- Lui, H., Wang, H., DeLong, C., Fowke, L.C., Crosby, W.L., and Fobert, P.R. (2000). The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant J. 21, 379–385. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Hurwitz, J., and Massagué, J. (1995). Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature 375, 159–161. [DOI] [PubMed] [Google Scholar]
- Magyar, Z., Mészáros, T., Miskolczi, P., Deák, M., Fehér, A., Brown, S., Kondorosi, E., Athanasiadis, A., Pongor, S., Bilgin, M., Bakó, L., Koncz, C., and Dudits, D. (1997). Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K. (1993). Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127–130. [DOI] [PubMed] [Google Scholar]
- McLeod, M., Stein, M., and Beach, D. (1987). The product of the mei3+ gene, expressed under control of the mating-type locus, in-duces meiosis and sporulation in fission yeast. EMBO J. 6, 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShea, A., Samuel, T., Eppel, J.-T., Galloway, D.A., and Funk, J.O. (2000). Identification of CIP-1-associated regulator of cyclin B (CARB), a novel p21-binding protein acting in the G2 phase of the cell cycle. J. Biol. Chem. 275, 23181–23186. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, V., De Veylder, L., Van Montagu, M., and Inzé, D. (1999). Cyclin-dependent kinases and cell division in higher plants: The nexus. Plant Cell 11, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nakayama, K., Ishida, N., Shirane, M., Inomata, A., Inoue, T., Shishido, N., Horii, I., Loh, D.Y., and Nakayama, K.-i. (1996). Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85, 707–720. [DOI] [PubMed] [Google Scholar]
- Nourse, J., Firpo, E., Flanagan, W.M., Coats, S., Polyak, K., Lee, M.-H., Massague, J., Crabtree, G.R., and Roberts, J.M. (1994). Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature 372, 570–573. [DOI] [PubMed] [Google Scholar]
- Peters, J.-M. (1998). SCF and APC: The yin and yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 10, 759–768. [DOI] [PubMed] [Google Scholar]
- Pines, J. (1994). Protein kinases and cell cycle control. Semin. Cell Biol. 5, 399–408. [DOI] [PubMed] [Google Scholar]
- Polyak, K., Lee, M.-H., Erdjument-Bromage, H., Koff, A., Roberts, J.M., Tempst, P., and Massagué, J. (1994). Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66. [DOI] [PubMed] [Google Scholar]
- Porceddu, A., De Veylder, L., Hayles, J., Van Montagu, M., Inzé, D., and Mironov, V. (1999). Mutational analysis of two Arabidopsis thaliana cyclin-dependent kinases in fission yeast. FEBS Lett. 446, 182–188. [DOI] [PubMed] [Google Scholar]
- Pyke, K.A., Marrison, J.L., and Leech, R.M. (1991). Temporal and spatial development of the cells of the expanding first leaf of Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 42, 1407–1416. [Google Scholar]
- Rogers, S., Wells, R., and Rechsteiner, M. (1986). Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 234, 364–368. [DOI] [PubMed] [Google Scholar]
- Russo, A.A., Jeffrey, P.D., Patten, A.K., Massagué, J., and Pavletich, N.P. (1996). Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A–Cdk2 complex. Nature 382, 325–331. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Baumann, S., Mattes, R., and Steinbiss, H.H. (1995). Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. BioTechniques 19, 196–200. [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (1995). Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9, 1149–1163. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (1999). CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Smith, M.L., Chen, I.-T., Zhan, Q., Bae, I., Chen, C.-Y., Gilmer, T.M., Kastan, M.B., O'Connor, P.M., and Fornace, A.J., Jr. (1994). Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science 266, 1376–1380. [DOI] [PubMed] [Google Scholar]
- Tomoda, K., Kubota, Y., and Kato, J.-y. (1999). Degradation of the cyclin-dependent kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398, 160–165. [DOI] [PubMed] [Google Scholar]
- Toyoshima, H., and Hunter, T. (1994). p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78, 67–74. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens–mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens, M., and Clarke, J. (1998). Leaf development in Arabidopsis. Plant Physiol. Biochem. 36, 47–60. [Google Scholar]
- Vlach, J., Hennecke, S., and Amati, B. (1997). Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip21. EMBO J. 16, 5334–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Fowke, L.C., and Crosby, W.L. (1997). A plant cyclin–dependent kinase inhibitor gene. Nature 386, 451–452. [DOI] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin–dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. [DOI] [PubMed] [Google Scholar]
- Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L.C. (2000). Expression of the plant cyclin–dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24, 613–623. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Wong, C., Liu, D., Finegold, M., Harper, J.W., and Elledge, S.J. (1999). p21CIP1 and p57KIP2 control muscle differentiation at the myogenin step. Genes Dev. 13, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]